Abstract
Dehydrogenase/reductase (SDR family) X-linked (DHRSX) is a novel human gene without any substantial functional annotation and was initially cloned and identified in our laboratory. In this study, we present evidence that it encodes a non-classical secretory protein and promotes starvation induced autophagy. Using the Baf.A1 assay and N-terminal sequencing, we showed that DHRSX is secreted in a non-classical form. We expressed and purified a recombinant human GST-DHRSX fusion protein. Functional studies revealed that HeLa and U2OS cells overexpressing DHRSX or treated with the GST-DHRSX fusion protein exhibited higher levels of starvation-induced autophagy, resulting in increased endogenous LC3-II levels, a punctate GFP-LC3 distribution, and structures associated with autophagy, with a lower accumulation of autophagy substrates such as p62 and polyQ80. Accordingly, knockdown of endogenous DHRSX through specific siRNAs reduced LC3-II levels obviously in U2OS cells induced by starvation. Collectively, these results demonstrate that DHRSX is a novel non-classical secretory protein involved in the positive regulation of starvation induced autophagy and provide a new avenue for research on this protein family and autophagy regulation.
Keywords: Autophagy, DHRSX, Non-classical secretory protein, Novel gene
Introduction
The rapid development of functional genomics has led to the identification of a large number of novel genes whose sequences have been determined but whose functions remain unclear. Using bioinformatics techniques and high-throughput screening, we have already identified several novel genes, such as NSA2 (Nop seven-associated 2) 1, CMTM3 (CKLF-like MARVEL transmembrane domain-containing member 3) 2 and TM9SF1 (transmembrane 9 superfamily member 1) 3, that are associated with cellular proliferation, death or autophagy, respectively.
Dehydrogenase/reductase (SDR family) X-linked (DHRSX) encodes a putative secretory protein belonging to the short-chain dehydrogenase/reductase (SDR) superfamily, which contains several thousand members as NAD(P)(H)-dependent oxidoreductases, sharing sequence motifs and displaying similar mechanisms 4. Studies have shown that this family plays an important role in multiple physiological or pathological processes. For example, HEP27 (encoded by gene DHRS2), identified initially in growth-arrested human hepatoblastoma HepG2 cells, inhibits Mdm2 and Stabilizes p53 5, 6 and exhibits a protective role against apoptosis induced by oxidative stress 7. retSDR1, also known as DHRS3, is a regulator of vitamin A metabolism and was reported as one of the three candidate genes that were significantly overexpressed in favorable neuroblastoma 8, 9. DHRS9, another SDR member with retinol dehydrogenase activity mediates Epstein-Barr virus lytic infection induced retinoic acid-responsive gene expression 10, 11. However, to date, many of the SDR members lack biochemical characterization and substantial functional annotation 12, 13. What is more, no studies have provided evidence supporting the involvement of these proteins in autophagy.
Autophagy is a highly conserved and dynamic process in eukaryotes in which the cytoplasm component, including excess or aberrant organelles, is sequestered into double-membrane autophagosomes and subsequently delivered to lysosomes for the breakdown and eventual recycling of macromolecules 14. When cells encounter environmental stresses, such as nutrient starvation, hypoxia, oxidative stress, etc, the level of autophagy can be dramatically augmented 15 and nutrient deprivation through Earle's balanced salt solution (EBSS), a starvation medium, is often used to induced autophagy 16. Many useful and convenient methods can be used to monitor autophagy, such as electron microscopy, LC3 western blotting, fluorescence microscopy and measuring autophagic substrate degradation 17. An increasing number of biological functions have been shown to be affected by autophagy, including cell survival 18, inflammation 19, gut homeostasis 20, tumor regulation 21 and diabetes 22; however, the detailed mechanisms underlying autophagy regulation remain to be revealed.
In the present study, we showed that the overexpression of DHRSX or treatment with the GST-DHRSX fusion protein increased the levels of EBSS induced autophagy in HeLa and U2OS cells, suggesting that this novel protein might be involved in the regulation of starvation-induced autophagy.
Materials and methods
Cell culture and reagents
HeLa, U2OS and HEK293T cells were obtained from the American Type Culture Collection and cultured in DMEM containing 10% fetal bovine serum (HyClone, Logan, UT, USA) supplemented with 100 mg/mL penicillin-streptomycin in 5% CO2 at 37℃. HeLa cells stably transfected with the GFP-LC3 construct (HeLa-GFP-LC3) were a kind gift from Professor Yingyu Chen in our laboratory. All other cell lines used for RT-PCR were maintained in our laboratory. siRNAs against DHRSX were synthesize by GenePharma (Shanghai, China). The sense and anti-sense sequences of specific siRNAs were as follows. DHRSX-homo-523, 5' GACCAACCUUCUCUUGGAUTT 3' and 5' AUCCAAGAGAAGGUUGGUCTT 3'; DHRSX-homo-988, 5' GCAGCUGUGGUCUAAGAGUTT 3' and 5' ACUCUUAGACCACAGCUGCTT 3'. Opti-MEM and Lipofectamine™ 2000 were purchased from Life Technologies (USA). The transfections were performed according to the manufacturer's instructions. Earle's balanced salt solution (EBSS) was obtained from M&C Gene Technology (Beijing, China). Chloroquine (CQ, Sigma; 100 mM stock) was dissolved in H2O. Antibodies against LC3I/II, p62, AKT, mTOR or Beclin1 were a gift of Professor Yingyu Chen. The antibody against β-actin was purchased from Sungene Biotech (Tianjin, China). Rabbit anti-DHRSX polyclonal antibodies were generated using recombinant human DHRSX protein, followed by affinity purification and validation through ELISA and immunoblot analysis. IRDyeTM700 or IRDyeTM800-labeled secondary antibodies were obtained from Odyssey (Lincoln, NE, USA). The polyQ80-luciferase and polyQ19-luciferase constructs were a gift from Professor Yingyu Chen. The Dual Luciferase Reporter System was purchased from Promega (Madison, WI, USA).
Reverse transcription PCR and real-time quantitative PCR
The normal human tissue cDNA libraries were obtained from Clontech (Mountain View, CA, USA) and the cDNA libraries of tumor cell lines were prepared in our laboratory. Total RNA was isolated using TRIzol reagent (Life Technologies, USA), and reverse transcription was performed using the RevertAid First Strand cDNA Synthesis Kit (Fermentas, Canada) according to the manufacturer's instructions. Real-time PCR was performed using the “Power SYBR Green PCR Master Mix” (Applied Biosystems, Forster City, CA, USA). The following specific forward and reverse primer sequences were used: DHRSX, 5'-CGGCTGGTTGCTTTTCAA-3' and 5'-TCTCTTTCTCGTTGTATAGGTAATGG-3'; and GAPDH, 5'-TCCACTGGCGTCTTCACC-3' and 5'-GGCAGAGATGATGACCCTTTT-3'.
N-terminal sequencing
The DHRSX-myc-his eukaryotic protein with a 6-his tag at C-terminal was purified from the supernatants of HEK293T cells overexpressing that protein using nickel affinity chromatography, followed by separation by SDS-PAGE and transfer onto a PVDF membrane in CAPS buffer. Subsequently, the membrane was stained with Coomassie R-250, and the specific band of interest was subjected to N-terminal sequencing (ABI PROCISETM494).
Expression and purification of recombinant GST-DHRSX fusion protein
pGEX-4T-3-DHRSX-positive Transetta (DE3) cells (TransGen Biotech, Beijing, China) were cultured to the logarithmic growth phase (OD600 = 0.6), and subsequently, IPTG was added at a final concentration of 0.01 mM. The cells were then cultured for an additional 8 h, after which they were collected and resuspended in PBS, followed by supersonic lysis in an ice bath for 500 s intermittently. The resulting lysate was centrifuged at 13,000 ×g for 20 min, and the supernatant was purified with Glutathione Sepharose 4B (GE, Fairfield, CT, USA) according to the manufacturer's instructions and dialyzed in PBS. The products were analyzed by SDS-PAGE and western blotting.
Western blot analysis
The cells were harvested and lysed in RIPA buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, and 1% Triton X-100) containing protease inhibitors. After incubation on ice for 20 min, the insoluble materials were removed through centrifugation, followed by protein quantification using the BCA kit (Pierce, Rockford, IL, USA). Total proteins were resolved by SDS-PAGE, followed by transfer to nitrocellulose membranes and sequential incubation with specific primary and secondary antibodies. The proteins were visualized using the Odyssey Infrared Imager System (LI-COR Biosciences, Lincoln, NE, USA), and the gray value of the indicated band was quantified using Odyssey software.
Confocal microscopy and electron microscopy
The cells were treated with 50 ng/ml GST or GST-DHRSX fusion protein for 24 h before autophagy induction in EBSS containing 25 μM chloroquine (CQ) for 1 h. Subsequently, the cells were rinsed with PBS and fixed in 4% paraformaldehyde for 20 min, and the GFP-LC3 dots were detected by confocal microscopy (Leica TCS SP5 Confocal System, Germany). Electron microscopy was performed as previously described 23.
Luciferase reporter assay
The cells were cotransfected with polyQ80-luciferase and DHRSX or the empty vector using Lipofectamine 2000 reagent according to the manufacturer's instructions. The same procedure was used for the cotransfection of polyQ19-luciferase. After 48 h, autophagy was induced in EBSS for 1 h. Subsequently, the luciferase activity in the whole-cell lysates was analyzed using the Luciferase Reporter System. The polyQ80-luciferase activity was normalized to that of polyQ19-luciferase.
Statistical analysis. Significant differences between groups were tested using an unpaired two-tailed t test. The symbols ''*'', ''**'' and “***” indicate statistical significance at P < 0.05, P < 0.01, and P < 0.001, respectively.
Results
Bioinformatics analysis and expression pattern of DHRSX
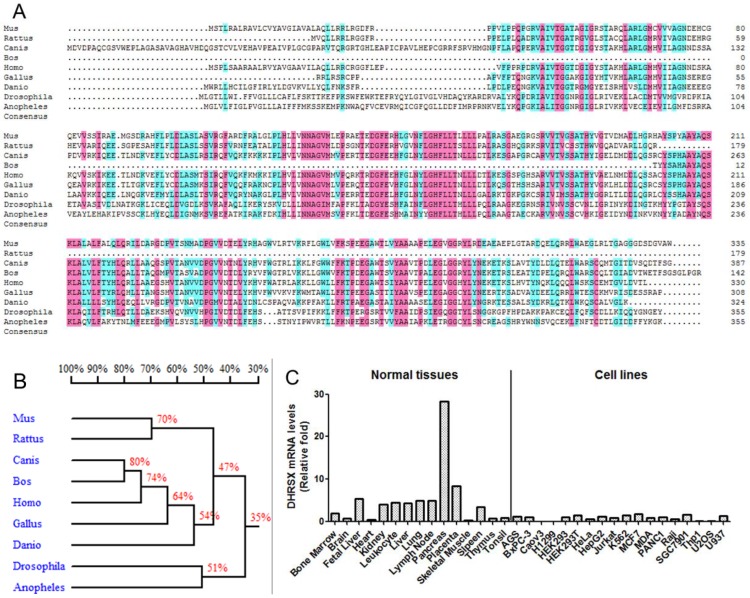
We initially performed a basic bioinformatics analysis of DHRSX. According to the NCBI database (http://www.ncbi.nlm.nih.gov/), this gene is located on chromosomes Xp22.33 and Yp11.2 and comprises 7 exons and 6 introns. The alignment of the protein sequences from different organisms, using DNAMAN software, showed that DHRSX is a highly conserved protein in humans, mice, rats, cows, dogs, chickens, fruit flies, zebrafish, and mosquitoes (Fig. 1A, 1B). An analysis of DHRSX expression through real-time qPCR using a panel of normal human tissues and cell lines revealed that DHRSX is expressed in a wide spectrum of tissues, with high levels of expression detected in the pancreas; notably, in a series of cell lines, the DHRSX expression levels were obviously decreased (Fig. 1C).
Fig 1.
Bioinformatics analysis and the expression pattern of DHRSX. (A) The protein sequence alignment using DNAMAN software revealed significant similarities among the DHRSX proteins from different organisms. Pink and azure represent homology levels of 100% and 75%, respectively. (B) Homology tree of DHRSX from different species. The identity of the amino acid sequences between different species was indicated as a percentage. (C) Expression pattern of DHRSX using real-time qPCR in a panel of normal human tissues and cell lines.
DHRSX is a novel non-classical secretory protein
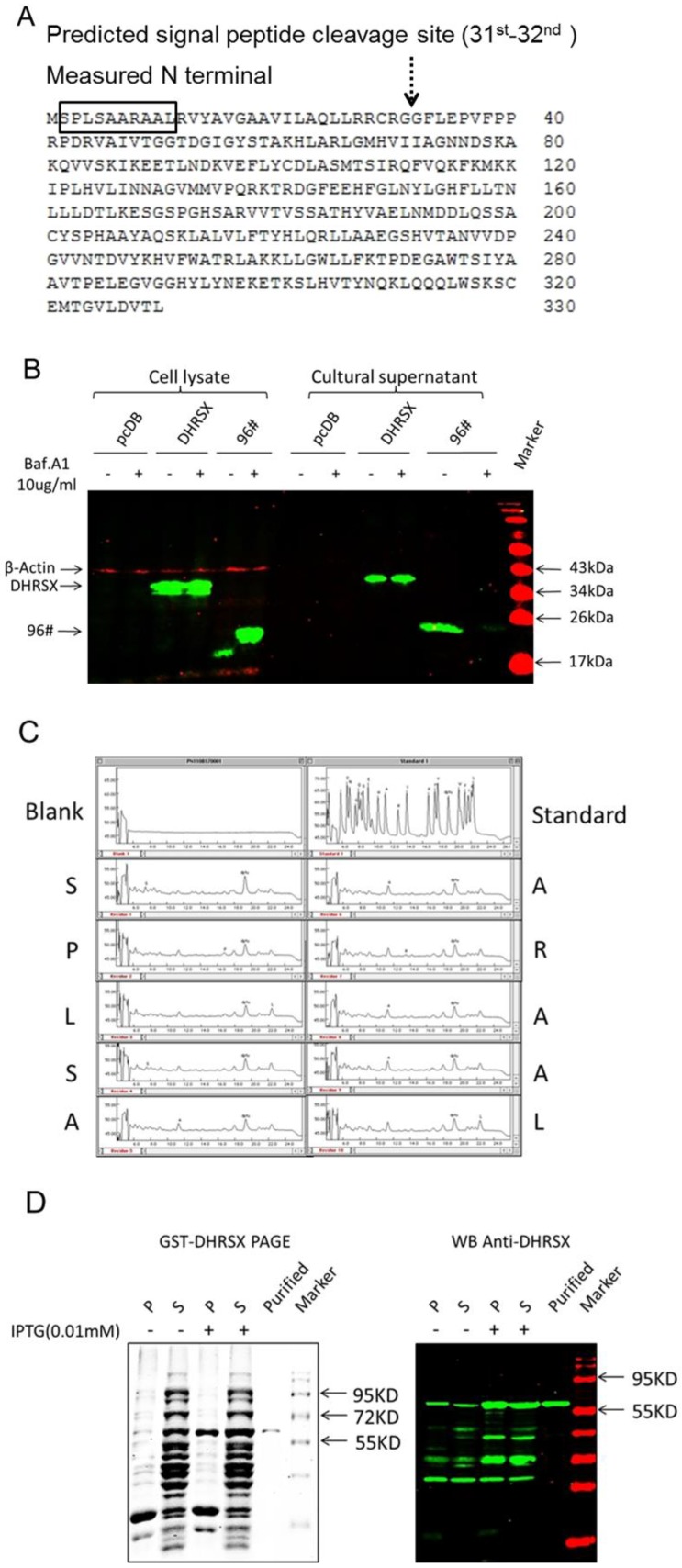
The SignalP software (http://www.cbs.dtu.dk/services/SignalP/) analysis predicted that DHRSX contained a potential signal peptide with a putative cleavage site between amino acids 31 and 32 (Fig. 2A), and the analysis using PSORT II (http://psort.hgc.jp/) suggested that DHRSX was an extracellular protein, showing a high prediction score of 19.0 (data not shown). Then, we experimentally characterized the secretion behavior of DHRSX. The western blot analysis demonstrated that DHRSX-myc-his eukaryotic protein, with a 6-his tag at C-terminal, could be detected in the supernatant of HEK293T cells overexpressing this protein, but DHRSX secretion was not suppressed by Baf.A1, a classical secretion inhibitor 24 (Fig. 2B). Furthermore, N-terminal sequencing demonstrated that the first 10 amino acids of DHRSX-myc-his protein were identical to the N-terminal sequence of the full-length DHRSX (Fig. 2A, 2C), indicating that DHRSX is secreted as a full-length protein. These data suggest that DHRSX is indeed secreted but in a non-classical form 25.
Fig 2.
Identification of the non-classical secretion of DHRSX-myc-his and preparation of the GST-DHRSX fusion protein. (A, C) N-terminal sequencing demonstrated that the first 10 amino acids of DHRSX-myc-his eukaryotic protein, with a 6-his tag at C-terminal, were identical to the N-terminal sequence of the full-length DHRSX, indicated with a black box, suggesting that this protein was secreted in its full-length form, although a potential signal peptide with a putative cleavage site between amino acids 31 and 32, indicated with a black dotted arrow, was predicted using SignalP software. (B) Western blot using anti-his as the primary antibody showed that DHRSX-myc-his was detectable in the supernatant, after enrichment by nickel affinity chromatography, and this secretion could not be inhibited using 10 μg/ml Baf.A1, an inhibitor of classical secretion. pcDB served as a negative control. 96#, a confirmed classical secretory protein, was used as a positive control for Baf.A1 inhibition. (D) The results of SDS-PAGE (left) and western blot analysis (right) using a rabbit anti-DHRSX polyclonal antibody as the primary antibody, whose specificity has been previously identified (data not shown), demonstrated the purity and specificity of the GST-DHRSX fusion protein before and after purification with Glutathione Sepharose 4B. “P” and “S”, respectively, indicate the precipitate and supernatant from the E. coli cell lysate induced with or without 0.01 mM IPTG.
To facilitate further functional studies, we constructed the human DHRSX prokaryotic expression construct pGEX-4T-3-DHRSX, and subsequently expressed and purified the recombinant human GST-DHRSX fusion protein, which was identified as a high-quality protein using SDS-PAGE and western blot analysis (Fig. 2D).
DHRSX promotes autophagic flux induced through starvation in HeLa and U2OS cells
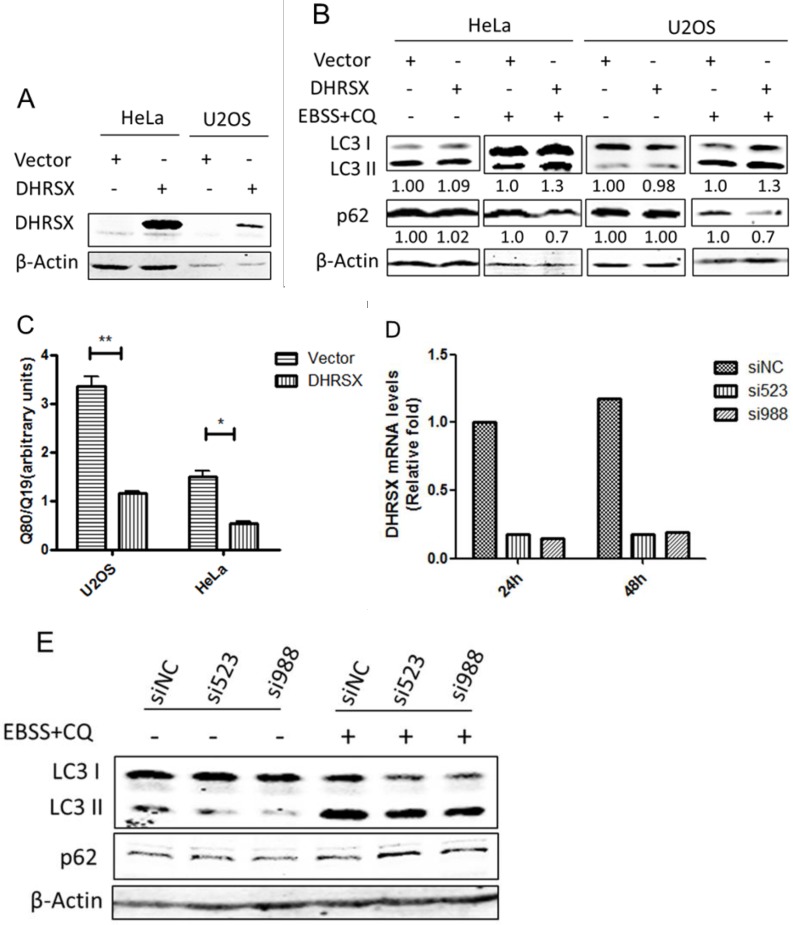
To investigate the functional involvement of DHRSX in autophagy, we overexpressed this protein in HeLa and U2OS cells (Fig. 3A). Interestingly, the overexpression of DHRSX alone did not affect the levels of LC3 II or the selective autophagy substrate p62, while with the induction of autophagy using Earle's balanced salt solution (EBSS) in combination with chloroquine (CQ), a well-known lysosomotropic agent serving as an inhibitor of autophagy flux at the late stage 26, forced DHRSX expression resulted in increased LC3 II expression and decreased p62 accumulation compared with the negative control (Fig. 3B). To confirm the positive effect of DHRSX on autophagy, we monitored the accumulation of polyglutamine (polyQ80), another autophagy substrate, to directly assess the progression of autophagic flux. The results showed that the ectopic expression of DHRSX resulted in lower polyQ80-luciferase activities in both HeLa and U2OS cells treated with EBSS for 1 h (Fig. 3C), suggesting that the degradation of autolysosomes was increased. Accordingly, knockdown of endogenous DHRSX through specific siRNAs reduced LC3-II levels obviously and increased the amount of p62 slightly in U2OS cells induced by starvation (Fig. 3D, 3E).
Fig 3.
Overexpressed DHRSX increased LC3 II expression and reduced autophagy substrate accumulation in HeLa and U2OS cells after starvation for 1 h. (A) Anti-DHRSX was used as a control for DHRSX overexpression in HeLa and U2OS cells. β-Actin was used as a loading control, and the empty vector served as a negative control. (B) The ectopic expression of DHRSX for 48 h increased LC3 II and reduced p62 protein expression in HeLa and U2OS cells after starvation in EBSS containing 25 μM chloroquine (CQ) for 1 h. The ratios of the corresponding protein to β-actin expression normalized to the empty control were shown below the western blots. (C) PolyQ80-luciferase activity normalized to that of polyQ19-luciferase was remarkably decreased compared with the negative control in HeLa and U2OS cells transfected with DHRSX for 48 h, followed by treatment in EBSS for 1 h. (D) Real-time quantitative PCR was used to identify the silencing efficiency of the siRNAs against DHRSX with GAPDH as an internal reference. (E) After knockdown of endogenous DHRSX with the indicated siRNAs for 48 h and another 1 h for starvation, LC3 II levels were obviously reduced than that of the control siRNA, while the p62 protein was slightly increased.
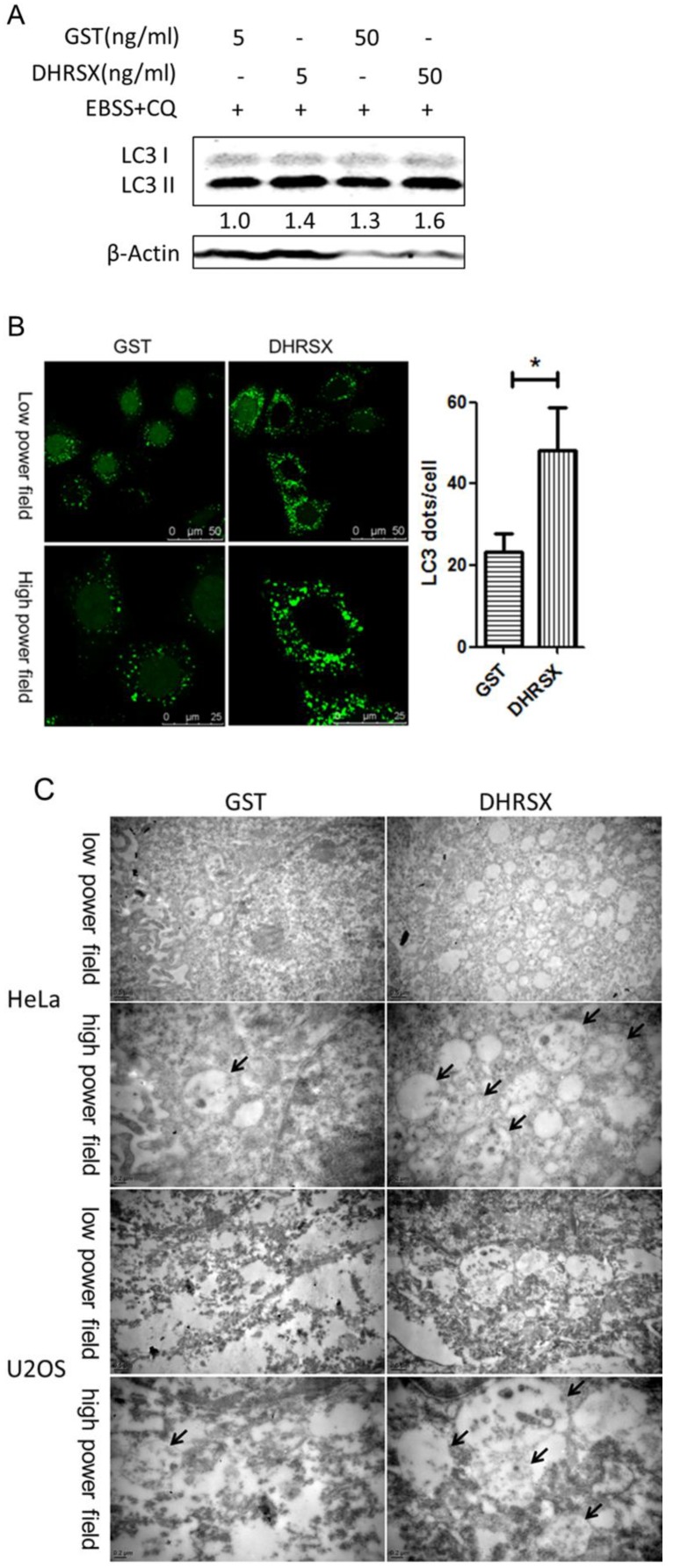
In HeLa cells treated with the recombinant protein GST-DHRSX, the results were consistent with those of the DHRSX overexpression experiments, showing an increased amount of LC3 II after autophagy induction for 1 h (Fig. 4A). Moreover, confocal microscopy revealed that GFP-LC3 punctuation was markedly increased in stably transfected Hela-GFP-LC3 cells treated with GST-DHRSX, followed by autophagy induction in EBSS containing CQ for 1 h (Fig. 4B). Electron microscopy showed that after the induction of autophagy in EBSS containing CQ for 1 h, HeLa and U2OS cells treated with GST-DHRSX generated more autophagosomes and autolysosomes than cells treated with the GST control protein (Fig. 4C).
Fig 4.
Recombinant DHRSX protein increased the formation of autophagic structures in starvation-induced HeLa and U2OS cells. (A) HeLa cells treated with GST-DHRSX or GST as a control for 24 h showed an increased amount of LC3 II after autophagy induction in EBSS containing 25 μM chloroquine for 1 h. (B) GFP-LC3 expression in stably transfected Hela-GFP-LC3 cells treated with 50 ng/ml GST-DHRSX or GST as a control for 24 h, followed by autophagy induction for 1 h, was determined using confocal microscopy. The number of GFP-LC3 puncta in at least 50 cells was shown on the right of the representative images. (C) Representative transmission electron micrographs of HeLa and U2OS cells after treatment with 50 ng/ml GST-DHRSX or GST as a negative control for 24 h, followed by autophagy induction for 1 h. The black arrows indicate typical autophagy structures.
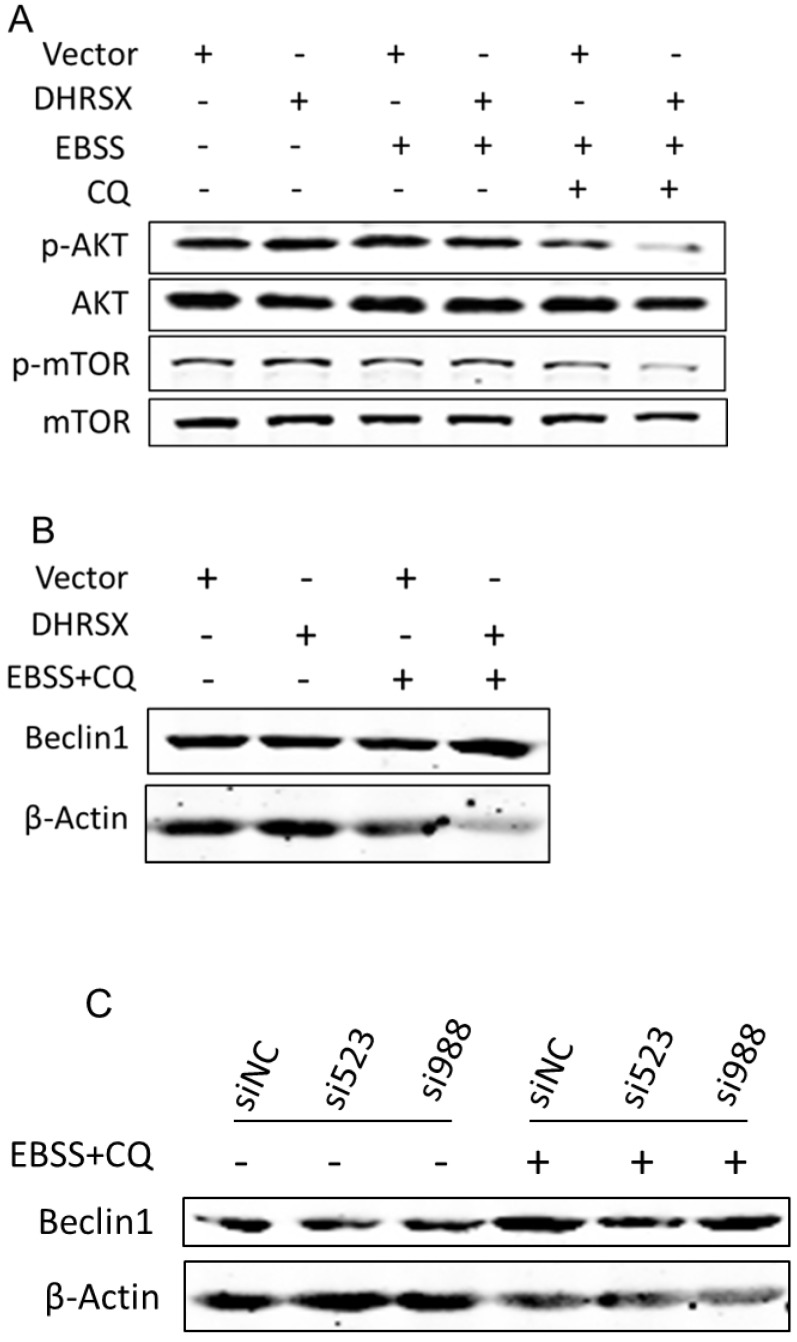
To clarify the underlying mechanism, we detected some essential autophagy regulators. Studies have shown that the AKT (PKB)/mTOR (mammaliam target of rapamycin) axis is a vital pathway regulating autophagy 15. Our analyses revealed that ectopic DHRSX expression in U2OS cells treated with EBSS containing CQ significantly reduced the phosphorylation of AKT and mTOR (Fig. 5A). It was early known that Beclin1 was a necessary regulator for autophagy and was found to facilitate autophagy induced by serum and amino-acid deprivation 27. In our investigation, the amount of Beclin1 was increased obviously in starvation induced U2OS cells overexpressing DHRSX (Fig. 5B). What's more, knockdown of endogenous DHRSX via specific siRNAs resulted in less Beclin1 protein (Fig. 5C).
Fig 5.
Ectopic DHRSX lead to down-regulation of phosphorylation AKT/mTOR and up-regulation of Beclin1 protein during starvation induced autophagy. The phosphorylation of AKT and mTOR was significantly reduced (A) while the amount of Beclin1 was increased obviously (B) in starvation induced U2OS cells overexpressing DHRSX. (C) Knockdown of endogenous DHRSX via indicated siRNAs resulted in less Beclin1 protein in U2OS cells after treatment with EBSS containing 25 μM chloroquine for 1 h.
Taken together, these results provided the first demonstration of an obvious role for the novel non-classical secretory protein DHRSX in promoting starvation induced autophagy and indicated that the facilitation of DHRSX in autophagy was associated with the down-regulation of AKT/mTOR phosphorylation together with the up-regulation of Beclin1.
Discussion
Although the sequence of DHRSX has been identified, no experimental studies concerning its function are available. In the present study, we provide the first systematic investigation of the secretory behavior and function of DHRSX.
The results of basic bioinformatics analysis indicate that DHRSX is a highly conserved protein that is expressed in many different species, suggesting that this gene might be involved in some conserved biological processes. Moreover, the expression of DHRSX in normal tissues and cell lines should be further investigated.
Studies have shown that unconventional secretory proteins lack signal peptides and their export from cells is not affected by brefeldin A, an inhibitor of the classical secretory pathway 28. Interestingly, the SignalP software predicted a potential signal peptide cleavage site in this protein, although the results of N-terminal sequencing showed that secreted DHRSX contained the complete predicted signal peptide sequence, consistent with results that its secretion could not be suppressed using the classical secretion inhibitor Baf.A1. What's more, the actual molecular weight of DHRSX (about 38 kDa) was much closer to its full-length form (36.4 kDa) than the predicted form with the potential signal peptide cut (33kDa). All the results above indicated that DHRSX was secreted in an unconventional pathway. Further studies will reveal the detailed secretion mechanism.
Autophagy is a highly regulated process and under the control of a large number of molecules and multiple signaling pathways. To characterize the involvement of DHRSX in autophagy, we overexpressed DHRSX or treated cells with the recombinant DHRSX protein. We observed increased LC3 II expression and autophagic structure formation in DHRSX group compared with the negative control. In addition, we detected lower levels of selective autophagy substrates p62 and polyQ80, whose total cellular expression levels were inversely correlated with autophagic activity 17.
There are good examples that a secreted protein could promote autophagy, such as TNF-α (Tumor Necrosis Factor) 29 and MIF (Macrophage migration inhibitory factor) 30. Then, how DHRSX augmented starvation induced autophagy was further studied. mTOR (mammaliam target of rapamycin) is a highly conserved serine/threonine protein kinase and a master regulator of autophagy serving as a central sensor of growth factors, nutrient signals and engery status 31. mTOR integrates upstream activating signals that inhibit autophagy through the class I PI3K-Akt (Akt, also known as PKB) pathway. In a word, PI3K/Akt/ mTOR acts as a pivotal axis that negatively regulates autophagy 15. Beclin 1, another crucial regulator of autophagy, can intervene at many major steps in autophagic pathways through the interaction with specific Beclin 1-binding proteins. Though its role in autophagy is complicated, in more cases, Beclin 1 acts as a positive regulator 32. In our study, we found that the phosphorylation of AKT and mTOR was significantly reduced while the amount of Beclin1 was increased obviously in starvation induced U2OS cells overexpressing DHRSX. Importantly, knockdown of endogenous DHRSX via specific siRNAs resulted in less Beclin1 protein.
These results were exactly consistent with those previously reported. Nevertheless, what's puzzling is just that there existed no obvious changes in these molecules without starvation induction. Maybe, it reflected that it was not the initiation but the process that DHRSX took part in. In short, these results indicated that the inhibition of phosphorylation of AKT/mTOR and up-regulation of Beclin1 might account for the positive effect of DHRSX on autophagy. Further studies will include more in-depth mechanism research.
In summary, in the present study, we performed the first systematic analysis of the functional involvement of the novel non-classical secretory protein DHRSX in promoting starvation-induced autophagy. These findings provide a new direction for research on this protein family and autophagy regulation.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (30971483; 81172001).
References
- 1.Zhang H, Ma X, Shi T. et al. NSA2, a novel nucleolus protein regulates cell proliferation and cell cycle. Biochem Bioph Res Co. 2010;391:651–58. doi: 10.1016/j.bbrc.2009.11.114. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Li J, Cui Y. et al. CMTM3, located at the critical tumor suppressor locus 16q22.1, is silenced by CpG methylation in carcinomas and inhibits tumor cell growth through inducing apoptosis. Cancer Res. 2009;69:5194–201. doi: 10.1158/0008-5472.CAN-08-3694. [DOI] [PubMed] [Google Scholar]
- 3.He P, Peng Z, Luo Y. et al. High-throughput functional screening for autophagy-related genes and identification of TM9SF1 as an autophagosome-inducing gene. Autophagy. 2009;5:52–60. doi: 10.4161/auto.5.1.7247. [DOI] [PubMed] [Google Scholar]
- 4.Kavanagh KL, Jörnvall H, Persson B. et al. The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65:3895–906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DONADEL G, GARZELLI C, FRANK R. et al. Identification of a novel nuclear protein synthesized in growth-arrested human hepatoblastoma HepG2 cells. Eur J Biochem. 1991;195:723–29. doi: 10.1111/j.1432-1033.1991.tb15759.x. [DOI] [PubMed] [Google Scholar]
- 6.Deisenroth C, Thorner AR, Enomoto T. et al. Mitochondrial HEP27 Is a c-Myb Target Gene That Inhibits Mdm2 and Stabilizes p53. Mol Cell Biol. 2010;30:3981–93. doi: 10.1128/MCB.01284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monge M, Colas E, Doll A. et al. Proteomic approach to ETV5 during endometrial carcinoma invasion reveals a link to oxidative stress. Carcinogenesis. 2009;30:1288–97. doi: 10.1093/carcin/bgp119. [DOI] [PubMed] [Google Scholar]
- 8.Cerignoli F, Guo X, Cardinali B. et al. retSDR1, a Short-Chain Retinol Dehydrogenase/Reductase, Is Retinoic Acid-inducible and Frequently Deleted in Human Neuroblastoma Cell Lines. Cancer Res. 2002;62:1196–204. [PubMed] [Google Scholar]
- 9.Kamei N, Hiyama K, Yamaoka H. et al. Evaluation of genes identified by microarray analysis in favorable neuroblastoma. Pediatr Surg Int. 2009;25:931–37. doi: 10.1007/s00383-009-2448-1. [DOI] [PubMed] [Google Scholar]
- 10.Rexer BN, Ong DE. A Novel Short-Chain Alcohol Dehydrogenase from Rats with Retinol Dehydrogenase Activity, Cyclically Expressed in Uterine Epithelium. Biol Reprod. 2002;67:1555–64. doi: 10.1095/biolreprod.102.007021. [DOI] [PubMed] [Google Scholar]
- 11.Jones RJ, Dickerson S, Bhende PM. et al. Epstein-Barr Virus Lytic Infection Induces Retinoic Acid-responsive Genes through Induction of a Retinol-metabolizing Enzyme, DHRS9. J Biol Chem. 2007;282:8317–24. doi: 10.1074/jbc.M608667200. [DOI] [PubMed] [Google Scholar]
- 12.Keller B, Volkmann A, Wilckens T. et al. Bioinformatic identification and characterization of new members of short-chain dehydrogenase/reductase superfamily. Mol Cell Endocrinol. 2006;248:56–60. doi: 10.1016/j.mce.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 13.Bray JE, Marsden BD, Oppermann U. The human short-chain dehydrogenase/reductase (SDR) superfamily: A bioinformatics summary. Chem-Biol Interact. 2009;178:99–109. doi: 10.1016/j.cbi.2008.10.058. [DOI] [PubMed] [Google Scholar]
- 14.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–52. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–31. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng S, Wu Y, Chen B. et al. Impaired autophagy due to constitutive mTOR activation sensitizes TSC2-null cells to cell death under stress. Autophagy. 2011;7:1173–86. doi: 10.4161/auto.7.10.16681. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N, Yoshimori T, Levine B. Methods in Mammalian Autophagy Research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiang L, Wu C, Ming M. et al. Autophagy Controls p38 Activation to Promote Cell Survival under Genotoxic Stress. J Biol Chem. 2013;288:1603–11. doi: 10.1074/jbc.M112.415224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen ML, Yi L, Jin X. et al. Resveratrol attenuates vascular endothelial inflammation by inducing autophagy through the cAMP signaling pathway. Autophagy. 2013;9:2033–45. doi: 10.4161/auto.26336. [DOI] [PubMed] [Google Scholar]
- 20.Randall-Demllo S, Chieppa M, Eri R. Intestinal Epithelium and Autophagy: Partners in Gut Homeostasis. Front Immunol. 2013;4:301. doi: 10.3389/fimmu.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu B, Wen X, Cheng Y. Survival or death: disequilibrating the oncogenic and tumor suppressive autophagy in cancer. Cell Death Dis. 2013;4:e892. doi: 10.1038/cddis.2013.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson CM, Magnaudeix A, Yardin C. et al. Autophagy Dysfunction and its Link to Alzheimer's Disease and Type II Diabetes Mellitus. CNS Neurol Disord Drug Targets. 2014;13:226–46. doi: 10.2174/18715273113126660146. [DOI] [PubMed] [Google Scholar]
- 23.Yu C, Wang L, Lv B. et al. TMEM74, a lysosome and autophagosome protein, regulates autophagy. Biochem Bioph Res Co. 2008;369:622–29. doi: 10.1016/j.bbrc.2008.02.055. [DOI] [PubMed] [Google Scholar]
- 24.Lippincott-Schwartz J, Yuan LC, Bonifacino JS. et al. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–13. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nickel W. The mystery of nonclassical protein secretion. Eur J Biochem. 2003;270:2109–19. doi: 10.1046/j.1432-1033.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 26.Solomon VR, Lee H. Chloroquine and its analogs: A new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol. 2009;625:220–33. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 27.Liang XH, Jackson S, Seaman M. et al. Induction of autophagy and inhibition of tumorigenesis by beclin1. Nature. 1999;402:672–76. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 28.Nickel W. Pathways of unconventional protein secretion. Curr Opin Biotech. 2010;21:621–26. doi: 10.1016/j.copbio.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Djavaheri-Mergny M, Amelotti M, Mathieu J. et al. NF-kB Activation Represses Tumor Necrosis Factor- -induced Autophagy. J Biol Chem. 2006;281:30373–82. doi: 10.1074/jbc.M602097200. [DOI] [PubMed] [Google Scholar]
- 30.Chuang Y, Su W, Lei H. et al. Macrophage Migration Inhibitory Factor Induces Autophagy via Reactive Oxygen Species Generation. PLoS ONE. 2012;7:e37613. doi: 10.1371/journal.pone.0037613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12:1509–18. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 32.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]