Abstract
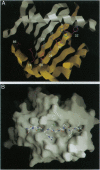
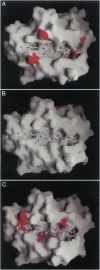
The binding of invariant chain to major histocompatibility complex (MHC) proteins is an important step in processing of MHC class II proteins and in antigen presentation. The question of how invariant chain can bind to all MHC class II proteins is central to understanding these processes. We have employed molecular modeling to predict the structure of class II-associated invariant chain peptide (CLIP)-MHC protein complexes and to ask whether the predicted mode of association could be general across all MHC class II proteins. CLIP fits identically into the MHC class II alleles HLA-DR3, I-Ak, I-Au, and I-Ad, with a consistent pattern of hydrogen bonds, contacts, and hydrophobic burial and without bad contacts. Our model predicts the burial of CLIP residues Met-91 and Met-99 in the deep P1 and P9 anchor pockets and other detailed interactions, which we have compared with available data. The predicted pattern of I-A allele-specific effects on CLIP binding is very similar to that observed experimentally by alanine-scanning mutations of CLIP. Together, these results indicate that CLIP may bind in a single, general way across products of MHC class II alleles.
Full text
PDF
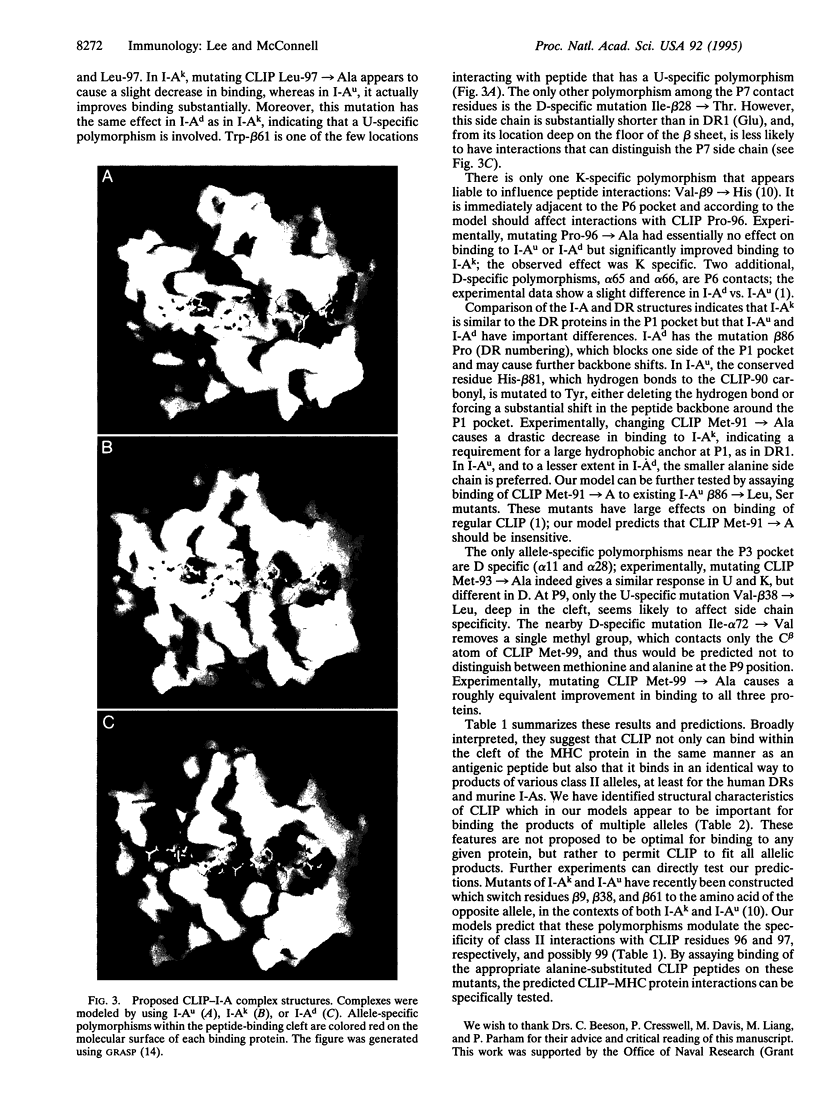

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Demotz S., Barbey C., Corradin G., Amoroso A., Lanzavecchia A. The set of naturally processed peptides displayed by DR molecules is tuned by polymorphism of residue 86. Eur J Immunol. 1993 Feb;23(2):425–432. doi: 10.1002/eji.1830230219. [DOI] [PubMed] [Google Scholar]
- Fremont D. H., Matsumura M., Stura E. A., Peterson P. A., Wilson I. A. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992 Aug 14;257(5072):919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- Garrett T. P., Saper M. A., Bjorkman P. J., Strominger J. L., Wiley D. C. Specificity pockets for the side chains of peptide antigens in HLA-Aw68. Nature. 1989 Dec 7;342(6250):692–696. doi: 10.1038/342692a0. [DOI] [PubMed] [Google Scholar]
- Gautam A. M., Pearson C., Quinn V., McDevitt H. O., Milburn P. J. Binding of an invariant-chain peptide, CLIP, to I-A major histocompatibility complex class II molecules. Proc Natl Acad Sci U S A. 1995 Jan 3;92(1):335–339. doi: 10.1073/pnas.92.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Levitt M. Accurate prediction of the stability and activity effects of site-directed mutagenesis on a protein core. Nature. 1991 Aug 1;352(6334):448–451. doi: 10.1038/352448a0. [DOI] [PubMed] [Google Scholar]
- Lee C. Predicting protein mutant energetics by self-consistent ensemble optimization. J Mol Biol. 1994 Feb 25;236(3):918–939. doi: 10.1006/jmbi.1994.1198. [DOI] [PubMed] [Google Scholar]
- Lee C., Subbiah S. Prediction of protein side-chain conformation by packing optimization. J Mol Biol. 1991 Jan 20;217(2):373–388. doi: 10.1016/0022-2836(91)90550-p. [DOI] [PubMed] [Google Scholar]
- Malcherek G., Gnau V., Jung G., Rammensee H. G., Melms A. Supermotifs enable natural invariant chain-derived peptides to interact with many major histocompatibility complex-class II molecules. J Exp Med. 1995 Feb 1;181(2):527–536. doi: 10.1084/jem.181.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Nash D. K., Eckels D. D. Differential effect of polymorphism at HLA-DR1 beta-chain positions 85 and 86 on binding and recognition of DR1-restricted antigenic peptides. J Immunol. 1993 Mar 1;150(5):1813–1821. [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Sette A., Southwood S., Miller J., Appella E. Binding of major histocompatibility complex class II to the invariant chain-derived peptide, CLIP, is regulated by allelic polymorphism in class II. J Exp Med. 1995 Feb 1;181(2):677–683. doi: 10.1084/jem.181.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern L. J., Brown J. H., Jardetzky T. S., Gorga J. C., Urban R. G., Strominger J. L., Wiley D. C. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994 Mar 17;368(6468):215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- Tate K. M., Lee C., Edelman S., Carswell-Crumpton C., Liblau R., Jones P. P. Interactions among polymorphic and conserved residues in MHC class II proteins affect MHC-peptide conformation and T cell recognition. Int Immunol. 1995 May;7(5):747–761. doi: 10.1093/intimm/7.5.747. [DOI] [PubMed] [Google Scholar]
- Zhang W., Young A. C., Imarai M., Nathenson S. G., Sacchettini J. C. Crystal structure of the major histocompatibility complex class I H-2Kb molecule containing a single viral peptide: implications for peptide binding and T-cell receptor recognition. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8403–8407. doi: 10.1073/pnas.89.17.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]