Abstract
Gastrointestinal (GI) cancers remain one of the most common malignancies and are the second common cause of cancer deaths worldwide. The limited effectiveness of therapy for patients with advanced stage and recurrent disease is a reflection of an incomplete understanding of the molecular basis of GI carcinogenesis. Major advancements have improved our understanding of pathology and pathogenesis of GI cancers, but high mortality rates, unfavorable prognosis and lack of clinical predictive biomarkers provide an impetus to investigate new sensitive and specific diagnostic and prognostic markers for GI cancers. MicroRNAs (miRNAs) are short (19–24 nucleotides) noncoding RNA molecules that regulate gene expression at the posttranscriptional level thus playing an important role in modulating various biological processes including, but not limited, to developmental processes, proliferation, apoptosis, metabolism, differentiation, epithelial-mechenchymal transition and are involved in the initiation and progression of various human cancers. Unique miRNA expression profiles have been observed in various cancer types at different stages, suggesting their potential as diagnostic and prognostic biomarkers. Due to their tumor-specific and tissue-specific expression profiles, stability, robust clinical assays for detection in serum as well as in formalin-fixed tissue samples, miRNAs have emerged as attractive candidates for diagnostic and prognostic applications. This review summarizes recent research supporting the utility of miRNAs as novel diagnostic and prognostic tools for GI cancers.
Keywords: Gastrointestinal cancers, Diagnosis, Prognosis, miRNAs
Introduction
Gastrointestinal cancers refer to malignant conditions of the gastrointestinal tract (GI tract) and digestive tract associated organs including the esophagus, stomach, biliary system, pancreas, small intestine, large intestine, rectum and anus. The GI cancers are collectively the major cause of cancer related morbidity and mortality worldwide [1]. Prognosis of patients with GI cancers remains largely unsatisfactory due to loco-regional recurrence [1]. Despite improvements, the current treatment strategies including surgery, radiotherapy (RT) and/or chemotherapy (CT) have marginally improved the curative expectations and the quality of life of patients; however, the effectiveness of these new tools depends largely on the stage in which tumors can be detected. Therefore, these cancers can be managed by developing biomarkers, which could enhance early detection, improve patient stratification and therapy response prediction resulting in a more favorable disease outcome. Currently, the most important conventional prognostic factors for survival of GI patients are histological tumor grade and tumor stage at the time of diagnosis, including depth of tumor invasion and involvement of regional lymph nodes. In addition to these clinicopathological parameters, biomarkers are being intensively sought and validated for GI cancers. However, low sensitivity and specificity of available biomarkers limit their utility particularly in screening for early stages or in distinguishing aggressive and indolent tumors [2]. Currently, the relative abundance of several proteins is used as an indicator for diagnostic and prognostic potential. However, complexity of protein composition, diversity of posttranslational modifications, low relative abundance of many proteins and difficulties in developing suitable high-affinity detection agents hinder the development of new protein-based biomarkers. This has prompted investigators to identify more specific and sensitive novel markers, which can supplement or complement existing detection methods for better diagnosis and management of lethal GI cancers.
In recent years, miRNAs present in the body fluids such as plasma, serum, urine, saliva, etc. and tissues have been utilized as biomarkers in various diseases including many cancers [3]. These miRNAs are short noncoding RNA molecules of approximately 19–24 nucleotides (nt), involved in post-transcriptional regulation of gene expression. miRNAs bind to the 3’ untranslated region of mRNA, which leads to either translational repression, or mRNA degradation initiated by miRNA guided rapid deadenylation [4]. They act as master regulators for many important biological processes including ontogeny, cell proliferation, apoptosis, migration, differentiation, metabolism, stress, viral infection, cancer initiation and progression and drug resistance [5–8]. Although hundreds of miRNAs have been shown to be deregulated in cancers, the group of miRNAs actually playing any pathogenic role in cancer has not yet been fully defined [9]. However, recent high throughput screening studies have documented several miRNA expression signatures as promising biomarkers in a wide array of human cancers that are potentially capable of predicting favorable prognosis or exhibit association with tumor development, progression and response to therapy [10–12]. In cancers, approximately half of the deregulated miRNAs have been located near fragile sites, in loci associated with loss of heterozygosity or with DNA amplification, and in common breakpoints [13]. Being physiologically important and deregulated in several malignancies, miRNAs are considered to carry important information about the pathophysiological status of cancer patients.
While the majority of miRNAs are intracellular, in recent years extracellular circulating miRNAs have been detected in a variety of biological fluids, such as blood, serum and plasma, saliva, urine, breast milk, seminal plasma, tears, amniotic fluid, colostrum, bronchial lavage, cerebrospinal fluid, peritoneal fluid, and pleural fluid [14–17]. Although Lawrie et al identified miRNAs from serum of diffused B cell lymphoma patients; it remained unknown whether the miRNAs detected originated from tumor cells or from nonmalignant cell types [18]. The best possible sources of these circulating miRNAs may include not only apoptosis and necrosis of circulating and primary tumor cells, but also immune cells and other blood cells [19]. However, Chen et al., showed different serum miRNA expression profiles among the cancer and the healthy controls suggesting the presence of tumor-specific miRNAs in serum and plasma [20]. It is possible that circulatory miRNAs predominantly originate from apoptotic and necrotic tumor cells and reflect pathophysiology of the underlying disease, thus serving as useful biomarkers to monitor the clinical course of tumors. Initially the stability of miRNAs in body fluids was debatable; however recent studies indicate that circulating miRNAs are present in extracellular vesicles including exosomes, microvesicles and apoptotic bodies, which provide protection from nucleases present abundantly in the body fluids [21, 22]. In addition to vesicle bound miRNAs in body fluids, miRNAs also bind to high density and low density lipoproteins and RNA-binding proteins Agonaute 2 (Ago 1) and Agonaute 2 (Ago2).
Overall, tissue specific expression of miRNAs, ease of access in the cell-free body fluids, remarkable stability, sensitive and inexpensive detection supports their potential as disease biomarkers [23–25]. Therefore miRNAs are considered to be attractive candidates as diagnostic, prognostic and predictive biomarkers [26]. Furthermore, a single miRNA can affect several cellular processes, and therefore, successful targeting of miRNAs can potentially provide novel therapeutic avenues to combat malignancies. In this review article, we provide an updated overview of literature and summarize the current knowledge about the diagnostic and prognostic applications of miRNAs in GI cancers.
Esophageal Cancer
Esophageal cancer is the 3rd most common type of cancer among the GI cancers and 6th leading cause of cancer related deaths. In the United States, about 17,990 new cases and 15,210 deaths were estimated in 2013 [27]. The epidemiology of esophageal cancer has changed markedly over the past several decades in the United States. Until the 1970s, squamous cell carcinoma was the most prevalent type of esophageal cancer (90–95%). However due to the lifestyle changes, the incidence of adenocarcinoma has increased intensely in the last two decades [28]. Numerous molecular and histological changes were associated in the multistage conversion of normal squamous epithelium to Barrett’s esophagus, low grade and high grade dysplasia and frank adenocarcinoma. Specifically, esophageal adenocarcinoma (EAC) is the most common aggressive tumor that arises from the Barrett’s esophagus and Barrett’s metaplasia [29]. Hence Barrett’s esophagus is the pre-neoplastic condition suitable for identifying and predicting the candidate biomarkers for early detection and prognostic evaluation. Several studies have highlighted the importance of miRNAs involved during the progression of esophageal cancer [10, 30].
Altered expression of miRNAs during the development of esophageal tumors has been thoroughly investigated during the last decade [30]. Wijnhoven et al [31] reported deregulation of 44 miRNAs in the columnar gastric and squamous esophageal epithelium. Furthermore, real time PCR analysis revealed miR-21, miR-143, miR-145, miR-194 and miR-215 to be significantly upregulated in columnar epithelium as compared to normal esophageal squamous tissues. Maru et al [32] demonstrated miRNA-196a as a potential marker with dual role in growth acceleration and anti-apoptotic function during the progression from normal esophagus-toBarrett’s metaplasia- to dysplasia- to full blown esophageal cancer. Recently, a genome-wide PCR based profiling of 754 miRNAs was evaluated in the sequential stages of esophageal cancer and oncogenic miRNAs such as miR-21, miR-25, and miR-223 and tumor suppressor miRNAs such as miR-205, miR-203, let-7c, miR-133a and miR-133b were found to be aberrantly expressed in Barrett’s esophagus and EAC specimens [33]. Tumor suppressor effects of miR-133a have been shown in variety of cancers. Restoration of miR-133a in cancer cells inhibited proliferation, migration, invasion and resulted in G2/S arrest hence validating the tumor suppressive function of miR-133a in cancer cells [33]. Similar tumor suppressor effects have been documented for miR-133b using various cell culture experiments. In silico analysis revealed EGFR and FGFR1 as the predictive targets of 133a and 133b suggesting that these two miRNA’s were suggested as potential biomarkers and therapeutic targets for esophageal cancer [33]. Also, a significant down-regulation of miR-375 was exclusively observed in adenocarcinoma tissues indicating this miRNAs as a promising biomarker which can be interpreted with histological grading as well [33]. Other expression profiling studies using next generation sequencing (NGS) in a paired normal vs EAC tissues reveled 26 miRNAs to be frequently deregulated in EAC. Out of 26 deregulated miRNAs, miR-31 and miRNA-31* were extensively identified in the high grade dysplasia and adenocarcinoma specimens [34]. In addition, miR-375 showed remarkable down regulation in EAC [34]. Thus, miRNAs are critical molecular markers that can potentially aid in discriminating the patients who are at high risk of developing adenocarcinoma from the pre-neoplastic lesions of Barrett’s esophagus.
Despite aggressive treatment, the clinical outcome remains poor in patients with esophageal cancer. The dismal prognosis of esophageal cancer is mainly attributed to the asymptomatic nature of the disease and late presentation of patients with advanced disease stages i.e mostly detected only after metastasis, making it difficult for surgical removal procedures [35]. Overexpression of miRNA-21 in esophageal tumors was associated with positive lymph nodes and decreased survival of esophageal cancer patients [36]. Significant decrease in cell proliferation and invasion were observed when anti-miR- 21 was overexpressed in human esophageal squamous cell carcinoma cell lines (TE6, TE8, TE10, TE11, TE12, TE14, KYSE30) [36]. In addition, differential expression of miR-375 expression was inversely correlated with pathological stages, metastasis and disease free survival (DFS) of esophageal cancer patients. Importantly, expression of miR-375 was found to regulate genes associated with chemotherapy resistance [37]. Another miRNA miR-143 was found to be associated with tumor recurrence and invasion in esophageal cancer [38]. Liu et al documented the tumor suppressor role of cluster of two miRNAs, miR-143 and miR-145 in esophageal cancer and observed a significant association between risk of esophageal cancer development with silencing of these miRNAs [39]. Several lines of evidence have established that 5q region of the genomic DNA is more frequently lost or deleted in esophageal cancer and human miR-145 and miR-143 were predicted to be associated with 5q33 genomic region [40]. Taken together, miR-145 and miR-143 might be candidate miRNAs for better prognosis of esophageal cancer [39].
Gastric Cancer
Despite three decades of progress in cancer treatment, gastric cancer (GC) remains a significant health problem and second leading cause of death worldwide [27]. Gastric cancer incidence varies dramatically with geographic locations with highest in Eastern Asia (Japan), Eastern Europe, South America and central Asia (Middle East) and lowest in North America [28]. In the USA, GC is the second most common cancer among the GI malignancies with approximately 21,600 new cases and 10,990 deaths estimated to occur in 2013 [27] The asymptomatic and late onset of gastric cancer practically nullifies medical interventions at the diagnostic stage. However, sequential evolution of histological changes from normal gastric epithelium to gastritis to gastric metaplasia to dysplasia to carcinoma provides a window of opportunity for new detection strategies and newer biomarkers to prevent GC advancement [41]. Therefore, there is a critical need for an early diagnosis based on established markers and alterations in signaling pathways that predispose the gastric epithelial cells for neoplastic transformation.
The involvement of oncogenic and tumor suppressor miRNA in GC is well established [42]. Gastric juice miRNAs are promising biomarkers to detect GC at an earlier stage and several studies have reported the presence of miR-200c, miR-150, miR-342 and miR-378 in the gastric juice as potential candidates for screening gastric cancer patients at earlier stage [43, 44]. Another study using gastric juice from patients with GC, gastric ulcers, gastritis and normal healthy subjects demonstrated significantly lower expression of miR-129-1-3p and miR-129-2-3p in GC patients as comparison with healthy controls [45]. In addition to gastric juice, recently Li et al., reported plasma miRNA-199a-3p as a potential diagnostic marker for early stage GC [46]. A serum based genome wide analysis of patients with GC and healthy controls indicated a unique miRNA signature (miR-1, miR-20a, miR-27a, miR-34 and miR-423-5p) as a novel diagnostic marker for GC [47]. Validation of several miRNAs in a cohort of 60 GC patient and 60 healthy serum samples identified over expression of miR-21 and miR-223 in patients with Helicobacter pylori (H. pylori) infection as compared with GC patients without H.pylori infection [48]. Thus, H. pylori infection which is one of the risk factors for GC can also influence the miRNA expression profile in the patient serum. In addition, serum miR-21, miR-146a and miR-148a were also recognized not only as promising biomarkers but are also useful in prediction of lymph node metastasis of GC [49].
In addition to their utility as diagnostic biomarkers, miRNAs can also be used as clinical outcome predictors. Teng et al reported the over expression Lin28 as predictive indicator for locally advanced GC patients undergoing neoadjuvant chemotherapy [50]. In addition, down regulation of miR-206 or overexpression miR-200c are associated with poor prognosis of GC patients [44, 51]. Lower expression of miR-125a-5p is also regarded as independent prognostic indicator for GC [52].
Hepatocellular Carcinoma
Hepatocellular carcinoma (HCC) is the lethal malignancy and the 3rd leading cause of cancer-related deaths worldwide [53, 54]. HCC has extremely poor prognosis with five year survival rate of less than 9% [55]. Therefore, exploration of unexplored molecular mechanisms of HCC will help us to understand its pathogenesis, identify early diagnostic, prognostic markers and therapeutic targets for this lethal cancer.
Unique miRNA expression profiles have been associated with the development, progression and therapy response of hepatocellular carcinoma [56]. Expression profiling of miRNAs in 24 HCC patient tissue samples identified significant upregulation of miR-18, and miR-224 and downregulation of miR-199a*, miR-199a, miR-195, miR-200a and miR-125a compared to their adjacent normal tissues [57]. Several other reports also documented significant down regulation of miR-139, miR-145, miR-214, miR-21, 221, 222, 34a miR-148a, miR-124a-2 and Let-7c and upregulation of miR-9, miR-21, miR-25, miR-137, miR-151, miR-155, miR-182, miR-183, miR-338, miR-219-1, miR-207, miR-185, miR-186 miR-216, miR-221, miR-222, miR-224 and miR-301 [58–60] in HCC tissues and cell lines. A signature of 20 miRNAs was found to differentiate primary HCC from non-metastatic tumors [58]; whereas a set of 40 miRNAs precisely differentiated HCC from cirrhotic tissues. A significant association of miR-222 overexpression with advanced stage and shorter disease-free survival of HCC patients was also observed [61]. Multivariate analysis of upregulated set of miRNAs (miR-155, miR-15a & b, miR-432, miR-486-3p and miR-30b) also showed a significant association with recurrence-free survival in HCC patients [62]. Similarly, expression of miR-15b negatively correlated with risk of HCC recurrence [63]. In yet another study, miR-29c and miR-140-5p were found to be downregulated in HCC patients and associated with poor prognosis [64, 65], suggesting their tumor suppressor role in HCC and indicating their utility as valuable biomarkers for prognosis of HCC. Furthermore, down regulation of miR-140-5p in HCC tissue samples was associated with multiple nodules, differentiation, capsular formation, vein invasion, and shorter overall and disease-free survival of HCC patients [65]. Murakami et al reported over expression of miR-18b in HCC patients which associated with shorter relapse-free survival [66]. Furthermore, significant over expression of miR-18b in poorly differentiated HCC was observed [66]. Abnormal expression of miRNAs has also been associated with drug resistance resulting in poor prognosis and reduced patient survival [67, 68]. In addition to their diagnostic and prognostic relevance, expression of miRNAs like miR-26 in HCC patients have been used as an indicator for responsiveness to Interferon (IFN) treatment [56, 59].
Gallbladder Cancer
Gallbladder cancer (GBC) is the 5th most common gastrointestinal cancer [69] with expected 6000 new cases in 2013 as per the estimates of American Cancer Society. Due to late stage diagnosis, extensive hepatic, lymph-node and vascular metastases, GBC is associated with extremely poor prognosis with overall five-year survival rate of <5% [69, 70]. Chronic inflammation of the gallbladder (Cholecystitis) and gallbladder calcification (Cholelithiasis) are usually considered as conspicuous causative precursors of GBC [71]. Over the years advances have been made to improve the diagnostic and therapeutic modalities (radiation and conventional chemotherapy) to target GBC, yet the prognostic scenario of this lethal malignancy remains as grim as ever [72]. While distant metastases often preclude surgical resection, the extremely high recurrence rates even after complete resection lead to marginal improvement in the overall survival [69]. This necessitates urgent efforts for the identification of reliable diagnostic, prognostic tumor markers and novel cellular targets for curative therapeutic approaches. So far the role of miRNAs in GBC is not very well documented. However, recently a significant overexpression of miR-155 was reported in GBC when compared to normal gall bladder and pancreatobiliary maljunction cases [70]. Up-regulation of miR-155 in GBC cases correlated with poor prognosis of the GBC patients compared to the patients with low miR-155 expression [70]. Further, the study showed the involvement of this aberrantly expressed miRNA in proliferation, aggressive behavior with increased lymph node metastasis and vessel invasion [70]. This study supports the potential of miR-155 as a molecular target in GBC therapy particularly for inhibition of lymph node metastasis and as a prognostic biomarker for GBCs [70]. Moreover, the utility of increased miR-155 expression for distinguishing GBC from benign diseases of gall bladder such as Xanthogranulomatous cholecystitis was also suggested [70]. GBC-specific upregulation with no effect on expression during inflammatory conditions associated with pancreatobiliary maljunction and possibility of its stable detection in serum and/or bile mark it as a prospective novel diagnostic marker for GBC [70]. Molecular studies and clinicopathological analysis of GBCs revealed strong association of aberrantly over-expressed miR-20a with growth, distant metastasis and poor prognosis [73]. It was demonstrated that expression of miR-20a is induced by increased levels of TGF-β1 in GBC that leads to epithelial to mesenchymal transition (EMT) mediated through inhibition of Smad7 transcription and activation of β-catenin signaling pathway [73]. In addition to the above mentioned miRNAs with oncogenic function, Jin et al. have demonstrated that, miR-34a acts as a tumor suppressor in GBC and decreased expression of miR-34a was associated with poor prognosis of GBC patients [74]. Overexpression of miR-34a resulted in inhibition of colony forming ability of CD44+CD133+ GBC tumor stem like cells in vitro. Furthermore, overexpression of miR-34a resulted in reduced xenograft tumors [74].
Various single nucleotide polymorphisms (SNPs) in miRNA genes lead to variations in miRNA expression resulting in diverse functions and, therefore, may represent ideal candidate biomarkers for cancer prognosis. A recent North-India population-based case-control study of GBC explored the function of common single nucleotide polymorphisms (SNPs) in pre-microRNA genes as potential biomarkers for GBC susceptibility [75]. Patients carrying more than two variant alleles due to hsa-miR-146a G>C polymorphism (rs2910164), hsa-mir-196a2 C>T (rs11614913) polymorphism and hsa-mir-499 T>C (rs3746444) polymorphism were associated with increased overall risk of developing GBC with borderline significance. This risk was further accentuated in patients with gallstone disease [75].
Pancreatic Cancer
Pancreatic cancer (PC) is the 4th leading cause of cancer related deaths in the USA with less than 6% five-year survival [27]. Despite extensive efforts in understanding the molecular pathology, PC remains one of the most aggressive forms of cancer due to insufficient knowledge of PC pathogenesis, resistance to conventional therapy, the lack of early diagnostic and predictive markers. The other reason being the late clinical presentation of the disease; approximately 80% of newly diagnosed patients have cancers already metastasized to adjacent organs, for which no curative therapy is currently available. Therefore, better management of PC necessitates the discovery of early diagnostic and predictive markers.
Several studies over the past decade have revealed differential expression of miRNAs in normal pancreas, pre-malignant lesions (PanINs) and pancreatic ductal adenocarcinoma (PDAC) and therefore support diagnostic and prognostic utility of miRNAs [76]. Several miRNA expression profiling studies have revealed PDAC-specific overexpression of miRNAs (miR-376a, miR-301, miR-132 and miR-212) as compared to normal or benign adjacent pancreas [77, 78]. However, Zhang S et al demonstrated downregulation of miR-132 in cancer compared to normal and benign tissues [79]. A recent study has reported the downregulation of KRAS targeting miR-96 in PC as compared to normal tissues and demonstrated that the overexpression of miR-96 in PC cells resulted in inhibition of cell proliferation, invasion, induced apoptosis and reduced tumor growth [80]. Ikenaga et al. reported overexpression of miR-203 in PC compared to normal pancreas and chronic pancreatitis; and found it to be an independent predictor of poor prognosis in cases with no residual tumors [81]. Expression of miR-216 and 217 and the lack of miR-133a was found to be the only characteristic of the normal pancreatic tissues [82]. Several studies have documented the lower expression of let-7 in PC [83, 84]. A miRNA microarray analysis revealed differential expression of 25 (21 upregulated and 4 downregulated) miRNA distinguishing PDAC from chronic pancreatitis and normal pancreas [82]. Among the 25 upregulated miRNAs, 6 miRNAs namely miR30a-3p, miR-105, miR-127, miR-187, miR-452, and miR-518a-2 were found to be better prognostic indicators in patients with lymph node positive disease. A similar study indicated that a subset of 20 miRNAs could discriminate between PDAC, chronic pancreatitis and normal pancreas [85]. Whereas this study observed overexpression of miR-31, miR-145, and miR-146a in PC cases, another study by Papaconstantinou et al, reported downregulation of these miRNAs in PC samples compared to normal pancreas [86]. One possible explanation for this discrepancy might be that the latter study used micro dissected PDAC tissues, whereas the former used the whole PC tissue and most of the time, PC tissues consists 90% of desmoplasia. A similar study by Schultz et al., reported 32 differentially expressed miRNAs between PDAC and chronic pancreatitis, among which miR-148a, miR-196b, miR-196a and miR-205 were already found deregulated in previous other studies [87]. In addition to these studies, over expression of miR-103 and miR-107, and decreased expression of miR-155 were useful discriminators of tumors from the normal pancreas [88]. Munding et al., identified 78 deregulated miRNAs and out of these only miR-135b performed best in discriminating PDAC from Chronic pancreatitis [89]. miR-21 over expression has been shown in 79% of PDACs and correlated with worse prognosis of the patients [90]. However, no significant correlation of miRNA-21 overexpression was observed with tumor size, differentiation, nodal status or patient T-stage [91]. Another study recently showed upregulation of miR-21, miR-155, miR-210, miR-221 and miR-222 and downregulation of miR-31, miR-122, miR-145 and miR-146a in PCs compared with normal samples of cancer-free individuals [86].
Traditionally, diagnosis of PC is most often done by tissue biopsies and ultrasound guided fine needle aspiration. However, detection of serum and plasma miRNAs can be used as a non-invasive method to determine the progression and extent of metastasis. To this end, Wang et al studied the plasma miRNA expression profile of four miRNAs (miR-21, miR-210, miR-155, and miR-196a) in PC patients and observed the differential upregulation of miR-155 and miR-196a, distinguished PC patients from normal healthy controls with a sensitivity of 64% and a specificity of 89%, suggesting that plasma miRNA profiling can be a sensitive and specific blood-based biomarker assay for PC [92]. The expression of miR-18a was also significantly higher in PC samples compared to healthy controls [93]. Similarly, differential upregulation of miR-2 has also been observed in the sera of PDAC patients compared to healthy controls [94]. In addition to plasma/serum, miR-196a and miR-217 are over expressed and detectable in the fine needle aspirates (FNA), and used for diagnosis of PDAC and in discriminating them from chronic pancreatitis [95]. Higher expression of miR-451 and miR-486-5p, and lower expression of let-7c, let-7d, let-7f, and miR-200c in the FNA samples of PC patients was also reported recently [83, 96].
Not only have miRNAs been used as diagnostic markers, they have also been analyzed as prognostic indicators for PC. The first such analysis was done by Bloomston et al., who compared the miRNA expression profile of lymph node positive pateints who succumbed to the disease within 2 years and those that survived beyond 2 years. The study reported the association of disease outcome with deregulation of miR-30a-3p, miR-105, miR-127, miR-187, miR-452, and miR-518a-2 [82]. The same study reported the association of poor survival with over expression of 2 miRNAs (miR-196a-2 and miR-219) [median, 14.3 months (95% confidence interval, 12.4–16.2) vs. 26.5 months (95% confidence interval, 23.4–29.6); P = 009] [82]. In addition, miR-155, miR-203, miR-210, miR-222, miR-203, miR-21 and miR-221 overexpression has also been significantly associated with increased risk of death compared to patients with reduced expression of these miRNAs [81, 96–98]. Using high-throughput Toray’s 3D Gene™-miRNA chip, Giovannetti et al., identified miRNA-211 as a single prognostic indicator in resected PDACs [99]. Recently, Schultz et al., observed a significant correlation between overexpression of 2 miRNAs (miR-212 and miR-675) and low expression of miR-148a, miR-187, and let-7g with shorter overall survival (OS) [87, 96, 98]. After calculating prognostic index (PI) for these five miRNAs, the median survival was observed to be 1.09 years [95% Confidence Interval (CI), 0.98–1.43] for PI > median PI compared to 2.23 years (CI 1.84–4.36) for PI < median. This association was independent of various clinicopathological characteristics of the patient like age, gender, KRAS mutation status, tumor stage, American Society of Anesthesiologists (ASA) score, and differentiation of tumor. Using a cohort of 48 PDAC patient samples, high expression of miR-21 and down regulation of miR-30d was also associated with overall poor survival [100]. The miR-200 and Let-7 downregulation has also been inversely associated with worsening tumor and patient survival in other studies [83, 83, 96, 96, 98]. From the Let-7 family, in particular a significant downregulation of Let-7b has been observed in human PC cell lines and tissue and associated with PC pathogenesis, apoptosis, and cell growth by targeting several target genes, thereby functioning as a tumor suppressor [83, 98].
Intestinal cancers
A recent study examined the miRNA profile in a cohort of well differentiated neuroendocrine small intestinal tumors, and found that five miRNAs were up-regulated (miR-96, -182, -183, -196a and miRNA-200a), while four were down-regulated (miR-31, -129-5p, -133a, -215) [101]. Since these malignancies are very rare, much needs to be unraveled regarding the precise role of miRNAs in the progression of this cancer.
Despite being the second leading cause of cancer related deaths in the United States [27], most early detection methods for colorectal cancer (CRC) are invasive and/or expensive. The miRNA profile is altered in the tumor tissues, serum as well as in feces of individuals with CRC and several studies have indicated that these deregulated miRNAs could potentially be used both as diagnostic and prognostic indicators of the disease. Several miRNAs including miR-429, miR-17-3p, miR-29a, miR-221, miR-92a and miR-21 have been characterized as possible prognostic markers of CRC [102–106], while miR-150 and miR-126 are being proposed as predictive markers of response to chemotherapy [107]. More specifically, over expression of miR-21 in CRC has been associated with: poor response to adjuvant therapy and more rapid recurrence in stage III patients [108], development of distant metastasis [109], reduced disease-free interval [110], reduced disease free survival in stage II colon cancer patients [111] and worse overall survival [112]. miR-31 was upregulated in CRC and associated with advanced stage and/or worse outcomes [113–115]. In addition, overexpression of miR-141, miR-200c [116] and miR-203 [117, 118] in tumors predicted poor prognosis, either alone or in combination with other miRNAs [119].
Twenty miRNAs were found to be altered in the serum of CRC patients [120]. Based on the miRNA profile prevalent in stage I–II and stage IV patients, it was possible to distinguish CRC patients from a control group [120]. Another study used Real Time PCR to analyze miRNA expression in both CRC tissue and adjacent normal tissue and a panel of colorectal cancer cell lines. It was observed that 13 miRNAs were present at significantly different levels, the most dramatically downregulated were miR-133b and miR-145, while miR-31 (which was also associated with the stage of the cancer), miR-96, miR-135b and miR-183, were significantly up-regulated [121]. Further, higher expression of miR-720 has been reported in CRC tissue compared to normal colon epithelial tissues [121, 122]. Faltejskova et al., independently observed significant downregulation of miR-215, miR-375, miR-378 and miR-422a, and upregulation of miR-135b in CRC tumor tissues compared to adjacent normal tissues [123]. A significant correlation of miR-215 and miR-422a expression with clinical stage was observed [123]. A recent study by Zhang et al., identified 35 differentially expressed miRNAs in stage II colon cancer patients compared to adjacent normal mucosa [124]. Cluster of six miRNAs including miR-21-5p, miR-20a-5p, miR-103a-3p, miR-106b-5p, miR-143-5p, and miR-215 successfully categorized patients into high-risk and low-risk groups with large differences in 5-year disease-free survival. Yet another study looked at miRNA levels in the fecal samples obtained from both normal volunteers and individuals with colon adenomas and CRC. It was found that miR-21 and miR-106a were elevated in fecal samples from CRC and adenoma patients [125]. It was also demonstrated that certain miRNAs can have prognostic value. For instance, miR-29c expression was found to be decreased in UICC stage II and II patients that have an early relapse in comparison with those exhibiting no early relapse [126]. Further, miR-29c was found to inhibit proliferation and migration as well as cause an accumulation of the G1 and G2 population. Serum miR-29c was observed in these early relapse patients, thus indicating a possible role for this miRNA as a prognostic tool [126]. As in other malignancies, the pattern of expression of miRNAs has been shown to vary during the progression of colon cancer. A panel of miRNAs was shown to contribute to tumor development. The miRNAs miR-10b, miR-210 and miR-708 were found to distinguish primary colon cancer and metastases [127]. The same study identified alterations in a set of interconnected miRNA regulatory networks that governed various cellular processes, including epithelial to mesenchymal transition (EMT). The miRNA miR-10b was found to be linked to patient survival when expressed in metastases [127]. A recent study by Shivapurkar et al using serum samples from CRC patients reported significant downregulation of miR-103 and upregulation of miR-596 [128]. Furthermore, the same study reported down-regulated miR-103 in blood samples from CRC patients with disease recurrence compared to recurrence-free patients.
Individual miRNAs have been found to affect disease progression, lending credence to the notion that targeting these miRNAs could be a viable therapeutic strategy in the future. The miRNAs miR-143 and miR-145 were found to act in an opposing fashion, either inhibiting or aiding disease progression [129]. Both miRNAs were found to have a nearly mutually exclusive target gene set. The miR-143 miRNA was found to target DNA methyltrasferase 3A in CRC, thereby acting as a tumor suppressor miRNA. Conversely, miR-145 was found to be an oncogenic miRNA, targeting mRNAs involved in cell cycle (MYC and CCND2) and neuregulin (MYC and ADAM17) pathways [129]. The miRNA miR-133a was found to be a critical tumor suppressor miRNA, acting in part by regulating levels of the p53 protein [130], by targeting the 3’ UTR of the ring finger and FYVE-like domain containing E3 ubiquitin ligase protein (RFFL). This miRNA was found to be down-regulated in primary colon cancer in comparison to surrounding normal tissue.
The KRAS and TP53 mutations are present in a significant proportion of CRC patients (5–20%), and a recent study has indicated that the miRNA let-7a regulates KRAS through TP53 in the KRAS mutant HCT116 cells [131]. miRNAs have been found to up-regulate other proteins aiding in CRC progression, such as Fascin1 (FSCN1), which is regulated by the miR-451 via the inhibition of AMPK1 and subsequent activation of mTORC1 [132]. Likewise, the important cell cycle regulatory molecule, CDC25A, was found to be regulated by miR-21 [133]. The reduced levels of the miR-224 and the passenger strand of miR-221 were found to reduce the levels of the metastasis suppressor Maspin, thereby promoting the metastasis of CRC [134]. It has also been suggested that the loss of DICER1 function (involved in miRNA biogenesis) imparts cancer stem cell-like properties to CRC cells, that impacts the biogenesis of several critical miRNAs such as miR-34a and miR-126 [135].
Conclusions and future perspectives
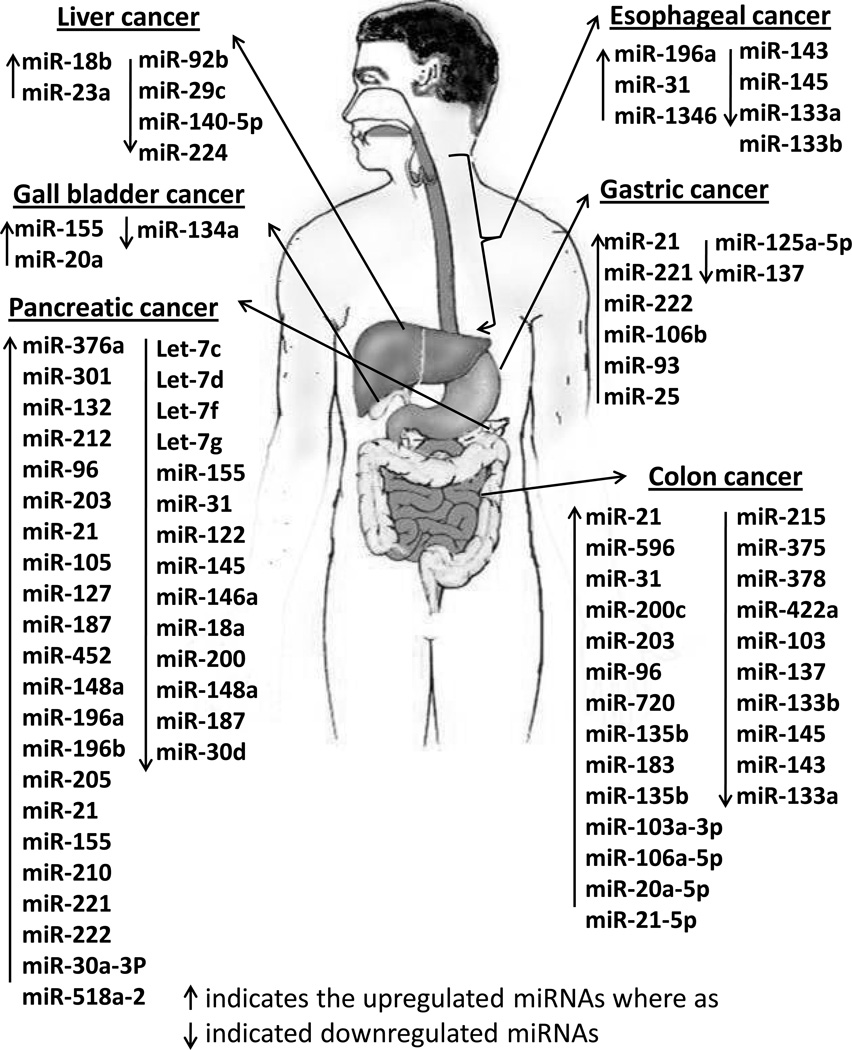
miRNAs play an intrinsic role in the cancer progression, from the initiation of the disease (by regulating levels of critical genes such as KRAS and p53) to the advanced metastatic stages of the malignancy. A complex, finely tuned network of miRNAs govern crucial processes such as EMT. The level of miRNAs in feces and serum reflect changes in miRNA expression patterns in the tumor tisses and may be exploited as non-invasive diagnostic and prognostic tools. Given that altering the level of a single miRNA can trigger a cascade of signaling events culminating in a generalized increase/decrease in proliferation, apoptosis, cell growth etc. effective miRNA targeting strategies could yield dramatic results, potentially altering the course of the disease. However, current miRNA targeting technology is hampered by several practical difficulties, primarily, inefficient targeting to the disease-affected organ. Should these significant obstacles be overcome, miRNAs could well be used as prognostic and diagnostic tools as well as viable therapeutic targets. Overall, differentially expressed miRNAs in GI cancers and GI tract associated organs are summarized in Table 1 and Table 2 respectively. These differentially expressed miRNAs are also shown in Figure 1.
Table 1.
Differentially expressed miRNAs in gastrointestinal (GI) tract cancers.
| miRNAs | Type of Cancer | Status | Identified miRNA Targets | Ref. |
|---|---|---|---|---|
| miR-205 | Esophageal cancer | Up | ZEB1 and ZEB2 | [34] |
| miR-31 | Esophageal cancer | Up | KSR2, EMP1 RGS4 | [35] |
| miR-21 | Esophageal cancer | Up | cdc25A, PTEN | [33–34, 37] |
| miR-143, miR-145, miR-133a, miR-133b | Esophageal cancer | Down | FSCN1 | [33–34, 40] |
| miR-375 | Esophageal cancer | Down | JAK2 and c-MYC | [34] |
| miR-143 | Esophageal cancer | Down | DNMT3A | [40] |
| miR-21 and miR-20a | Gastric cancer | Up | PTEN | [36, 40, 46] |
| miR-129-1-3p, miR-129-2-3p | Gastric cancer | Down | CP110, TOCA1, ABLIM1,SOX2 | [46] |
| miR-199a-3p | Gastric cancer | Up | CD44 | [47] |
| miR-223 | Gastric cancer | Up | SH2B3, CDR2, CARM1 | [40] |
| miR-146a | Gastric cancer | Up | SOCS1 | [50] |
| miR1-148a | Gastric cancer | Up | DNMT3B | [50] |
| miR-206 | Gastric cancer | Down | BCL2, BDNF | [44–45, 52] |
| miR-200c | Gastric cancer | Up | TrkB | [44–45, 52] |
| miR-21 | Colon cancer | Up | cdc25A, PTEN, E2F, MYC, NFKB, β-Catenin | [108–112,125] |
| miR-596 | Colon cancer | Up | LGALS3BP | [128] |
| miR-31 | Colon cancer | Up | KSR2, EMP1 RGS4 | [113–115] |
| miR-200c | Colon cancer | Up | ZEB, CDH1, PLS1, LSR, EPS8L2 | [116] |
| miR-203 | Colon cancer | Up | p63, SOCS3), c-jun (AP-1) | [106,118] |
| miR-96 | Colon cancer | Up | FOXA3a, FOXO1, RAD51, REV1 | [121] |
| miR-720 | Colon cancer | Up | TWIST | [121–122] |
| miR-135b | Colon cancer | Up | LATS2, β-TrCP, NDR2 and LZTS1 | [121,123] |
| miR-106b-5p | Colon cancer | Up | RBL1 RBL2 and CASP8 | [124] |
| miR-133b | Colon cancer | Down | (MCL-1), BCL2L2, c-Met FSCN1 | [121] |
| miR-143 | Colon cancer | Down | DNMT3A | [121] |
| miR-215 | Colon cancer | Down | MDM2, TYMS and SIP1/ZEB2 | [123] |
| miR-378 | Colon cancer | Down | IGF1R | [123] |
| miR-442a | Colon cancer | Down | CYP7A1 | [123] |
| miR-29c | Colon cancer | Down | SIRT1 | [126] |
| miR-103 | Colon cancer | Down | DAPK and KLF4 | [128] |
| miR-145 | Colon cancer | Down | MYC, CCND2, ADAM17 | [125] |
| miR-375 | Colon cancer | Down | p53 | [123] |
Table 2.
Differentially expressed miRNAs in cancers of GI tract associated organs.
| miRNAs | Type of Cancer | Status | Identified miRNA Targets | Ref. |
|---|---|---|---|---|
| miR-155 | Gall bladder Cancer | Up | HDAC1, SHIP1 | [70] |
| miR-20a | Gall bladder Cancer | Up | BCL2, PTEN, STAT3, VEGFA, WEE1 | [73] |
| miR-34a | Gall bladder Cancer | Down | NOTCH1, CDC25, CDK6, Bcl2, HDAC1 | [74] |
| miR-18b | Hepatic Cancer | Up | TNRC6B | [67] |
| miR-23a | Hepatic Cancer | Up | TOP1 | [60] |
| miR-29c | Hepatic Cancer | Down | SIRT1 | [65] |
| miR-140-5p | Hepatic Cancer | Down | TGFBR1 and FGF9 | [66] |
| miR-224 | Hepatic Cancer | Down | PPP2R1B | [57,59] |
| miR-376a miR-301 miR-212 miR-96 miR-21 |
Pancreatic cancer | Up | CDK2, AGO2 FOXF2, BCC3, PTEN, Col2A1 Rb, PTCH1 FOXA3a, FOXO1, RAD51, REV1 PTEN, P53, NF-κB,TGFBR2, Tropomyosin 1 |
[78–79,81–82, 87, 120,100] |
| miR-105 miR-187 |
Pancreatic cancer | Up | TLR2, CDK6 DAB2 |
[83, 88, 97] |
| miR-148a miR-196b miR-196a miR-205 |
Pancreatic cancer | Up | DNMT3B, ROCK1 HOXA9/MEIS1, FAS ANXA1 PTEN, PHLPP2 |
[88] |
| miR-155 miR-210 miR-221 miR-222 |
Pancreatic cancer | Up | HDAC1, SHIP1 E2F3, EFNA3, GIT2, MNT, ZNF462, EGF3 E2F, MYC, NFKB, β-Catenin FOS, FOXO3, PTEN, TKB1 |
[87] |
| miRNA-211 | Pancreatic cancer | Up | CHOP/GADD153 | [99] |
| miR-196a | Pancreatic cancer | Up | HOXA9/MEIS1, FAS | [93] |
| miR-18a | Pancreatic cancer | Down | KRAS, PIAS3 | [94] |
| Let-7b, let-7c, let-7d and let-7f miR-200c |
Pancreatic cancer | Down | KRAS, NRAS ZEB, CDH1, PLS1, LSR, EPS8L2 |
[84, 97, 100–101] |
| let-7g | Pancreatic cancer | Down | Collagen type I α2 | [88, 97] |
| miR-30d | Pancreatic cancer | Down | EZH2, SMAD1, BECN1 | [100] |
| Let-7 miR-155 |
Pancreatic cancer | Down | RAS, HMG2A, cdc34 APC, FOXO3, MECP2, RUNX2, SMAD1 |
[84, 85, 89] |
| miR-122 miR-145 miR-146a |
Pancreatic cancer | Down | CUX1, CAT1 SOX9, ADAD17 TRAF6 |
[87] |
| miR-200 | Pancreatic cancer | Down | FAP | [84, 97] |
Figure 1.
Important differentially expressed miRNAs in GI malignancies and GI tract-associated organs. These include cancers from esophagus, stomach liver, pancreas, colon, and gall bladder. Upregulated miRNAs are shown in red color whereas the down regulated miRNAs are shown in black color.
Despite significant advances in diagnosis and treatment, morbidity and mortality rates of GI malignancies have remained unchanged over the past decade. The poor diagnostic and prognostic performance of currently used biomarkers mandates identification and evaluation of novel biomarkers with increased sensitivity and specificity. Since their discovery, miRNAs have been assigned important roles and act as master regulators of various physiological processes like metabolism, cell differentiation, stress etc. However, miRNA deregulation contributes to tumorigenesis by augmenting several cellular processes like cell proliferation, migration and invasion. Not only miRNAs have the potential to serve as diagnostic and prognostic markers, they are also being considered as biomarkers for predicting the response to therapy in several malignancies. Their high stability in circulation and formalin fixed tissue samples and relatively inexpensive detection methods suitable for high-throughput analysis makes miRNAs promising candidates for ex vivo detection. Due to their tissue and disease stage specific expression, miRNA not only can identify the presence or absence of tumors, they can also determine the primary organ or tissue affected. In addition they can also identify the clinical and pathological stage of the disease. Furthermore, miRNAs can predict risk of progression, relapse, and metastasis, and help to evaluate possible clinical scenarios in relation to the therapy response.
Even though miRNAs have been used as diagnostic and prognostic markers, we still have only a nascent understanding of the role of miRNAs in cancer in general and still face challenges in the research field of tumor-related miRNAs. Use of different methodologies in the miRNA profiling studies have limited the comparability of data and makes it difficult to identify a consistent miRNA signature for diagnosis and prognosis of GI cancers. However, the rapid pace of improvement in miRNA expression profiling technologies and given that new miRNAs continue to be discovered it is likely that future studies will identify useful expression signatures that may be even more informative for predicting the behavior of GI cancers. Most published studies do not account for the confounding influence of various pathological factors such as hypoxia, infection and cytotoxic treatment [136]. Therefore, to improve the reliability, quality assurance and validity of miRNA signatures, it is recommended to use standardized and more stringent protocols for collecting and analyzing samples. This approach will lead to reduction in inter-study discrepancies and provide highly reproducible results. The clinical validity of these candidate miRNA signatures should be determined using large independent cohorts in multi-centric studies. In combination with the use of more robust platforms, more appropriate bio-computational and statistical analyses should be used to identify candidate miRNA signatures. In addition, use of miRNA signatures along with already established biomarkers for GI cancers may help to better predict the disease outcome on indidual patients. Presence of miRNAs in exosomes, microvesicles and/or apoptotic bodies can have different functions during the development of cancer. Much of the current literature describing serum/plasma based miRNA expression profiles does not describe the sub-types of circulating miRNAs, suggesting that future studies concerning circulating miRNAs for diagnostic and prognostic purposes should be focused on the type of circulating miRNAs present in body fluids.
Genetically engineered mouse (GEM) models have been designed to recapitulates histopathological alterations found in human cancers. Integrated human and mouse gene expression analysis of tumor have revealed important similarities [137]. Furthermore, similar miRNA expression profile between mouse p48-Cre/KrasG12D and human PC patients has been reported [138]. Therefore, similar future studies should also be designed using relevant GEM models of other GI cancers to identify pathologically important miRNAs.
Acknowledgments
This work was supported in part by the grants from National Institutes of Health (RO1 CA133774, EDRN UO1CA111294, SPORE P50 CA 127297, R21 CA156037, P20 RR021937 and U54 CA163120).
Footnotes
Conflicts of Interest: There are no potential conflicts of interest involved with this work.
References
- 1.Marusawa H, Jenkins BJ. Inflammation and gastrointestinal cancer: An overview. Cancer Lett. 2013;10 doi: 10.1016/j.canlet.2013.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Lumachi F, Brandes AA, Ermani M, et al. Sensitivity of serum tumor markers CEA and CA 15-3 in breast cancer recurrences and correlation with different prognostic factors. Anticancer Res. 2000;20:4751–4755. [PubMed] [Google Scholar]
- 3.Tricoli JV, Jacobson JW. MicroRNA: Potential for Cancer Detection, Diagnosis, and Prognosis. Cancer Res. 2007;67:4553–4555. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- 4.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol. 2008;18:103–110. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 8.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 9.Grady WM, Tewari M. The next thing in prognostic molecular markers: microRNA signatures of cancer. Gut. 2010;59:706–708. doi: 10.1136/gut.2009.200022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Jay C, Nemunaitis J, Chen P, et al. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 2007;26:293–300. doi: 10.1089/dna.2006.0554. [DOI] [PubMed] [Google Scholar]
- 12.Yu SL, Chen HY, Yang PC, et al. Unique MicroRNA signature and clinical outcome of cancers. DNA Cell Biol. 2007;26:283–292. doi: 10.1089/dna.2006.0555. [DOI] [PubMed] [Google Scholar]
- 13.Bullrich F, Fujii H, Calin G, et al. Characterization of the 13q14 tumor suppressor locus in CLL: identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res. 2001;61:6640–6648. [PubMed] [Google Scholar]
- 14.Park NJ, Zhou H, Elashoff D, et al. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanke M, Hoefig K, Merz H, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 16.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber JA, Baxter DH, Zhang S, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrie CH, Chi J, Taylor S, et al. Expression of microRNAs in diffuse large B cell lymphoma is associated with immunophenotype, survival and transformation from follicular lymphoma. J Cell Mol Med. 2009;13:1248–1260. doi: 10.1111/j.1582-4934.2008.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanco-Calvo M, Calvo L, Figueroa A, et al. Circulating microRNAs: molecular microsensors in gastrointestinal cancer. Sensors (Basel) 2012;12:9349–9362. doi: 10.3390/s120709349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 21.Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 22.Zernecke A, Bidzhekov K, Noels H, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turchinovich A, Weiz L, Langheinz A, et al. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci. 2012;37:460–465. doi: 10.1016/j.tibs.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 28.Lee JH. Interventional gastroenterology: esophageal and pancreatic cancers. Semin Oncol. 2005;32:25–28. doi: 10.1053/j.seminoncol.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 29.Agarwal A, Polineni R, Hussein Z, et al. Role of epigenetic alterations in the pathogenesis of Barrett's esophagus and esophageal adenocarcinoma. Int J Clin Exp Pathol. 2012;5:382–396. [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Gu J, Wang KK, et al. MicroRNA expression signatures in Barrett's esophagus and esophageal adenocarcinoma. Clin Cancer Res. 2009;15:5744–5752. doi: 10.1158/1078-0432.CCR-09-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wijnhoven BP, Hussey DJ, Watson DI, et al. MicroRNA profiling of Barrett's oesophagus and oesophageal adenocarcinoma. Br J Surg. 2010;97:853–861. doi: 10.1002/bjs.7000. [DOI] [PubMed] [Google Scholar]
- 32.Maru DM, Singh RR, Hannah C, et al. MicroRNA-196a is a potential marker of progression during Barrett's metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–1948. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X, Ajani JA, Gu J, et al. MicroRNA expression signatures during malignant progression from Barrett's esophagus to esophageal adenocarcinoma. Cancer Prev Res (Phila) 2013;6:196–205. doi: 10.1158/1940-6207.CAPR-12-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leidner RS, Ravi L, Leahy P, et al. The microRNAs, MiR-31 and MiR-375, as candidate markers in Barrett's esophageal carcinogenesis. Genes Chromosomes Cancer. 2012;51:473–479. doi: 10.1002/gcc.21934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim TJ, Kim HY, Lee KW, et al. Multimodality assessment of esophageal cancer: preoperative staging and monitoring of response to therapy. Radiographics. 2009;29:403–421. doi: 10.1148/rg.292085106. [DOI] [PubMed] [Google Scholar]
- 36.Hiyoshi Y, Kamohara H, Karashima R, et al. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res. 2009;15:1915–1922. doi: 10.1158/1078-0432.CCR-08-2545. [DOI] [PubMed] [Google Scholar]
- 37.Kong KL, Kwong DL, Chan TH, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 38.Mayne GC, Hussey DJ, Watson DI. MicroRNAs and esophageal cancer--implications for pathogenesis and therapy. Curr Pharm Des. 2013;19:1211–1226. doi: 10.2174/138161213804805702. [DOI] [PubMed] [Google Scholar]
- 39.Liu R, Liao J, Yang M, et al. The cluster of miR-143 and miR-145 affects the risk for esophageal squamous cell carcinoma through co-regulating fascin homolog 1. PLoS One. 2012;7:e33987. doi: 10.1371/journal.pone.0033987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pack SD, Karkera JD, Zhuang Z, et al. Molecular cytogenetic fingerprinting of esophageal squamous cell carcinoma by comparative genomic hybridization reveals a consistent pattern of chromosomal alterations. Genes Chromosomes Cancer. 1999;25:160–168. doi: 10.1002/(sici)1098-2264(199906)25:2<160::aid-gcc12>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 41.Carrasco G, Corvalan AH. Helicobacter pylori-Induced Chronic Gastritis and Assessing Risks for Gastric Cancer. Gastroenterol Res Pract. 2013;2013:393015. doi: 10.1155/2013/393015. Epub;%2013 Jul 29.: 393015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Link A, Kupcinskas J, Wex T, et al. Macro-role of microRNA in gastric cancer. Dig Dis. 2012;30:255–267. doi: 10.1159/000336919. [DOI] [PubMed] [Google Scholar]
- 43.Cai H, Yuan Y, Hao YF, et al. Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med Oncol. 2013;30:452–0452. doi: 10.1007/s12032-012-0452-0. [DOI] [PubMed] [Google Scholar]
- 44.Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med. 2012;10:186. doi: 10.1186/1479-5876-10-186. 186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X, Luo L, Wu Y, et al. Gastric juice miR-129 as a potential biomarker for screening gastric cancer. Med Oncol. 2013;30:365–0365. doi: 10.1007/s12032-012-0365-y. [DOI] [PubMed] [Google Scholar]
- 46.Li C, Li JF, Cai Q, et al. MiRNA-199A-3p: A potential circulating diagnostic biomarker for early gastric cancer. J Surg Oncol. 2013;108:89–92. doi: 10.1002/jso.23358. [DOI] [PubMed] [Google Scholar]
- 47.Liu R, Zhang C, Hu Z, et al. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784–791. doi: 10.1016/j.ejca.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 48.Li BS, Zhao YL, Guo G, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One. 2012;7:e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SY, Jeon TY, Choi CI, et al. Validation of Circulating miRNA Biomarkers for Predicting Lymph Node Metastasis in Gastric Cancer. J Mol Diagn. 2013;15:661–669. doi: 10.1016/j.jmoldx.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Teng RY, Zhou JC, Jiang ZN, et al. The relationship between Lin28 and the chemotherapy response of gastric cancer. Onco Targets Ther. 2013;6:1341–1345. doi: 10.2147/OTT.S45705. 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Q, Zhang C, Huang B, et al. Downregulation of microRNA-206 is a potent prognostic marker for patients with gastric cancer. Eur J Gastroenterol Hepatol. 2013;25:953–957. doi: 10.1097/MEG.0b013e32835ed691. [DOI] [PubMed] [Google Scholar]
- 52.Nishida N, Mimori K, Fabbri M, et al. MicroRNA-125A-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17:2725–2733. doi: 10.1158/1078-0432.CCR-10-2132. [DOI] [PubMed] [Google Scholar]
- 53.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics. 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 54.Schutte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma--epidemiological trends and risk factors. Dig Dis. 2009;27:80–92. doi: 10.1159/000218339. [DOI] [PubMed] [Google Scholar]
- 55.Sherman M. Hepatocellular carcinoma: epidemiology, risk factors, and screening. Semin Liver Dis. 2005;25:143–154. doi: 10.1055/s-2005-871194. [DOI] [PubMed] [Google Scholar]
- 56.Ji J, Shi J, Budhu A, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami Y, Yasuda T, Saigo K, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. %20. [DOI] [PubMed] [Google Scholar]
- 58.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y, Lee AT, Ma JZ, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitoR-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 60.Pineau P, Volinia S, McJunkin K, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong QW, Ching AK, Chan AW, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16:867–875. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 62.Huang YH, Lin KH, Chen HC, et al. Identification of postoperative prognostic microRNA predictors in hepatocellular carcinoma. PLoS One. 2012;7:e37188. doi: 10.1371/journal.pone.0037188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chung GE, Yoon JH, Myung SJ, et al. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection of hepatocellular carcinoma. Oncol Rep. 2010;23:113–119. [PubMed] [Google Scholar]
- 64.Bae HJ, Noh JH, Kim JK, et al. MicroRNA-29c functions as a tumor suppressor by direct targeting oncogenic SIRT1 in hepatocellular carcinoma. Oncogene. 2013;10 doi: 10.1038/onc.2013.216. [DOI] [PubMed] [Google Scholar]
- 65.Yang H, Fang F, Chang R, et al. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor beta receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205–217. doi: 10.1002/hep.26315. [DOI] [PubMed] [Google Scholar]
- 66.Murakami Y, Tamori A, Itami S, et al. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer. 2013;13:99–13. doi: 10.1186/1471-2407-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Takahashi S, Tasaka A, et al. Involvement of microRNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:565–575. doi: 10.1111/j.1440-1746.2012.07271.x. [DOI] [PubMed] [Google Scholar]
- 68.Zhuo L, Liu J, Wang B, et al. Differential miRNA expression profiles in hepatocellular carcinoma cells and drug-resistant sublines. Oncol Rep. 2013;29:555–562. doi: 10.3892/or.2012.2155. [DOI] [PubMed] [Google Scholar]
- 69.Lai CH, Lau WY. Gallbladder cancer--a comprehensive review. Surgeon. 2008;6:101–110. doi: 10.1016/s1479-666x(08)80073-x. [DOI] [PubMed] [Google Scholar]
- 70.Kono H, Nakamura M, Ohtsuka T, et al. High expression of microRNA-155 is associated with the aggressive malignant behavior of gallbladder carcinoma. Oncol Rep. 2013;30:17–24. doi: 10.3892/or.2013.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wistuba II, Gazdar AF. Gallbladder cancer: lessons from a rare tumour. Nat Rev Cancer. 2004;4:695–706. doi: 10.1038/nrc1429. [DOI] [PubMed] [Google Scholar]
- 72.Marsh RW, Alonzo M, Bajaj S, et al. Comprehensive review of the diagnosis and treatment of biliary tract cancer 2012. Part I: diagnosis-clinical staging and pathology. J Surg Oncol. 2012;106:332–338. doi: 10.1002/jso.23028. [DOI] [PubMed] [Google Scholar]
- 73.Chang Y, Liu C, Yang J, et al. miR-20a triggers metastasis of gallbladder carcinoma. J Hepatol. 2013;59:518–527. doi: 10.1016/j.jhep.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 74.Jin K, Xiang Y, Tang J, et al. miR-34 is associated with poor prognosis of patients with gallbladder cancer through regulating telomere length in tumor stem cells. Tumour Biol. 2013 doi: 10.1007/s13277-013-1207-z. [DOI] [PubMed] [Google Scholar]
- 75.Srivastava K, Srivastava A, Mittal B. Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J Hum Genet. 2010;55:495–499. doi: 10.1038/jhg.2010.54. [DOI] [PubMed] [Google Scholar]
- 76.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 77.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park JK, Henry JC, Jiang J, et al. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun. 2011;406:518–523. doi: 10.1016/j.bbrc.2011.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang S, Hao J, Xie F, et al. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- 80.Yu S, Lu Z, Liu C, et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 2010;70:6015–6025. doi: 10.1158/0008-5472.CAN-09-4531. [DOI] [PubMed] [Google Scholar]
- 81.Ikenaga N, Ohuchida K, Mizumoto K, et al. MicroRNA-203 expression as a new prognostic marker of pancreatic adenocarcinoma. Ann Surg Oncol. 2010;17:3120–3128. doi: 10.1245/s10434-010-1188-8. [DOI] [PubMed] [Google Scholar]
- 82.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 83.Torrisani J, Bournet B, du Rieu MC, et al. let-7 MicroRNA transfer in pancreatic cancer-derived cells inhibits in vitro cell proliferation but fails to alter tumor progression. Hum Gene Ther. 2009;20:831–844. doi: 10.1089/hum.2008.134. [DOI] [PubMed] [Google Scholar]
- 84.Watanabe S, Ueda Y, Akaboshi S, et al. HMGA2 maintains oncogenic RAS-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am J Pathol. 2009;174:854–868. doi: 10.2353/ajpath.2009.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schultz NA, Werner J, Willenbrock H, et al. MicroRNA expression profiles associated with pancreatic adenocarcinoma and ampullary adenocarcinoma. Mod Pathol. 2012;25:1609–1622. doi: 10.1038/modpathol.2012.122. [DOI] [PubMed] [Google Scholar]
- 86.Papaconstantinou IG, Manta A, Gazouli M, et al. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013;42:67–71. doi: 10.1097/MPA.0b013e3182592ba7. [DOI] [PubMed] [Google Scholar]
- 87.Schultz NA, Andersen KK, Roslind A, et al. Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer--five microRNAs in a prognostic index. World J Surg. 2012;36:2699–2707. doi: 10.1007/s00268-012-1705-y. [DOI] [PubMed] [Google Scholar]
- 88.Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 89.Munding JB, Adai AT, Maghnouj A, et al. Global microRNA expression profiling of microdissected tissues identifies miR-135b as a novel biomarker for pancreatic ductal adenocarcinoma. Int J Cancer. 2012;131:E86–E95. doi: 10.1002/ijc.26466. [DOI] [PubMed] [Google Scholar]
- 90.Dillhoff M, Liu J, Frankel W, et al. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dillhoff M, Wojcik SE, Bloomston M. MicroRNAs in solid tumors. J Surg Res. 2009;154:349–354. doi: 10.1016/j.jss.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morimura R, Komatsu S, Ichikawa D, et al. Novel diagnostic value of circulating miR- 18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;105:1733–1740. doi: 10.1038/bjc.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ali S, Almhanna K, Chen W, et al. Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am J Transl Res. 2010;3:28–47. [PMC free article] [PubMed] [Google Scholar]
- 95.Matthaei H, Wylie D, Lloyd MB, et al. miRNA biomarkers in cyst fluid augment the diagnosis and management of pancreatic cysts. Clin Cancer Res. 2012;18:4713–4724. doi: 10.1158/1078-0432.CCR-12-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ali S, Saleh H, Sethi S, et al. MicroRNA profiling of diagnostic needle aspirates from patients with pancreatic cancer. Br J Cancer. 2012;107:1354–1360. doi: 10.1038/bjc.2012.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Greither T, Grochola LF, Udelnow A, et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 98.Li Y, VandenBoom TG, Kong D, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Giovannetti E, van d V, Funel N, et al. High-throughput microRNA (miRNAs) arrays unravel the prognostic role of MiR-211 in pancreatic cancer. PLoS One. 2012;7:e49145. doi: 10.1371/journal.pone.0049145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jamieson NB, Morran DC, Morton JP, et al. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin Cancer Res. 2012;18:534–545. doi: 10.1158/1078-0432.CCR-11-0679. [DOI] [PubMed] [Google Scholar]
- 101.Li SC, Essaghir A, Martijn C, et al. Global microRNA profiling of well-differentiated small intestinal neuroendocrine tumors. Mod Pathol. 2013;26:685–696. doi: 10.1038/modpathol.2012.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Corte H, Manceau G, Blons H, et al. MicroRNA and colorectal cancer. Dig Liver Dis. 2012;44:195–200. doi: 10.1016/j.dld.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 103.Luo X, Burwinkel B, Tao S, et al. MicroRNA signatures: novel biomarker for colorectal cancer? Cancer Epidemiol Biomarkers Prev. 2011;20:1272–1286. doi: 10.1158/1055-9965.EPI-11-0035. [DOI] [PubMed] [Google Scholar]
- 104.Madhavan D, Cuk K, Burwinkel B, et al. Cancer diagnosis and prognosis decoded by blood-based circulating microRNA signatures. Front Genet. 2013;4:116. doi: 10.3389/fgene.2013.00116. eCollection;%2013.: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mazeh H, Levy Y, Mizrahi I, et al. Differentiating benign from malignant thyroid nodules using micro ribonucleic acid amplification in residual cells obtained by fine needle aspiration biopsy. J Surg Res. 2013;180:216–221. doi: 10.1016/j.jss.2012.04.051. [DOI] [PubMed] [Google Scholar]
- 106.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma Y, Zhang P, Wang F, et al. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 2012;3:1291. doi: 10.1038/ncomms2276. 1291. [DOI] [PubMed] [Google Scholar]
- 108.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 110.Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet. 2010;200:154–160. doi: 10.1016/j.cancergencyto.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 111.Nielsen BS, Jorgensen S, Fog JU, et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011;28:27–38. doi: 10.1007/s10585-010-9355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shibuya H, Iinuma H, Shimada R, et al. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–320. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 113.Chang KH, Miller N, Kheirelseid EA, et al. MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis. 2011;26:1415–1422. doi: 10.1007/s00384-011-1279-4. [DOI] [PubMed] [Google Scholar]
- 114.Schee K, Boye K, Abrahamsen TW, et al. Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer. 2012;12:505. doi: 10.1186/1471-2407-12-505. 505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang CJ, Stratmann J, Zhou ZG, et al. Suppression of microRNA-31 increases sensitivity to 5-FU at an early stage, and affects cell migration and invasion in HCT-116 colon cancer cells. BMC Cancer. 2010;10:616. doi: 10.1186/1471-2407-10-616. 616-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xi Y, Formentini A, Chien M, et al. Prognostic Values of microRNAs in Colorectal Cancer. Biomark Insights. 2006;2:113–121. 113–121. [PMC free article] [PubMed] [Google Scholar]
- 117.Bovell LC, Shanmugam C, Putcha BD, et al. The prognostic value of microRNAs varies with patient race/ethnicity and stage of colorectal cancer. Clin Cancer Res. 2013;19:3955–3965. doi: 10.1158/1078-0432.CCR-12-3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mazeh H, Mizrahi I, Ilyayev N, et al. The Diagnostic and Prognostic Role of microRNA in Colorectal Cancer - a Comprehensive review. J Cancer. 2013;4:281–295. doi: 10.7150/jca.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vickers MM, Bar J, Gorn-Hondermann I, et al. Stage-dependent differential expression of microRNAs in colorectal cancer: potential role as markers of metastatic disease. Clin Exp Metastasis. 2012;29:123–132. doi: 10.1007/s10585-011-9435-3. [DOI] [PubMed] [Google Scholar]
- 120.Hofsli E, Sjursen W, Prestvik WS, et al. Identification of serum microRNA profiles in colon cancer. Br J Cancer. 2013;108:1712–1719. doi: 10.1038/bjc.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bandres E, Cubedo E, Agirre X, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;19:5–29. doi: 10.1186/1476-4598-5-29. 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ragusa M, Statello L, Maugeri M, et al. Specific alterations of the microRNA transcriptome and global network structure in colorectal cancer after treatment with MAPK/ERK inhibitors. J Mol Med (Berl) 2012;90:1421–1438. doi: 10.1007/s00109-012-0918-8. [DOI] [PubMed] [Google Scholar]
- 123.Faltejskova P, Svoboda M, Srutova K, et al. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med. 2012;16:2655–2666. doi: 10.1111/j.1582-4934.2012.01579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang JX, Song W, Chen ZH, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 125.Link A, Balaguer F, Shen Y, et al. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766–1774. doi: 10.1158/1055-9965.EPI-10-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang IP, Tsai HL, Huang CW, et al. The functional significance of microRNA-29c in patients with colorectal cancer: a potential circulating biomarker for predicting early relapse. PLoS One. 2013;8:e66842. doi: 10.1371/journal.pone.0066842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pizzini S, Bisognin A, Mandruzzato S, et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics. 2013;14:589. doi: 10.1186/1471-2164-14-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shivapurkar N, Weiner LM, Marshall JL, et al. Recurrence of early stage colon cancer predicted by expression pattern of circulating microRNAs. PLoS One. 2014;9:e84686. doi: 10.1371/journal.pone.0084686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Arndt GM, Dossey L, Cullen LM, et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer. 2009;20(9):374. doi: 10.1186/1471-2407-9-374. 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dong Y, Zhao J, Wu CW, et al. Tumor Suppressor Functions of miR-133a in Colorectal Cancer. Mol Cancer Res. 2013;11:1051–1060. doi: 10.1158/1541-7786.MCR-13-0061. [DOI] [PubMed] [Google Scholar]
- 131.Luu C, Heinrich EL, Duldulao M, et al. TP53 and let-7a micro-RNA regulate K-Ras activity in HCT116 colorectal cancer cells. PLoS One. 2013;8:e70604. doi: 10.1371/journal.pone.0070604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen MB, Wei MX, Han JY, et al. MicroRNA-451 regulates AMPK/mTORC1 signaling and fascin1 expression in HT-29 colorectal cancer cells. Cell Signal. 2013;10 doi: 10.1016/j.cellsig.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 133.Wang P, Zou F, Zhang X, et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yuan K, Xie K, Fox J, et al. Decreased Levels of miR-224 and the Passenger Strand of miR-221 Increase MBD2, Suppressing Maspin and Promoting Colorectal Tumor Growth and Metastasis in Mice. Gastroenterology. 2013;145:853–864. doi: 10.1053/j.gastro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Iliou MS, da Silva-Diz V, Carmona FJ, et al. Impaired DICER1 function promotes stemness and metastasis in colon cancer. Oncogene. 2013;10 doi: 10.1038/onc.2013.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang J, Wang Q, Liu H, et al. MicroRNA expression and its implication for the diagnosis and therapeutic strategies of gastric cancer. Cancer Lett. 2010;297:137–143. doi: 10.1016/j.canlet.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 137.Desai KV, Xiao N, Wang W, et al. Initiating oncogenic event determines geneexpression patterns of human breast cancer models. Proc Natl Acad Sci U S A. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.LaConti JJ, Shivapurkar N, Preet A, et al. Tissue and serum microRNAs in the Kras(G12D) transgenic animal model and in patients with pancreatic cancer. PLoS One. 2011;6:e20687. doi: 10.1371/journal.pone.0020687. [DOI] [PMC free article] [PubMed] [Google Scholar]