Abstract
Cells continually adjust their gene expression profiles in order to adapt to the availability of nutrients. Glucose is a major regulator of pxancreatic β-cell function and cell growth. However, the mechanism of β-cell adaptation to high levels of glucose remains uncertain. To identify the specific targets responsible for adaptation to high levels of glucose, the differentially expressed genes from primary rat islets treated with 3.3 and 16.7 mmol/l glucose for 24 h were detected by DNA microarray. The results revealed that the expression levels of genes that encode enzymes required for de novo cholesterol biosynthesis [3-hydroxy-3-methylglutaryl-CoA synthase 1 (Hmgcs1), 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr), mevalonate (diphospho) decarboxylase (Mvd), isopentenyl-diphosphate δ-isomerase 1 (Idi1), squalene epoxidase (Sqle) and 7-dehydrocholesterol reductase (Dhcr7)] were significantly increased in islets treated with high levels of glucose compared with those in the islets treated with lower glucose levels. Quantitative polymerase chain reaction further confirmed that glucose stimulated the expression levels of these genes in a dose- and time-dependent manner. A similar result was obtained in islets isolated from rats subjected to 12, 24, 48 and 72 h of continuous glucose infusion. It has previously been recognized that cholesterol homeostasis is important for β-cell function. The present study provides, to the best of our knowledge, the first evidence for the involvement of the de novo cholesterol biosynthesis pathway in the adaptation of rat islets to high levels of glucose in vitro and in vivo.
Keywords: high glucose, pancreatic β-cell, nutrition adaptation, cholesterol biosynthesis
Introduction
As a major global chronic health problem, type 2 diabetes affects 336 million people worldwide (1). Type 2 diabetes develops when pancreatic β cells fail to secrete enough insulin in the face of insulin resistance or increased insulin demand, most likely due to β-cell exhaustion (2). Therefore, it is critical to reveal the mechanism of β-cell compensation and prevent the progression of β-cell decompensation for the long-term management of the disease.
Cells continually adjust their gene expression profiles in order to adapt to the availability of nutrients such as glucose (3,4). The majority of nonproliferating, differentiated cells depend on the efficiency of ATP production through oxidative phosphorylation to maintain their integrity. These cells metabolize glucose to pyruvate through glycolysis following which they oxidize the majority of the pyruvate to CO2 through the tricarboxylic acid (TCA) cycle in the mitochondria (5). In lipogenic tissues or rapidly proliferating cells, citrate, generated by the TCA cycle from glucose, is preferentially exported from the mitochondria to the cytosol and cleaved by ATP citrate lyase to produce cytosolic acetyl coenzyme A (acetyl-CoA) (6–8), which is a vital building block for the de novo biosynthesis of fatty acids and cholesterol. Previous studies have reported that islets from mice with a specific inactivation of the ATP-binding cassette transporter 1 (Abca1), a cellular cholesterol transporter, in their β cells, demonstrated altered cholesterol homeostasis and impaired insulin secretion (9–11). This suggests that abnormality of the cholesterol metabolism may contribute to the impaired β-cell function in diabetes. However, as glucose is a major regulator of pancreatic β-cell function and the main source of precursors (including acetyl-CoA) for cholesterol synthesis, whether high levels of glucose regulate the expression levels of genes responsible for de novo cholesterol biosynthesis in islets has not yet been validated.
Materials and methods
Materials
RPMI-1640 medium, fetal bovine serum and other culture reagents were obtained from Gibco Life Technologies (Grand Island, NY, USA). The cell culture plates were purchased from Nalge Nunc International (Roskilde, Denmark). Collagenase type XI was purchased from Sigma (St. Louis, MO, USA). The anti-3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr) antibody was purchased from Abcam (Cambridge, MA, USA). Anti-rabbit IgG conjugated with horseradish peroxidase was obtained from Cell Signaling Technology, Inc. (Beverly, MA, USA).
Rat infusions
The animal treatment was reviewed and approved by the Animal Care Committee of Shanghai Jiao Tong University School of Medicine (Shanghai, China). Male Sprague Dawley rats (Shanghai Laboratory Animal Center, Shanghai, China), weighing 250–300 g, were housed under a controlled temperature (21°C) and a 12 h light-dark cycle with unrestricted access to water and a standard laboratory diet. The animals were randomly divided into two groups, 4 rats in each group, receiving either saline or glucose, with heparin (40 U/ml). The infusion technique was similar to that described by Bonner-Weir et al (12). Under general anesthesia, indwelling catheters were inserted into the right jugular vein. The catheters were tunneled subcutaneously and exteriorized at the base of the neck. Following a recovery period of three to five days, the rats were infused (via the jugular vein catheter at 2 ml/h) with either 0.45% saline or 50% glucose in 0.45% saline, for 12, 24, 48 and 72 h. Animals were allowed access to food and water ad libitum during the infusion period. Upon completion of the infusion, the animals were sacrificed and their islets were isolated. The isolated islets were stored at −80°C with TRIzol (Invitrogen Life Technologies, Carlsbad, CA, USA) until RNA isolation.
Islet isolation and treatment
Islets of Langerhans were isolated from male Sprague Dawley rats by an in situ infusion of the pancreas with collagenase, and separated by density gradient centrifugation (13). The concentration of collagenase type XI was 0.5 mg/ml. Isolated rat islets were transferred to 6-well plates (300 islets per well) and cultured for 6, 12, 24, 48 or 72 h in RPMI-1640 medium containing 3.3, 8.3, 11.1 or 16.7 mmol/l glucose, supplemented with 0.25% bovine serum albumin (BSA) at 37°C and with 5% CO2.
RNA sample preparation and array hybridization
Total RNA was extracted from isolated islets using TRIzol according to the manufacturer’s instructions. Sample labeling and array hybridization were performed according to the instructions for the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies, Inc., Santa Clara, CA, USA). The total RNA from each sample was linearly amplified and labeled with Cy3-CTP (Agilent Technologies, Inc., Santa Clara, CA, USA). The labeled cRNAs were purified using an RNeasy Mini kit (Qiagen, Hilden, Germany). The concentration and specific activity (pmol Cy3/μg cRNA) of the labeled cRNAs were measured by NanoDrop (ND-1000; Thermo Scientific, Wilmington, DE, USA). A total of 1 μg of each labeled cRNA was fragmented by adding 11 μl of 10X blocking agent and 2.2 μl of 25X fragmentation buffer (both from Agilent Technologies, Inc.,). The mixture was heated at 60°C for 30 min and 55 μl 2X gene expression (GE) hybridization buffer (Agilent Technologies, Inc.,) was added to dilute the labeled cRNA. A total of 100 μl of the hybridization solution was dispensed into a gasket slide and assembled to the gene expression microarray slide. The slides were incubated for 17 h at 65°C in an Agilent Hybridization Oven (Agilent Technologies, Inc.). The hybridized arrays were washed and scanned using the Agilent DNA Microarray Scanner system (part no. G2565BA; Agilent Technologies, Inc.).
Microarray data analysis
Agilent Feature Extraction software (version 11.0.1.1; Agilent Technologies, Inc.) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX software package (version 11.5.1; Agilent Technologies, Inc.). Differentially expressed genes were identified through volcano plot filtering. Gene ontology (GO) and pathway analyses were performed using the standard enrichment computation method.
Quantitative polymerase chain reaction (qPCR)
The total RNA isolated from the islets was reverse-transcribed using a Promega Reverse Transcription kit (Promega Corporation, Madison, WI, USA). In order to quantify the transcript abundance of the genes of interest, qPCR was performed using SYBR Green Premix Ex Taq (Takara Bio, Inc., Shiga, Japan) with a 7300 Real-Time PCR machine (Applied Biosystems, Foster City, CA, USA). The relative expression levels were normalized to 18S mRNA levels in each sample. The sequences of the specific primers that were used were as follows: 3-hydroxy-3-methylglutaryl-CoA synthase 1 (Hmgcs1), 5′-GTCCCTCCACAAATGACCAC-3′ (forward), 3′-ATG ACAGCCGACTCAGGTTC-5′ (reverse); Hmgcr, 5′-TGCTGCTTTGGCTGTATGTC-3′ (forward), 3′-TGAGCG TGAACAAGAACCAG-5′ (reverse); mevalonate kinase (Mvk), 5′-TGGAGCAACTGGAGAAGCTG-3′ (forward), 3′-ATGTCCAGGCTTGGGAGTGT-5′ (reverse); phosphomevalonate kinase (Pmvk), 5′-TCAGCTGTAGGCCTGGTG AA-3′ (forward), 3′-TGCTCCTTGAGTGGACCAGA-5′ (reverse); mevalonate (diphospho) decarboxylase (Mvd), 5′-GAAAACTTCGCTGGCTACGG-3′ (forward) and 3′-CAACCCCTTTCTCCAGATGC-5′ (reverse); isopentenyl diphosphate δ-isomerase 1 (Idi1), 5′-CACTGG CAGGAGTGATTGGA-3′ (forward), 3′-TTGCTGGCATTG ATTTCAGG-5′ (reverse); farnesyl diphosphate farnesyl transferase 1 (Fdft1), 5′-ACATGGCCATCAGTGTGGAG-3′ (forward), 3′-AATTCTGCCATCCCACATCC-5′ (reverse); squalene epoxidase (Sqle), 5′-TGGTGGAGGAATGACAGT CG-3′ (forward), 3′-AAGCAAGCTTTTCGGAGCTG-5′ (reverse); 7-dehydrocholesterol reductase (Dhcr7), 5′-CGC TTCCAAAGCCAAGAATC-3′ (forward), 3′-ACACAA TGAACGGTGCGAAG-5′ (reverse); sterol regulatory element binding protein 1 (Srebp1), 5′-CATCCGCTT CTTACAGCACA-3′ (forward), 3′-TCATGCCCTCCA TAGACACA-5′ (reverse); 18S, 5′-CACGGGTGACGGGGA ATCAG-3′ (forward), 3′-CGGGTCGGGAGTGGGTAA TTTG-5′ (reverse).
Western blot analysis
The cultured islets in the 6-well plates were washed twice with ice-cold phosphate-buffered saline (PBS) and immediately placed into a lysis buffer containing 25 mmol/l 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES; pH 7.4), 1% Nonidet P-40, 100 mmol/l NaCl, 2% glycerol, 5 mmol/l NaF, 1 mmol/l ethylenediamine tetraacetic acid (EDTA), 1 mmol/l Na3VO4, 1 mmol/l sodium pyrophosphate (NaPPi), 1 mmol/l phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 5 μg/ml leupeptin and 5 μg/ml pepstatin. Lysates were centrifuged at 14,000 × g for 10 min at 4°C. The protein concentration of the extracts was determined according to the Bradford method, using BSA as the standard. Samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on 8% polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF)-Plus membranes (Bio-Rad, Hercules, CA, USA). The transferred membranes were incubated with anti-Hmgcr antibody at 1:1,000 dilution overnight at 4°C. Primary antibodies were detected with donkey anti-rabbit IgG conjugated with horseradish peroxidase at 1:2,000 for 1 h at room temperature. The blotted membrane was developed with enhanced chemiluminescence (ECL) Advance (Cell Signaling Technology, Inc., Boston, MA, USA) and imaged with a LAS-4000 Super CCD Remote Control Science Imaging system (Fujifilm, Tokyo, Japan).
Statistical analysis
Data are presented as mean ± standard error of the mean. Comparisons were performed using analysis of variance (ANOVA) for multiple groups or the Student’s t-test for two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Genes involved in cholesterol biosynthesis are upregulated in rat islets exposed to high levels of glucose
To identify the specific targets responsible for the adaptation to high levels of glucose, the differentially expressed genes from primary rat islets treated with 3.3 and 16.7 mM glucose for 24 h were detected by a whole-genome DNA microarray. Among a total of 44,000 probe sets, 894 genes were upregulated by high levels of glucose, revealing changes of ≥2-fold. GO analysis was used to identify the underlying biological themes in the genes in response to treatment with high levels of glucose. As shown in Table I, seven genes, termed in the gene ontology as ‘cholesterol biosynthetic process’ (GO: 0006695), were significantly upregulated in group treated with high levels of glucose. The qPCR results for Hmgcs1, Hmgcr, Mvd, Idi1, Sqle and Dhcr7 confirmed the genomic analyses, whereas those for Fdft1 did not (Fig. 1). Hmgcr, the rate-limiting enzyme in cholesterol biosynthesis, catalyzes the synthesis of mevalonate from hydroxymethyl glutaric acid acyl coenzymeA. Western blot analysis revealed increased protein levels of Hmgcr (Fig. 1). When combined, these data demonstrate that high levels of glucose markedly increased the expression levels of various genes involved in cholesterol biosynthesis in rat islets.
Table I.
Increased expression folds of cholesterol biosynthesis-associated genes in isolated rat islets incubated with 16.7 compared with 3.3 mmol/l glucose as demonstrated by microarray analysis.
| Gene symbol | Gene name | Gene function | mRNA fold change |
|---|---|---|---|
| Hmgcs1 | 3-Hydroxy-3-methylglutaryl- CoA synthase 1 | Catalyzes the conversion of (S)-3-hydroxy-3-methylglutaryl-CoA and CoA to acetyl-CoA, acetoacetyl-CoA and H2O | 2.494 |
| Hmgcr | 3-Hydroxy-3-methylglutaryl- CoA reductase | Enzyme involved in mevalonate synthesis | 2.675 |
| Mvd | Mevalonate (diphospho) decarboxylase | Enzyme that catalyzes the conversion of mevalonate pyrophosphate to isopentenyl pyrophosphate | 2.044 |
| Idi1 | Isopentenyl-diphosphate δ-isomerase 1 | Catalyzes the interconversion of isopentenyl diphosphate to dimethylallyl diphosphate | 5.411 |
| Fdft1 | Farnesyl-diphosphate farnesyl transferase 1 | Catalyzes the conversion of trans-farnesyl diphosphate to squalene | 2.279 |
| Sqle | Squalene epoxidase | Enzyme that catalyzes sterol biosynthesis | 2.591 |
| Dhcr7 | 7-Dehydrocholesterol reductase | Catalyzes the reduction of 7-dehydrocholesterol and is involved in cholesterol biosynthesis | 2.139 |
| Srebp1 | Sterol regulatory element binding protein 1 | Transcription factor: binds to the sterol regulatory element 1 | 2.289 |
Figure 1.
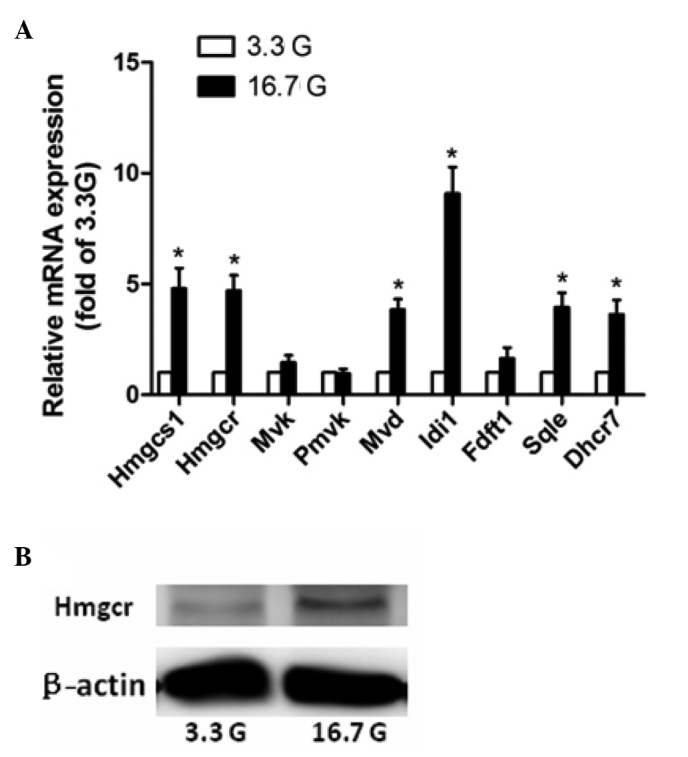
Effect of high levels of glucose on the expression levels of genes involved in cholesterol biosynthesis in rat islets in vitro. Isolated islets were incubated at 3.3 (3.3G) or 16.7 (16.7G) mmol/l glucose for 24 h. (A) The mRNA expression levels of nine genes involved in cholesterol biosynthesis were detected by quantitative polymerase chain reaction (qPCR). (B) The expression level of the 3-hydroxy-3-methylglutaryl-coenzyme A reductase (Hmgcr) protein was detected by western blot analysis. Data are expressed as mean ± standard error of the mean for three separate experiments. *P<0.05 vs. 3.3 mmol/l glucose treatment. A representative blot from three independent experiments is shown. All three experiments revealed similar results. Hmgcs1, 3-hydroxy-3-methylglutaryl-CoA synthase 1; Mvk, mevalonate kinase; Pmvk, phosphomevalonate kinase; Mvd, mevalonate (diphospho) decarboxylase; Idi1, isopentenyl-diphosphate δ-isomerase 1; Fdft1, farnesyl-diphosphate farnesyl transferase 1; Sqle, squalene epoxidase; Dhcr7, 7-dehydrocholesterol reductase.
Cholesterol biosynthetic gene expression levels are increased in the islets from rats infused with high levels of glucose
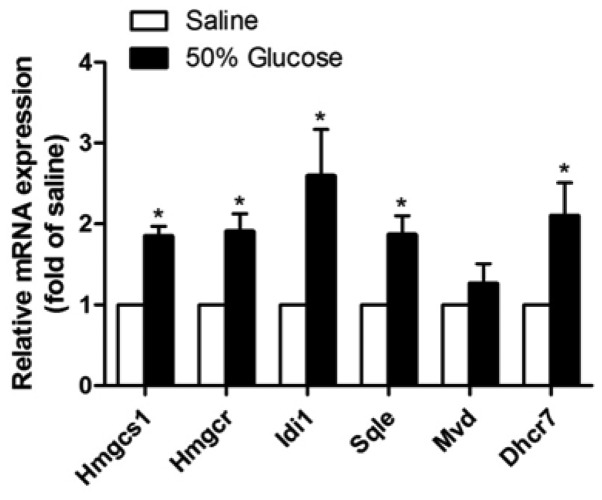
To further investigate the effect of high levels of glucose on cholesterol biosynthetic gene expression levels in vivo, the present study used a continuous glucose-infusion model as previously described (12). Infusion with 50% glucose (2 ml/h) for 24 h causes a significant increase in the plasma glucose level in rats (14,15). Consistent with the in vitro results of the current study, Hmgcs1, Hmgcr, Idi1, Sqle and Dhcr7 mRNA expression levels were significantly higher in islets isolated from rats infused with high levels of glucose for 24 h compared with those in the saline-infused control rats (Fig. 2). Furthermore, the expression level of the Idi1 gene revealed the highest increase (259±58%, P<0.05) among all genes tested, in accordance with in vitro results.
Figure 2.
Infusion with high levels of glucose increases the expression levels of cholesterol biosynthesis genes in rat islets in vivo. Sprague Dawley rats were infused with 50% glucose or saline for 24 h (n=5). The islets were isolated and the expression levels of mRNA were detected by quantitative polymerase chain reaction (qPCR). Data are expressed as mean ± standard error of the mean. *P<0.05 vs. saline treatment. Hmgcs1, 3-hydroxy-3-methylglutaryl-CoA synthase 1; Hmgcr, 3-hydroxy-3-methylglutaryl-CoA reductase; Idi1, isopentenyl-diphosphate δ-isomerase 1; Sqle, squalene epoxidase; Mvd, mevalonate (diphospho) decarboxylase; Dhcr7, 7-dehydrocholesterol reductase.
Glucose increases the expression levels of cholesterol biosynthetic genes in islets in a dose-dependent manner
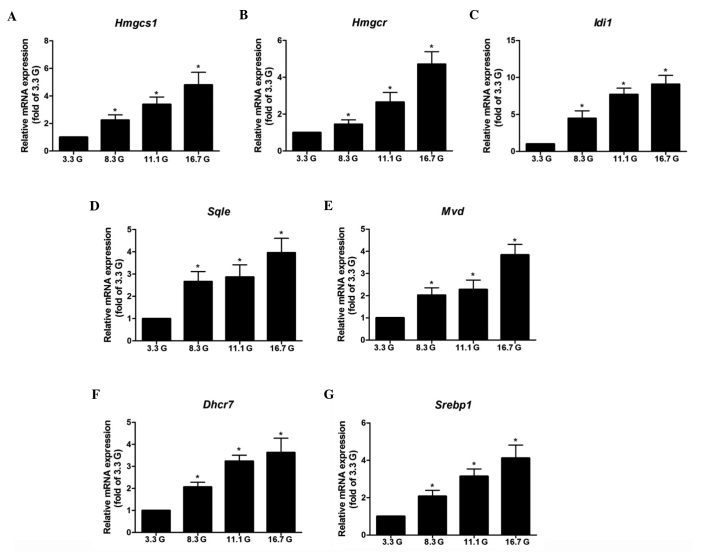
Isolated rat islets were incubated in 3.3, 8.3, 11.1 and 16.7 mmol/l glucose for 24 h. The qPCR results demonstrated that glucose increased the mRNA expression levels of Hmgcs1, Hmgcr, Mvd, Idi1, Sqle, Dhcr7 and Srebp1 in a dose-dependent manner, with notable effects observed at the concentration of 8.3 mmol/l (Fig. 3). The expression level of the of Srebp1 gene, which functions as a transcription factor for the gene expression of cholesterol biosynthetic enzymes, was also assessed. The qPCR results revealed that glucose markedly enhanced the expression level of Srebp1 in the same pattern as its target genes (Fig. 3).
Figure 3.
Dose-dependent effects of high levels of glucose on the expression levels of genes involved in cholesterol biosynthesis in rat islets. Following isolation, the rat islets were incubated with 3.3, 8.3, 11.1 or 16.7 mmol/l glucose and the expression levels of mRNA of (A) 3-hydroxy-3-methylglutaryl-CoA synthase 1 (Hmgcs1), (B) 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr), (C) isopentenyl-diphosphate δ-isomerase 1 (Idi1), (D) squalene epoxidase (Sqle), (E) mevalonate (diphospho) decarboxylase (Mvd), (F) 7-dehydrocholesterol reductase (Dhcr7) and (G) sterol regulatory element-binding protein 1 (Srebp 1) were detected by quantitative polymerase chain reaction (qPCR). Data are presented as mean ± standard error of the mean. *P<0.05 vs. 3.3 mmol/l glucose treatment.
Time course for the expression of cholesterol biosynthetic genes in islets exposed to high levels of glucose in vitro or in vivo
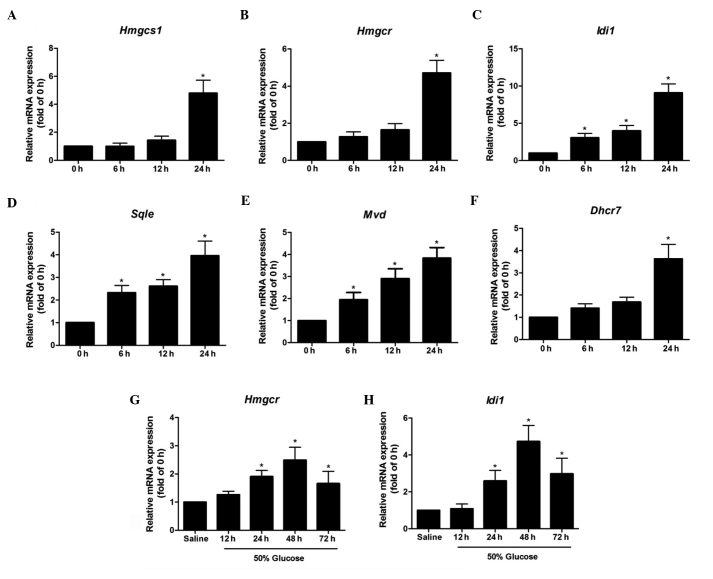
The time-dependent effects of high levels of glucose on the expression levels of cholesterol biosynthetic genes in isolated rat islets were detected (Fig. 4). In islets exposed to 16.7 mmol/l glucose for 6 and 12 h, the mRNA levels of Hmgcs1, Hmgcr and Dhcr7 remained comparable with those at 0 h, and increased significantly at 24 h by 380±92, 371±68, and 263±65%, respectively (P<0.05). However, the gene expression levels of Sqle and Mvd were increased ~2-fold by high levels of glucose at 6 h, and increased markedly with the lengthening of the exposure time. The Idi1 gene exhibited the most notable change in expression level among the genes tested, increasing by 206±59% at 6 h and 808±119% at 24 h (P<0.05). In islets isolated from 50% glucose-infused rats for 12 h, the mRNA level of Hmgcr remained unchanged when compared with that of the saline control group; however, it significantly increased with the lengthening of the infusion time to 24 h, and reached a peak level after 48 h (Fig. 4). The expression level pattern of the Idi1 gene was similar to that of Hmgcr in islets from glucose-infused rats (Fig. 4).
Figure 4.
Time course for the expression of cholesterol biosynthetic genes in islets exposed to high levels of glucose in vitro or in vivo. Following isolation, the rat islets were incubated with 16.7 mmol/l glucose for the indicated time and the expression levels of mRNA of (A) 3-hydroxy-3-methylglutaryl-CoA synthase 1 (Hmgcs1), (B) 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr), (C) isopentenyl-diphosphate δ-isomerase 1 (Idi1), (D) squalene epoxidase (Sqle), (E) mevalonate (diphospho) decarboxylase (Mvd) and (F) 7-dehydrocholesterol reductase (Dhcr7) were detected by quantitative polymerase chain reaction (qPCR). After the Sprague Dawley rats were infused with 50% glucose for the indicated time, the islets were isolated for analysis by qPCR to determine the mRNA expression levels of (G) Hmgcr and (H) Idi1. Data are expressed as mean ± standard error of the mean for three separate experiments. *P<0.05 vs. 0 h or saline treatment.
Discussion
As a main source for precursors (including acetyl-CoA), glucose has notable long-term actions on the de novo biosynthesis of lipids (16). Numerous studies investigating the link between chronic high levels of glucose and pancreatic β-cell lipid biosynthesis have focused on fatty acids (FAs). Chronic high levels of glucose markedly enhance the gene expression levels of acetyl-CoA carboxylase and fatty acid synthase and induce de novo fatty acid synthesis in pancreatic β cells (17–19). In the present study, the expression levels of genes involved in de novo cholesterol biosynthesis (Hmgcs1, Hmgcr, Mvd, Idi1, Sqle and Dhcr7) were significantly increased in islets treated with high levels of glucose. These results were further confirmed in islets isolated from rats subjected to 12, 24, 48 and 72 h continuous glucose infusion. This suggests that high levels of glucose may enhance cholesterol biosynthesis in the islets through increasing the gene expression levels of associated enzymes.
Cholesterol is synthesized via a cascade of enzymatic reactions known as the mevalonate pathway, which involves >20 enzymes from several subcellular compartments (20). This series of reactions is primarily regulated by a rate-limiting step involving the conversion of hydroxylmethylglutaryl-coenzyme A (HMG-CoA) into mevalonate. The rate-limiting reduction of HMG-CoA to mevalonate is an important regulatory step in cholesterol synthesis (21). The results of the present study revealed that high levels of glucose enhanced the gene expression levels of Hmgcr in rat islets in a dose-dependent manner when the cells were exposed to them for 24 h, and exposure to high levels of glucose also increased the protein levels of the gene. The qPCR results from glucose-infused rat islets confirmed this effect of high levels of glucose in vivo. Hmgcr acts early in the cholesterol synthesis pathway, with >20 subsequent enzymes required to produce cholesterol. The process of their regulation remains largely unstudied. The current study has provided the first published evidence, to the best of our knowledge, that high levels of glucose significantly increase the gene expression levels of Mvd, Idi1, Sqle and Dhcr7 in rat islets; these genes encode enzymes that serve as flux-controlling points in the cholesterol synthesis process beyond Hmgcr.
It has long been known that chronic high levels of glucose stimulate β-cell proliferation (12,22–26); however, the mechanism underlying the role of glucose in these events remains to be fully determined. Cell proliferation requires nutrients, energy and biosynthetic activity to duplicate all the macromolecular components during each passage through the cell cycle (27). The cholesterol content and rate of cholesterol biosynthesis are elevated in proliferating normal tissues and a reduction in cholesterol biosynthesis inhibits cell growth (28). This suggests the presence of a link between cell proliferation and the cholesterol biosynthetic pathway. The mevalonate pathway is also a crucial biochemical process for the generation of other key metabolic end products. There is an expanding list of intermediates that are known to be involved in cholesterol synthesis, and which have been credited with regulatory functions in the control of cell growth (28). Farnesyl diphosphate and other phosphorylated products of the mevalonate pathway are essential to the post-translational processing and physiological function of small G proteins, nuclear lamins and growth factor receptors (29–30), which are crucially involved in the regulation of proliferation. In addition, the expression levels of the steroidogenic acute regulatory protein (StAR) and steroid-5-α-reductase (Srd5a1), involved in the synthesis of steroid hormones from cholesterol, were observed to be increased by treatment with high levels of glucose in the DNA microarray data in the present study (data not shown). The regulatory mechanism of these genes by glucose and the possible role of these intermediates in glucose-stimulated β-cell proliferation requires further investigation.
In conclusion, based on the microarray analysis and qPCR validation, the present study identified a set of genes encoding cholesterol biosynthetic enzymes that were induced by high levels of glucose in rat islets. The current results provide evidence that enhanced cholesterol biosynthesis may play a role in the adaptive changes of β cells to high levels of glucose in vivo and in vitro.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (30600294, 81170720 and 81270910), Natural Science Foundation of Shanghai (11ZR1429500), Science and Technology Funds from Pudong New Area (PKJ2012-Y07), and the Academic Leaders Training Program of Pudong Health Bureau of Shanghai (PWRd2011-01).
Abbreviations
- TCA
tricarboxylic acid
- BSA
bovine serum albumin
- Hmgcs1
3-hydroxy-3-methylglutaryl-CoA synthase 1
- Hmgcr
3-hydroxy-3-methylglutaryl-CoA reductase
- Mvk
mevalonate kinase
- Pmvk
phosphomevalonate kinase
- Mvd
mevalonate (diphospho) decarboxylase
- Idi1
isopentenyl-diphosphate δ-isomerase 1
- Fdft1
farnesyl diphosphate farnesyl transferase 1
- Dhcr7
7-dehydrocholesterol reductase
- Sqle
squalene epoxidase
- Srebp1
sterol regulatory element binding protein 1
- HMG-CoA
hydroxymethylglutaryl-coenzyme A
References
- 1.Ashcroft FM, Rorsman P. Diabetes mellitus and the β cell: the last ten years. Cell. 2012;148:1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y, Taylor SI, Tan SL, Sonenberg N. When translation meets metabolism: multiple links to diabetes. Endocr Rev. 2003;24:91–101. doi: 10.1210/er.2002-0018. [DOI] [PubMed] [Google Scholar]
- 3.Ganapathy V, Thangaraju M, Prasad PD. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol Ther. 2009;121:29–40. doi: 10.1016/j.pharmthera.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Fiaschi T, Marini A, Giannoni E, Taddei ML, Gandellini P, De Donatis A, Lanciotti M, Serni S, Cirri P, Chiarugi P. Reciprocal metabolic reprogramming through lactate shuttle coordinately influences tumor-stroma interplay. Cancer Res. 2012;72:5130–5140. doi: 10.1158/0008-5472.CAN-12-1949. [DOI] [PubMed] [Google Scholar]
- 5.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chypre M, Zaidi N, Smans K. ATP-citrate lyase: a mini-review. Biochem Biophys Res Commun. 2012;422:1–4. doi: 10.1016/j.bbrc.2012.04.144. [DOI] [PubMed] [Google Scholar]
- 8.Beigneux AP, Kosinski C, Gavino B, Horton JD, Skarnes WC, Young SG. ATP-citrate lyase deficiency in the mouse. J Biol Chem. 2004;279:9557–9564. doi: 10.1074/jbc.M310512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunham LR, Kruit JK, Pape TD, Timmins JM, Reuwer AQ, Vasanji Z, et al. β-cell ABCA1 influences insulin secretion, glucose homeostasis and response to thiazolidinedione treatment. Nat Med. 2007;13:340–347. doi: 10.1038/nm1546. [DOI] [PubMed] [Google Scholar]
- 10.Kruit JK, Wijesekara N, Fox JE, Dai XQ, Brunham LR, Searle GJ, et al. Islet cholesterol accumulation due to loss of ABCA1 leads to impaired exocytosis of insulin granules. Diabetes. 2011;60:3186–3196. doi: 10.2337/db11-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wijesekara N, Zhang LH, Kang MH, Abraham T, Bhattacharjee A, Warnock GL, Verchere CB, Hayden MR. miR-33a modulates ABCA1 expression, cholesterol accumulation, and insulin secretion in pancreatic islets. Diabetes. 2012;61:653–658. doi: 10.2337/db11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonner-Weir S, Deery D, Leahy JL, Weir GC. Compensatory growth of pancreatic β-cells in adult rats after short-term glucose infusion. Diabetes. 1989;38:49–53. doi: 10.2337/diab.38.1.49. [DOI] [PubMed] [Google Scholar]
- 13.Kinasiewicz A, Juszczak M, Pachecka J, Fiedor P. Pancreatic islets isolation using different protocols with in situ flushing and intraductal collagenase injection. Physiol Res. 2004;53:327–333. [PubMed] [Google Scholar]
- 14.Zhang H, Li W, Wang Q, Wang X, Li F, Zhang C, et al. Glucose-mediated repression of menin promotes pancreatic β-cell proliferation. Endocrinology. 2012;153:602–611. doi: 10.1210/en.2011-1460. [DOI] [PubMed] [Google Scholar]
- 15.Ammon HP, Bacher M, Brändle WF, Waheed A, Roenfeldt M, el-Sayed ME, Ahmed AA, Wahl MA. Effect of forty-eight-hour glucose infusion into rats on islet ion fluxes, ATP/ADP ratio and redox ratios of pyridine nucleotides. J Endocrinol. 1998;156:583–590. doi: 10.1677/joe.0.1560583. [DOI] [PubMed] [Google Scholar]
- 16.Dashty M. A quick look at biochemistry: carbohydrate metabolism. Clin Biochem. 2013;46:1339–1352. doi: 10.1016/j.clinbiochem.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 17.Roche E, Farfari S, Witters LA, Assimacopoulos-Jeannet F, Thumelin S, Brun T, Corkey BE, Saha AK, Prentki M. Long-term exposure of β-INS cells to high glucose concentrations increases anaplerosis, lipogenesis, and lipogenic gene expression. Diabetes. 1998;47:1086–1094. doi: 10.2337/diabetes.47.7.1086. [DOI] [PubMed] [Google Scholar]
- 18.Berne C. The metabolism of lipids in mouse pancreatic islets: the biosynthesis of triacylglycerols and phospholipids. Biochem J. 1975;152:667–673. doi: 10.1042/bj1520667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diraison F, Ravier MA, Richards SK, Smith RM, Shimano H, Rutter GA. SREBP1 is required for the induction by glucose of pancreatic β-cell genes involved in glucose sensing. J Lipid Res. 2008;49:814–822. doi: 10.1194/jlr.M700533-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharpe LJ, Brown AJ. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) J Biol Chem. 2013;288:18707–18715. doi: 10.1074/jbc.R113.479808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medina MW, Krauss RM. The role of HMGCR alternative splicing in statin efficacy. Trends Cardiovasc Med. 2009;19:173–177. doi: 10.1016/j.tcm.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swenne I. Effects of aging on the regenerative capacity of the pancreatic B-cell of the rat. Diabetes. 1983;32:14–19. doi: 10.2337/diab.32.1.14. [DOI] [PubMed] [Google Scholar]
- 23.Swenne I. Glucose-stimulated DNA replication of the pancreatic islets during the development of the rat fetus. Effects of nutrients, growth hormone, and triiodothyronine. Diabetes. 1985;34:803–807. doi: 10.2337/diab.34.8.803. [DOI] [PubMed] [Google Scholar]
- 24.Chick WL. β-cell replication in rat pancreatic monolayer cultures. Effects of glucose, tolbutamide, glucocorticoid, growth hormone and glucagons. Diabetes. 1973;22:687–693. doi: 10.2337/diab.22.9.687. [DOI] [PubMed] [Google Scholar]
- 25.Paris M, Bernard-Kargar C, Berthault MF, Bouwens L, Ktorza A. Specific and combined effects of insulin and glucose on functional pancreatic β-cell mass in vivo in adult rats. Endocrinology. 2003;144:2717–2727. doi: 10.1210/en.2002-221112. [DOI] [PubMed] [Google Scholar]
- 26.Heit JJ, Karnik SK, Kim SK. Intrinsic regulators of pancreatic β-cell proliferation. Annu Rev Cell Dev Biol. 2006;22:311–338. doi: 10.1146/annurev.cellbio.22.010305.104425. [DOI] [PubMed] [Google Scholar]
- 27.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Rao KN. The significance of the cholesterol biosynthetic pathway in cell growth and carcinogenesis (review) Anticancer Res. 1995;15:309–314. [PubMed] [Google Scholar]
- 29.Thurnher M, Gruenbacher G, Nussbaumer O. Regulation of mevalonate metabolism in cancer and immune cells. Biochim Biophys Acta. 2013;1831:1009–1015. doi: 10.1016/j.bbalip.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Mo H, Elson CE. Studies of the isoprenoid-mediated inhibition of mevalonate synthesis applied to cancer chemotherapy and chemoprevention. Exp Biol Med (Maywood) 2004;229:567–585. doi: 10.1177/153537020422900701. [DOI] [PubMed] [Google Scholar]