Abstract
Interfacing synthetic materials with biomacromolecules provides new systems for biological applications. We report the creation of a reversible multivalent supramolecular "zipper" recognition motif between gold nanoparticles and proteins. In this assembly, carboxylate-functionalized nanoparticles interact strongly with oligohistidine tags. This interaction can be tuned through His-tag length, and offers unique binding profiles based on the pH and electrolyte concentration of the medium.
Introduction
Tailoring molecular recognition between synthetic materials and biomolecules provides a versatile strategy for creating bioconjugate systems.1 A variety of supramolecular approaches have been devised to interface synthetic and biological systems for diverse applications.2 However, using these systems in physiological environments such as is challenging, where high concentrations of proteins and other biomolecules compete for interaction.
Co-engineering of biomolecules and synthetic materials and provides a strategy for generating high affinity and reversible molecular interactions.3 Inspiration for this codesign can be obtained from Nature: naturally occurring molecular zippers, including duplex DNA4 and leucine zippers5 exhibit robust multivalent reversible interactions in intracellular conditions. Microtubules polymerize and de-polymerize through the formation of specific molecular zippers.6 This multivalent motif7 has been used to create synthetic molecular duplexes8 through non-covalent interactions including electrostatic interactions,9 hydrogen bonding,10 π–π interactions,11 and van der Waals forces to generate zippers.12
Multivalency is a key structural prerequisite for zipper motifs. Nanomaterials offers molecular scaffolds that can be engineered to present multivalent recognition elements.13 Gold nanoparticles (AuNPs) provide a particularly versatile platform for biomolecular recognition,14 and have been interfaced with proteins for a wide variety of applications.15 The AuNP surface can be readily engineered to feature recognition elements. Additionally, AuNPs can be generated with sizes commensurate to proteins, providing surface complementarity for recognition while maintaining effective biological function.16
The metal ion-mediated oligohistidine-nitrilotriacetate recognition motif has been widely employed to capture proteins using nanomaterials.17 We hypothesized that the oligohistidine cationic tail18 used in this strategy could be employed as a zipper component for interaction with nanomaterials. In this report, we demonstrate a reversible molecular zipper between His-tagged proteins and carboxylate functionalized AuNPs. This zipper exhibits high affinity binding in physiologically relevant environments, including serum conditions. The system is also environmentally responsive, with binding dictated by solution pH. This new recognition motif presents opportunities for engineering specific molecular interactions between synthetic and biomolecules.
Results and discussion
The host nanoparticle was provided by AuNPs (2nm core diameter) functionalized with anionic ligands (AuNP-COOH) that can interact with proteins without denaturation.19 We next explored the interaction of these inherently multivalent carboxylate particles with a family of His-tagged green fluorescent proteins (GFP)20 (Fig. 1). We cloned and purified three eGFP21 variants carrying different length of N-terminal His-tags: one His (1×His-GFP), six His (6×His-GFP), and twelve His (12×His-GFP) to determine the required number of interactions, These proteins were all anionic, with predicted pI values of 5.8, 6.1, and 6.5, respectively.
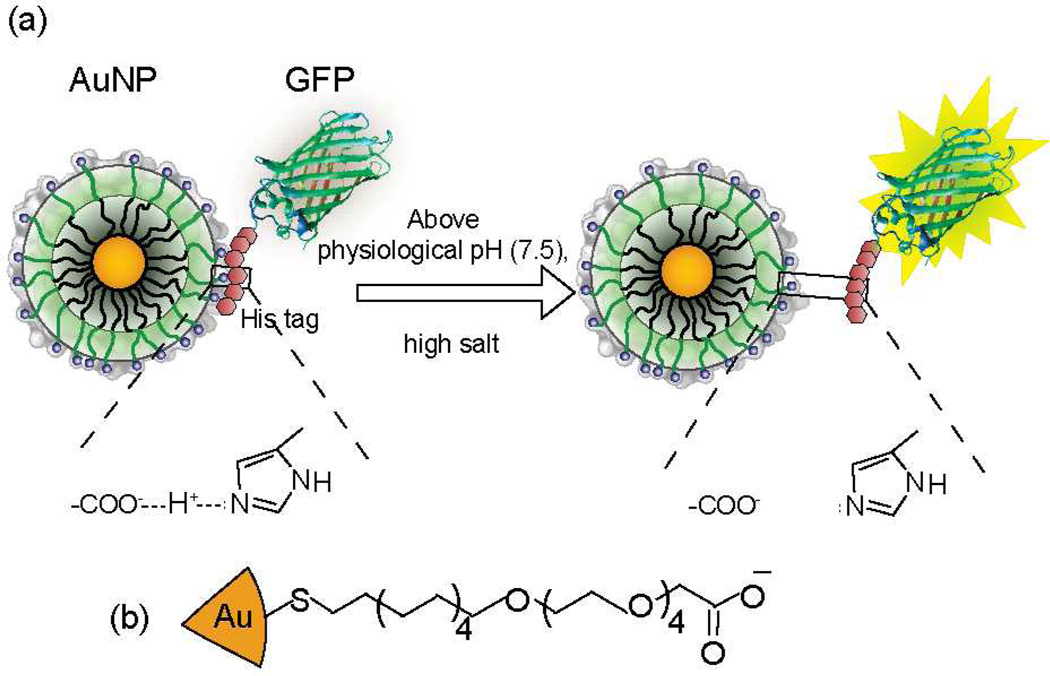
Fig. 1.
(a) Zipper formation between AuNP-COOH and N-terminus oligohistidine-tagged GFPs through carboxylate-histidine interaction (b) The chemical structure of 2 nm gold core naoparticle AuNP-COOH.
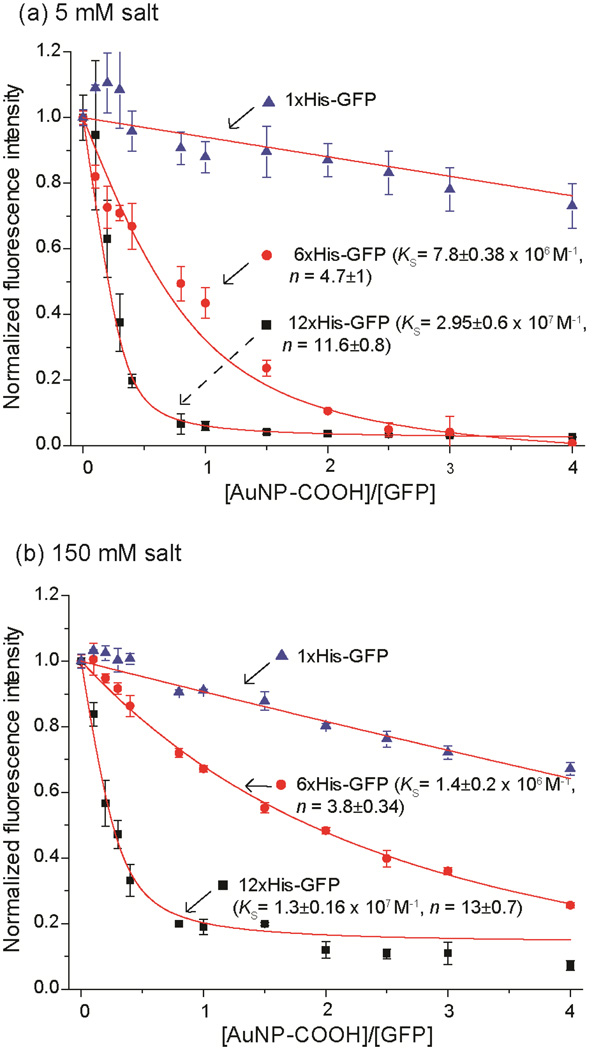
The binding efficiency of AuNP-COOH with the His-tagged GFPs was quantified through fluorescence titration,22 utilizing the quenching properties of the AuNP.23 At low ionic strength (5 mM phosphate buffer, PB) AuNP-COOH bound both 12×His-GFP and 6×His-GFP with high affinity (Fig. 2a). The binding constant (KS) values for 12×His-GFP (Ks= 2.95 ±0.6 × 107 M−1) was ~3-fold higher than that of 6×His-GFP (Ks= 7.8 ±0.38 × 106 M−1), indicating that multivalency is crucial for zipper formation. Interestingly, more GFPs bound to each nanoparticle for 12×His-GFP (n= 11.6 ±0.8) than for 6×His-GFP (n= 4.7 ±1), potentially due to decreased secondary repulsion between the anionic GFPs.24 No observable binding was observed with 1×His-GFP, demonstrating that specific zipper formation was required for interaction.
Fig. 2.
The interaction of AuNP-COOH with His-tagged GFP variants. Fluorescence (λex=475 nm, λem=510 nm) titrations between nanoparticles and GFPs (100 nM) in (a) 5 mM phosphate buffer (PB), and (b) PBS buffer (150 mM NaCl in 5 mM PB) at pH 7.4. The complex association constant (KS) and the binding stoichiometry (n) were determined using previously reported method.21
The pragmatic use of non-covalent bioconjugates requires high affinity interactions at physiological ionic strength. In previous studies, electrostatic interactions between nanoparticles and proteins were fully disrupted at quite low salt concentrations, typically 10–50 mM salt.25 In contrast, high binding affinities were observed between AuNP-COOH and both 12×His-GFP (Ks= 1.3 ±0.16 × 107 M−1), and 6×His-GFP (Ks= 1.4 ±0.2 × 106 M−1) in PBS buffer (150 mM NaCl in 5 mM PB, pH 7.4) (Fig. 2b). Notably, a larger n value was observed for 12×His-GFP, similar to the one at low (5 mM) electrolyte concentration.
Reversible zipper formation at physiologically relevant conditions
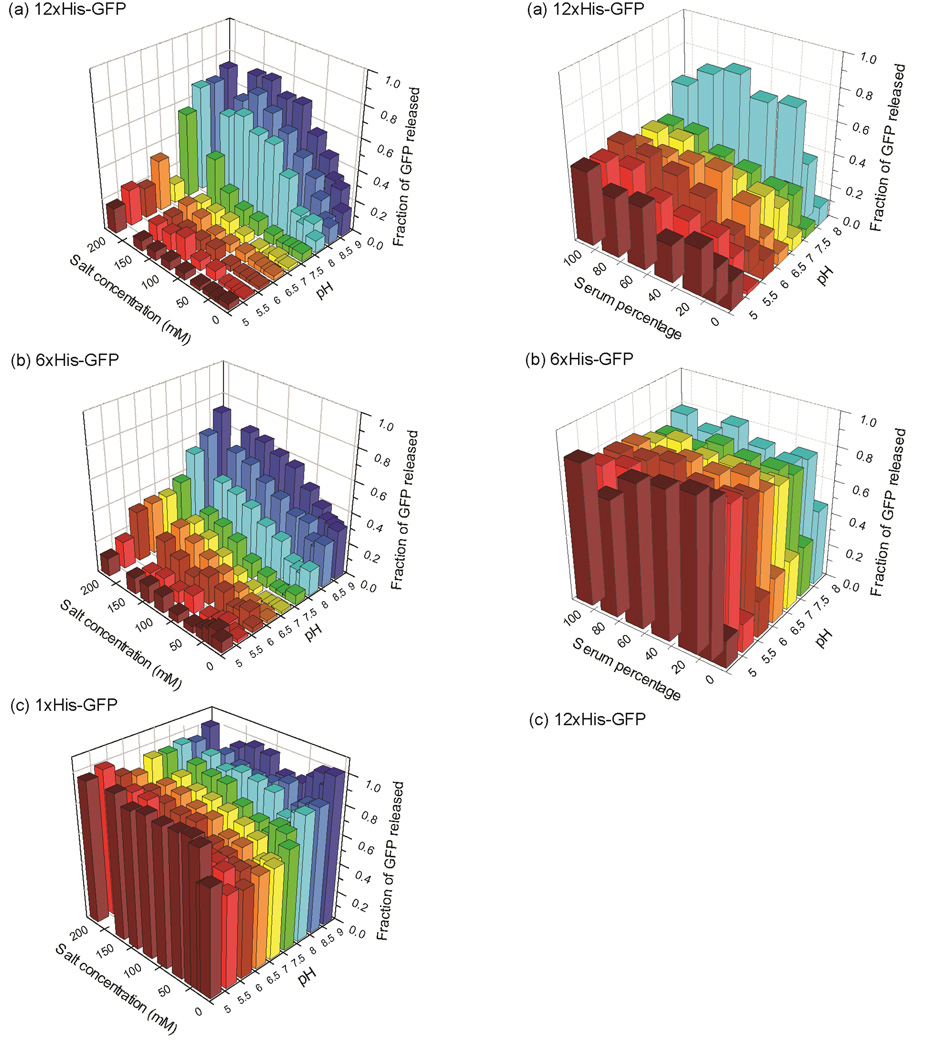
One of the key advantages of supramolecular bioconjugates is their ability to respond to environmental changes. pH is an important biological parameter. For example, normal tissues have a pH of 7.4, while tumor tissues have lower pH (~6 to 7).26 Additionally, pH decreases through the endosomal/lysosomal pathways inside cells, reaching a pH of ~4.8.27 In our system, the histidine tag in GFPs offers a potentially pH-switchable recognition scaffold. To explore this possibility, we investigated the pH and ionic strength dependent reversibility of the carboxylate-histidine zipper formation. Both 12×His-GFP and 6×His-GFP interacted strongly with AuNP-COOH below pH ~7.5 at physiological salt concentration (PBS). Significantly, above pH ~7.5 the carboxylate-histidine zipper disassembled, releasing the GFP from the nanoparticles surface (Fig. 3a and 3b). As expected, 1×His-GFP did not interact with nanoparticles at any condition (Fig. 3c). Taken together, these studies demonstrated the pH response of the zipper motif.
Fig. 3.
Responsiveness of the carboxylate-histidine zipper towards pH and salt concentration. Fluorescence titrations between 400 nM of AuNP-COOH and 100 nM of (a) 12×His-GFP, (b) 6×His-GFP, and (c) 1×His-GFP were performed parametrically varied pH and salt (NaCl) concentrations in 5 mM PB. The intensity of GFP released from nanoparticles was normalized against the intensity of free GFP.
Reversible zipper formation in serum conditions
In vivo applications including protein and gene delivery require specific and reversible interactions between synthetic carrier materials and the cargo molecules in serum.28 Serum presents a complex competitive chemical environment featuring a high (~1 mM) concentration of protein,29 making it challenging to engineer effective recognition motifs. We parametrically investigated the serum concentration and pH dependent reversibility of the carboxylate-histidine zipper. At pH <7.5 and at 10% serum (cell culture condition), 12×His-GFP exhibited a high affinity binding towards AuNP-COOH (Fig. 4a). Significantly, in 55% serum condition (in vivo condition) at pH 7.5 there was substantial binding between AuNP-COOH and 12×His-GFP (Fig. 4c). While the binding isotherm is complex, considerable binding was observed at high nanomolar concentrations. In contrast, 6×His-GFP did not bind with AuNP-COOH at any serum condition under investigation (Fig. 4b), indicating that a high degree of multivalency is crucial for carboxylate-histidine zipper formation in complex biological environments.
Fig. 4.
Reversible carboxylate-histidine zipper formation between AuNP-COOH and (a) 12×His-GFP, and (b) 6×His-GFP at serum conditions. 400 nM of AuNP-COOH was titrated against 100 nM of His-tagged GFPs varying the serum percentage and pH at 150 mM salt (1×PBS) concentration. (c) Fluorescence titrations between AuNP-COOH and 12×His-GFP (100 nM) at 55% serum condition, pH 7.4.
Conclusions
In summary, we have tailored a molecular zipper based on multivalent carboxylate-histidine interactions through co-engineering of the AuNP surface and proteins. The carboxylate-histidine zipper exhibited high affinity interactions under physiologically relevant conditions that were pH responsive, making these systems attractive starting points for delivery and imaging applications. In a broaer context, these studies demonstrate how co-engineering of biomolecules and nanoparticles can be used to generate bioconjugates with new and useful properties..
Experimental section
Materials and methods
Cloning and over expression of green fluorescent proteins (GFPs)
Genetic engineering manipulation and protein expression were done according to standard protocols. (a) To generate 1×His-GFP, a constitutive expression vector (pUCCB-ntH6-eGFP) was purchased from Addgene (plasmid id- 32557).30 For the sake of purification, a 6×His tag was placed on the N-terminus of 1×His-GFP, upstream of a thrombin cleavage site. (b) 6×His-GFP expression vector (pET21-d-GFP) was obtained from Novagen. (c) 12×His-GFP was generated by incorporating twelve histidines in the N-terminus of GFP. Briefly, using GFP as the template, PCR was performed with the following primers. Subsequently, the PCR product was digested (using BamHI and HindIII restriction enzymes) and inserted into pQE80 vector, downstream of nucleotides for six histidine tag to construct pQE80-12×His-GFP expression vector. Successful cloning was confirmed by DNA sequencing. Forward primer: 5’- ACGATGGATCCCACCATCACCAT -3’ Reverse primer: 5’- GTGACAAGCTTTTACTTGTACAGCTC -3’
To produce recombinant proteins, plasmids carrying 1×His-GFP, 6×His-GFP, or 12×His-GFP was transformed into Escherichia coli BL21(DE3) strain. A transformed colony was picked up to grow small cultures in 50 mL 2×YT media at 37 °C for overnight. The following day, 15 mL of grown culture was inoculated into one liter 2×YT media and allowed to grow at 37 °C until OD reaches 0.6. At this point, the protein expression was induced by adding isopropyl-b-D-thiogalactopyranoside (IPTG; 1 mM final concentration) at 25 °C. After 16 hours of induction, the cells were harvested and the pellets were lysed using a microfluidizer. His-tagged fluorescent proteins were purified from the lysed supernatant using HisPur cobalt columns. The integrity and the purity of native protein were determined by 12% SDS-PAGE gel.
1×His-GFP was cleaved from its 6×His tag using thrombin-agarose beads (Thrombin CleanCleave™ Kit, Sigma-Aldrich) as described in the instruction manual. After the cleavage, 1×His-GFP was passed through a HisPur cobalt column to remove the cleaved 6×His tag. Further, the residual 6×His was removed by a 10KD-MWCO (molecular weight cut off) filter.
Synthesis and characterization of nanoparticles
Carboxylate functionalized gold nanoparticles (AuNP-COOH) were synthesized according to a previous report.31 Briefly, Brust-Schiffrin two-phase synthesis was used to synthesize pentanethiol-coated AuNPs with core diameter ~2 nm.32 The Murray place-exchange method was followed to obtain AuNP-COOH.33 The monolayer protected nanoparticles were re-dispersed in water. The excess ligand/pentanethiol were removed by dialysis using a 10,000 MWCO snake-skin membrane. The final concentration was measured by UV spectroscopy at 502 nm. To assess their quality, the nanoparticles were characterized by Zeta potential (surface charge), Dynamic Light Scattering (DLS) (hydrodynamic radius), and Transmission Electron Microscopy (TEM) (core size) as shown in Fig. S1.
Fluorescence titration
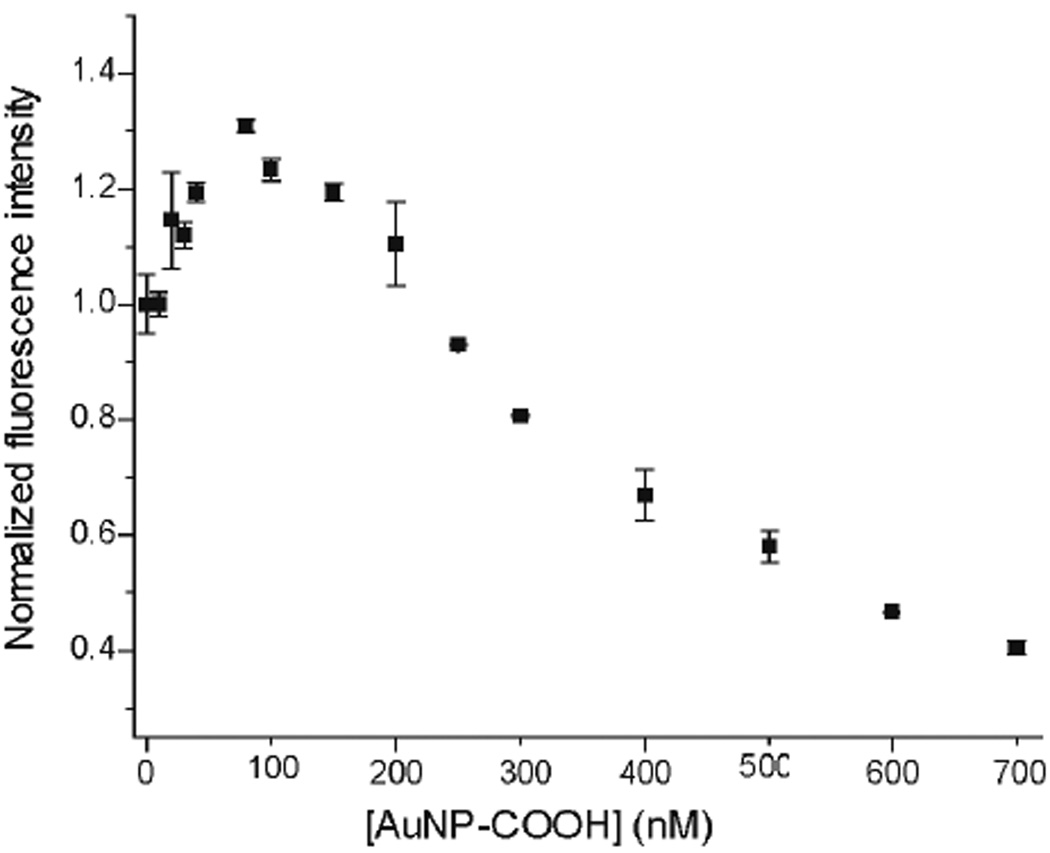
Fluorescence titration experiments between nanoparticles and GFPs were carried out as described previously.34 Briefly, the change of fluorescence intensity of GFPs at 510 nm was measured with an excitation wavelength of 475 nm at various concentrations of nanoparticles from 0 to 400 nM on a Molecular Devices SpectraMax M3 microplate reader (at 25 °C). Quenching of fluorescence intensity arising from 100 nM GFP was observed with increasing nanoparticle concentration. Nonlinear least-squares curve fitting analysis was carried out to estimate the binding constant (KS) and association stoichiometry (n, [GFP]/[AuNP-COOH]) using a one site binding model.21
For the pH and salt dependent interactions (fluorescence titrations) between nanoparticles and GFPs, the concentration of GFP chosen was 100 nM for each study. The concentrations of AuNP-COOH used for the titrations were 400 nM. The fluorescence intensity for each study was normalized against the intensity of GFP without nanoparticles at their respective pH and salt (NaCl in 5 mM PB) concentration. The titrations were carried out in triplicates, and repeated at least twice with different batches of nanoparticles.
Similar fluorescence titrations were performed for the serum concentration and pH dependent interactions between AuNP-COOH and His-tagged GFPs. Both the nanoparticle (400 nM) and GFP (100 nM) concentrations were kept fixed, varying the serum percentage and pH of the solutions. In a typical experiment, AuNP-COOH/GFP complexes were made first, incubated at dark for 10 minutes, then the required serum amount was added to the complexes, followed by immediate shaking for 30 seconds. Fluorescence reading was taken after 30 minutes of incubation.
Supplementary Material
Acknowledgements
This research was funded by NIH (GM077173 and EB014277), and NSF instrumentation (MRSEC, DMR-0820506).
Footnotes
Electronic Supplementary Information (ESI) available: Nanoparticle characterizations- i.e. Zeta potential, DLS, and TEM and GFP sequence. See DOI: 10.1039/b000000x/
Notes and references
- 1.(a) Lehn JM. Science. 2002;295:2400. doi: 10.1126/science.1071063. [DOI] [PubMed] [Google Scholar]; (b) Zhang J, Landry MP, Barone PW, Kim JH, Lin S, Ulissi ZW, Lin D, Mu B, Boghossian AA, Hilmer AJ, Rwei A, Hinckley AC, Kruss S, Shandell MA, Nair N, Blake S, Sen F, Sen S, Croy RG, Li D, Yum K, Ahn JH, Jin H, Heller DA, Essigmann JM, Blankschtein D, Strano MS. Nat. Nanotechnol. 2013;8:959. doi: 10.1038/nnano.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Ostuni E, Yan L, Whitesides GM. Colloids Surf. B. 1999;15:3. [Google Scholar]; (b) Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Nature. 2000;405:665. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]; (c) Seeman NC, Belcher AM. Proc. Natl. Acad. Sci. U S A. 2002;99:6451. doi: 10.1073/pnas.221458298. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Niemeyer CM. Angew. Chem. Int. Ed. 2001;40:4128. doi: 10.1002/1521-3773(20011119)40:22<4128::AID-ANIE4128>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]; (e) Cooper W, Waters ML. Curr. Opin. Chem. Biol. 2005;9:627. doi: 10.1016/j.cbpa.2005.10.015. [DOI] [PubMed] [Google Scholar]; (e) Chen I, Ting AY. Curr. Opin. Biotechnol. 2005;16:35. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]; (f) Mout R, Moyano DF, Rana S, Rotello VM. Chem. Soc. Rev. 2012;41:2539. doi: 10.1039/c2cs15294k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostiainen MA, Hiekkataipale P, Laiho A, Lemieux V, Seitsonen J, Ruokolainen J, Ceci P. Nat. Nanotechnol. 2013;8:52. doi: 10.1038/nnano.2012.220. [DOI] [PubMed] [Google Scholar]
- 4.Weaver RF. Molecular Biology. 5th ed. McGraw-Hill; 2011. [Google Scholar]
- 5.(a) Nir E, Kleinermanns K, de Vries MS. Nature. 2000;408:949. doi: 10.1038/35050053. [DOI] [PubMed] [Google Scholar]; (b) Landschulz WH, Johnson PF, McKnight SL. Science. 1988;240:1759. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]; (c) Munro OQ, du Toit K, Drewes SE, Crouch NR, Mulholland DA. New J. Chem. 2006;30:197. [Google Scholar]
- 6.(a) Kikkawa M, Metlagel Z. Cell. 2006;127:1302. doi: 10.1016/j.cell.2006.12.009. [DOI] [PubMed] [Google Scholar]; (b) Sandblad L, Busch KE, Tittmann P, Gross H, Brunner D, Hoenger A. Cell. 2006;127:1415. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Levine PM, Carberry TP, Holub JM, Kirshenbaum K. Med. Chem. Commun. 2013;4:493. [Google Scholar]
- 8.(a) Bisson AP, Carver FJ, Hunter CA, Walthot JP. J. Am. Chem. Soc. 1994;116:10292. [Google Scholar]; (b) Zeng H, Yang X, Flowers RA, Gong B. J. Am. Chem. Soc. 2002;124:2903. doi: 10.1021/ja010701b. [DOI] [PubMed] [Google Scholar]; (c) Yang XW, Hua FJ, Yamato K, Ruckenstein E, Gong B, Kim W, Ryu CY. Angew. Chem. Int. Ed. 2004;43:6471. doi: 10.1002/anie.200460472. [DOI] [PubMed] [Google Scholar]; (d) Wu L, McElheny D, Huang R, Keiderling TA. Biochemistry. 2009;48:10362. doi: 10.1021/bi901249d. [DOI] [PubMed] [Google Scholar]; (e) Hwang S, Hilty C. J. Phys. Chem. B. 2011;115:15355. doi: 10.1021/jp206405b. [DOI] [PubMed] [Google Scholar]; (f) Culik RM, Jo H, DeGrado WF, Gai F. J. Am. Chem. Soc. 2012;134:8026. doi: 10.1021/ja301681v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diss ML, Kennan AJ. Org. Lett. 2008;10:3797. doi: 10.1021/ol801461a. [DOI] [PubMed] [Google Scholar]
- 10.Spencer R, Chen KH, Manuel G, Nowick JS. Eur. J. Org. Chem. 2013;3523 [Google Scholar]
- 11.Waters ML. Biopolymers. 2004;76:435. doi: 10.1002/bip.20144. [DOI] [PubMed] [Google Scholar]
- 12.(a) Bisson AP, Carver FJ, Eggleston DS, Haltiwanger RC, Hunter CA, Livingstone DL, McCabe JF, Rotger C, Rowan AE. J. Am. Chem. Soc. 2000;122:8856. [Google Scholar]; (b) Kim HW, Jung J, Han M, Lim S, Tamada K, Hara M, Kawai M, Kim Y, Kuk Y. J. Am. Chem. Soc. 2011;133:9236. doi: 10.1021/ja2031486. [DOI] [PubMed] [Google Scholar]
- 13.(a) Rana S, Bajaj A, Mout R, Rotello VM. Adv. Drug. Deliv. Rev. 2012;64:200. doi: 10.1016/j.addr.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Drechsler U, Erdogan B, Rotello VM. Chemistry. 2004;10:5570. doi: 10.1002/chem.200306076. [DOI] [PubMed] [Google Scholar]; (c) Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Angew. Chem. Int. Ed. Engl. 2010;49:3280. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel MC, Astruc D. Chem. Rev. 2004;104:293. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 15.(a) Walkey CD, Chan WC. Chem. Soc. Rev. 2012;41:2780. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]; (b) Mout R, Rotello VM. Isr. J. Chem. 2013;53:521. [Google Scholar]
- 16.Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 17.(a) Abad JM, Mertens SF, Pita M, Fernández VM, Schiffrin DJ. J. Am. Chem. Soc. 2005;127:5689. doi: 10.1021/ja042717i. [DOI] [PubMed] [Google Scholar]; (b) De M, Rana S, Rotello VM. Macromol. Biosci. 2009;9:174. doi: 10.1002/mabi.200800289. [DOI] [PubMed] [Google Scholar]; (c) Boeneman K, Mei BC, Dennis AM, Bao G, Mattoussi JRH, Medintz IL. J. Am. Chem. Soc. 2009;131:3828. doi: 10.1021/ja809721j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marti DN, Bosshard HR. J. Mol. Biol. 2003;330:621. doi: 10.1016/s0022-2836(03)00623-5. [DOI] [PubMed] [Google Scholar]
- 19.(a) Bayraktar H, You CC, Rotello VM, Knapp MJ. J. Am. Chem. Soc. 2007;129:2732. doi: 10.1021/ja067497i. [DOI] [PubMed] [Google Scholar]; (b) Fischer NO, Verma A, Goodman CM, Simard JM, Rotello VM. J. Am. Chem. Soc. 2003;125:13387. doi: 10.1021/ja0352505. [DOI] [PubMed] [Google Scholar]; (c) Hong R, Emrick T, Rotello VM. J. Am. Chem. Soc. 2004;126:13572. doi: 10.1021/ja0461163. [DOI] [PubMed] [Google Scholar]
- 20.Tsien RY. Annu. Rev. Biochem. 1998;67:509. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 21.Cormack BP, Valdivia RH, Falkow S. Gene. 1996;173:33. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 22.You CC, De M, Han G, Rotello VM. J. Am. Chem. Soc. 2005;127:12873. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]
- 23.(a) Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, Rotello VM. Chem. Sci. 2010;1:134. [Google Scholar]; (b) De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UH, Rotello VM. Nat. Chem. 2009;1:461. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torrens F, Castellano G. J. Cheminform. 2010;2(Suppl 1):P12. [Google Scholar]
- 25.(a) Wang L, Wang H, Yuan L, Yang W, Wu Z, Chen H. J. Mater. Chem. 2011;21:13920. [Google Scholar]; (b) Peng ZG, Hidajat K, Uddin MS. J. Colloid Interface Sci. 2004;271:277. doi: 10.1016/j.jcis.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 26.(a) Tannock IF, Rotin D. Cancer Res. 1989;49:4373. [PubMed] [Google Scholar]; (b) Helmlinger G, Yuan F, Dellian M, Jain RK. Nat. Med. 1997;3:177. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 27.Sorkin A, Von Zastrow M. Nat. Rev. Mol. Cell Biol. 2002;3:600. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 28.Tang R, Kim CS, Solfiell DJ, Rana S, Mout R, Velázquez-Delgado EM, Chompoosor A, Jeong Y, Yan B, Zhu ZJ, Kim C, Hardy JA, Rotello VM. ACS Nano. 2013;7:6667. doi: 10.1021/nn402753y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG. Mol. Cell Proteomics. 2002;1:947. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 30.Vick JE, Johnson ET, Choudhary S, Bloch SE, Lopez-Gallego F, Srivastava P, Tikh IB, Wawrzyn GT, Schmidt-Dannert C. Appl. Microbiol. Biotechnol. 2011;92:1275. doi: 10.1007/s00253-011-3633-4. [DOI] [PubMed] [Google Scholar]
- 31.Hong R, Emrick T, Rotello VM. J. Am. Chem. Soc. 2004;126:13572. doi: 10.1021/ja0461163. [DOI] [PubMed] [Google Scholar]
- 32.(a) Kanaras AG, Kamounah FS, Schaumburg K, Kiely CJ, Brust M. Chem. Commun. 2002;20:2294. doi: 10.1039/b207838b. [DOI] [PubMed] [Google Scholar]; (b) Brust M, Walker M, Bethell D, Schiffrin DJ, Whyman R. J. Chem. Soc. Chem. 1994;801 [Google Scholar]
- 33.Templeton AC, Wuelfing MP, Murray RW. Acc. Chem. Res. 2000;33:27. doi: 10.1021/ar9602664. [DOI] [PubMed] [Google Scholar]
- 34.Rana S, Singla AK, Bajaj A, Elci SG, Miranda OR, Mout R, Yan B, Jirik FR, Rotello VM. ACS Nano. 2012;6:8233. doi: 10.1021/nn302917e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.