Abstract
Microalgae tend to accumulate lipids as an energy storage material in the specific organelle, oleosomes. Current studies have demonstrated that lipids derived from microalgal oleosomes are a promising source of biofuels, while the oleosome formation mechanism has not been fully elucidated. Oleosome-associated proteins have been identified from several microalgae to elucidate the fundamental mechanisms of oleosome formation, although understanding their functions is still in infancy. Recently, we discovered a diatom-oleosome-associated-protein 1 (DOAP1) from the oleaginous diatom, Fistulifera solaris JPCC DA0580. The DOAP1 sequence implied that this protein might be transported into the endoplasmic reticulum (ER) due to the signal sequence. To ensure this, we fused the signal sequence to green fluorescence protein. The fusion protein distributed around the chloroplast as like a meshwork membrane structure, indicating the ER localization. This result suggests that DOAP1 could firstly localize at the ER, then move to the oleosomes. This study also demonstrated that the DOAP1 signal sequence allowed recombinant proteins to be specifically expressed in the ER of the oleaginous diatom. It would be a useful technique for engineering the lipid synthesis pathways existing in the ER, and finally controlling the biofuel quality.
Keywords: signal sequence, endoplasmic reticulum, diatom oleosome-associated protein, Fistulifera solaris JPCC DA0580, marine oleaginous diatom
1. Introduction
With an increased demand for a sustainable energy supply, biofuel production has attracted much attention. Microalgal biodiesel production has been researched to meet such demand due to its advantageous features (e.g., global carbon dioxide fixation, no competition for food, much higher biomass yield than higher plants, and oil accumulation at a high level inside the cells) [1]. Several oleaginous microalgae can accumulate triacylglycerol (TAG) in high level as a form of the oleosome (also known as oil body), and such promising oil producers have been intensively studied to understand the TAG biosynthesis [2,3,4,5,6].
A current trend in this field is genome and transcriptome analyses to determine the active synthesis pathways for fatty acids and TAG in the target oil-producing organisms [7,8,9,10,11], while proteome analysis has also been launched to identify the proteins closely attached around the oleosomes. The proteomic approach is expected to identify the novel protein machineries directly participating in the oleosome formation, which conventional pathway analysis can hardly address. It leads to the elucidation of the biological mechanism for oleosome development, and can provide promising targets of genetic engineering for the purpose of oil production improvements. However, the oleosome-associated proteins have been studied in only a few microalgae [12,13,14,15,16,17,18,19].
Among such rare examples, we have focused on Fistulifera solaris JPCC DA0580, an oleaginous marine diatom screened from our marine microalgal culture collection [17]. Beneficial features of this strain for practical biodiesel production include a high growth rate, high lipid content (up to 60%, w/w), a low unsaturation degree of the accumulated lipids, as well as ease of mass cultivation [20,21,22,23]. Recently, the proteome analysis for this diatom identified two oleosome-associated proteins, G12504 and G16188 (formerly g4301 and g6574, respectively) [17]. The GFP-fusion experiment demonstrated that G16118 (tentative potassium channel) showed a broad subcellular distribution including the oleosome. In contrast, G12504 (containing a quinonprotein alcohol dehydrogenase-like domain) exhibited a strict localization only on the oleosomes, implying that this protein could play a role for oleosome formation, and contain a specific signal sequence directing the proteins onto the oleosomes. This notion led us to further research the sequence features of this diatom-oleosome-associated protein, G12504 (referred to as DOAP1 in this study) in detail, and encouraged us to estimate the routing mechanism of this protein into oleosome-targeting. Particularly we focused on the N-terminal signal sequence which primarily governs the localization of the nuclear-encoded polypeptides within the cell organelles [24].
In this study, we carefully determined the N-terminal signal sequence of DOAP1, and fused it with GFP in order to examine the initial localization of this oleosome-associated protein. Fluorescent microscopy of the fusion protein revealed that the signal sequence of DOAP1 could transport proteins into endoplasmic reticulum (ER) of F. solaris, implying the initial localization of DOAP1 at the ER and subsequent transportation to the oleosomes. Additionally, this study also means the success in specific expression of the recombinant protein into the oleaginous microalgal ER where important lipid synthesis reactions occur [25]. It would be useful for future metabolic engineering for improvement of biofuel quality.
2. Results
2.1. Characterization of Doap1 Gene Structure
Comparison between the genomic and cDNA sequences revealed that doap1 gene includes one internal intron. RNA-seq data (partially published [25,26]) supported the transcribed region with 1977 bp (Supplementary Figure S1). TATA-box candidate sequences were found upstream of the transcribed region, indicating the presence of a promoter for doap1 gene. It would be reasonable to consider that translation of DOAP1 starts from the most forward start codon in the RNA-seq supporting region, thus the start codon was predicted to locate 93 bp-downstream from the transcription initiation site (Supplementary Figure S1).
The coding region is estimated to produce a polypeptide with 562 amino acid residues (~59.0 kDa, Supplementary Figure S2). Sequence features of DOAP1 were examined with the SignalP [27] and InterProScan algorisms, and it was predicted that DOAP1 contains an N-terminal signal sequence (ranging from M1 to A19), as well as quinonprotein alcohol dehydrogenase-like superfamily (IPR011047, ranging from Q18 to P184). Subcellular localization of DOAP1 was predicted with a series of bioinformatics tools we have utilized [17,25], and as a result ER-localization was assumed. A proline knot motif was found in the plant oleosome-associated proteins, and demonstrated to work as a specific signal for oleosome-targeting in plants (oleosins) [28,29]. In the case of microalgal oleosome-associated proteins found to date, the proline knot motif is not present, but proline-rich hydrophobic domain is contained [19]. Similarly, DOAP1 is less likely to have the proline knot-like motif, while a proline-rich region (Supplementary Figure S2) and a highly hydrophobic region [17] individually exist at the C-terminus. BLAST screening revealed that Phaeodactylum tricornutum, a pennate diatom same with F. solaris, is the only organism which has an ortholog of this protein according to the present national center for biotechnology information (NCBI) database, while its function remains unknown.
2.2. GFP Expression in the Transformants
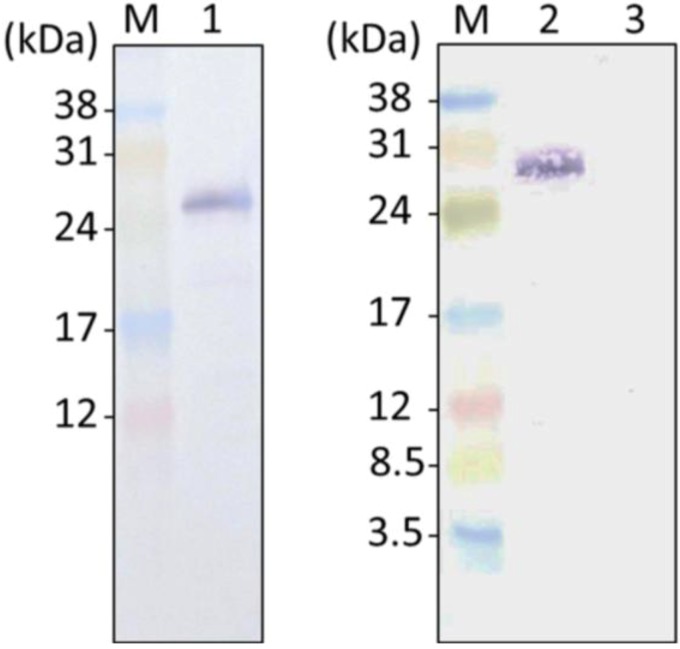
To confirm the initial subcellular localization of DOAP1, we attempted to express the fusion protein of DOAP1 signal sequence and GFP (SDOAP1-GFP). Although the DOAP1-coding region was assumed as mentioned above, we determined to fuse the DNA fragment ranging from the tentative promoter region to the predicted coding region of the N-terminal 57 amino acid of DOAP1 (including the signal sequence and a part of mature DOAP1, see also Supplementary Figures S1 and S2) with green fluorescence protein (gfp) gene in order to ensure the actual native signal sequence can be expressed. It should be noted that our previous study also utilized the same DNA region to express the full length of DOAP1 fused with GFP [17]. The constructed expression vector was introduced into F. solaris cells. As-prepared transformants were subjected to Western blotting to confirm whether the fusion protein SDOAP1-GFP was produced in the cells. A specific band was visualized in the transformant sample using anti-GFP antibody (Figure 1). The detected protein was larger than the neat GFP produced in the F. solaris transformants (Supplementary Figure S3), suggesting the successful expression of the target fusion protein. Furthermore, its size was smaller than the intact protein corded (approximately 33 kDa); this could be caused by the cleavage of the signal peptide after transportation [30]. As a negative control experiment, wild type cells were also examined, and no signal was detected.
Figure 1.
Green fluorescence protein (GFP) detection with Western blotting from F. solaris JPCC DA0580 transformants expressing neat GFP (Lane 1); SDOAP1-GFP (Lane 2); and wild-type cells (Lane 3). Lane M represents molecular marker.
2.3. ER-Targeting of SDOAP1-GFP
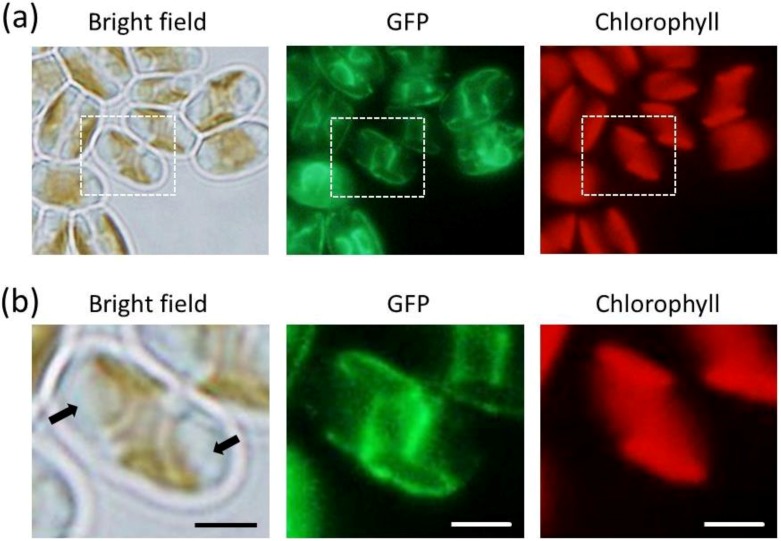
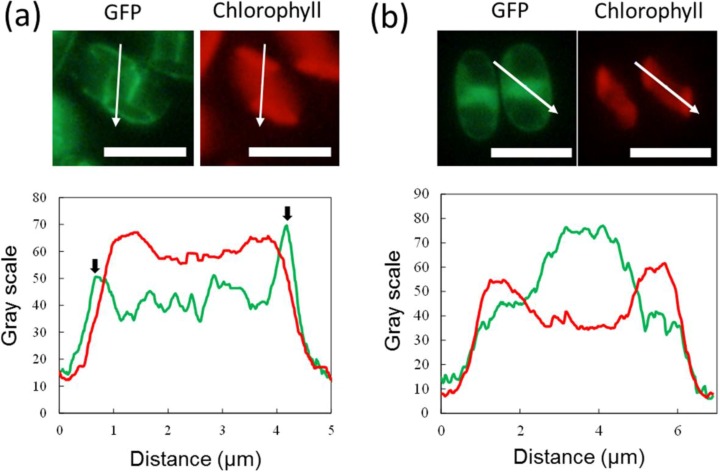
To examine whether the DOAP1 signal sequence directs proteins to specific organelles, the cells expressing SDOAP1-GFP were observed using a fluorescent microscope. The intense fluorescence was observed around the chloroplast, as well as central cellular region (Figure 2). This fluorescence distribution was obviously different from that of the fusion protein consisted of full length of DOAP1 and GFP, which strictly localized at the oleosomes [17]. When GFP and chlorophyll distribution was spatially profiled, it was demonstrated that the peak of GFP fluorescence was outside of the chlorophyll (Figure 3a). This feature was reproducibly confirmed in several cells (Supplementary Figure S4), suggesting that SDOAP1-GFP localizes outside the chloroplast. In the case of the transformants expressing the neat GFP, only the central cellular region emitted significant fluorescence, suggesting the expression at the cytoplasm (Figure 3b). When the cells were stained with the Hoechst dye, the nucleus was demonstrated to localize at the center of the cell, and surrounded by the GFP emission (Supplementary Figure S5).
Figure 2.
Microscopy studies on F. solaris JPCC DA0580 transformants expressing SDOAP1-GFP. (a) Bright field and fluorescent images of the transformants; (b) Magnified images of the square regions in (a). Black arrows represent oleosomes. (scale bar = 2 μm).
Figure 3.
Fluorescence profiling on the microscopic images of F. solaris JPCC DA0580 expressing SDOAP1-GFP (a) and neat GFP (b). Fluorescent signals of GFP and chlorophyll along the white arrows in the images are shown in green and red lines, respectively. Fluorescent peaks of GFP outside of the chlorophyll fluorescent region are shown with the black arrows. (scale bar = 5 μm).
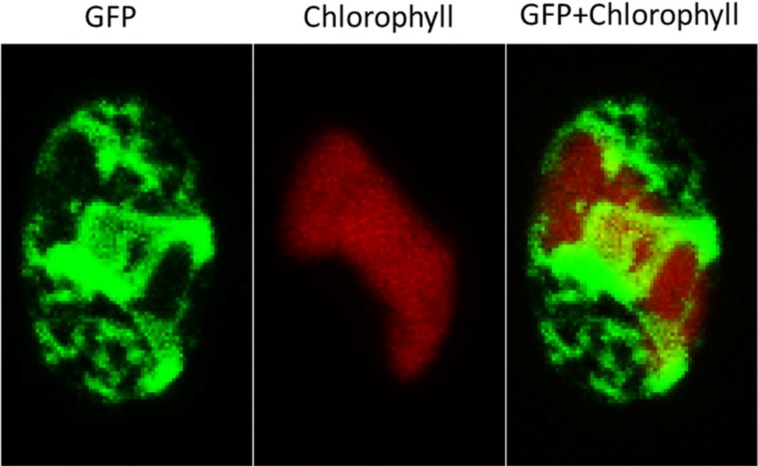
Confocal microscopy reconstituted three-dimensional (3D) arrangement of SDOAP1-GFP in the cells. GFP fluorescence delineated the network of membranes thought to represent the ER (Figure 4 and Supplementary Figure S6). The fluorescent network extended through the cell, and surrounded the chloroplast. This 3D arrangement closely resembles that observed in P. tricornutum expressing GFP at the ER [24]. When F. solaris was stained with an ER-specific dye, a similar feature was observed (Supplementary Figure S7).
Figure 4.
Confocal microscopic images of F. solaris JPCC DA0580 transformant expressing SDOAP1-GFP.
3. Discussion
DOAP1 has been identified an oleosome-associated protein through our proteomic research. It was predicted to have 506 amino acids in length previously [17]. However, recently obtained RNA-seq data allowed us to find the N-terminal extension, which we have fortunately introduced in the expression vector in the previous study [17] but did not consider that this extra sequence may be a part of the coding region. By SignalP algorism, this sequence is predicted to be an ER-targeting signal sequence which can be recognized by a Sec61 complex for transition across the ER membrane [31].
Next, to test whether this tentative signal sequence can route proteins into the ER of F. solaris, we constructed the expression vector of SDOAP1-GFP, where the signal sequence and a part of mature DOAP1 (M1–M57 in the Supplementary Figure S2) were together fused at the N-terminus of GFP. Western blotting for the F. solaris transformants harboring the SDOAP1-GFP expression gene confirmed substantial expression of the target protein, as well as the cleavage of the signal sequence. Although the cleavage site is likely to be the point between A19 and Q20 according to the SignalP algorism, actual cleavage site still remains unclear, and can be elucidated with N-terminal sequencing of the cleaved protein [24]. In vivo localization of SDOAP1-GFP in F. solaris was observed with fluorescent microscopy. As a result, intensive green fluorescence was detected surrounding the chloroplast and nucleus (Figure 2 and Figure 3, and Supplementary Figure S4 and S5). Confocal microscopic analysis further revealed the meshwork-like structure showing GFP fluorescence around the chloroplast, suggesting that the target fusion protein was transported into the ER. It has been widely accepted that diatoms have complex chloroplasts which are surrounded by four membranes owing to their evolutional history with secondary symbiosis [32]. The innermost and second innermost membranes are actually corresponding to the two membranes of the primary chloroplast. The space between the second outermost membrane and the outermost membrane represents the ER (also known as chloroplast-ER; CER). Organelle-specific targeting by recombinant proteins in diatoms has been studied in a pennate diatom, P. tricornutum [24,30,33,34]. ER-targeting has been achieved by fusing the specific signal presequence at the N-terminus of proteins, while the inner chloroplast-targeting needed an extra transit sequence-like domain following the ER signal. In the case of ER-targeting by GFP, meshwork of green fluorescence was observed around the chloroplast as well as the nucleus in P. tricornutum [24,30]; the GFP distribution was similar with that observed in this study. These previous studies also support that the GFP distribution in this study represented the ER-targeting of SDOAP1-GFP. ER-staining with the specific dye further supported this notion.
When DOAP1 with its full length was labeled with GFP, fluorescence was only observed on the oleosomes [17]. In contrast, the truncated DOAP1 including the signal sequence and the following partial mature protein sequence (Supplementary Figure S2) directed the GFP into the ER, and little fluorescence was observed from the oleosomes (usually two oleosomes exist in a F. solaris cell at the polar position). These results suggest that DOAP1 could localize in the ER at first due to the N-terminal signal sequence, then it is transported onto the oleosomes. Specific signals for targeting oleosome (e.g., proline knot) is not yet identified in DOAP1, thus the transportation mechanism from the ER to oleosomes still remains unknown. We assume that the proline-rich region at the C-terminus of DOAP1 might be a possible candidate of oleosome-targeting signal because similar feature was found in lipid droplet surface protein (LDSP) in other microalga, Nannochloropsis sp. [19]. Incidentally SignalP did not detect ER-signal sequence from LDSP. In order to specify the oleosome-targeting sequence in DOAP1, the GFP fusion experiment with various truncated forms of DOAP1 will be performed in the near future. The protein transportation from the ER to oleosomes also implies the direct interaction between these two organelles, otherwise DOAP1 cannot eventually move to the oleosomes. This notion is consistent with our previous study, in which the oleosome membrane was hypothesized to be derived from the ER membrane [17].
Another significance of this study is establishment of the method for ER-specific recombinant protein expression in the oleaginous microalga. At the ER, many critical reactions with regard to the biofuel productivity and quality take place. For instance, acyl-chain desaturation and elongation would be performed at ER [25], and the acyl-chain length and desaturation degree directly affects the resultant biodiesel fuel quality [35]. Engineering these metabolic pathways by transporting heterogeneous enzymes could be a promising approach to control the fuel quality derived from F. solaris, and the DOAP1 signal sequence could achieve the transportation by just fusing it at the N-terminus of the target proteins.
4. Experimental Section
4.1. Culture Conditions
The marine diatom, F. solaris JPCC DA0580, was isolated from the junction of the Sumiyo and Yakugachi Rivers, in Kagoshima, Japan (28°15'N, 129°24'E) [21]. F. solaris was cultured in the f/2 medium [36] (75 mg NaNO3, 6 mg Na2HPO4·2H2O, 0.5 μg vitamin B12, 0.5 μg biotin, 100 μg Thiamine HCl, 10 mg Na2SiO3·9H2O, 4.4 mg Na2-EDTA, 3.16 mg FeCl3·6H2O, 12 μg CoSO4·5H2O, 21 μg ZnSO4·7H2O, 0.18 mg MnCl2·4H2O, 70 μg CuSO4·5H2O, and 7 μg Na2MoO4·2H2O) dissolved per liter of artificial seawater. Transformant cells were incubated in the f/2 medium with antibiotics G418 (500 μg/mL). Cultures were aerated with sterile air at 25°C under 140 μmol/m2/s of continuous illumination.
4.2. Characterization of Nucleotide and Protein Sequences
DNA sequence neighboring the tentative doap1 (g12504) gene was retrieved from our domestic database of F. solaris whole genome sequence (Supplementary Figure S1) [37]. cDNA sequence and RNA-seq data regarding doap1 gene has already been obtained in our previous studies [17,25]. TATA-boxes upstream of doap1 gene were predicted with the polymerase II promoter function of GENETYX ver.10. Protein features were analyzed with SignalP [27], InterProScan and BLAST.
4.3. Vector Construction and Transformation
The expression vector, pSP-DOAP1GFP/GAPDH, for full length of DOAP1 (formerly g4301) fused with GFP, was constructed in our previous study [17], in which the predicted coding region (without intron) and its up-stream sequence (638 bp) were synthesized (Integrated DNA Technologies, Inc., Coraville, IA, USA), and inserted between the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) promoter derived from F. solaris and gfp gene. Transcription is terminated by the fucoxanthin chlorophyll a/c-binding protein A (fcpA) terminator derived from P. tricornutum [38]. In this study, the DNA fragment including a part of doap1 gene was amplified by polymerase chain reaction (PCR) using the primer pair (5′-ATGTTCCCTGGGCATTCGTG-3′ and 5′-CTTGTCTCCCGACAACAAGATG-3′) and pSP-DOAP1GFP/GAPDH as a template. The amplified fragment was inserted between the same promoter and GFP gene. The constructed plasmid was referred to as pSP-SDOAP1GFP/GAPDH.
Transformation of F. solaris was performed by microparticle bombardment using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) as described previously [17,38].
4.4. Western Blotting
F. solaris transformants (1 × 107 cells) were collected by centrifugation, washed with water, suspended in 100 μL of 1% (w/v) sodium dodecyl sulfate (SDS) in aqueous solution and boiled for 10 min. After centrifugation, supernatant was collected, and SDS sample buffer was added (final concentration of 62.5 mM Tris-HCl, pH 6.8, 5% 2-mercaptoethanol, 2% SDS, 5% sucrose, and 0.002% bromophenol blue). Denatured proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using a 12.5% (w/v) gel, and transferred to a polyvinylidene difluoride membrane. GFP was then detected using alkaline phosphatase (ALP)-labeled anti-GFP antibody (Rockland immunochemicals Inc., Gilbertsville, PA, USA, 1/5000 dilution from stock in PBS containing 0.05% Tween 20). BCIP/NBT-Blue (Sigma, St. Louis, MO, USA) was used as the ALP substrate for visualization.
4.5. Fluorescent Microscopy and Image Analysis
Transformant cells were observed using a fluorescent microscope BX51 (Olympus Corporation, Tokyo, Japan); a NIBA filter set for GFP, a WIG filter set for chlorophyll and a WU filter set for Hoechst 33342 fluorescence, respectively. Confocal microscopy was performed with Fluoview FV1000 (Olympus Corporation, Tokyo, Japan). Hoechst staining was conducted by adding Hoechst 33342 (Invitrogen, Eugene, OR, USA) to the cell culture at 1:50 volume ratio (final concentration = 200 μg/mL). The images obtained were analyzed with Image J program. In order to display the grayscale of the GFP and chlorophyll fluorescence, the fluorescence images obtained were converted into 8-bit black-and-white images, and then plot profiling was performed.
5. Conclusions
The fusion experiment with GFP and the signal sequence of the oleosome-associated protein DOAP1 revealed that DOAP1 contains the signal sequence targeting the ER. This result suggests that DOAP1 could initially be transported into the ER with the aid of its signal sequence, and subsequently transported onto the oleosomes. This new finding implies the interaction between the ER membrane and oleosome. Protein targeting to the ER achieved in this study is also useful for engineering the lipid synthesis pathway in F. solaris because key reactions for lipid synthesis including elongation and desaturation of acyl chains occur in the ER. This could contribute to improvement of biodiesel quality derived from F. solaris.
Acknowledgments
This study was supported by the Japan Science and Technology Agency (JST) and CREST.
Supplementary Files
Supplementary Information (PDF, 786 KB)
Author Contributions
T.T. designed overall experiments. Y. M. and Y. S. performed experiments and analyzed data. All authors, including Y.M., Y. S., T.Y., and T.T. participated in discussions and preparation of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Smith V.H., Sturm B.S., Denoyelles F.J., Billings S.A. The ecology of algal biodiesel production. Trends Ecol. Evol. 2010;25:301–309. doi: 10.1016/j.tree.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Liang Y., Sarkany N., Cui Y. Biomass and lipid productivities of Chlorella vulgaris under autotrophic, heterotrophic and mixotrophic growth conditions. Biotechnol. Lett. 2009;31:1043–1049. doi: 10.1007/s10529-009-9975-7. [DOI] [PubMed] [Google Scholar]
- 3.Mahapatra D.M., Chanakya H., Ramachandra T. Euglena sp. as a suitable source of lipids for potential use as biofuel and sustainable wastewater treatment. J. Appl. Phycol. 2013;25:855–865. doi: 10.1007/s10811-013-9979-5. [DOI] [Google Scholar]
- 4.Matsunaga T., Matsumoto M., Maeda Y., Sugiyama H., Sato R., Tanaka T. Characterization of marine microalga, Scenedesmus sp. strain JPCC GA0024 toward biofuel production. Biotechnol. Lett. 2009;31:1367–1372. doi: 10.1007/s10529-009-0029-y. [DOI] [PubMed] [Google Scholar]
- 5.Oh S.H., Han J.G., Kim Y., Ha J.H., Kim S.S., Jeong M.H., Jeong H.S., Kim N.Y., Cho J.S., Yoon W.B., et al. Lipid production in Porphyridium cruentum grown under different culture conditions. J. Biosci. Bioeng. 2009;108:429–434. doi: 10.1016/j.jbiosc.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Rodolfi L., Chini Zittelli G., Bassi N., Padovani G., Biondi N., Bonini G., Tredici M.R. Microalgae for oil: Strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 2009;102:100–112. doi: 10.1002/bit.22033. [DOI] [PubMed] [Google Scholar]
- 7.Corteggiani Carpinelli E., Telatin A., Vitulo N., Forcato C., D’Angelo M., Schiavon R., Vezzi A., Giacometti G.M., Morosinotto T., Valle G. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant. 2014;7:323–325. doi: 10.1093/mp/sst120. [DOI] [PubMed] [Google Scholar]
- 8.Guarnieri M.T., Nag A., Smolinski S.L., Darzins A., Seibert M., Pienkos P.T. Examination of triacylglycerol biosynthetic pathways via de novo transcriptomic and proteomic analyses in an unsequenced microalga. PLoS One. 2011;6:e25851. doi: 10.1371/journal.pone.0025851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radakovits R., Jinkerson R.E., Fuerstenberg S.I., Tae H., Settlage R.E., Boore J.L., Posewitz M.C. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat. Commun. 2012;3:686. doi: 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rismani-Yazdi H., Haznedaroglu B.Z., Hsin C., Peccia J. Transcriptomic analysis of the oleaginous microalga Neochloris oleoabundans reveals metabolic insights into triacylglyceride accumulation. Biotechnol. Biofuels. 2012;5:74. doi: 10.1186/1754-6834-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Alvarez S., Hicks L.M. Comprehensive comparison of iTRAQ and label-free LC-based quantitative proteomics approaches using two Chlamydomonas reinhardtii strains of interest for biofuels engineering. J. Proteome Res. 2012;11:487–501. doi: 10.1021/pr2008225. [DOI] [PubMed] [Google Scholar]
- 12.Davidi L., Katz A., Pick U. Characterization of major lipid droplet proteins from Dunaliella. Planta. 2012;236:19–33. doi: 10.1007/s00425-011-1585-7. [DOI] [PubMed] [Google Scholar]
- 13.Frandsen G.I., Mundy J., Tzen J.T. Oil bodies and their associated proteins, oleosin and caleosin. Physiol. Plant. 2001;112:301–307. doi: 10.1034/j.1399-3054.2001.1120301.x. [DOI] [PubMed] [Google Scholar]
- 14.Lin I., Jiang P.-L., Chen C.-S., Tzen J.T. A unique caleosin serving as the major integral protein in oil bodies isolated from Chlorella sp. cells cultured with limited nitrogen. Plant Physiol. Biochem. 2012;61:80–87. doi: 10.1016/j.plaphy.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Moellering E.R., Benning C. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot. Cell. 2010;9:97–106. doi: 10.1128/EC.00203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen H.M., Baudet M., Cuiné S., Adriano J.M., Barthe D., Billon E., Bruley C., Beisson F., Peltier G., Ferro M. Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: With focus on proteins involved in lipid metabolism. Proteomics. 2011;11:4266–4273. doi: 10.1002/pmic.201100114. [DOI] [PubMed] [Google Scholar]
- 17.Nojima D., Yoshino T., Maeda Y., Tanaka M., Nemoto M., Tanaka T. Proteomics analysis of oil body-associated proteins in the oleaginous diatom. J. Proteome Res. 2013;12:5293–5301. doi: 10.1021/pr4004085. [DOI] [PubMed] [Google Scholar]
- 18.Peled E., Leu S., Zarka A., Weiss M., Pick U., Khozin-Goldberg I., Boussiba S. Isolation of a novel oil globule protein from the green alga Haematococcus pluvialis (Chlorophyceae) Lipids. 2011;46:851–861. doi: 10.1007/s11745-011-3579-4. [DOI] [PubMed] [Google Scholar]
- 19.Vieler A., Brubaker S.B., Vick B., Benning C. A lipid droplet protein of Nannochloropsis with functions partially analogous to plant oleosins. Plant Physiol. 2012;158:1562–1569. doi: 10.1104/pp.111.193029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Y., Maeda Y., Yoshino T., Matsumoto M., Tanaka T. Profiling of fatty acid methyl esters from the oleaginous diatom Fistulifera sp. strain JPCC DA0580 under nutrition-sufficient and -deficient conditions. J. Appl. Phycol. 2014 doi: 10.1007/s10811-014-0265-y. [DOI] [Google Scholar]
- 21.Matsumoto M., Sugiyama H., Maeda Y., Sato R., Tanaka T., Matsunaga T. Marine diatom, Navicula sp. strain JPCC DA0580 and marine green alga, Chlorella sp. strain NKG400014 as potential sources for biodiesel production. Appl. Biochem. Biotechnol. 2010;161:483–490. doi: 10.1007/s12010-009-8766-x. [DOI] [PubMed] [Google Scholar]
- 22.Sato R., Maeda Y., Yoshino T., Tanaka T., Matsumoto M. Seasonal variation of biomass and oil production of the oleaginous diatom Fistulifera sp. in outdoor vertical bubble column and raceway-type bioreactors. J. Biosci. Bioeng. 2014;117:720–724. doi: 10.1016/j.jbiosc.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 23.Satoh A., Ichii K., Matsumoto M., Kubota C., Nemoto M., Tanaka M., Yoshino T., Matsunaga T., Tanaka T. A process design and productivity evaluation for oil production by indoor mass cultivation of a marine diatom, Fistulifera sp. JPCC DA0580. Bioresour. Technol. 2013;137:132–138. doi: 10.1016/j.biortech.2013.03.087. [DOI] [PubMed] [Google Scholar]
- 24.Apt K.E., Zaslavkaia L., Lippmeier J.C., Lang M., Kilian O., Wetherbee R., Grossman A.R., Kroth P.G. In vivo characterization of diatom multipartite plastid targeting signals. J. Cell Sci. 2002;115:4061–4069. doi: 10.1242/jcs.00092. [DOI] [PubMed] [Google Scholar]
- 25.Liang Y., Maeda Y., Sunaga Y., Muto M., Matsumoto M., Yoshino T., Tanaka T. Biosynthesis of polyunsaturated fatty acids in the oleaginous marine diatom Fistulifera sp. strain JPCC DA0580. Mar. Drugs. 2013;11:5008–5023. doi: 10.3390/md11125008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemoto M., Maeda Y., Muto M., Tanaka M., Yoshino T., Mayama S., Tanaka T. Identification of a frustule-associated protein of the marine pennate diatom Fistulifera sp. strain JPCC DA0580. Mar. Genomics. 2014 doi: 10.1016/j.margen.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 28.Abell B.M., Holbrook L.A., Abenes M., Murphy D.J., Hills M.J., Moloney M.M. Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell. 1997;9:1481–1493. doi: 10.1105/tpc.9.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang A.H. Oleosins and oil bodies in seeds and other organs. Plant Physiol. 1996;110:1055–1061. doi: 10.1104/pp.110.4.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilian O., Kroth P.G. Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 2005;41:175–183. doi: 10.1111/j.1365-313X.2004.02294.x. [DOI] [PubMed] [Google Scholar]
- 31.Stork S., Lau J., Moog D., Maier U.G. Three old and one new: Protein import into red algal-derived plastids surrounded by four membranes. Protoplasma. 2013;250:1013–1023. doi: 10.1007/s00709-013-0498-7. [DOI] [PubMed] [Google Scholar]
- 32.Gibbs S.P. The chloroplast endoplasmic reticulum: Structure, function, and evolutionary significance. Int. Rev. Cytol. 1981;72:49–99. doi: 10.1016/S0074-7696(08)61194-8. [DOI] [Google Scholar]
- 33.Gruber A., Vugrinec S., Hempel F., Gould S.B., Maier U.G., Kroth P.G. Protein targeting into complex diatom plastids: Functional characterisation of a specific targeting motif. Plant Mol. Biol. 2007;64:519–530. doi: 10.1007/s11103-007-9171-x. [DOI] [PubMed] [Google Scholar]
- 34.Peschke M., Moog D., Klingl A., Maier U.G., Hempel F. Evidence for glycoprotein transport into complex plastids. Proc. Natl. Acad. Sci. USA. 2013;110:10860–10865. doi: 10.1073/pnas.1301945110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knothe G. Dependence of biodiesel fuel properties on the structure of fatty acid alkyl esters. Fuel Process. Technol. 2005;86:1059–1070. [Google Scholar]
- 36.Guillard R.R., Ryther J.H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka T., Maeda Y., Veluchamy A., Tanaka M., Bowler C., Muto M., Sunaga Y., Tanaka M., Yoshino T., Taniguchi T., et al. Genome and transcriptome analyses of theoleaginous diatom Fistulifera sp. reveals the oil accumulation mechanism. 2014. to be submitted for publication.
- 38.Muto M., Fukuda Y., Nemoto M., Yoshino T., Matsunaga T., Tanaka T. Establishment of a genetic transformation system for the marine pennate diatom Fistulifera sp. strain JPCC DA0580—A high triglyceride producer. Mar. Biotechnol. 2013;15:48–55. doi: 10.1007/s10126-012-9457-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information (PDF, 786 KB)