Abstract
More than any other species, humans form social ties to individuals who are neither kin nor mates, and these ties tend to be with similar people. Here, we show that this similarity extends to genotypes. Across the whole genome, friends’ genotypes at the single nucleotide polymorphism level tend to be positively correlated (homophilic). In fact, the increase in similarity relative to strangers is at the level of fourth cousins. However, certain genotypes are also negatively correlated (heterophilic) in friends. And the degree of correlation in genotypes can be used to create a “friendship score” that predicts the existence of friendship ties in a hold-out sample. A focused gene-set analysis indicates that some of the overall correlation in genotypes can be explained by specific systems; for example, an olfactory gene set is homophilic and an immune system gene set is heterophilic, suggesting that these systems may play a role in the formation or maintenance of friendship ties. Friends may be a kind of “functional kin.” Finally, homophilic genotypes exhibit significantly higher measures of positive selection, suggesting that, on average, they may yield a synergistic fitness advantage that has been helping to drive recent human evolution.
Keywords: genetics, social networks, kinship detection
Human social interactions, and the networks they give rise to, show striking structural regularities (1, 2), even when comparing modernized networks with those in hunter–gatherer societies (3). Indeed, friendship is a fundamental characteristic of human beings (3–5), and genes are known to play a role in the formation (6), attributes (7), and structures (8) of friendship ties. Social ties also evince homophily, the tendency of people to form connections with phenotypically similar others (9). Evolutionary models suggest that homophily can evolve under a wide range of conditions if there is a fitness advantage to same-type interactions (10, 11). And candidate gene studies (12, 13) have recently identified one gene variant that exhibits positive correlation or similarity between friends (homophily) and another variant that exhibits negative correlation or dissimilarity (heterophily). It remains unclear, however, whether this phenomenon extends to multiple genotypes across the whole genome, and it is not known what role genotypic correlation may have played in human evolution.
There are (at least) four possible reasons that friends may exhibit homophily in their genotypes (12). First, correlation in genotypes may be a trivial by-product of the tendency of people to make friends with geographically proximate or ethnoracially similar individuals who also tend to share the same ancestry. Thus, it is important to use strict controls for population stratification in tests of genetic correlation (below, we rely on the widely used principal-components method to control for ancestry). Second, people may actively choose and retain friends of a similar genotype or they may avoid or terminate friendships with people who have different genotypes (“birds of a feather flock together”). This process may take place via a variety of mechanisms; for example, although it is unlikely that people would observe the actual genotypes of others around them, they can observe and prefer certain phenotypes, and these may obviously be influenced by specific genotypes. It is well known that people prefer to associate with others they resemble phenotypically (9), but what is not known is how this observation translates to the single-nucleotide polymorphism (SNP) level. Third, people may actively choose particular environments, and, in those environments, they may be more likely to encounter people with similar phenotypes influenced by specific genotypes. If people then choose friends from within these environments (even at random), it would tend to generate correlated genotypes. Fourth, people may be chosen by third parties or otherwise selected into environments or circumstances where they then come into contact with similar people. These four reasons are not mutually exclusive, of course, and they may operate in parallel; two people may become friends through both active choice of each other and active or passive choice of a convivial environment.
In contrast, there are fewer reasons that friends may exhibit heterophily in their genotypes (12). For example, heterophily is not likely to arise by population stratification, nor by a simple process of people choosing, or being drawn to, the same environment for the same reason. Instead, there are two other processes that might be at work. First, certain environments might foster interactions between individuals with dissimilar traits. Second, people may actively choose to befriend people of a different type (“opposites attract”). In fact, such a phenomenon has been proposed for reproductive relationships, and some experiments suggest that men and women may choose partners with opposite immune system genotypes (14, 15).
Importantly, all of these processes may be at work simultaneously, and humans may select friends and environments based on a wide variety of traits, some of which result in advantages when homophily is present (synergy) and others of which may yield advantages to heterophily (complementarity or specialization) (3, 11). The people to whom we are connected provide important capabilities, from the ability to ward off infections, to the ability to transmit or exploit useful information, to the ability to reciprocate cooperative exchanges. Consequently, the fitness advantage of some gene variants might be influenced by their parallel presence or absence in other individuals to whom a person is connected.
Evolutionary models show that preferences for both homophily and heterophily can evolve depending on the relative fitness advantages of genotypic similarity and dissimilarity on given traits (10). However, these models also show that homophily evolves under a much wider variety of conditions than heterophily—even when the fitness advantage to dissimilarity exceeds the fitness advantage to similarity (10). The reason is that it is less costly to find and successfully interact with a similar partner in a population of similar individuals than it is to find and successfully interact with a dissimilar partner in a population of dissimilar individuals. For an intuition regarding this observation, consider populations at fixation. For populations with an advantage to homophily, all individuals have the same trait at fixation, and so they all will gain the advantage in every interaction. In contrast, for populations with an advantage to heterophily, some individuals have one trait and some have another, meaning there are still likely to be at least some same-type encounters in the population that do not yield the advantage to dissimilar-type interactions. These theoretical models thus suggest that we should find more genotypes that are positively correlated between friends than negatively correlated and that we should, on average, expect friends to exhibit greater genetic similarity across the genome as a whole (10).
If homophily generally contributes to evolutionary fitness across a wide variety of traits, then we would also expect to see signs of positive selection for genes that exhibit positive correlation between friends. If so, it would suggest that our capacity to make friends with unrelated strangers may have played a role in human evolution. This capacity to form friendships and this preference for homophily [which is also seen in other social animals such as dolphins (16) and primates (17)] may possibly reflect the extended workings of a kin detection system (18) such that genetically similar (but unrelated) friends are a kind of “functional kin.” Humans may—when choosing friends from among individuals who are strictly not related to them—come to choose individuals who do, after all, resemble them on a genotypic level.
Here, we conduct, to our knowledge, the first genome-wide analysis of correlation in genotypes between friends. We emphasize that we are not conducting a genome-wide association study (GWAS) of a propensity to be friendly (or some similar complex social trait); rather, we are using GWAS techniques to identify certain theorized patterns (10) across the whole genome. Using data from the Framingham Heart Study, we analyzed 466,608 (unimputed) SNPs in 1,932 unique subjects who are in one or more of 1,367 friendship pairs (see SI Appendix for data construction and summary). The data we used (which we have uploaded to a shared data repository at www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000153.v6.p5) are exceedingly scarce; we know of no other dataset of any significant size that has information on both friendship ties and common genetic variants across the whole genome (SI Appendix). As a check against false positives, beyond the other procedures described below, we also performed a split-sample replication study. We also emphasize that, as in other whole-genome investigations with circumscribed samples (19, 20), our interest is not in any particular SNP, but rather in the pattern across the whole genome.
To assess general, overall homophily within pairs of friends, we calculated the kinship coefficient (21) (the probability that two alleles sampled at random from two individuals are identical by state), a measure that is equal to half the relatedness measure used in genome-wide complex trait analysis (GCTA) approaches (22) (although the pairs of friends here are not actually related). Positive values for this measure indicate that genotypes are positively correlated, and negative values indicate that two individuals are not related and, in fact, tend to have opposite genotypes. To measure heterophily, we calculated the empirical probability that two individuals have opposite genotypes at a given SNP, measured by the proportion of SNPs for which neither allele is identical by state.
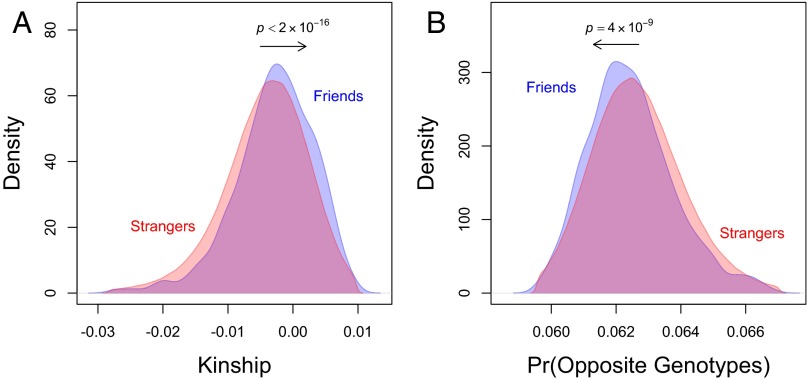
For comparison, we also calculated these measures for all nonkin “stranger” pairs using the same set of 1,932 subjects who are in the friends sample. After removing kin (who can, of course, be identified using genotyping) and after removing pairs who had a social relationship (i.e., friends, spouses, etc.), we identified 1,196,429 stranger pairs (SI Appendix). Fig. 1A shows that the distribution of kinship coefficients for friends is shifted right relative to the strangers. A simple difference-in-means test suggests that friends tend to be significantly more genetically “related” than strangers (+0.0014, P < 2 × 10−16), and, as a benchmark, the size of the difference roughly corresponds to the kinship coefficient we would expect for fourth cousins (0.0010). This difference cannot be explained by the ancestral composition of the sample or by cryptic relatedness because the same people are used in both the friends and strangers samples (the only thing that differs is the set of relationships between them); and we emphasize again that we can be sure these pairs of friends are not, in fact, distant cousins because they are strictly unrelated and there is no identity by descent. Meanwhile, Fig. 1B shows that friends also tend to have fewer SNPs where the genotypes are exactly opposite (–0.0002, P = 4 × 10−9). Both of these results indicate that pairs of (strictly unrelated) friends generally tend to be more genetically homophilic than pairs of strangers from the same population, but the weaker results for opposite genotypes suggest that this general tendency toward homophily may be obscuring a tendency for some specific parts of the genome to be heterophilic.
Fig. 1.
Friends exhibit significantly more homophily (positive correlation) than strangers in genome-wide measures. Overlapping density plots show that, compared with strangers, friends have (A) higher kinship coefficients and (B) lower proportions of opposite genotypes (SNPs for which neither allele is identical by state) in 1,367 friendship pairs and 1,196,429 stranger pairs observed in the same set of subjects (SI Appendix). On average, friends have a kinship coefficient that is +0.0014 greater than friends, a value that corresponds to the relatedness of fourth cousins. P values are from difference-in-means tests (SI Appendix).
The results so far do not control for population stratification because we wanted to characterize overall similarity. However, it is important to remember that some of the similarity in genotypes can be explained by simple assortment into relationships with people who have the same ancestral background. The Framingham Heart Study is composed of mostly whites (e.g., of Italian descent), so it is possible that a simple preference for ethnically similar others could explain the results in Fig. 1. However, in the following results, we applied strict controls for population stratification to ensure that any correlation we observed was not due to such a process.
To characterize the genotypes that are most likely to be homophilic or heterophilic, we conducted a GWAS regressing subject’s expected genotype on friend’s expected genotype for 1,468,013 common SNPs (minor allele frequency >0.10; see SI Appendix for imputation and regression details). For this GWAS analysis, we used both unimputed and imputed SNPs to improve power, but we emphasize, again, that our interest here is not in any particular SNP, but rather in the pattern across the whole genome.
Although the individuals in the Framingham Heart Study are almost all of European ancestry, population stratification has been shown to be a concern even in samples of European Americans (23). Relying on a widely used procedure to control for population stratification, we calculated the first 10 principal components of the subject–gene matrix with EIGENSTRAT (24). None of our subjects are classified as outliers, defined as individuals whose score is at least six SDs from the mean on one of the top 10 principal components. Nonetheless, consistent with past approaches (24), we included all 10 principal components for both the subject and the subject’s friend (20 variables in all) as controls for ancestry in each regression (SI Appendix).
To eliminate the possibility that the results are influenced by people tending to make friends with distant relatives, we use only the 907 friend pairs where kinship was ≤0 (recall that kinship can be less than zero when unrelated individuals tend to have negatively correlated genotypes). This procedure ensures that pairs of friends in the GWAS are not actually biologically related at all. It also allows us to set aside the remaining 458 pairs of friends for a split-sample replication analysis (discussed below). However, note that this procedure biases against finding homophilic SNPs because it means the average correlation between friends will be weakly negative.
Finally, we guarded against false positives by conducting an additional “strangers” GWAS for comparison with the “friends” GWAS. For the strangers analysis, we drew 907 random pairs from the stranger sample, and, to maintain comparability, we also restricted these stranger pairs to have a kinship ≤0 (SI Appendix). Importantly, both the friends GWAS and the strangers GWAS contained exactly the same people and genotypes—only the relationships between these people were different (friends vs. strangers).
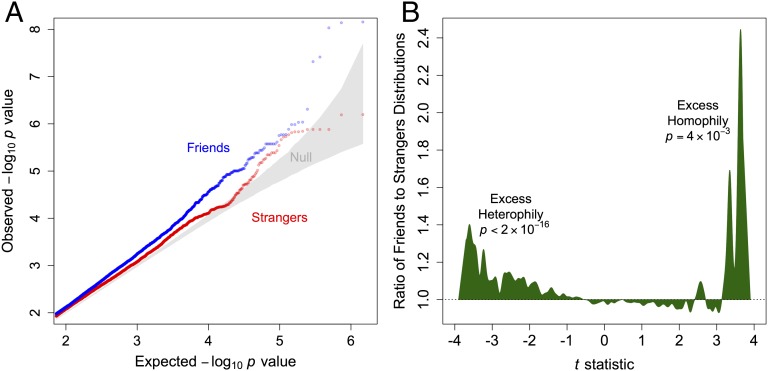
Fig. 2A shows QQ plots of observed versus expected P values for both GWASs. We would expect some variance inflation because of the restriction on the kinship coefficient to pairs that show no positive relatedness; the average correlation in genotypes resulting from this restriction is slightly negative (mean kinship = –0.003), which causes an excess number of markers to show negative correlation and low P values. To establish a baseline for this effect, we first measured the variance inflation factor in the strangers GWAS (λ = 1.020) and note in Fig. 2A that there is a slight upward shift that corroborates this tendency.
Fig. 2.
Friends exhibit significantly more homophily (positive correlation) and heterophily (negative correlation) than strangers in a genome-wide association study (GWAS) with strict controls for population stratification. (A) QQ plot of observed vs. expected P values from separate GWAS of genetic correlation shows more outliers for pairs of friends (blue) than pairs of strangers (red). Null distribution (gray) shows 95% confidence region for values possible due to chance. The strangers GWAS shows that some inflation is due to restricting observations to unrelated pairs of individuals, which causes genotypes to be negatively correlated on average. Over and above this baseline, the friends GWAS shows that friend pairs tend to have many markers that exhibit even lower P values, and this pattern is consistent with traits that are highly polygenic (25). (B) Distribution of t statistics in the friends GWAS divided by the distribution of t statistics in the strangers GWAS shows that friends tend to have both more heterophilic (negatively correlated) and also more homophilic (positively correlated) SNPs in the tails of the distribution. P values are from Kolmogorov–Smirnov tests (SI Appendix).
In contrast, the friends GWAS is shifted even higher and yields even lower P values than expected for many SNPs. In fact, the variance inflation for friends is more than double, at λ = 1.046, despite the fact that the two GWAS were generated using exactly the same regression-model specification. This shift is what we would expect if there were widespread low-level genetic correlation in friends across the genome, and it is consistent with recent work that shows that polygenic traits can generate inflation factors of these magnitudes (25). As supporting evidence for this interpretation, notice that Fig. 2A shows that there are many more outliers for the friends group than there are for the comparison stranger group, especially for P values less than 10−4. This result suggests that polygenic homophily and/or heterophily (rather than sample selection, population stratification, or model misspecification) accounts for at least some of the inflation and therefore that a relatively large number of SNPs are significantly correlated between pairs of friends (albeit each with probably small effects) across the whole genome.
To explore more fully this difference in results between the friends and strangers GWAS, in Fig. 2B we compare their t statistics to see whether the differences in P values are driven by homophily (positive correlation) or heterophily (negative correlation). The results show that the friends GWAS yields significantly more outliers than the comparison stranger group for both homophily (Kolmogorov–Smirnov test, P = 4 × 10−3) and heterophily (P < 2 × 10−16).
Although a few individual SNPs were genome-wide significant (SI Appendix), our interest is not in individual SNPs per se; and the homophily present across the whole genome, coupled with the evidence that friends exhibit both more genetic homophily and heterophily than strangers, suggests that there are many genes with low levels of correlation. In fact, we can use the measures of correlation from the friends GWAS to create a “friendship score” that can be used to predict whether two people are likely to be friends in a hold-out replication sample, based on the extent to which their genotypes resemble each other (SI Appendix). This replication sample contains 458 friend pairs and 458 stranger pairs that were not used to fit the GWAS models (SI Appendix). The results show that a one-standard-deviation change in the friendship score derived from the GWAS on the original friends sample increases the probability that a pair in the replication sample are friends by 6% (P = 2 × 10−4), and the score can explain ∼1.4% of the variance in the existence of friendship ties. This amount of variance is similar to the variance explained using the best currently available genetic scores for schizophrenia and bipolar disorder (0.4–3.2%) (26) and body-mass index (1.5%) (27). Although no other large datasets with fully genotyped friends exist at this time, we expect that a future GWAS on larger samples of friends might help to improve these friendship scores, boosting both efficiency and variance explained out of sample.
We expect that there are likely to be dozens and maybe even hundreds of genetic pathways that form the basis of correlation in specific genotypes, and our sample gives us enough power to detect a few of these pathways. We first conducted a gene-based association test of the likelihood that the set of SNPs within 50 kb of each of 17,413 genes exhibit (i) homophily or (ii) heterophily (SI Appendix). We then aggregated these results to conduct a gene-set analysis to determine whether the most significantly homophilic and heterophilic genes are overrepresented in any functional pathways documented in the KEGG and GOSlim databases (SI Appendix). In addition to examining the top 1% most homophilic and most heterophilic genes, we also examined the top 25% because highly polygenic traits may exhibit small differences across a large number of genes (28), and we expect homophily to be highly polygenic based on prior theoretical work (10).
Table 1 shows that three gene sets are significantly overrepresented in these analyses, after adjusting for multiple testing. In the 174 most homophilic genes (top 1%), we found that an olfactory transduction pathway is significantly overrepresented (P = 4 × 10−5, adjusted P = 0.009), suggesting that friends tend to have genotypes that yield similar senses of smell. When we increased the threshold to the top 25% of homophilic genes, we also found that the linoleic acid metabolism system is significantly overrepresented (P = 2 × 10−5, adjusted P = 0.005). For heterophilic genes, a gene set that characterizes certain immune system processes achieved significance (P = 5 × 10−4, adjusted P = 0.036), which suggests that friends have different genotypes for coping with infection, a previously hypothesized possibility (12). For comparison, we conducted the same gene-set analyses for strangers, and we did not find any significantly overrepresented gene sets (SI Appendix), suggesting that the procedure does not generate false positives. We cannot yet be sure whether this set of findings means that only a few biological systems are highly correlated or that many systems are weakly correlated; future analyses with larger samples may help to resolve this question.
Table 1.
Gene-set analysis shows three genome-wide significant gene sets that are overrepresented
| Friends results | |||
| Set 1 | Set 2 | Set 3 | |
| Correlation type | Homophily | Homophily | Heterophily |
| Set threshold | Top 1% | Top 25% | Top 25% |
| Gene set ID | KEGG hsa04740 | KEGG hsa00591 | GO 0002376 |
| Description | Olfactory transduction | Linoleic acid metabolism | Immune system process |
| No. in threshold set | 14 | 18 | 353 |
| Total no. measured | 355 | 29 | 1,198 |
| No. in gene set | 461 | 30 | 1,562 |
| Percent in threshold set | 3.9 | 62.1 | 29.5 |
| Percent measured | 77.0 | 96.7 | 76.7 |
| Z score | 5.05 | 4.64 | 3.55 |
| Fisher's test exact P | 4 x 10−5 | 2 x 10−5 | 5 x 10−4 |
| Benjamini–Hochberg adjusted P | 0.009 | 0.005 | 0.036 |
Friend pairs tend to have similar gene variants (homophily) in the olfactory and linoleic acid systems and different gene variants (heterophily) in the immune system. By comparison, the same analysis with stranger pairs shows no significant gene sets.
Although it is possible that these identified pathways, and other pathways yet to be identified, have played an important role in recent human evolution—and, indeed, prior work shows strong positive selection “for genes related to immune response, reproduction (especially spermatogensis), and sensory perception (especially olfaction)” (29)—the foregoing overrepresentation analysis does not address whether natural selection has generally favored genotypic homophily. To test the hypothesis that homophilic SNPs are generally under recent positive selection, we used the Composite of Multiple Signals (CMS) score (30). This score combines signals from several measures of positive selection to create a single value that indicates the likelihood that an SNP has been increasing in frequency due to selection pressure over the last 30,000 y (SI Appendix).
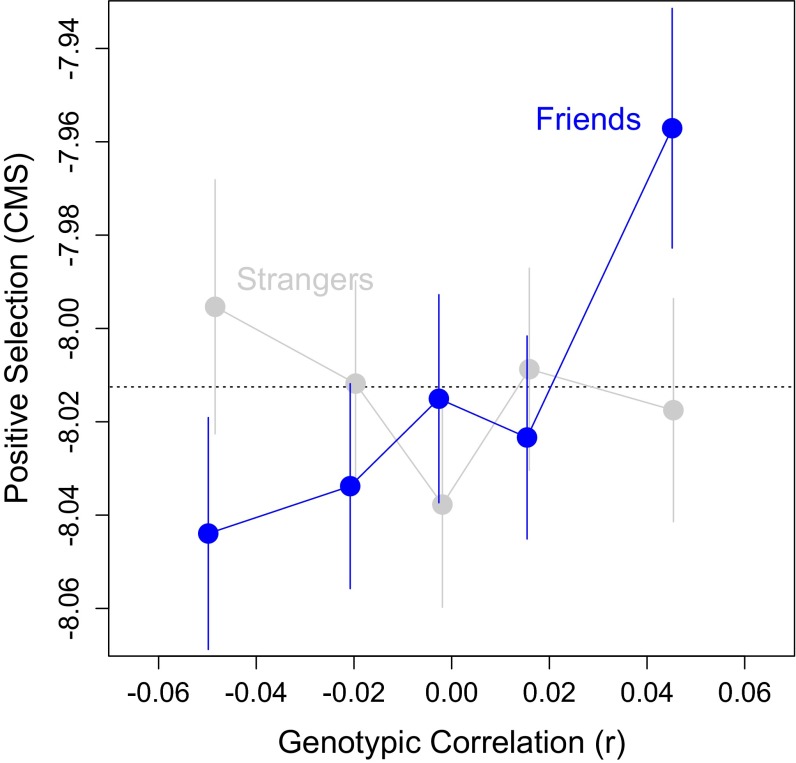
In Fig. 3, we show that, after correcting for correlated outcomes due to linkage disequilibrium and for varying precision in the GWAS estimates (SI Appendix), the top 20% most homophilic SNPs have significantly higher CMS scores than the other 80% (+0.07, SE 0.02, P = 0.003). For comparison, note that this effect is about half the size of the difference in CMS scores between intragenic and intergenic SNPs (+0.15, SE 0.02), which we would expect to be large given the functional role of variants within genes relative to those between genes. In contrast, Fig. 3 also shows that CMS scores are not significantly higher for the most homophilic SNPs in the strangers GWAS (–0.00, SE 0.02, P = 0.86). This observation suggests that the whole-genome regression model we used does not generate false positives.
Fig. 3.
Homophilic (positively correlated) SNPs are more likely to be under recent positive selection. Plot shows mean composite of multiple signals (CMS) score by SNP correlation quintile for friends (blue) and strangers (gray). Each quintile contains ∼293,600 SNPs. Vertical lines show the SEM corrected for correlated observations due to linkage disequilibrium (SI Appendix). For reference, the horizontal dotted line shows the mean CMS score.
Furthermore, we evaluated a model that fits the CMS score to the level of correlation in each SNP, allowing the linear relationship to be different for homophilic and heterophilic SNPs (SI Appendix). This model (which also serves as a robustness check) showed that there is a positive and significant relationship in the friends GWAS for homophilic SNPs (P = 0.03). As the level of positive correlation increases, so does the expected CMS score. There is no relationship for negatively correlated (heterophilic) SNPs (P = 0.63). And, for comparison, there is no relationship in the strangers GWAS between genetic correlation and positive selection for either homophily (P = 0.77) or heterophily (P = 0.28). In sum, it appears that, overall, across the whole genome, the genotypes humans tend to share in common with their friends are more likely to be under recent natural selection than other genotypes.
It is intriguing that genetic structure in human populations may result not only from the formation of reproductive unions, but also from the formation of friendship unions. This observation, in turn, has relevance for the idea of an evocative gene-environment correlation, proposed more than 30 y ago, which suggests that a person’s genes can lead one to seek out circumstances that are compatible with one’s genotype (31, 32). Our results suggest that these circumstances could include not only the physical environment but also the social environment, and therefore the genotypic constitution of one’s friends. As Tooby and Cosmides argue, “not only do individual humans have different reproductive values that can be estimated based on various cues they manifest, but they also have different association values” (11). People may seek out particular, convivial social environments that affect their fitness.
The existence of excess genetic similarity between friends is also relevant to the growing area of indirect genetic effects (33), wherein the phenotypic traits of focal individuals are influenced by the genomes of their neighbors, in a kind of “network epistasis.” (12) In fact, our results support the idea that humans might be seen as metagenomic not just with respect to the microbes within them (34), but also with respect to the humans around them. It may be useful to view a person’s genetic landscape as a summation of the genes within the individual and within the people surrounding the individual, just as in certain other organisms (33, 35).
Pairs of friends are, on average, as genetically similar to one another as fourth cousins, which seems noteworthy because this estimate is above and beyond mean ancestry and background relatedness. Acquiring friends who resemble oneself genotypically from among a group of strangers may reflect a number of processes, including the selection of particular friends or particular environments. Whatever its cause, however, the subtle process of genetic sorting in human social relationships might have an important effect on a number of other biological and social processes, from the spread of germs to the spread of information.
Insofar as the process involves the actual selection of friends, it may reflect the extended workings of some sort of kinship detector postulated in humans (18). One’s friends, in other words, may evince a kind of functional relatedness (identity by state)—and may perhaps do so especially for particular biological systems—rather than evincing an actual relatedness (identity by descent) as in the case of kin. Forming social ties to functional kin who perceive or cope with the environment in a similar way to oneself can result in both individuals benefiting from each other’s deliberately or accidentally created benefits (“positive externalities”); for example, if one individual builds a fire because he feels cold in the same circumstances as the other, both benefit (11). Genetic correlation between friends may even enhance the opportunity for natural selection to operate at the level of social groups established on a basis other than kinship; such associations have long been postulated in the theoretical evolutionary genetics literature, but there is little extant evidence (36, 37).
Kin recognition has been shown in many vertebrates (38), and it is important for stabilizing cooperation and promoting inclusive fitness benefits in some species (39). There is suggestive evidence for some sort of kin detection system in humans as well, such that, for each individual encountered, an unspecified system may compute and update a continuous measure of kinship that corresponds to the genetic relatedness of the self to the other individual (18). In part, this system would be driven by objectives such as behaving altruistically toward, and avoiding sexual relations with, kin. A number of mechanisms by which kin detection might take place have been proposed, including coresidence duration monitoring, perinatal association, and other cues, such as facial resemblance or odor. Cues of kinship may foster altruistic impulses and cooperative exchanges with individuals displaying those cues, and it is not hard to imagine that such a system might possibly be extended to preferential (active) friendship formation.
In this regard, our findings regarding homophily on certain olfactory system features are intriguing and supportive. There is evidence that olfaction plays a role in human (and other primate) kin recognition (40, 41) and even some suggestive evidence that people are able to distinguish friends from strangers based on blind odor tests (42, 43). The olfaction ontology in which we detect substantial homophily has some genes coding for odorant receptors; it is possible that individuals who smell things in the same way are drawn to similar environments where they interact with and befriend one another. Olfaction is also connected to other processes, such as emotional contagion and communication, and to the avoidance of inappropriate ingestions; these processes too may benefit from the synergistic presence of genotypically similar others.
The implications of the finding regarding homophily on genes related to linoleic acid metabolism are unclear. Linoleic acid is a precursor for substances involved in a broad range of important bodily processes (ranging from adipocyte function to bone formation to the regulation of gene expression) (44), and the component genes in the pathway are related to the metabolism of cholesterol, steroids, and various ingested substances, although it is intriguing that linoleic acid compounds might be used by moths as pheromones (45). Possibly, this pathway is related to the restrained consumption or the specific metabolism of various foodstuffs, traits for which homophily may be advantageous and heterophily self-injurious.
The observed heterophily on an immune system ontology has interesting implications. Prior work has provided evidence of an active process contributing to genetic heterophily between mates with respect to the avoidance of similar HLA haplotypes (14, 15, 46) (although these genes are not part of the present gene set). In the case of friends, there may also be advantages to complementarity rather than synergy when it comes to immune system function because surrounding oneself with others who are dissimilar to oneself in this regard may be an adaptive strategy. If one is already relatively resistant to a particular pathogen, it would be best to have friends who were resistant to different pathogens, thus mitigating the interpersonal spread of both. Genes affecting the immune system do not necessarily benefit from interpersonal ties to genotypically similar individuals.
It may be possible to use an approach similar to that outlined here, but with much larger samples of friendship pairs, and perhaps coupled with the addition of an equally large number of spousal pairs, to identify the genetic basis of kin detection. The extent to which friends and spouses resemble each other could itself be taken as a phenotype, and one could imagine doing a GWAS to isolate which regions of the genome contribute to our ability to pick suitable friends and mates.
Finally, the human evolutionary environment is not limited to the physical environment (sunshine, altitude) or biological environment (predators, pathogens) but also includes the social environment, which may itself be an evolutionary force (47). Our finding that positively correlated genotypes are under positive selection suggests that the genes of other people might modify the fitness advantages of one’s own genes, thus affecting the speed and outcome of evolution. In particular, communication—whether involving scent, sight, or sound—may be the key to this synergy. The human capacity to collaborate not only with kin but also with unrelated members of our species may have dramatically increased the potential gains from synergy, and this shift not only would favor interactions with generally similar partners, but also would affect the overall desire to search out such partners (10, 11). Therefore, it is possible that we evolved a predilection for homophily once we started to frequently interact socially with unrelated individuals. Such an effect would especially speed up the evolution of phenotypes that are intrinsically synergistic, and this observation may help shed light on the finding that evolution in humans is accelerating (48).
Supplementary Material
Acknowledgments
We thank Jason Boardman, David Cesarini, Chris Dawes, Jan-Emmanuel De Neve, Feng Fu, Erez Lieberman, Akihiro Nishi, Martin Nowak, David Rand, Pardis Sabeti, Zach Steinert-Threlkeld, Juan Ugalde, and Ajit Varki for helpful comments. We thank Shervin Tabrizi and Pardis Sabeti for sharing the CMS data with us. This work was supported by National Institute on Aging Grant P-01 AG031093, National Institute of General Medical Sciences Grant P-41 GM103504-03, and the Star Family Foundation. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract N01-HC-25195); funding for the genotyping of the data was provided by NHLBI Contract N02-HL-64278. Data was downloaded from National Institutes of Health Database of Genotypes and Phenotypes, project 780, with accession no. phs000153.v7.p6.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “In the Light of Evolution VIII: Darwinian Thinking in the Social Sciences,” held January 10–11, 2014, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS website at www.nasonline.org/ILE-Darwinian-Thinking.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400825111/-/DCSupplemental.
References
- 1.Borgatti SP, Mehra A, Brass DJ, Labianca G. Network analysis in the social sciences. Science. 2009;323(5916):892–895. doi: 10.1126/science.1165821. [DOI] [PubMed] [Google Scholar]
- 2.Barabási AL. Scale-free networks: A decade and beyond. Science. 2009;325(5939):412–413. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- 3.Apicella CL, Marlowe FW, Fowler JH, Christakis NA. Social networks and cooperation in hunter-gatherers. Nature. 2012;481(7382):497–501. doi: 10.1038/nature10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seyfarth RM, Cheney DL. The evolutionary origins of friendship. Annu Rev Psychol. 2012;63:153–177. doi: 10.1146/annurev-psych-120710-100337. [DOI] [PubMed] [Google Scholar]
- 5.Hruschka DJ. Friendship: Development, Ecology, and Evolution of a Relationship. Berkeley, CA: Univ of California Press; 2010. [Google Scholar]
- 6.Burt A. A mechanistic explanation of popularity: Genes, rule breaking, and evocative gene-environment correlations. J Pers Soc Psychol. 2009;96(4):783–794. doi: 10.1037/a0013702. [DOI] [PubMed] [Google Scholar]
- 7.Guo G. Genetic similarity shared by best friends among adolescents. Twin Res Hum Genet. 2006;9(1):113–121. doi: 10.1375/183242706776402920. [DOI] [PubMed] [Google Scholar]
- 8.Fowler JH, Dawes CT, Christakis NA. Model of genetic variation in human social networks. Proc Natl Acad Sci USA. 2009;106(6):1720–1724. doi: 10.1073/pnas.0806746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McPherson M, Smith-Lovin L, Cook JM. Birds of a feather: Homophily in social networks. Annu Rev Sociol. 2001;27:415–444. [Google Scholar]
- 10.Fu F, Nowak MA, Christakis NA, Fowler JH. The evolution of homophily. Sci Rep. 2012;2:845. doi: 10.1038/srep00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tooby J, Cosmides L. Friendship and the Banker’s Paradox: Other pathways to the evolution of adaptations for altruism. Proc Br Acad. 1996;88:119–143. [Google Scholar]
- 12.Fowler JH, Settle JE, Christakis NA. Correlated genotypes in friendship networks. Proc Natl Acad Sci USA. 2011;108(5):1993–1997. doi: 10.1073/pnas.1011687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boardman JD, Domingue BW, Fletcher JM. How social and genetic factors predict friendship networks. Proc Natl Acad Sci USA. 2012;109(43):17377–17381. doi: 10.1073/pnas.1208975109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wedekind C, Seebeck T, Bettens F, Paepke AJ. MHC-dependent mate preferences in humans. Proc Biol Sci. 1995;260(1359):245–249. doi: 10.1098/rspb.1995.0087. [DOI] [PubMed] [Google Scholar]
- 15.Wedekind C, Füri S. Body odour preferences in men and women: Do they aim for specific MHC combinations or simply heterozygosity? Proc Biol Sci. 1997;264(1387):1471–1479. doi: 10.1098/rspb.1997.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lusseau D, Newman MEJ. Identifying the role that animals play in their social networks. Proc Biol Sci. 2004;271(Suppl 6):S477–S481. doi: 10.1098/rsbl.2004.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brent LJH, Lehmann J, Ramos-Fernández G. Social network analysis in the study of nonhuman primates: A historical perspective. Am J Primatol. 2011;73(8):720–730. doi: 10.1002/ajp.20949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieberman D, Tooby J, Cosmides L. The architecture of human kin detection. Nature. 2007;445(7129):727–731. doi: 10.1038/nature05510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward LD, Kellis M. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science. 2012;337(6102):1675–1678. doi: 10.1126/science.1225057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell CD, et al. Demonstrating stratification in a European American population. Nat Genet. 2005;37(8):868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 24.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Yang J, et al. GIANT Consortium Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19(7):807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell SM, et al. International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speliotes EK, et al. MAGIC Procardis Consortium Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee PH, O’Dushlaine C, Thomas B, Purcell SM. INRICH: Interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28(13):1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312(5780):1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 30.Grossman SR, et al. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327(5967):883–886. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- 31.Kendler KS, Baker JH. Genetic influences on measures of the environment: A systematic review. Psychol Med. 2007;37(5):615–626. doi: 10.1017/S0033291706009524. [DOI] [PubMed] [Google Scholar]
- 32.Scarr S, McCartney K. How people make their own environments: A theory of genotype greater than environment effects. Child Dev. 1983;54(2):424–435. doi: 10.1111/j.1467-8624.1983.tb03884.x. [DOI] [PubMed] [Google Scholar]
- 33.Biscarini F, et al. Across-line SNP association study for direct and associative effects on feather damage in laying hens. Behav Genet. 2010;40(5):715–727. doi: 10.1007/s10519-010-9370-0. [DOI] [PubMed] [Google Scholar]
- 34.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature. 2010;467(7311):82–85. doi: 10.1038/nature09354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamilton WD. Innate social aptitudes of man: An approach from evolutionary genetics. In: Fox R, editor. Biosocial Anthropology. London: Malaby Press; 1975. pp. 133–153. [Google Scholar]
- 37.Smith JM. Group selection. Q Rev Biol. 1976;51:277–283. [Google Scholar]
- 38.Griffin AS, West SA. Kin discrimination and the benefit of helping in cooperatively breeding vertebrates. Science. 2003;302(5645):634–636. doi: 10.1126/science.1089402. [DOI] [PubMed] [Google Scholar]
- 39.Chaine AS, Schtickzelle N, Polard T, Huet M, Clobert J. Kin-based recognition and social aggregation in a ciliate. Evolution. 2010;64(5):1290–1300. doi: 10.1111/j.1558-5646.2009.00902.x. [DOI] [PubMed] [Google Scholar]
- 40.Porter RH. Olfaction and human kin recognition. Genetica. 1998-1999;104(3):259–263. doi: 10.1023/a:1026404319384. [DOI] [PubMed] [Google Scholar]
- 41.Boulet M, Charpentier MJE, Drea CM. Decoding an olfactory mechanism of kin recognition and inbreeding avoidance in a primate. BMC Evol Biol. 2009;9:281. doi: 10.1186/1471-2148-9-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsson SB, Barnard J, Turri L. Olfaction and identification of unrelated individuals: Examination of the mysteries of human odor recognition. J Chem Ecol. 2006;32(8):1635–1645. doi: 10.1007/s10886-006-9098-8. [DOI] [PubMed] [Google Scholar]
- 43.Weisfeld GE, Czilli T, Phillips KA, Gall JA, Lichtman CM. Possible olfaction-based mechanisms in human kin recognition and inbreeding avoidance. J Exp Child Psychol. 2003;85(3):279–295. doi: 10.1016/s0022-0965(03)00061-4. [DOI] [PubMed] [Google Scholar]
- 44.Belury MA. Dietary conjugated linoleic acid in health: physiological effects and mechanisms of action. Annu Rev Nutr. 2002;22:505–531. doi: 10.1146/annurev.nutr.22.021302.121842. [DOI] [PubMed] [Google Scholar]
- 45.Rule GS, Roelofs WL. Biosynthesis of sex pheromone components from linolenic acid arctiid moths. Arch Insect Biochem. 1989;12:89–97. [Google Scholar]
- 46.Ober C, et al. HLA and mate choice in humans. Am J Hum Genet. 1997;61(3):497–504. doi: 10.1086/515511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrmann E, Call J, Hernàndez-Lloreda MV, Hare B, Tomasello M. Humans have evolved specialized skills of social cognition: The cultural intelligence hypothesis. Science. 2007;317(5843):1360–1366. doi: 10.1126/science.1146282. [DOI] [PubMed] [Google Scholar]
- 48.Hawks J, Wang ET, Cochran GM, Harpending HC, Moyzis RK. Recent acceleration of human adaptive evolution. Proc Natl Acad Sci USA. 2007;104(52):20753–20758. doi: 10.1073/pnas.0707650104. [DOI] [PMC free article] [PubMed] [Google Scholar]