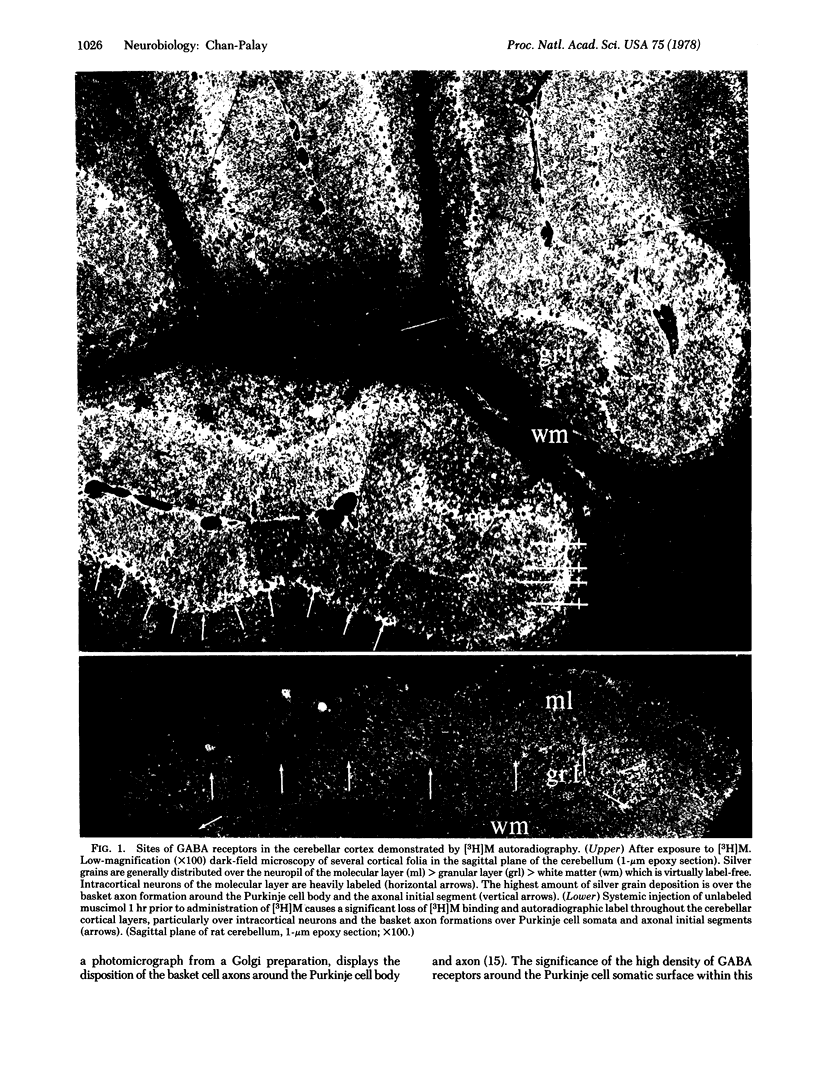
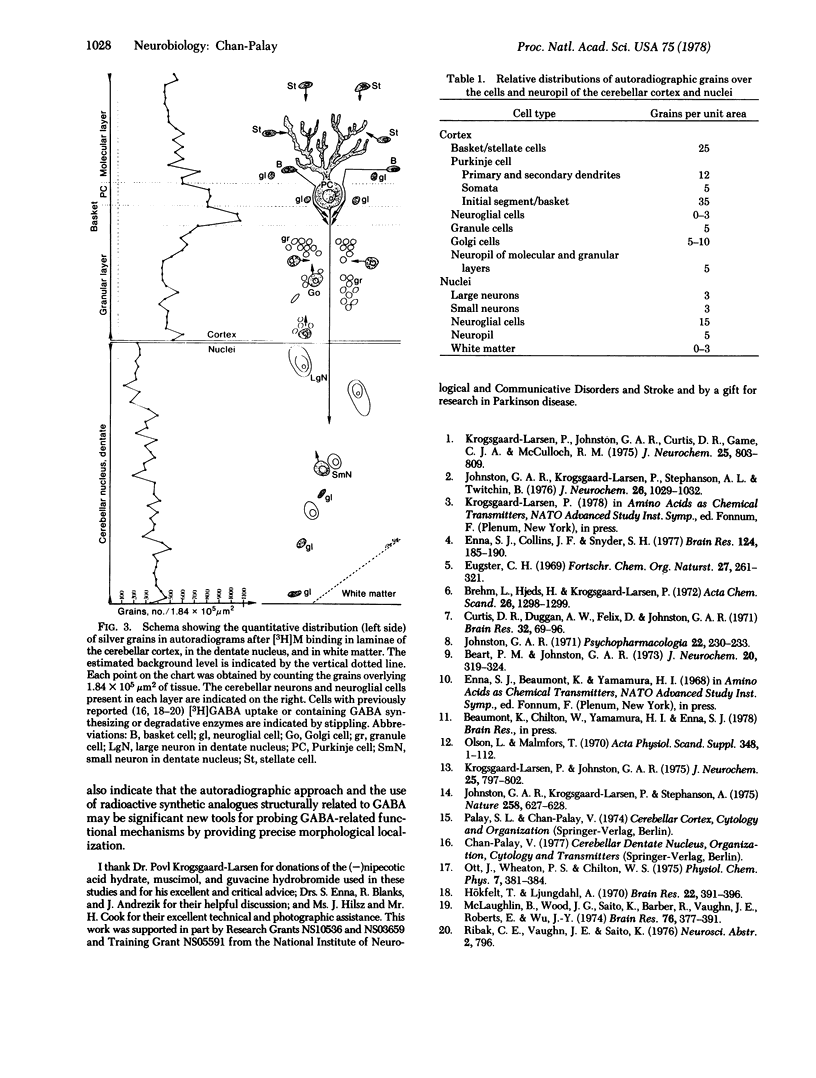
Abstract
Muscimol, a structural analogue and potent agonist of γ-aminobutyric acid (GABA), was used in its tritiated form for the autoradiographic localization of GABA receptors in the rat central nervous system. [3H]Muscimol ([3H]M) was incubated with brain slices or was injected intracortically or into intraocular brain transplants. As indicated by [3H]M binding and autoradiographic silver grains, GABA receptors display a laminar distribution over the cerebellar cortex, cerebral cortex, and hippocampus (in order of decreasing quantity); and a nonlaminar distribution in the caudate nucleus and substantia nigra. [3H]M binding was not affected by brief prior treatment of brain slices with (-)nipecotic acid or guvacine, two potent inhibitors of GABA uptake, indicating receptor binding specificity. Systemic administration of unlabeled muscimol interferred with binding of [3H]M binding subsequently administered in vitro, indicating that muscimol or a metabolite of it traverses the blood/brain barrier and binds to receptor sites, possibly in a manner competitive with [3H]M. [3H]M binding was greatest in the cerebellum. Quantitative analyses of the distribution of autoradiographic silver grains in the cerebellar cortex and dentate nucleus showed a general distribution of GABA receptors in the neuropil: molecular layer > granular layer > cerebellar nuclei > white matter. The highest binding of [3H]M occurred on the Purkinje cell somatic surface, in the basket axon formation surrounding the cell body and its axon initial segment, and somewhat less on basket and stellate cell somata. Neuroglial cells of the cortex have no [3H]M binding capacity; some glial cells in the cerebellar nuclei do. The role of glial cells in GABA uptake, metabolism, and GABA-receptor-mediated mechanisms remains to be clarified. The distribution of GABA receptors as indicated by [3H]M binding differs from the distribution of [3H]GABA uptake and GABA synthesizing and degradative sites.
Keywords: cerebellum, Purkinje cells, cerebrum, hippocampus, striato-nigral
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beart P. M., Johnston G. A. GABA uptake in rat brain slices: inhibition by GABA analogues and by various drugs. J Neurochem. 1973 Feb;20(2):319–324. doi: 10.1111/j.1471-4159.1973.tb12131.x. [DOI] [PubMed] [Google Scholar]
- Brehm L., Hjeds H., Krogsgaard-Larsen P. The structure of muscimol, a GABA analogue of restricted conformation. Acta Chem Scand. 1972;26(3):1298–1299. doi: 10.3891/acta.chem.scand.26-1298. [DOI] [PubMed] [Google Scholar]
- Curtis D. R., Duggan A. W., Felix D., Johnston G. A. Bicuculline, an antagonist of GABA and synaptic inhibition in the spinal cord of the cat. Brain Res. 1971 Sep 10;32(1):69–96. doi: 10.1016/0006-8993(71)90156-9. [DOI] [PubMed] [Google Scholar]
- Enna S. J., Collins J. F., Snyder S. H. Stereospecificity and structure--activity requirements of GABA receptor binding in rat brain. Brain Res. 1977 Mar 18;124(1):185–190. doi: 10.1016/0006-8993(77)90878-2. [DOI] [PubMed] [Google Scholar]
- Eugster C. H. Chemie der Wirkstoffe aus dem Fliegenpilz (Amanita muscaria) Fortschr Chem Org Naturst. 1969;27:261–321. [PubMed] [Google Scholar]
- Hökfelt T., Ljungdahl A. Cellular localization of labeled gamma-aminobutyric acid (3H-GABA) in rat cerebellar cortex: an autoradiographic study. Brain Res. 1970 Sep 16;22(3):391–396. doi: 10.1016/0006-8993(70)90480-4. [DOI] [PubMed] [Google Scholar]
- Johnston G. A., Krogsgaard-Larsen P., Stephanson A. L., Twitchin B. Inhibition of the uptake of GABA and related amino acids in rat brain slices by the optical isomers of nipecotic acid. J Neurochem. 1976 May;26(5):1029–1032. doi: 10.1111/j.1471-4159.1976.tb06488.x. [DOI] [PubMed] [Google Scholar]
- Johnston G. A., Krogsgaard-Larsen P., Stephanson A. Betel nut constituents as inhibitors of gamma-aminobutyric acid uptake. Nature. 1975 Dec 18;258(5536):627–628. doi: 10.1038/258627a0. [DOI] [PubMed] [Google Scholar]
- Johnston G. A. Muscimol and the uptake of -aminobutyric acid by rat brain slices. Psychopharmacologia. 1971;22(3):230–233. doi: 10.1007/BF00401785. [DOI] [PubMed] [Google Scholar]
- Krogsgaard-Larsen P., Johnston G. A., Curtis D. R., Game C. J., McCulloch R. M. Structure and biological activity of a series of conformationally restricted analogues of GABA. J Neurochem. 1975 Dec;25(6):803–809. doi: 10.1111/j.1471-4159.1975.tb04411.x. [DOI] [PubMed] [Google Scholar]
- Krogsgaard-Larsen P., Johnston G. A. Inhibition of GABA uptake in rat brain slices by nipecotic acid, various isoxazoles and related compounds. J Neurochem. 1975 Dec;25(6):797–802. doi: 10.1111/j.1471-4159.1975.tb04410.x. [DOI] [PubMed] [Google Scholar]
- McLaughlin B. J., Wood J. G., Saito K., Barber R., Vaughn J. E., Roberts E., Wu J. Y. The fine structural localization of glutamate decarboxylase in synaptic terminals of rodent cerebellum. Brain Res. 1974 Aug 23;76(3):377–391. doi: 10.1016/0006-8993(74)90815-4. [DOI] [PubMed] [Google Scholar]
- Olson L., Malmfors T. Growth characteristics of adrenergic nerves in the adult rat. Fluorescence histochemical and 3H-noradrenaline uptake studies using tissue transplantations to the anterior chamber of the eye. Acta Physiol Scand Suppl. 1970;348:1–112. [PubMed] [Google Scholar]
- Ott J., Wheaton P. S., Chilton W. S. Fate of muscimol in the mouse. Physiol Chem Phys. 1975;7(4):381–384. [PubMed] [Google Scholar]