Gata6 regulates differentiation, metabolism and survival of peritoneal macrophages.
Abstract
The transcription factor Gata6 regulates proliferation and differentiation of epithelial and endocrine cells and cancers. Among hematopoietic cells, Gata6 is expressed selectively in resident peritoneal macrophages. We thus examined whether the loss of Gata6 in the macrophage compartment affected peritoneal macrophages, using Lyz2-Cre x Gata6flox/flox mice to tackle this issue. In Lyz2-Cre x Gata6flox/flox mice, the resident peritoneal macrophage compartment, but not macrophages in other organs, was contracted, with only a third the normal number of macrophages remaining. Heightened rates of death explained the marked decrease in peritoneal macrophage observed. The metabolism of the remaining macrophages was skewed to favor oxidative phosphorylation and alternative activation markers were spontaneously and selectively induced in Gata6-deficient macrophages. Gene expression profiling revealed perturbed metabolic regulators, including aspartoacylase (Aspa), which facilitates generation of acetyl CoA. Mutant mice lacking functional Aspa phenocopied the higher propensity to death and led to a contraction of resident peritoneal macrophages. Thus, Gata6 regulates differentiation, metabolism, and survival of resident peritoneal macrophages.
Implicated in both tissue damage and repair, resident macrophages regulate numerous homeostatic, developmental, and host defense responses, rendering them therapeutic targets (Davies et al., 2013) where it might be desirable to selectively impact discrete macrophage populations. Recent studies confirm resident macrophages in adult mice sustain their own homeostasis through local proliferation, rather than replenishment from blood monocytes (Schulz et al., 2012; Hashimoto et al., 2013; Yona et al., 2013). Marked diversity in gene expression among macrophages residing in different organs suggests that distinct transcriptional programs may affect the maintenance of only single macrophage populations. For example, Spi-C is selectively expressed by red pulp macrophages, and it is essential for sustenance of this population, apparently due to its role in coordinating gene expression that facilitates handling of iron in macrophages that recycle aged erythrocytes (Kohyama et al., 2009; Haldar et al., 2014). Recently, we found that Gata6 is selectively expressed by resident peritoneal macrophages and predicted a set of peritoneal macrophage-specific genes that may be controlled by Gata6 in this population (Gautier et al., 2012). Subsequently, two studies identified a role for Gata6 in controlling the peritoneal macrophage pool (Okabe and Medzhitov, 2014; Rosas et al., 2014), including evidence that the ability of macrophages to proliferate during an inflammatory challenge was compromised (Rosas et al., 2014) and identification of retinoic acid as a signal that induces Gata6 in peritoneal macrophages (Okabe and Medzhitov, 2014). However, neither study explained what cellular events caused contraction of the macrophage pool within the peritoneum under resting conditions. Here, we show that apoptosis is induced in peritoneal macrophages in the absence of Gata6, at least in part because Gata6 expression either directly or indirectly supported expression of aspartoacylase (Aspa) that deacetylates N-acetyl aspartate. Mice bearing mutations in Aspa likewise displayed reduced macrophage counts concomitant with increased death. However, only the more global Gata6 deficiency, but not single deficiency in Aspa, resulted in a selective and seemingly cell autonomous polarization of peritoneal macrophages toward an alternatively activated phenotype. Collectively, these data delineate a role for Gata6 in regulating macrophage survival and activation state.
RESULTS AND DISCUSSION
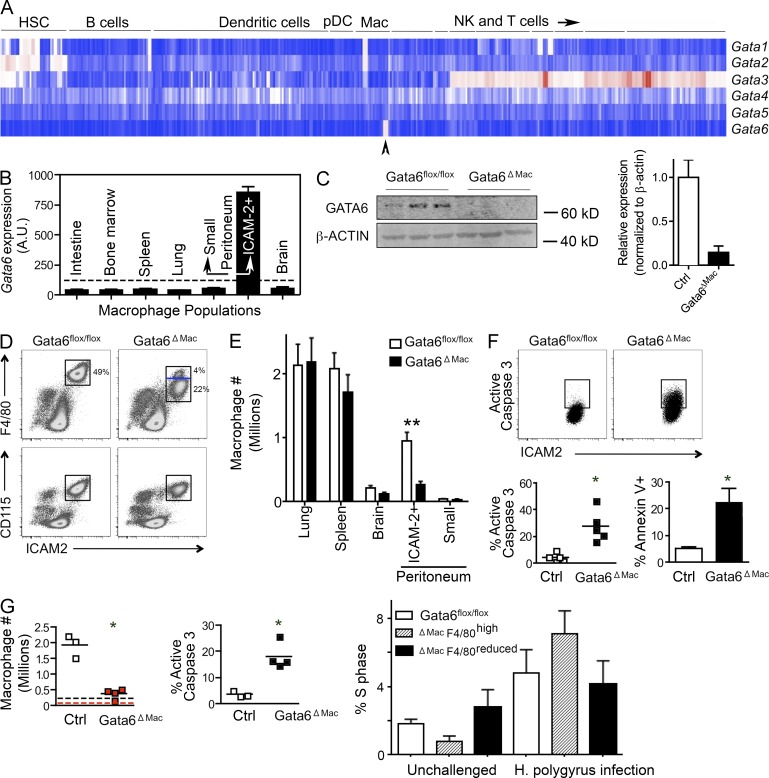
The transcription factor Gata6 was expressed only in F4/80hi peritoneal macrophages, corresponding with ICAM-2+ peritoneal macrophages (Gautier et al., 2012), within the entire hematopoietic system (Fig. 1, A and B). Thus, we crossed mice expressing the Cre recombinase downstream of the Lyz2 promoter, active in myeloid cells including macrophages (Clausen et al., 1999), with mice bearing floxed Gata6 alleles (Sodhi et al., 2006), generating mice specifically lacking Gata6 in macrophages (Gata6ΔMac) to be compared with controls bearing the floxed alleles in the absence of Cre recombinase (Gata6flox/flox; Fig. 1 C). Total cell numbers from the peritoneum were reduced by 25 ± 13%. Moreover, the frequency of F4/80hi ICAM-2+ was selectively reduced in Gata6ΔMac mice, to 54 ± 16% of control mice. Together, these changes led to a marked reduction in Gata6ΔMac peritoneal macrophages compared with control mice (Fig. 1 D). A second, minor CD115+ F4/80lo ICAM-2lo macrophage population residing in the peritoneum (Gautier et al., 2013), which did not express Gata6 (Fig. 1 B), was unaffected in Gata6ΔMac mice (Fig. 1 E). Macrophage frequency in other organs was unchanged (Fig. 1 E). Increased activated caspase 3 and Annexin V in Gata6ΔMac macrophages revealed augmented apoptosis (Fig. 1 F). Consistent with previous work (Jenkins et al., 2013), infection with the parasite Heligmosomoides polygyrus led to a greater than fivefold increase in peritoneal macrophages in control mice, as the ICAM-2+ resident macrophage population associated with Gata6 expression was induced to proliferate during infection (Fig. 1 G). Percentage of cycling macrophages was not significantly affected in Gata6ΔMac macrophages compared with controls before or after infection (Fig. 1 G). However, Gata6ΔMac macrophage numbers scarcely elevated above baseline numbers observed in uninfected control mice (Fig. 1 G) because of markedly elevated apoptosis (Fig. 1 G). Thus, F4/80hi ICAM-2+ resident peritoneal macrophages survival was selectively impaired in resting and parasite-challenged Gata6ΔMac mice.
Figure 1.
Gata6 deficiency decreases peritoneal macrophage density and induces apoptosis. (A) Expression of mRNA for the GATA family of transcription factors within the resting hematopoietic system profiled by the Immunological Genome Project. Arrowhead points to peritoneal macrophage. (B) Signal intensity of gene expression for Gata6 from array data comparing resident macrophages from multiple organs. Dotted line, cutoff representing positive expression after data normalization. (C) Immunoblot for Gata6 on sorted macrophages, in which all ICAM-2+ macrophages were collected from each genotype. (D) Gata6+ macrophages quantified after cell counts and gating during flow cytometric analysis on cells expressing F4/80, ICAM-2, and CD115. Blue line in gate delineates loss of F4/80 surface intensity in most Gata6ΔMac macrophages. (E) Macrophage counts in various organs are plotted. (F) Percent macrophages expressing active caspase 3 or Annexin V. (G) Peritoneal macrophages quantified in mice infected with H. polygyrus. Enumeration of these macrophages plotted in control Gata6flox/flox mice (black) or Gata6ΔMac mice (red); baselines for each strain shown as dotted line in same color. Percent macrophages positive for active caspase 3 after H. polygyrus infection are plotted, and S phase was analyzed in unchallenged and infected mice. Data are derived from 2–8 experiments, with 2–5 replicates per group, performed for each part of the figure. Means ± SEM are shown. *, P < 0.05; **, P < 0.001 relative to controls using two-tailed Student’s t tests. Statistical significance in S phase analysis is not depicted, but all S phase analyses are statistically significant (one-way ANOVA) comparing unchallenged control mice to infected mice, P < 0.05, but differences between genotypes in the same condition are not significant.
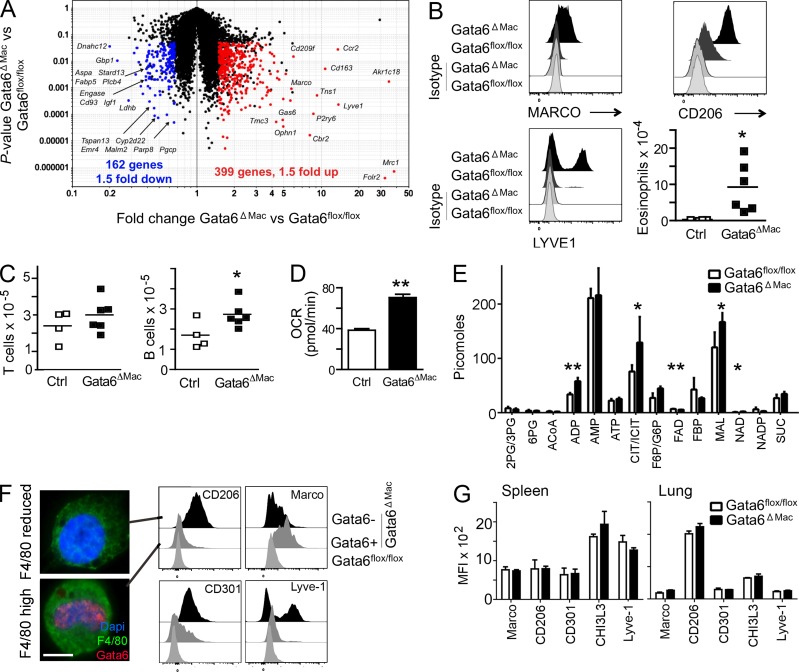
After flow cytometric cell sorting and gene expression analysis using whole mouse genome arrays, we found that notably elevated mRNA transcripts in Gata6ΔMac macrophages were those associated with alternative activation (Gordon and Martinez, 2010) of macrophages, including mRNA transcripts encoding CD163, LYVE-1, Arg1, Clec10a (CD301), Chi3L3, and CD206 (mannose receptor, Mrc1; Fig. 2 A and Table S1), as well as mRNA for MARCO, which is associated with innate macrophage activation (Mukhopadhyay et al., 2014). Increased cell surface levels of corresponding proteins were accordingly observed (Fig. 2 B). Functional evidence in support of alternative activation included a significant increase in peritoneal eosinophils (Fig. 2 B) associated with type 2 immune responses (Gause et al., 2013), along with expanded resident B1a lymphocytes, whereas T cell counts were unchanged (Fig. 2 C). Infiltrating monocytes and neutrophils were not found, ruling out generalized local inflammation. Gata6ΔMac macrophages metabolism was oriented to oxidative phosphorylation, another feature of alternative activation (Vats et al., 2006; Pearce and Pearce, 2013), as observed by increased oxygen consumption rates (OCR; Fig. 2 D). Profiling of metabolic intermediates from sorted control and Gata6ΔMac macrophages, respectively, revealed greater ADP (34 ± 3 vs. 58 ± 12 pmol/106 cells), citrate or isocitrate (CIT/ICIT; 76 ± 12 vs. 129 ± 46 pm/106 cells), NAD+ (1.1 ± 0.4 vs. 2.1 ± 0.5 pmol/106 cells), and malate (MAL; 120 ± 28 vs. 167 ± 23 pmol/106 cells) levels in Gata6ΔMac macrophages, with reduced amounts of FAD (6.9 ± 0.4 vs. 5.5 ± 0.5 pmol/106 cells), consistent with a more active citric acid cycle (Fig. 2 E).
Figure 2.
Gene expression changes and alternative activation of Gata6ΔMac macrophages. (A) Scatter plots depict mRNA transcripts significantly decreased (left, blue) or increased (right, red) in Gata6flox/flox (Ctrl) versus Gata6ΔMac mice. (B) Expression of macrophage polarization and activation markers and eosinophil counts in the peritoneum. (C) Peritoneal B1-a and T lymphocyte counts. (D) Oxygen consumption rates (OCR) of Gata6flox/flox and Gata6ΔMac mice. (E) Mass spectrometric analysis of metabolites. (F) As shown in Fig. 1 D (blue line in flow cytometry gate), Gata6ΔMac peritoneal macrophages were sorted into two populations based on a retention or reduction of the originally high levels of F4/80, then stained for nuclear Gata6. Bar, 5 µm. Various markers on Gata6flox/flox (light gray histograms) and Gata6ΔMac macrophages with preserved expression of Gata6 (Gata6+, from F4/80high gate; gray histograms) and Gata6ΔMac with efficient deletion of Gata6 (Gata6− from F4/80reduced gate; black histograms) were compared. (G) Analysis of similar markers on spleen or lung macrophages. Data in the figure summarize results from three or more independent experiments with two to five replicates per experimental group. Metabolic analysis was performed using five separate pools of sorted macrophages for each genotype. *, P < 0.05; **, P < 0.001 relative to controls, assessed using two-tailed Student’s t tests.
Deletion of Gata6 correlated with decreased F4/80 expression (Fig. 1 D, events below blue line in Gata6ΔMac gate), but a minority of macrophages, i.e., 19.9 ± 10.2% of remaining ICAM-2+ macrophages, retained the originally high levels of F4/80 (Fig. 1 D) and only these cells immunostained with Gata6 mAb (Fig. 2 F). Among flow-sorted Gata6ΔMac macrophages with higher F4/80, only 26 ± 7% of nuclei had deleted Gata6, whereas 99 ± 0.4% had deleted Gata6 among Gata6ΔMac macrophages with reduced F4/80. Thus, the persistence of some resident macrophages that remained Gata6+, clearly demarcated by the same high expression of surface F4/80 as in WT mice, allowed us to address whether alternative activation was confined to macrophages that lost Gata6. Indeed, resident peritoneal macrophages with efficient deletion of Gata6 (F4/80 reduced) expressed higher level of CD206, CD301, and Lyve-1 compared with F4/80high macrophages or macrophages from control mice (Fig. 2 F). However, the population of macrophages that did not delete Gata6 (F4/80high) were those that induced Marco (Fig. 2 F). Macrophages in the splenic red pulp, lung (Fig. 2 G), or brain (not depicted) did not increase alterative activation markers in Gata6ΔMac mice. These data, therefore, indicate that deletion of Gata6 renders peritoneal macrophages prone to alternative activation. They may be more sensitive to external stimuli that promote alternative activation or there may be a derepression of an alternative activation program in a cell autonomous manner.
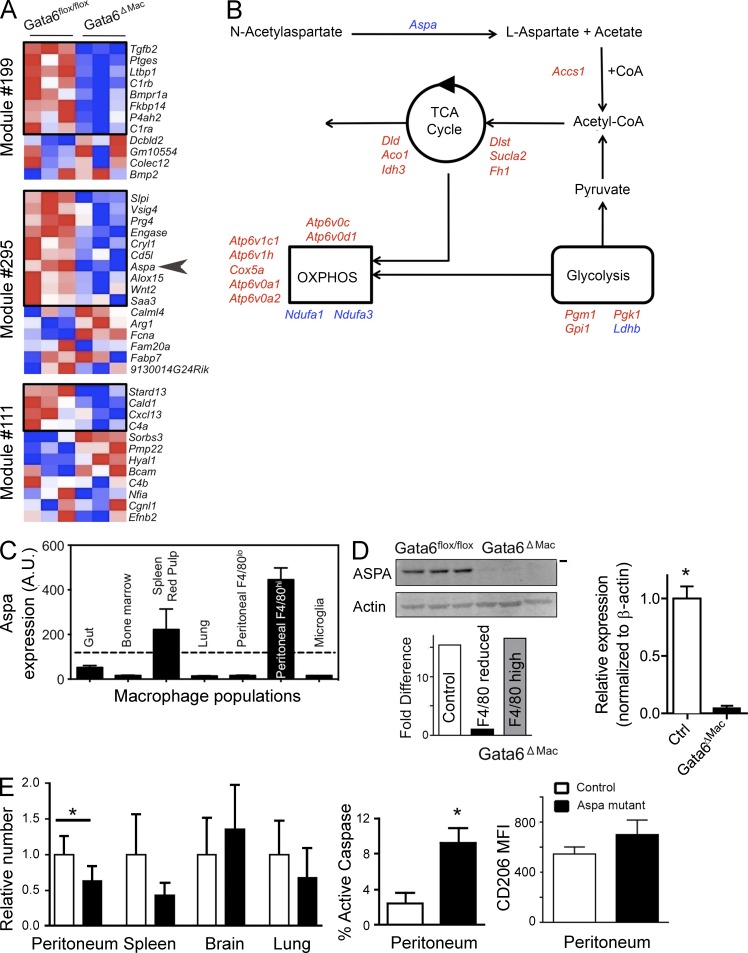
As mentioned, alternative activation has been linked to changes in cellular metabolic orientation (Vats et al., 2006; Pearce and Pearce, 2013). Furthermore, mRNA transcripts that distinguish peritoneal macrophages from other macrophages were especially those associated with lipid synthesis and signaling (Gautier et al., 2012). Several such transcripts were among those that were confined to modules of genes predicted by the Ontogenet algorithm to be regulated in peritoneal macrophages by Gata6 (Gautier et al., 2012). Most mRNA transcripts that were peritoneal macrophage-specific within these modules were indeed down-regulated in Gata6-deficient macrophages (Fig. 3 A). Others in the same modules not originally identified to be specific for peritoneal macrophages were notably up-regulated, including Arg1, which encodes arginase-1, an enzyme associated with canonical alternative activation (Fig. 3 A). Thus, we examined the expression of genes involved in metabolism and lipid synthesis. Whereas many metabolic genes were up-regulated, Aspa mRNA encoding a key enzyme at an early step in the pathway leading to acetyl CoA synthesis, aspartoacylase, was among the most down-regulated genes (Fig. 3 B). Furthermore, Aspa mRNA expression was highest in peritoneal macrophages and absent from other hematopoietic cells (http://www.immgen.org), including other macrophages, except for a more modest expression in red pulp macrophages (Fig. 3 C). Aspartoacylase is known to regulate lipid synthesis in the brain, and mutations in Aspa lead to Canavan disease characterized by defective synthesis of myelin (Traka et al., 2008). Its substrate N-acetylaspartate is the second-most abundant metabolite in the brain, being produced by neurons and used by oligodendrocytes to coordinate their differentiation, energy production, and lipid synthesis (Madhavarao and Namboodiri, 2006). We first confirmed that aspartoacylase protein was expressed by control peritoneal macrophages. It was reduced to nearly undetectable levels in Gata6ΔMac macrophages (Fig. 3 D), with its mRNA transcript selectively lost in the F4/80reduced population of macrophages that corresponded to those that deleted Gata6 (Fig. 3 D). To test whether loss of Aspa might be relevant to the loss of viable macrophages in the peritoneum, we quantified peritoneal macrophages in Nur7 mutant mice that bear a nonsense mutation in the Aspa (Traka et al., 2008).These mutants showed a 37% reduction in resident peritoneal macrophages (Fig. 3 E), along with enhanced apoptosis indicated by increased active caspase 3 (Fig. 3 E), although the extent of the loss in overall macrophage numbers was not as large as observed in Gata6ΔMac macrophages. As the mutant mice lack aspartoacylase wherever it is expressed, we cannot be sure that the increased death of peritoneal macrophages is due to inherent lack of aspartoacylase expression. However, it is interesting to note that Aspa mutant mice trended to reduced red pulp macrophages (P = 0.06), whereas macrophages in lung and brain, which do not express aspartoacylase, were not significantly decreased. Loss of expression of aspartoacylase did not lead to macrophage polarization, as shown by similar CD206 (Fig. 3 E) and other markers of alternative activation (not depicted) on the cell surface of peritoneal or other macrophages between mutant and control mice. We conclude that deficiency in Aspa increases peritoneal macrophage susceptibility to death, and partially accounts for the loss of peritoneal macrophages in Gata6ΔMac mice, but its loss is not sufficient to provoke alternative activation.
Figure 3.
Gata6 regulates aspartylcyclase expression that in turn affects peritoneal macrophage survival. (A) Heat map depicts pattern of expression of mRNA transcripts within modules earlier predicted to be controlled by Gata6. Transcripts within black boxes indicate those that are selectively expressed in peritoneal macrophages relative to other macrophages (Gautier et al., 2012). Arrowhead delineates Aspa. Data are compiled from three independent experiments. (B) Generalized schematic image of metabolic pathways and the expression of various mRNA transcripts in the pathways by Gata6ΔMac peritoneal macrophages. Red color depicts mRNA transcripts significantly up-regulated and blue depicts those significantly down-regulated. (C) Signal intensity of gene expression for Aspa from array data comparing resident macrophages from multiple organs. Dotted line, cutoff representing positive expression after data normalization. (D) Immunoblot for aspartoacylase (ASPA) in control Gata6flox/flox versus Gata6ΔMac mice. Each lane represents a distinct experiment in which pooled macrophages of the depicted genotype were sorted, with acquisition of all ICAM-2+ macrophages sorted from the respective genotypes. Bar on right represents 37-kD molecular weight marker. Real-time quantitative PCR from a single experiment from sorted macrophages pooled from four to seven mice (control vs. Gata6-deficient, respectively) was used; Aspa mRNA transcripts are shown as fold differences between sorted macrophage groups. (E) Macrophage counts in different organs and percent active caspase 3 in control versus Aspa (Nur7) mutant peritoneal macrophages. Far right bar graph shows CD206 MFI in peritoneal macrophages. Two experiments were conducted, with n = 9–10 for peritoneal analyses and n = 4–5 for other organs. *, P < 0.05 relative to control, determined by two-tailed Student’s t tests.
In conclusion, Gata6, selectively expressed in peritoneal macrophages, regulates peritoneal macrophage survival under homeostatic conditions. One of the underlying mechanisms supporting survival is the reliance of peritoneal macrophages on the enzyme aspartoacylase, but loss of this enzyme does not explain why surviving Gata6-deficient macrophages polarize themselves to an alternatively activated phenotype, whereas adjacent Gata6+ macrophages do not. Additional studies are needed to better understand the intrinsic regulation of polarization and tissue-specific macrophage differentiation. The Gata6-deficient peritoneal macrophage provides a compelling model system for such investigation.
MATERIALS AND METHODS
Mouse strains.
Gata6flox/flox mice on a mixed 129S1/SvImJ and CD-1 background were bred with Lyz2-Cre+/− on a C57BL/6 background to yield Cre+/− Gata6ΔMac mice and Cre−/− Gata6flox/flox littermate control mice. Nur7 mice bearing mutant Aspa alleles were on a C57BL/6J background and compared with C57BL/6J mice. All experimental procedures were approved by the Animal Studies Committee at Washington University in St. Louis.
Gene expression analysis.
RNA was amplified and hybridized on the Affymetrix Mouse Gene 1.0 ST array by the ImmGen Project consortium with double-sorted cell populations sorted directly into TRIzol (Life Technologies). These procedures followed a highly standardized protocol for data generation and documentation of quality control. A table listing QC data, replicate information, and batch information for each sample is also available on the ImmGen Project website. Data were analyzed with the GenePattern genomic analysis platform. Raw data were normalized with the robust multi-array algorithm, returning linear values between 10 and 20,000. A common threshold for positive expression at 95% confidence across the dataset was determined to be 120. Differences in gene expression signatures were identified and visualized with the Multiplot module of GenePattern. Probe sets were considered to have a difference in expression with a coefficient of variation of <0.5 and a p-value of ≥0.05 (Student’s t test). Data are deposited in GenBank under accession no. GSE37448 as part of the Immgen 2 dataset. Quantitative PCR was performed on ICAM-2+ peritoneal macrophages sorted from Gata6flox/flox or Gata6ΔMac mice in which F4/80high and F4/80reduced macrophages were distinctly sorted. 5′-TCCAAGGAATGAAAGTGGAGA-3′ was the sequence of the forward primer and 5′-TGCAATGGTTTCCAGTCTTG-3′ was the reverse primer.
Peritoneal macrophage quantification and characterization.
Two methods were used to quantify peritoneal macrophages. Total peritoneal cells were counted in a hemacytometer or using an automated cell counter. Then this number was multiplied by the percent of macrophages stained for CD115 (AFS98; eBioscience), ICAM-2 (3C4; BioLegend) and F4/80 (BM8, eBioscience) by flow cytometry. These antibodies allow us to distinguish the minor and major peritoneal macrophage populations (Gautier et al., 2012), with only the major macrophage population expressing ICAM-2 (Gautier et al., 2012). Alternatively, peritoneal cells obtained by lavage were incubated with FITC-conjugated anti–ICAM-2 mAb, and then a manual count of the fraction of ICAM-2+ cells was made and multiplied by the total number of peritoneal cells. Both methods yielded similar results. Other reagents and mAbs used in flow cytometry were Annexin V (Miltenyi Biotec) and antibodies against Ki67 (B56; BD), active caspase 3 (C92-605; BD), pH3 (D2C8; Cell Signaling technology), TIMD4 (RMT4-54; eBioscience), CD206 (MR5D3; Serotec), LYVE-1 (Abcam), Siglec F (E50-2440; BD), CD11b (M1/70; eBioscience), CD5 (53–7.3; BD), B220 (RA3-6B2; eBioscience), ly 6C/G (RB6-8C5; eBioscience), Ly6-G (1A8; BD), TCRb (H57-597; eBioscience), MHC-II (M5/114.15.2; eBioscience), and MARCO (ED31; Serotec).
H. polygyrus infection.
Mice were infected orally with 200 infectious larvae of H. polygyrus and euthanized 8 d later for collection of peritoneal lavage and enumeration of macrophages.
Cell cycle analysis.
Propidium iodide staining was used to analyze the cell cycle in peritoneal macrophages. A barcoding approach was used. Peritoneal cells stained for PE-conjugated CD11b were mixed 9:1 with a second collection of peritoneal cells from the same mouse separately stained with FITC-conjugated ICAM-2. Statistically doublets or larger aggregates could be CD11b+ ICAM-2− or CD11b+ ICAM-2+, but doublets that were ICAM-2+ CD11b− would be a vast minority of doublets. Thus, ICAM-2+ CD11b− macrophages from the mixture were gated and propidium iodide intensity was plotted on a linear scale.
Immunoblots.
FACS-sorted cells were homogenized in lysis buffer containing protease inhibitors. Protein extracts were run on Criterion gels (Bio-Rad Laboratories) and blotted onto nitrocellulose membranes. After blocking, immunoblots were incubated with primary Abs against Gata6 (D61E4; Cell Signaling Technology) or aspartoacylase (GeneTex) and β-actin (Cell Signaling Technology). Blots were then incubated with fluorescent secondary Abs and proteins were detected using the fluorescence-based Odyssey Infrared Imaging System (LI-COR Biosciences).
Metabolomics.
FACS-sorted macrophages (106 cells) were pelleted and rapidly washed (<10 s) with a mass spectrometry–compatible buffer (150 mM ammonium acetate solution) to prevent the presence of sodium and phosphate in the residue and limit interference with LC-MS analyses. After a second step of centrifugation, dry pellets were immediately frozen in liquid nitrogen to quench metabolism according to the University of Michigan Molecular Phenotyping Core facility’s instructions. Samples were shipped on dry ice to the Molecular Phenotyping Core facility where metabolites were extracted by exposing the cells to a chilled mixture of 80% methanol, 10% chloroform, and 10% water. Glycolytic and citric acid metabolites were then analyzed by the Molecular Phenotyping Core facility using LC-MS.
Statistical analysis.
The statistical significance of differences in mean values was analyzed with the unpaired, two-tailed Student’s t test or ANOVA for multiple comparisons. P values < 0.05 were considered statistically significant. Errors shown in bar graphs and mentioned in text refer to standard deviations.
Supplemental material.
Table S1 summarizes up-regulated mRNA transcripts in Gata6ΔMac macrophages compared to Gata6flox/flox macrophages. Table S2 summarizes down-regulated mRNA transcripts in Gata6ΔMac macrophages compared to Gata6flox/flox macrophages. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20140570/DC1.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant AI049653 to G. J. R. and utilized Core Services supported by a National Institutes of Health (NIH) grant (DK089503) to the University of Michigan. J.W.W. was supported by NIH training grant (T32DK007296).
The authors have no competing financial interests related to this work.
Footnotes
Abbreviations used:
- Aspa
- aspartoacylase
- Gata6ΔMac
- Lyz2-Cre x Gata6flox/flox
References
- Clausen, B.E., Burkhardt C., Reith W., Renkawitz R., and Förster I.. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8:265–277 10.1023/A:1008942828960 [DOI] [PubMed] [Google Scholar]
- Davies, L.C., Jenkins S.J., Allen J.E., and Taylor P.R.. 2013. Tissue-resident macrophages. Nat. Immunol. 14:986–995 10.1038/ni.2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause, W.C., Wynn T.A., and Allen J.E.. 2013. Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat. Rev. Immunol. 13:607–614 10.1038/nri3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., et al. . Immunological Genome Consortium. 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118–1128 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, E.L., Ivanov S., Lesnik P., and Randolph G.J.. 2013. Local apoptosis mediates clearance of macrophages from resolving inflammation in mice. Blood. 122:2714–2722 10.1182/blood-2013-01-478206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, S., and Martinez F.O.. 2010. Alternative activation of macrophages: mechanism and functions. Immunity. 32:593–604 10.1016/j.immuni.2010.05.007 [DOI] [PubMed] [Google Scholar]
- Haldar, M., Kohyama M., So A.Y., Kc W., Wu X., Briseño C.G., Satpathy A.T., Kretzer N.M., Arase H., Rajasekaran N.S., et al. . 2014. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 156:1223–1234 10.1016/j.cell.2014.01.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D., et al. . 2013. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 38:792–804 10.1016/j.immuni.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, S.J., Ruckerl D., Thomas G.D., Hewitson J.P., Duncan S., Brombacher F., Maizels R.M., Hume D.A., and Allen J.E.. 2013. IL-4 directly signals tissue-resident macrophages to proliferate beyond homeostatic levels controlled by CSF-1. J. Exp. Med. 210:2477–2491 10.1084/jem.20121999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama, M., Ise W., Edelson B.T., Wilker P.R., Hildner K., Mejia C., Frazier W.A., Murphy T.L., and Murphy K.M.. 2009. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 457:318–321 10.1038/nature07472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavarao, C.N., and Namboodiri A.M.. 2006. NAA synthesis and functional roles. Adv. Exp. Med. Biol. 576:49–66, discussion :361–363 10.1007/0-387-30172-0_4 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, S., Ramadass A.S., Akoulitchev A., and Gordon S.. 2014. Formation of distinct chromatin conformation signatures epigenetically regulate macrophage activation. Int. Immunopharmacol. 18:7–11 10.1016/j.intimp.2013.10.024 [DOI] [PubMed] [Google Scholar]
- Okabe, Y., and Medzhitov R.. 2014. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 157:832–844 10.1016/j.cell.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, E.L., and Pearce E.J.. 2013. Metabolic pathways in immune cell activation and quiescence. Immunity. 38:633–643 10.1016/j.immuni.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas, M., Davies L.C., Giles P.J., Liao C.T., Kharfan B., Stone T.C., O’Donnell V.B., Fraser D.J., Jones S.A., and Taylor P.R.. 2014. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 344:645–648 10.1126/science.1251414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S.E., Pollard J.W., et al. . 2012. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 336:86–90 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Sodhi, C.P., Li J., and Duncan S.A.. 2006. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC Dev. Biol. 6:19 10.1186/1471-213X-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka, M., Wollmann R.L., Cerda S.R., Dugas J., Barres B.A., and Popko B.. 2008. Nur7 is a nonsense mutation in the mouse aspartoacylase gene that causes spongy degeneration of the CNS. J. Neurosci. 28:11537–11549 10.1523/JNEUROSCI.1490-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vats, D., Mukundan L., Odegaard J.I., Zhang L., Smith K.L., Morel C.R., Wagner R.A., Greaves D.R., Murray P.J., and Chawla A.. 2006. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 4:13–24 10.1016/j.cmet.2006.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona, S., Kim K.W., Wolf Y., Mildner A., Varol D., Breker M., Strauss-Ayali D., Viukov S., Guilliams M., Misharin A., et al. . 2013. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 38:79–91 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.