Abstract
This unit describes the treatment of laboratory mice with the mutagen N-ethyl-N-nitrosourea (ENU) to achieve very highly induced rates of mutation throughout the genome. Further, it describes several popular mating schemes designed to produce animals displaying phenotypes associated with the induced mutations.
Keywords: alkylating agent, spermatogonial stem cells, per locus mutation frequency, phenotype-driven mutagenesis, genome sequencing
BASIC PROTOCOL
Many practical aspects of the murine system, such as the existence of inbred lines, moderate generation time, ease of experimental manipulation, and extensive bioinformatic annotation of the genome make it a useful system for investigation of mammalian biology. An additional factor that has elevated the mouse to its present importance as a model system is the ability to directly modify the genomic DNA sequence in a number of ways. This is usually done either by injection of DNA sequences into fertilized oocytes, the specific alteration of a locus by homologous recombination in embryonic stem cells, or most recently, utilizing a suite of genome editing tools that are rapidly changing the field (Li et al., 2011; Wang et al., 2013; Yang et al., 2013). While transgenic and knockout approaches are excellent for a genotype-driven query, they are less ideally suited to testing the genetic basis of a specific phenotype of interest (e.g., craniofacial abnormalities, skeletal defects). This is because a genotype-driven approach requires prior knowledge (or at least expectation) of the role of a gene of interest in a phenotype to be analyzed. Furthermore, knockout approaches generally result in complete abrogation of gene function, resulting in a null phenotype; partial loss of function is less readily obtained. However, hypomorphic (reduced function) phenotypes can be significantly different than the null phenotype and can be informative regarding the role of a specific gene in developmental or pathological processes.
The identification of genes responsible for spontaneous or induced mutations by positional cloning and/or direct sequencing are elements of a phenotype-driven strategy. This approach, while by no means trivial, has become markedly easier with the development of dense maps of genetic markers, strain-specific polymorphisms, the availability of a complete genome sequence, multiple bioinformatics resources such as expression databases, (Ng et al., 2009; Finger et al,. 2011; Harding et al., 2011), and affordable whole exome/whole genome sequencing (Fairfield et al., 2011; Leshchiner et al., 2012; Sheridan et al,. 2007). Given the potential of this approach to uncover gene effects that may have been unexpected, there is considerable interest in developing novel mutants lines for a wide variety of phenotypes—e.g., those affecting clinically relevant parameters such as cardiovascular or neurological function, and those affecting basic biological processes such as embryonic development and organogenesis.
This has led to a revival of interest in methods of efficient mutagenesis, such as treatment with the chemical N-ethyl-N-nitrosourea (ENU). The utility of this reagent for generating mutations in the mouse was clearly shown by Russell et al. (1979); however, because it was known to result in single nucleotide changes that were perceived as difficult to identify, it was originally employed by only a small cohort of intrepid investigators (Bode, 1984; Shedlovsky et al., 1986). As the tools for genomic analysis and positional cloning have been refined, the potential of ENU mutagenesis has become increasingly appreciated, and it has been employed both in wide-ranging screens involving large numbers of mice (Hrabe de Angelis, 2000; Nolan, 2000) and in smaller efforts focused on specific biological questions (Herron et al., 2002; Zarbalis et al., 2004; Huangfu and Anderson, 2005; Stottmann et al., 2011; Ha et al., 2013). Results of these efforts confirm the efficiency of the approach for producing novel mutations with interesting phenotypes. Other studies have illustrated how this strategy can inform interactions with a gene or disease of interest (Matera et al., 2008; Buchovecky et al., 2013). Thus, it is now feasible to apply to a mammalian system the same analytical approaches that have been historically so productive for the investigation of other model organisms such as yeast, C. elegans, and Drosophila.
There are a number of components for any mutagenesis experiment, including treatment regimens, husbandry issues, phenotype analysis, mutation mapping, and ultimately gene identification and validation. Clearly the most important step is the successful mutagenesis of mice, a detailed protocol of which is presented below
Materials
95% nondenatured ethanol
N-ethyl-N-nitrosourea (ENU) in a 100-ml serum containment bottle (store at 0°C until use)
Phosphate/citrate buffer (see recipe)
6- to 8-week-old male mice
Inactivating solution (see recipe)
1-, 10-, and 60-ml syringes with Luer-locks
16- and 27-G needles
CAUTION: N-ethyl-N-nitrosourea (ENU) is a hazardous chemical which is mutagenic, teratogenic, and carcinogenic. Thus, several important measures should be undertaken to ensure the safety of the operator when using this protocol. (1) The crystalline form of the chemical should always stay in the serum bottle containment vessel supplied by the manufacturer. (2) All manipulations of the agent that involve the dissolving, titration, application to animals, and holding of animals for a 24 hr period after application should be performed in a chemical fume hood. (3) The operator should wear chemical-resistant gloves whenever the agent is handled. (4) All spills and all materials coming into contact with the agent should be decontaminated with inactivating solution before disposal or cleaning and reuse.
-
1
Inject 10 ml of 95% nondenatured ethanol into the 100-ml serum bottle containing ENU using a 10-ml Luer-lock syringe and 16-G needle. Gently agitate the ethanolic ENU suspension until it dissolves completely. Facilitate the process by using the warmth from the hands to warm the containment vessel.
The final solution should be yellow and completely clear.Ethanol can be purchased either denatured or nondenatured. The denatured form has substances added that render it toxic and is therefore unsuitable for this protocol.ENU is available from Sigma in 1-g amounts sealed in a 100-ml serum bottle. An IsoPac also comes with the bottle for temperature control.It is advisable to use a fresh IsoPac for each ENU treatment if following a fractionated dose regimen. -
2
Using a 60-ml syringe and 16-G needle, inject 90 ml phosphate/citrate buffer in two 45-ml aliquots, pausing at 5- to 10-ml intervals to allow the pressure within the containment vessel to escape back into the injection syringe.
-
3
After adding all of the diluent, mix thoroughly by gentle agitation of the containment vessel.
Determine ENU concentration spectrophotometrically
-
4
Using a 1-ml Luer-lock syringe fitted with a 27-gauge needle, aspirate at least 200 μl ENU stock solution from the containment vessel. Transfer 200 μl into a 1.5-ml disposable microcentrifuge tube for spectroscopic analysis.
-
5
Dilute the ENU aliquot to 1000 μl with phosphate/citrate buffer (1:5 dilution) and transfer to a disposable cuvette.
-
6
Determine the OD398 nm of this diluted aliquot relative to a blank containing a 1:50 dilution of 95% ethanol in phosphate/citrate buffer in a disposable cuvette.
Typical OD398 readings will be between 1.3 and 1.5. -
7
Calculate the concentration of the ENU in the solution by dividing the OD398 by 0.72 and multiplying by 5 (to account for the dilution factor).
This calculation is based upon the observation that a 1 mg/ml solution gives an OD398 of 0.72 (Shedlovsky et al., 1986).Most manufacturer supplied bottles prove to have the 98% to 99% ENU that is reported on the label, but occasional preparations have been assayed at less; therefore, it is important to check and ensure that the desired dosage will be delivered. -
8
Perform a wavelength scan from 350 to 450 nm.
Most wavelength scans will reveal a somewhat broad peak centered at OD398 with some very minor shoulders both above (415 nm) and below (385 nm) the peak.
Inject animals
-
9
Calculate the necessary amount of ENU stock solution for injection using the following equation:
where W is the weight of the animal in grams, Y is the desired dose in micrograms ENU per gram body weight, X is the ENU stock concentration in milligrams per milliliter, and Z is the injection volume in milliliters.
The injection volume should be in the range of 0.2 to 1 ml. Volumes less than this may be difficult to inject accurately and volumes greater than this may result in an unacceptably high level of ethanol toxicity.See Critical Parameters and Troubleshooting for a discussion about choosing the appropriate dose. -
10
Inject 6- to 8-week old male mice with the ENU solution (see Critical Parameters). Select an injection site along the ventral midline of the animal to be treated, caudal to the rib cage, but rostral to the urinary bladder. Insert the injection needle through the abdominal skin and peritoneum. Inject the desired volume while observing the injection site.
The occurrence of a subcutaneous blister indicates that the needle is not being inserted at sufficient depth to penetrate the peritoneum. -
11
Clean all spills and soak all equipment coming into contact with ENU with the inactivating solution.
-
12
Inactivate remaining ENU stock solution in the following manner:
Inject 1-ml aliquots of inactivating solution into the containment vessel with a 1-ml Luer-lock syringe and 27-G needle.
Allow the pressure resulting from inactivation to escape into the injection syringe.
-
Leave an injection needle in the injection port of the containment vessel to allow gas to escape into the fume hood while the mice are also being kept in the hood.
Although fresh ENU is recommended for fractionated injections, it is also possible to store the ENU stock solution for several weeks at –20°C (Nolan et al., 1997), but it is important to recheck the concentration and purity by the above spectroscopic methods (steps 4 to 8).
-
11
Keep mice in an efficient chemical hood for at least 24 hr before moving them back into the mouse rooms. Spray cage bedding with inactivating solution before discarding.
Reagents and Solutions
Use sterile deionized, distilled water in all recipes and protocol steps.
Inactivating solution
0.1 M NaOH (4 g/liter)
20% (w/v) sodium thiosulfate (200 g/liter)
-
Store up to several months at room temperature
This solution, also known as alkaline thiosulfate, does not require sterilization.
Phosphate/citrate buffer
100 mM sodium phosphate (14.2 g dibasic salt/liter)
50 mM sodium citrate (14.7 g dihydrate salt/liter)
Adjust to pH 5.0
Store indefinitely if sterilized; otherwise use within several days
COMMENTARY
Background Information
N-ethyl-N-nitrosourea (ENU) mutagenesis of the mouse is used when there is a need for highly efficient induction of point mutations randomly distributed throughout the germline. ENU induces the highest mouse-germline mutation frequency of any known chemical or physical agent. In addition, it can be used in almost any mouse strain, although differences have been noted in the ability of different strains to withstand different mutagenesis regimens (Justice et al., 2000).
Because the typical mutagenic lesion of ENU is a single base change, it is capable of producing all of the different classes of mutations (i.e., hypo-, hyper-, neo-, nulli-, and antimorphs); however, this characteristic represents a potential disadvantage, because the identification of single base changes in the mouse genome still poses a significant challenge to rapid molecular characterization. Additionally, because of the logistics of mouse mutagenesis, most often only a single allele of any particular mutant locus is obtained. Thus, the causal role of a gene carrying a sequence change may require confirmation using an independent functional assay. Recently, multiple large scale efforts have been combined with the express goal of creating an ES cell line carrying a null mutation for each gene in the mouse genome (International Mouse Knockout Consortium, 2007). This makes obtaining alleles for complementation analyses much more tractable for the average researcher as cell lines can be ordered and chimeras produced by a core facility or commercially with no requirement for individual expertise in mouse transgenesis. The genome editing tools recently introduced have even further simplified the process of generating an engineered null (Li et al., 2011; Wang et al., 2013; Yang et al., 2013).
At present there is no comparable system for phenotype-driven analysis of mouse mutations. Transgenic insertional mutagenesis represents an alternative that has the virtue of facilitating a more rapid characterization of the mutagenized locus, but is substantially less efficient and can result in significant chromosomal deletions and rearrangements. Efforts to adapt transposable elements for use as mutagenic reagents have worked well for somatic mutagenesis, especially as applied for cancer gene discovery (Dupuy et al., 2005), but have been less productive for other phenotype-driven analyses.
An alternative approach is to use genotype-driven analysis, such as homologous recombination in ES cells, gene-trapping, or genome editing to generate mutant mice whose phenotype can be examined. While this has the advantage that the substantial task of positional cloning is not required, it represents a qualitatively different approach. Further, it is far less likely to uncover the hypomorphic phenotypes that are routinely obtained using ENU mutagenesis. Furthermore, the use of ENU mutagenesis does not preclude a genotype-driven approach. Several investigators have reported success identifying multiple mutant alleles of specific genes by direct sequencing of ENU-mutagenized mice or ES cell clones (Shedlovsky et al., 1993; Rinchik and Carpenter, 1999; Rajaraman et al., 2002; Vivian et al., 2002; Sakuraba et al.,2005). In addition, ENU mutagenized gametes have been stored and can be sequenced for mutation(s) in a gene(s) of interest (http://www.har.mrc.ac.uk/services/enu-dna-archive).
ENU
ENU is a member of the alkylating agent class of mutagens. It does not require metabolic activation to become mutagenic and, although its use in mouse mutagenesis began over 30 years ago (Russell et al., 1979), it remains the most potent mutagen yet discovered for the mouse germline. ENU has a low Swain-Scott substrate constant, which is a measure of the sensitivity of a substrate molecule to the nucleophilicity of a reagent molecule. Mutagens with low Swain-Scott substrate constants tend toward the following attributes. First, they employ a two-step SN1 mechanism (Shibuya and Morimoto, 1993). Second, they tend to be more active at alkylating oxygen molecules of the nitrogenous bases in DNA (especially O4-thymine, O2-thymine, and O2-cytosine). Third and finally, they tend to be mutagenic in the premeiotic stages of germ cell development (Vogel and Natarjan, 1995). Of reported mutations, 82% of those induced by ENU are AT to TA transversions and AT to GC transitions. These characteristic lesions are thought to arise from the aberrant base pairing of O4-thymine and O2-thymine, respectively (Justice, 2000).
Breeding Schemes
A variety of breeding schemes can be utilized in an ENU mutagenesis experiment (Fig. x.x.1). A one-generation breeding scheme can be used to produce animals displaying both autosomal and X-linked dominantly inherited traits. Two-generation schemes can be used in conjunction with mutations affecting a gene of interest, to isolate an allelic series or identify interacting genes. They are also useful in conjunction with deletion mutations, to isolate mutations prelocalized to the deletion interval, and to produce animals displaying X-linked recessive traits. Three-generation pedigree screens are used to produce animals displaying autosomal recessive traits. Typically these utilize a backcross of G2 females to their G1 parental males in order to increase the likelihood of mutation detection.
Figure 1.
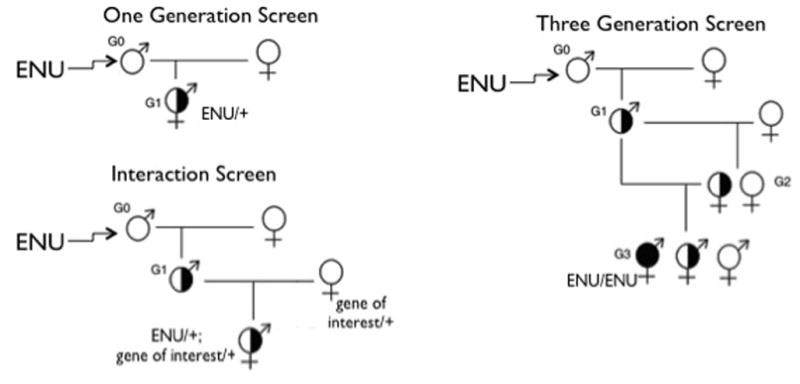
Breeding schemes for producing animals displaying mutant phenotypes.
Recently, specialized crosses have been used to more finely target specific phenotypes. An example is the use of “sensitized strains.” In this experiment a line carrying a specific mutation or heritable trait is used in mutagenesis experiments to identify loci that enhance or suppress a particular phenotype (Matera et al., 2008; Daxinger et al., 2013, Buchovecky et al., 2013). Mutagenesis has also been carried out on transgenic reporter mice, facilitating sensitive queries of specific developmental pathways (e.g. Dwyer et al., 2011; Ha et al., 2013). Finally, crosses with mice carrying inversions of specific regions (balancer chromosomes) have proven an effective means to permit the rapid localization of the subset of mutations that fall within the marked interval (Kile et al., 2003).
Applications
ENU mutagenesis can be used in a variety of different approaches. Perhaps the most effective has been the identification of novel alleles important in embryonic organogenesis (e.g. Herron et al., 2002; Huangfu and Anderson, 2005). This is likely due to the robust gene interaction network required for proper development as well as relatively little developmental homeostasis. Other successful applications of ENU mutagenesis have been to metabolic (Aigner et al., 2009), immunological (Hoebe and Beutler, 2008), and cellular phenotypes (Shima et al., 2003),
Mutation Mapping & Identification
The recent advances in sequencing technology and cost have significantly reduced the difficulty previously associated with mapping and sequence identification of causal mutations. The identification of a large number of SNPs associated with various mouse strains (e.g., MGI SNP database) have allowed a number of custom mapping panels to be created to allow initial mapping of mutations (e.g., Moran et al., 2006; Mega-MUGA panels). The genomic sequencing revolution now allows analysis of the entire coding sequence, “exome,” of a pool of mutants or exon-capture based on the map position at reasonable cost (e.g., Sheridan et al., 2011). Theoretically, whole genome sequencing could preclude the need to do any mapping by genetic outcross but the large number of mutations likely to be ascertained leads to a large challenge for verifying the single causal mutation (see below). The costs of next-generation sequencing can be somewhat alleviated by combining several mutant genomes into one sequencing experiment. The individual samples can be indexed to facilitate a post hoc analysis as necessary. The increasingly efficient costs of higher-coverage sequencing are making this an ever-more promising method of cloning. Extracting an ENU induced mutation from the large next-generation sequencing dataset is, in many cases, less difficult than other next-generation bioinformatic analyses. Most often the phenotypes are recessive, simplifying interpretation of the sequencing files as one desires only bases where the bioinformatics pipelines “calls” the read as “homozygous variant.” Furthermore, in many cases the mutant phenotypes are severe and not likely to simply be benign polymorphisms. As increasing numbers of mouse strains are being deeply sequenced, these natural variation (SNPs) of the species are become well-known and searchable (http://www.sanger.ac.uk/resources/mouse/), and be easily incorporated into an analysis of exome data to rule out some variants.
Validation
The application of next-generation sequencing, however, can dramatically increase the number of “variants,” or candidate mutations in the mutants of interest. These changes will represent the true mutation causing the phenotype, other non-causal ENU induced mutations, previously unknown strain specific polymorphisms, and spontaneous mutations arising independently. Some of these can be ruled by testing more phenotypic mouse mutants and looking for the mutation(s) held in common by all mutants. Depending on the design of the mutagenesis breeding scheme, genetic background can be used to determine which regions of the genome are from the mutagenized strain, and therefore must contain the mutation. This can be imputed from the genomic sequencing or determined with high-density SNP panels.
Fortunately, many options exist for generating a de novo allele of the gene of interest (see above –knockout mouse repositories and direct genome editing approaches). Whether a conditional allele or a null allele, this can be used in a complementation assay to demonstrate the candidate mutation is the truly causal mutation. In these assays, animals carrying both the ENU allele and the targeted allele of the mutated gene should have a phenotype matching or very close to, the ENU-generated phenotype. Some mutations will be predicted to code for a null allele and this can be verified in the laboratory. If, for example, a mutation shortens the transcript significantly or might result in an unstable protein, an antibody can used for immunoblotting to test for those effects in mutant tissue lysates. An ENU-induced predicted protein truncation and a Western immunoblot showed a smaller protein in mutants would be convincing evidence the appropriate gene and mutation have been identified.
Critical Parameters and Troubleshooting
ENU Dose
The single most critical parameter with respect to successful outcome is the ENU dose. This requires attention to accuracy in the determination of ENU stock concentration and in delivery. An excessively high dose will result in the permanent sterilization of the treated male (i.e., the spermatogonial stem cells fail to repopulate the testes), while a low dose will result in a low mutation frequency. It has been clearly shown that a fractionated-dose protocol is highly effective for generating mutations (Hitotsumachi et al., 1985), although some investigators continue to have success with single-dose regimens.
Mouse Strain
It has also been clearly shown that there is strain-dependent variation of ENU sensitivity (Davis et al., 1999; Justice et al., 2000). There are useful tabulations of effective treatment regimens (Hitotsumachi et al., 1985; Shedlovsky et al., 1986), but the logistics of a mutagenesis protocol generally preclude a rigorous determination of an optimal dose. Given this, and the time frame required to assess whether treatment was effective, one approach is to empirically determine the optimal dose protocol for the strain of interest by exposing animals to a range of doses. In this strategy, one uses a standard dose tolerated by most strains (e.g., 3 × 90 or 3 × 100 micrograms ENU per gram body weight), and includes additional cohorts of mice treated at higher and lower doses.
Population Size
One issue that cannot be readily addressed is the number of mice that must be examined to identify a novel allele of interest. This is because it is impossible to know the size of the target population—i.e., how many loci exist that, when mutated to a null or hypomorphic state, will result in the phenotype being examined. Moreover, it is impossible to know when all causal alleles leading to the phenotype of interest have been identified. Taking into account the variation in the induced mutation frequency from gene to gene and from mouse strain to mouse strain, and assuming an effective treatment, the average frequency of induced mutations at any specific locus is 1 mutation induced for every 1000 gametes recovered from ENU-treated animals. This varies as a function of a variety of factors. For example, mutagenic efficiency has a direct relationship to gene size and to the AT-richness of the gene. It is also important to sample enough progeny from a treated male to examine a representative population (suggested number: 30 to 50 gametes per male); however, because the germ line may be reconstituted by a limited number of spermatogonial stem cells, over-sampling often leads to repeat isolations.
Phenotype Assay
The ultimate success of a mutagenesis experiment is also contingent on the utilization of a suitable assay. This must be amenable to relatively high throughput analysis, given the potentially large numbers of mice that it will be necessary to examine. It is also desirable that the assay has low normal variation in the strain that is being tested. If this is not the case, it is likely that many apparent mutants will prove to be outliers of the normal phenotypic variation. Finally, it is important to evaluate the variation in the phenotype that is being assessed in different strain combinations to determine whether these introduce modifiers that increase phenotypic variability. This is because, in order to map the mutated locus, it will ultimately be necessary to score the phenotype in intercross or backcross progeny.
Animal Husbandry
Although many inbred mouse lines are readily available from commercial breeders, most efforts at mouse ENU mutagenesis will benefit from a mouse colony protocol that provides animals of various types. The following list provides an example of animals to have on hand:
Young males (8 to 10 weeks old) for treatment with mutagen. Males are used exclusively for the mutagenesis because ENU has been shown to be significantly more mutagenic toward male than female gametes (Russell et al., 1979). The 8 to 10 week age range is chosen because this covers the time when male mice are first becoming fertile. Since the overall time required to produce mutants can be long (see Time Considerations), it is important to begin the mutagenesis as early as possible in the reproductive life of the animal to ensure that a sufficient number of offspring are recovered to thoroughly sample the mutant gamete repertoire.
Fertile females as sentinel females to reveal when mutagenized males recover fertility.
Fertile females of specific inbred lines to cross to mutagenized males after they recover fertility.
A good source of information for mouse colony establishment and maintenance is Silver (1995).
Time Considerations
The time required to produce mutants will vary considerably depending the genetic screen used, the time required for treated animals to recover fertility, and the phenotypic assay. What follows is an estimate for a three-generation genetic screen to detect a trait in third generation neonates. More time would be needed to detect a trait in third generation adults. Less time would be needed to do a one or a two-generation screen.
Males being mutagenized must be 6 to 8 weeks old. The most common fractionated-dose treatment regimen requires 3 weeks. The recovery of fertility by mutagenized males will take a minimum of 10 to 15 weeks and some strains can take >30 weeks. When fertility is recovered, it is necessary to produce the desired 30 to 50 cohort of G1 offspring from each mutagenized male (cycling with two females per week would produce 30 to 50 animals in about 4 weeks). These will reach fertility in 6 weeks and G2 litters can be generated in 3 weeks. The female progeny require 6 weeks to reach fertility. G3 offspring from crosses between G2 and G1 animals can be generated in 3 weeks. Thus, a minimum of 40 to 50 weeks can be expected to elapse between the time of original ENU mutagenesis and the generation of the first G3 progeny that can be evaluated for recessive mutations.
Acknowledgments
The work in this article was funded by NIH grant nos. R01HD36404 and R01MH081187.
Footnotes
Internet Resources
Mammalian Genetics Unit, Harwell, U.K. ENU Mutagensis Web site.
http://www.informatics.jax.org/expression.shtml
Mouse Gene Expression Database (GXD) hosted by the Mouse Genome Informatics program at The Jackson Laboratory
The Allen Brain Atlas Project.
The GenitoUrinary Molecular Anatomy Project
http://www.informatics.jax.org/javawi2/servlet/WIFetch?page=snpQF
Mouse SNP Query Database from MGI
http://www.har.mrc.ac.uk/services/enu-dna-archive
Harwell Mouse ENU DNA Archive
http://csbio.unc.edu/CCstatus/Media/MegaMUGAFlyer.pdf
A recently developed high density SNP panel
http://www.sanger.ac.uk/resources/mouse/
Sanger Institute Mouse Genomics – deep sequencing of multiple inbred mouse strains
Key References
Fairfield et al., 201. See above.
Good evidence for the ability of exome sequencing to identify ENU mutations.
Justice, M.J. et al., 2000. See above.
The most complete collection of data pertaining to the sensitivity of mouse strains to ENU treatment.
Weber, J.S., Salinger, A., and Justice, M.J. 2000. Optimal N-ethyl-N-nitrosourea (ENU) doses for inbred strains. Genesis 26:230–233.
Further useful information about the dose-response characteristics of mouse inbred lines to ENU treatment.
Vivian, J.L. et al., 2002. See above.
A recent example of how ENU mutagenesis can be applied to embryonic stem cells in culture.
Contributor Information
Rolf Stottmann, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH.
David R. Beier, Center for Developmental Biology and Regenerative Medicine, Seattle Children’s Research Institute
Literature Cited
- Aigner B, Rathkolb B, Klaften M, Sedlmeier R, Klempt M, Wagner S, Michel D, Mayer U, Klopstock T, de Angelis MH, Wolf E. Generation of N-ethyl-N-nitrosourea-induced mouse mutants with deviations in plasma enzyme activities as novel organ-specific disease models. Experimental Physiology. 2009;94:412–21. doi: 10.1113/expphysiol.2008.045864. [DOI] [PubMed] [Google Scholar]
- Bode VC. Ethylnitrosourea mutagenesis and the isolation of mutant alleles for specific genes located in the T region of mouse chromosome 17. Genetics. 1984;108:457–470. doi: 10.1093/genetics/108.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchovecky CM, Turley SD, Brown HM, Kyle SM, McDonald JG, Liu B, Pieper AA, Huang W, Katz DM, Russell DW, Shendure J, Justice MJ. A suppressor screen in Mecp2 mutant mice implicates cholesterol metabolism in Rett syndrome. Nat Genetics. 2013;45:1013–1020. doi: 10.1038/ng.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Woychik RP, Justice MJ. Effective chemical mutagenesis in FVB/N mice requires low doses of ethylnitrosourea. Mammalian Genome. 1999;10:308–310. doi: 10.1007/s003359900992. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Harten SK, Oey H, Epp T, Isbel L, Huang E, Whitelaw N, Apedaile A, Sorolla A, Yong J, et al. An ENU mutagenesis screen identifies novel and known genes involved in epigenetic processes in the mouse. Genome Biol. 2013;14:R96. doi: 10.1186/gb-2013-14-9-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfield H, Gilbert GJ, Barter M, Corrigan RR, Curtain M, Ding Y, D’Ascenzo M, Gerhardt DJ, He C, Huang W, Richmond T, Rowe L, Probst FJ, Bergstrom DE, Murray SA, Bult C, Richardson J, Kile BT, Gut I, Hager J, Sigurdsson S, Mauceli E, Di Palma F, Lindblad-Toh K, Cunningham ML, Cox TC, Justice MJ, Spector MS, Lowe SW, Albert T, Donahue LR, Jeddeloh J, Shendure J, Reinholdt LG. Mutation discovery in mice by whole exome sequencing. Genome Biology. 2011;14:R86. doi: 10.1186/gb-2011-12-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger JH, Smith CM, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): update. 2011. Nucleic Acids Research. 2011;39:D835–41. doi: 10.1093/nar/gkq1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Stottmann RW, Furley AJ, Beier DR. A Forward Genetic Screen in Mice Identifies Mutants with Abnormal Cortical Patterning. Cereb Cortex. 2013 Aug 22; doi: 10.1093/cercor/bht209. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitotsumachi S, Carpenter DA, Russell WL. Dose-repetition increases the effectiveness of N-ethyl-N-nitrosourea in mouse spermatogonia. Proc Natl Acad Sci USA. 1985;82:6619–6621. doi: 10.1073/pnas.82.19.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SD, Armit C, Armstrong J, Brennan J, Cheng Y, Haggarty B, Houghton D, Lloyd-MacGilp S, Pi X, Roochun Y, Sharghi M, Tindal C, McMahon AP, Gottesman B, Little MH, Georgas K, Aronow BJ, Potter SS, Brunskill EW, Southard-Smith EM, Mendelsohn C, Baldock RA, Davies JA, Davidson D. The GUDMAP database--an online resource for genitourinary research. Development. 2011;138:2845–53. doi: 10.1242/dev.063594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe K, Beutler B. Forward genetic analysis of TLR-signaling pathways: an evaluation. Adv Drug Deliv Review. 2008;60:824–9. doi: 10.1016/j.addr.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Hrabé de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, Reis A, Richter T, Alessandrini F, Jakob T, Fuchs E, Kolb H, Kremmer E, Schaeble K, Rollinski B, Roscher A, Peters C, Meitinger T, Strom T, Steckler T, Holsboer F, Klopstock T, Gekeler F, Schindewolf C, Jung T, Avraham K, Behrendt H, Ring J, Zimmer A, Schughart K, Pfeffer K, Wolf E, Balling R. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nature Genetics. 2000;25:444–7. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- Herron BJ, Lu W, Rao C, Liu S, Peters H, Bronson RT, Justice MJ, McDonald JD, Beier DR. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nature Genetics. 2002;30:185–9. doi: 10.1038/ng812. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;12:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Justice MJ . International Mouse Knockout Consortium. Mutagenesis of the mouse germline. In: Jackson I, Abbot C, editors. Mouse Genetics and Transgenics: A practical Approach. Oxford University Press; Oxford: 2000. pp. 185–216. [Google Scholar]
- Justice MJ, Carpenter DA, Favor J, Neuhauser-Klaus A, Hrabé de Angelis M, Soewarto D, Moser A, Miller D, Chapman V, Weber JS, Rinchik EM, Hunsicker PR, Russell WL, Bode VC. Effects of ENU dosage on mouse strains. Mammalian Genome. 2000;11:484–488. doi: 10.1007/s003350010094. [DOI] [PubMed] [Google Scholar]
- Kile BT, Hentges KE, Clark AT, Nakamura H, Salinger AP, Liu B, Box N, Stockton DW, Johnson RL, Behringer RR, Bradley A, Justice MJ. Functional genetic analysis of mouse chromosome 11. Nature. 2003;425:81–6. doi: 10.1038/nature01865. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Leshchiner I, Alexa K, Kelsey P, Adzhubei I, Austin-Tse CA, Cooney JD, Anderson H, King MJ, Stottmann RW, Garnaas MK, Ha S, Drummond IA, Paw BH, North TE, Beier DR, Goessling W, Sunyaev SR. Mutation mapping and identification by whole-genome sequencing. Genome Res. 2012;22:1541–8. doi: 10.1101/gr.135541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, Murphy SL, Finn JD, Khazi FR, Zhou S, Paschon DE, Rebar EJ, Bushman FD, Gregory PD, Holmes MC, High KA. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;26:217–21. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera I, Watkins-Chow DE, Loftus SK, Hou L, Incao A, Silver DL, Rivas C, Elliott EC, Baxter LL, Pavan WJ. A sensitized mutagenesis screen identifies Gli3 as a modifier of Sox10 neurocristopathy. Hum Mol Genet. 2008;15:2118–31. doi: 10.1093/hmg/ddn110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JL, Bolton AD, Tran PV, Brown A, Dwyer ND, Manning DK, Bjork BC, Li C, Montgomery K, Siepka SM, Vitaterna MH, Takahashi JS, Wiltshire T, Kwiatkowski DJ, Kucherlapati R, Beier DR. Utilization of a whole genome SNP panel for efficient genetic mapping in the mouse. Genome Research. 2006;16:436–40. doi: 10.1101/gr.4563306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Bernard A, Lau C, Overly CC, Dong HW, Kuan C, Pathak S, Sunkin SM, Dang C, Bohland JW, Bokil H, Mitra PP, Puelles L, Hohmann J, Anderson DJ, Lein ES, Jones AR, Hawrylycz M. An anatomic gene expression atlas of the adult mouse brain. Nature Neuroscience. 2009;12:356–62. doi: 10.1038/nn.2281. [DOI] [PubMed] [Google Scholar]
- Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, Washbourne R, Hough T, Greenaway S, Hewitt M, Liu X, McCormack S, Pickford K, Selley R, Wells C, Tymowska-Lalanne Z, Roby P, Glenister P, Thornton C, Thaung C, Stevenson JA, Arkell R, Mburu P, Hardisty R, Kiernan A, Erven A, Steel KP, Voegeling S, Guenet JL, Nickols C, Sadri R, Nasse M, Isaacs A, Davies K, Browne M, Fisher EM, Martin J, Rastan S, Brown SD, Hunter J. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nature Genetics. 2000;25:440–3. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- Nolan PM, Kapfhamer D, Bucan M. Random mutagenesis screen for dominant behavioral mutations in mice. Methods. 1997;13:379–395. doi: 10.1006/meth.1997.0545. [DOI] [PubMed] [Google Scholar]
- Rajaraman S, Davis WS, Mahakali-Zama A, Evans HK, Russell LB, Bedell MA. Genetics. 2002;162:341–353. doi: 10.1093/genetics/162.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell WL, Kelly EM, Hunsicker PR, Bangham JW, Maddux SC, Phipps EL. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc Natl Acad Sci USA. 1979;76:5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Sezutsu H, Takahasi KR, Tsuchihashi K, Ichikawa R, Fujimoto N, Kaneko S, Nakai Y, Uchiyama M, Goda N, Motoi R, Ikeda A, Karashima Y, Inoue M, Kaneda H, Masuya H, Minowa O, Noguchi H, Toyoda A, Sakaki Y, Wakana S, Noda T, Shiroishi T, Gondo Y. Molecular characterization of ENU mouse mutagenesis and archives. Biochem Biophys Res Commun. 2005;336:609–16. doi: 10.1016/j.bbrc.2005.08.134. [DOI] [PubMed] [Google Scholar]
- Shedlovsky A, Guenet J-L, Johnson LL, Dove WF. Induction of recessive lethal mutations in the T/t – H-2 region of the mouse genome by a point mutagen. Genet Res Camb. 1986;47:135–142. doi: 10.1017/s0016672300022977. [DOI] [PubMed] [Google Scholar]
- Shedlovsky A, McDonald JD, Symula D, Dove WF. Mouse models for human PKU. Genetics. 1993;134:1205–1210. doi: 10.1093/genetics/134.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan R, Lampe K, Shanmukhappa SK, Putnam P, Keddache M, Divanovic S, Bezerra J, Hoebe K. Lampe1: An ENU-Germline Mutation Causing Spontaneous Hepatosteatosis Identified through Targeted Exon-Enrichment and Next-Generation Sequencing. PLoS One. 2007;6:e21979. doi: 10.1371/journal.pone.0021979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Morimoto K. A review of the genotoxicity of 1-ethyl-1-nitrosourea. Mutat Res. 1993;297:3–38. doi: 10.1016/0165-1110(93)90005-8. [DOI] [PubMed] [Google Scholar]
- Shima N, Hartford SA, Duffy T, Wilson LA, Schimenti KJ, Schimenti JC. Phenotype-based identification of mouse chromosome instability mutants. Genetics. 2003;163:1031–40. doi: 10.1093/genetics/163.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver LM. Mouse Genetics: Concepts and Applications. Oxford University Press; New York: 1995. [Google Scholar]
- Stottmann RW, Moran JL, Turbe-Doan A, Driver E, Kelley M, Beier DR. Focusing forward genetics: a tripartite ENU screen for neurodevelopmental mutations in the mouse. Genetics. 2011;188:615–24. doi: 10.1534/genetics.111.126862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian JL, Chen Y, Yee D, Schneider E, Magnuson T. An allelic series of mutations in Smad2 and Smad4 identfied in a genotype-based screen of N-ethyl-N-nitrosourea-mutagenized mouse embryonic stem cells. Proc Natl Acad Sci USA. 2002;99:15542–15547. doi: 10.1073/pnas.242474199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel EW, Natarjan AT. DNA damage and repair in somatic and germ line tissues. Mutat Res. 1995;330:183–208. doi: 10.1016/0027-5107(95)00040-p. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–9. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarbalis K, May SR, Shen Y, Ekker M, Rubenstein JL, Peterson AS. A focused and efficient genetic screening strategy in the mouse: identification of mutations that disrupt cortical development. PLoS Biology. 2004;2:E219. doi: 10.1371/journal.pbio.0020219. [DOI] [PMC free article] [PubMed] [Google Scholar]


