A novel MAIT cell antagonist, Ac-6-FP, stabilizes MR1 and can inhibit MAIT cell activation with the flexible TCR β-chain serving to fine-tune the affinity of the TCR for antigen-MR1 complexes.
Abstract
Mucosal-associated invariant T (MAIT) cells express an invariant T cell receptor (TCR) α-chain (TRAV1-2 joined to TRAJ33, TRAJ20, or TRAJ12 in humans), which pairs with an array of TCR β-chains. MAIT TCRs can bind folate- and riboflavin-based metabolites restricted by the major histocompatibility complex (MHC)-related class I−like molecule, MR1. However, the impact of MAIT TCR and MR1-ligand heterogeneity on MAIT cell biology is unclear. We show how a previously uncharacterized MR1 ligand, acetyl-6-formylpterin (Ac-6-FP), markedly stabilized MR1, potently up-regulated MR1 cell surface expression, and inhibited MAIT cell activation. These enhanced properties of Ac-6-FP were attributable to structural alterations in MR1 that subsequently affected MAIT TCR recognition via conformational changes within the complementarity-determining region (CDR) 3β loop. Analysis of seven TRBV6-1+ MAIT TCRs demonstrated how CDR3β hypervariability impacted on MAIT TCR recognition by altering TCR flexibility and contacts with MR1 and the Ag itself. Ternary structures of TRBV6-1, TRBV6-4, and TRBV20+ MAIT TCRs in complex with MR1 bound to a potent riboflavin-based antigen (Ag) showed how variations in TRBV gene usage exclusively impacted on MR1 contacts within a consensus MAIT TCR-MR1 footprint. Moreover, differential TRAJ gene usage was readily accommodated within a conserved MAIT TCR-MR1-Ag docking mode. Collectively, MAIT TCR heterogeneity can fine-tune MR1 recognition in an Ag-dependent manner, thereby modulating MAIT cell recognition.
Mucosal-associated invariant T (MAIT) cells are an evolutionarily conserved (Tilloy et al., 1999; Huang et al., 2009) innate-like population of T cells that are very abundant in humans (Porcelli et al., 1993; Tilloy et al., 1999; Reantragoon et al., 2012). MAIT cells have been implicated in protective and aberrant immunity, but their specific function remains obscure (Illés et al., 2004; Croxford et al., 2006; Peterfalvi et al., 2008; Godfrey et al., 2010; Gold et al., 2010; Le Bourhis et al., 2010; Miyazaki et al., 2011; Chua et al., 2012; Eckle et al., 2013; Gold and Lewinsohn, 2013; Meierovics et al., 2013; Birkinshaw et al., 2014; Serriari et al., 2014). Once activated via their αβ TCR, MAIT cells rapidly secrete an array of cytokines (Kawachi et al., 2006; Dusseaux et al., 2011; Tang et al., 2013). Unlike the classical MHC-restricted T lymphocytes, MAIT cells typically express an invariant TCR α-chain paired with one of a selected group of TCR β-chains, with the MAIT TCR being highly conserved across mammals (Tilloy et al., 1999; Huang et al., 2009). In humans, the TCR α-chain comprises the TRAV1-2 gene that combines with the TRAJ33 gene segment, with limited nonnucleotide (N) additions/deletions at the TRAV1-2-TRAJ33 junction. In mice, the MAIT TCR repertoire uses the orthologous TCR α-chain (TRAV1-TRAJ33). In addition, the human MAIT TCR α-chain repertoire also includes smaller subsets containing TRAJ20 and TRAJ12 gene segments paired with the TRAV1-2 gene (Reantragoon et al., 2013). Although the human MAIT TCR β-chain repertoire was considered to mainly be comprised of TRBV20, TRBV6-1, and TRBV6-4 genes (Tilloy et al., 1999), MR1-tetramer based studies have demonstrated that the MAIT TCR β-chain repertoire is more diverse (Reantragoon et al., 2013). Moreover, the MAIT TCR β-chain is typified by a hypervariable complementarity-determining region (CDR) 3β loop (Tilloy et al., 1999; Reantragoon et al., 2013). The semiinvariant nature of the MAIT TCR resonates with the repertoire diversity of type I NKT TCRs (Borg et al., 2007; Mallevaey et al., 2009; Pellicci et al., 2009; Patel et al., 2011; Rossjohn et al., 2012). However, although we have a growing understanding of how the NKT TCR repertoire can interact with a range of CD1d-restricted, lipid-based Ags, the molecular basis of MAIT TCR α- and β-chain heterogeneity, CDR3β hypervariability, and MR1-ligand diversity on MAIT cell function is unknown.
The MAIT TCR is restricted by the MHC class I–related molecule MR1 (Treiner et al., 2003; Huang et al., 2005; Huang et al., 2008). MR1 is a monomorphic Ag-presenting molecule that is highly conserved across mammals (Tsukamoto et al., 2013). Although the MR1 transcript is expressed widely (Hashimoto et al., 1995; Riegert et al., 1998), cell surface expression of MR1 is very low/absent, thereby indicating that other factors, including Ag supply, can determine the level of MR1 that egresses to the cell membrane (Huang et al., 2008; Chua et al., 2011). Recently, it has been established that MR1 can bind vitamin B–based precursors and derivatives that originate from folic acid (vitamin B9) and riboflavin (vitamin B2) biosynthesis (Kjer-Nielsen et al., 2012). Specifically, MR1 can present 6-formylpterin (6-FP), a naturally occurring photo-degradation product of folic acid, and a series of ribityllumazines, including 6,7-dimethyl-8-d-ribityllumazine (RL-6,7-DiMe), 6-methyl-7-hydroxy-8-d-ribityllumazine (RL-6-Me-7-OH; Kjer-Nielsen et al., 2012; Patel et al., 2013), 5-(2-oxoethylideneamino)-6-d-ribitylaminouracil (5-OE-RU), and 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU; Corbett et al., 2014). The MR1 Ag-binding cleft is ideally disposed to bind to these small organic metabolites, with the ligands being closely sequestered by an aromatic cradle within MR1, whereupon some of the ligands can form a covalent bond (Schiff base) with MR1 (Kjer-Nielsen et al., 2012). Although the 6-FP ligand up-regulated MR1 cell surface expression, it did not stimulate MAIT cells (Kjer-Nielsen et al., 2012). However, the ribityllumazines were stimulatory, with the extent of MAIT cell potency varying markedly, such that synthetic rRL-6-CH2OH (reduced 6-hydroxymethyl-8-d-ribityllumazine) is ∼1,000 times more potent than the weak agonists RL-6,7-DiMe and RL-6-Me-7-OH (Kjer-Nielsen et al., 2012). We recently traced the high potency of rRL-6-CH2OH to MR1 trapping and presentation of a relatively unstable pyrimidine-based transitory derivative (5-OP-RU), with similar high potency also found for a homologue with one carbon less (5-OE-RU) but no activity for another homologue with one extra carbon, 5-(1-methyl-2-oxopropylideneamino)-6-d-ribitylaminouracil (5-MOP-RU; Corbett et al., 2014). Thus, the MR1 cleft, although having a penchant for small molecule uracil derivatives, nonetheless has sufficient plasticity to accommodate a range of chemical entities, but the nature and relative impact of molecular determinants on MR1 function remains underexplored.
We recently provided insight into how the human TRAV1-2-TRAJ33-TRBV6-1+ MAIT TCR recognized human MR1 presenting 6-FP, RL-6-Me-7-OH (Patel et al., 2013) and two short lived adducts, 5-OE-RU and 5-OP-RU, arising from reaction of glyoxal and methylglyoxal, respectively, with 5-amino-6-d-ribitylaminouracil (5-A-RU), an intermediate in bacterial riboflavin synthesis (Corbett et al., 2014). Regardless of the stimulatory properties of the ligand, the MAIT TCR docked in a conserved manner with MR1 (Patel et al., 2013; Corbett et al., 2014), with this consensus docking mode being observed in a humanized bovine MR1-MAIT TCR ternary complex with undefined Ags bound (López-Sagaseta et al., 2013). The important role of the MAIT TCR α-chain was underpinned by the marked effect some residues had on MR1-Ag recognition. In contrast, the germline-encoded regions of the TCR β-chain appeared less ideally disposed to interact with MR1, which was consistent with an alanine-scanning mutagenesis study on the human MAIT TCR TRBV6-1 encoded region (Reantragoon et al., 2012). Nevertheless, the CDR3β loop interacted extensively with MR1, being situated directly above the Ag itself, thereby suggesting that CDR3β loop hypervariability may impact on MAIT TCR recognition in an Ag-dependent manner. MR1 tetramers loaded (Reantragoon et al., 2013) or refolded (Corbett et al., 2014) with the transitory intermediates stained the majority of MAIT cells, suggesting that MAIT cells, regardless of their MAIT TCR repertoire, are tailored to interact with potent Ags, in a manner analogous to how the CD1d-restricted ligand α-galactosylceramide represents the universal potent ligand for NKT cells (Matsuda et al., 2000). However, it is unknown whether less potent MAIT cell Ags are recognized differentially on account of variations in the MAIT TCR repertoire, as observed in NKT cell recognition of weak agonists (Pellicci et al., 2009; Matulis et al., 2010; Wun et al., 2011).
In this study, we address the impact of MAIT TCR heterogeneity on recognition of different MR1-restricted ligands. Although the MAIT TCR heterogeneity converged onto a common docking footprint on MR1, we show that TCR β-chain usage can modulate MAIT TCR recognition and fine-tune the response in an Ag-dependent manner.
RESULTS
A novel MAIT cell inhibitory ligand
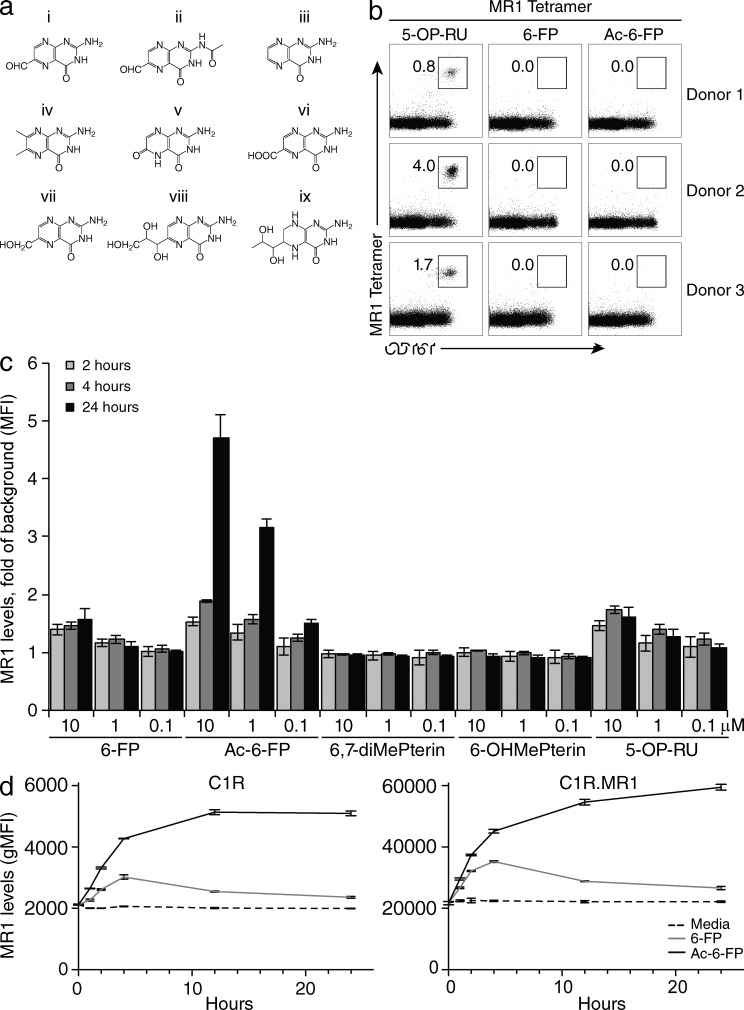
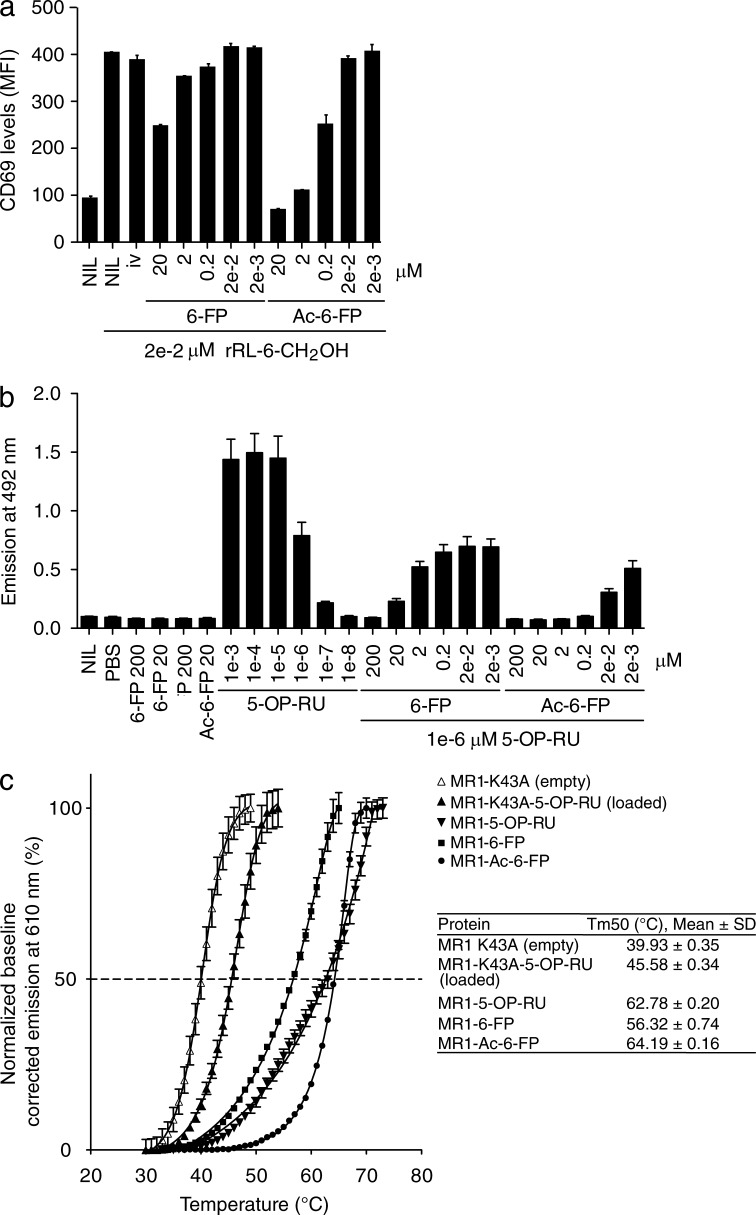
We previously established that 6-FP could up-regulate MR1 cell surface expression on C1R.MR1 cells without activating MAIT cells or Jurkat cells transduced with a MAIT TCR (Jurkat.MAIT; Kjer-Nielsen et al., 2012). Instead, 6-FP was a weak competitive antagonist of MAIT cell activation (Patel et al., 2013). Thus, we evaluated whether analogues of 6-FP could competitively inhibit MAIT cell recognition more potently. Although the compounds 6,7-dimethylpterin, 6-hydroxymethylpterin, pterin, l-monapterin, tetrahydrobiopterin, xanthopterin, and pterin-6-carboxylic acid (Fig. 1 a) could not be refolded with MR1 or increase MR1 cell surface expression, acetyl-6-FP (Ac-6-FP) readily refolded with MR1 as judged by gel filtration profile and cell staining with the MR1-reactive mAb 26.5 (not depicted). Analogous to MR1-6-FP tetramers, MR1–Ac-6-FP tetramers did not stain human PBMCs (Fig. 1 b), which is indicative of Ac-6-FP not representing a stimulating ligand for MAIT cells. Ac-6-FP differs from 6-FP by the presence of an acetyl group (Fig. 1 a), yet it still possesses a formyl group at the 6-position and thus was anticipated to form a Schiff base when bound to MR1. Ac-6-FP caused much greater surface up-regulation of MR1 when compared with 6-FP or other pterin-based compounds (Fig. 1 c). Interestingly, both ligands induced increased MR1 expression, but the up-regulation induced by 6-FP was slower and less pronounced than that caused by Ac-6-FP. Intriguingly, the effect of 6-FP was also transient, whereas it was maintained for at least 24 h with Ac-6-FP (Fig. 1 d). Ac-6-FP, although nonstimulatory for MAIT cells, nevertheless strongly inhibited Jurkat.MAIT activation in response to an agonist ligand, being a 100 fold more potent inhibitor than 6-FP in cell-based assays, in which CD69 up-regulation and IL-2 production were used as functional readouts (Fig. 2, a and b). To formally test the apparent stabilizing effect of Ac-6-FP, we undertook thermostability assays on the refolded MR1–Ag complex, and compared these values to the half maximum melting temperatures (Tm50) of recombinant MR1-6-FP, MR1–5-OP-RU, MR1-K43A-5-OP-RU, and MR1-K43A (empty; Fig. 2 c). The MR1-K43A protein, in which the Lys43 had been mutated to alanine to prevent Schiff base formation with any ligands (Reantragoon et al., 2013) was refolded in the absence of any ligands. This MR1-K43A empty molecule had a Tm50 of 40°C and served as the baseline to assess the impact of the effect of Schiff base formation and the presence of ligand on MR1 stability. Notably, although MR1-K43A-5-OP-RU exhibited a moderately increased stability (Tm50 46°C) to that of MR1-K43A, MR1–5-OP-RU had a markedly increased Tm50 value of 63°C. Thus, the ability of 5-OP-RU to form a Schiff base with MR1 resulted in a 17°C increase in MR1 stability. In contrast, the Tm50 of MR1-6-FP was 56°C, indicating that the stabilizing effect of the Schiff base formation is also dependent on the nature of the bound ligand. Interestingly, MR1–Ac-6-FP exhibited a Tm50 value of 64°C, suggesting that the presence of the acetyl group in Ac-6-FP led to an 8°C increase in stability compared with MR1-6-FP. The higher stability of MR1–Ac-6-FP was consistent with Ac-6-FP being a more potent inhibitor than 6-FP and having a prolonged effect on cell surface expression of MR1.
Figure 1.
MR1 tetramer staining and MR1 up-regulation by 6-FP and Ac-6-FP. (a) Chemical structures of MR1-restricted pterin ligands (6-FP [6-FP, i] and acetyl-6-FP [Ac-6-FP, ii]) and pterin-based compounds that did not bind to MR1 (pterin [iii], 6,7-dimethylpterin [6,7-diMePterin, iv], xanthopterin [v], pterin-6-carboxylic acid [vi], 6-hydroxymethylpterin [6-OHMePterin, vii], l-monapterin [viii], tetrahydrobiopterin [ix]). (b) Human PBMCs were stained with CD3- and CD161-specific mAbs and human tetrameric MR1–5-OP-RU, MR1-6-FP, or MR1–Ac-6-FP, respectively. Live, CD3+ cells were considered for CD161 (x axis) and tetramer staining (y axis), displaying dot plots and percentages of cells within boxed regions. Displayed are the results of three representative of 6 healthy blood donors tested. These experiments were performed twice using two independent batches of tetramer and yielding similar results. (c) MR1 expression levels on C1R.MR1 cells upon incubation with 6-FP, Ac-6-FP, 6,7-dimethylpterin, 6-hydroxymethylpterin, and 5-OP-RU added in indicated doses and for indicated time spans. Cells were stained with anti-MR1 mAb 26.5. Data shows MFI fold of background of MR1 expression levels, mean ± SEM of three independent experiments done in triplicates. These experiments were performed three times, yielding similar results and a representative experiment is shown. (d) Comparison of MR1 expression levels on C1R and C1R.MR1 cells upon incubation with 10 µM 6-FP or Ac-6-FP, or without ligand (media), for different time periods. Cells were stained with anti-MR1 mAb 26.5. Data shows untransformed geometric MFI (gMFI) of MR1 expression levels for each time point. Mean ± SEM of triplicates. These experiments were performed twice, yielding similar results and a representative experiment is shown.
Figure 2.
MAIT cell inhibitory ligands and MR1 stability. (a) Inhibition of Jurkat.MAIT (original WT, clone A-F7) cell activation by 6-FP or Ac-6-FP. 6-FP, Ac-6-FP (or controls – 6,7-diMePterin or no ligand) were added to C1R.MR1 cells at the indicated concentrations for 1 h before addition of Jurkat.MAIT cells and 0.02 µM synthetic rRL-6-CH2OH. Data shows MFI of CD69 expression levels for gated Jurkat.MAIT cells. Mean ± SEM of triplicates. These experiments were performed twice, yielding similar results and a representative experiment is shown. (b) Inhibition of Jurkat76.MAIT (original WT TCR, clone A-F7) cell activation by 6-FP or Ac-6-FP. 6-FP, Ac-6-FP (or controls, PBS or no ligand) were added to C1R.MR1 cells at the indicated concentrations and co-incubated with Jurkat76.MAIT cells and 0.02 µM synthetic rRL-6-CH2OH. Data shows emission at 492 nm correlating with IL-2 production. These experiments were performed three times, showing mean ± SD. (c) Thermostability of soluble MR1-ligand by fluorescence-based thermal shift assay. Shown is baseline-corrected, normalized emission at 610 nm plotted against temperature. Mean ± SEM of triplicate samples, and nonlinear curve-fits. The half maximum melt point (Tm50) is indicated as a dashed line. Displayed is a representative of three independent experiments yielding similar results. The table summarizes the data of three independent experiments, each in triplicate.
MAIT TCR-MR1–Ac-6-FP interaction
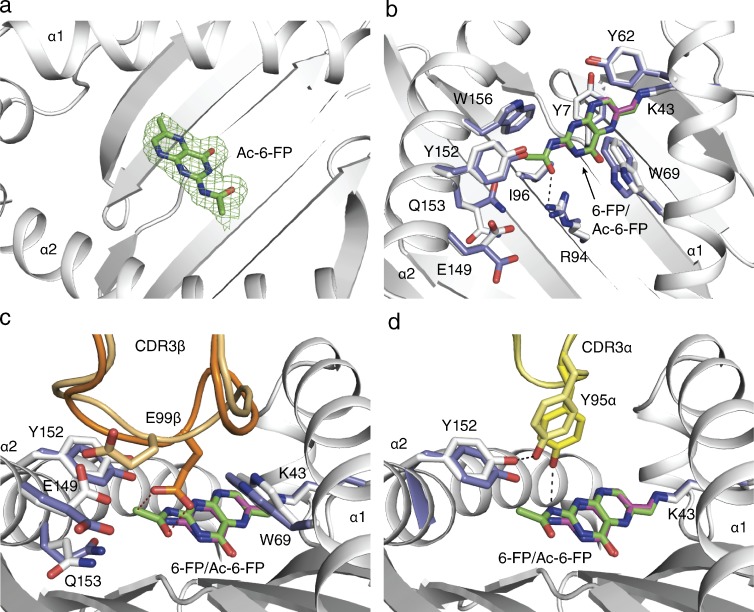
To understand the impact of the Ac-6-FP ligand, we expressed and refolded the TRBV6-1+ MAIT TCR (original WT; clone A-F7; Tilloy et al., 1999), added it to refolded MR1–Ac-6-FP, crystallized the ternary complex, and solved its structure to 2.0 Å resolution to an Rfac and Rfree of 18.0 and 22.0%, respectively (Table S1). The electron density at the MAIT TCR-MR1-Ag interface, and for the ligand itself, was unambiguous (Fig. 3 a), thereby permitting a direct comparison to the corresponding MAIT TCR-MR1-6-FP structure.
Figure 3.
MAIT TCR recognition of MR1–Ac-6-FP. (a) Electron density of Ac-6-FP in omit and final 2Fo-Fc maps of the ternary complex MR1–Ac-6-FP-MAIT original WT (clone A-F7) TCR. Electron density in mesh format, the ligand in ball and stick, MR1 in ribbon representation in white, a Fo-Fc omit map contoured at 3σ (green). (b–d) Structural basis for Ac-6-FP recognition by MR1 and original WT (clone A-F7) MAIT TCR compared with the previously published MR1-6-FP original WT (clone A-F7) MAIT TCR structure (PDB accession no. 4L4T). (b) Superposition of Ac-6-FP and 6-FP and MR1 induced conformational changes. (c) CDR3β conformational changes induced by Ac-6-FP. (d) Contacts between Ac-6-FP and Tyr95α from the CDR3α loop. MR1-6-FP residues are shown in white, pale yellow, and pale orange for the CDR3α and CDR3β loops, respectively, and the 6-FP ligand is shown in cyan. Equivalent residues from the MR1–Ac-6-FP shown in slate, yellow, and orange for the CDR3α and CDR3β loops, respectively and Ac-6-FP in green with contacts between TCR and Ac-6-FP indicated by dashed lines.
The Ac-6-FP ligand was bound within the MR1 aromatic cradle in the same orientation to that observed with 6-FP, thereby indicating that the inhibitory property of Ac-6-FP was not attributable to differences manifested by altered Ac-6-FP docking within MR1 (Fig. 3 b). As such, the formyl group of Ac-6-FP formed a Schiff base with Lys43, whereas the pterin ring was sandwiched between Tyr7, Tyr62, Trp69, and Trp156 (Fig. 3 b). The acetyl moiety formed van der Waals (vdw) interactions with Tyr152, Trp156, and Ile96, with the carbonyl moiety hydrogen bonding to Arg94, which is located at the base of the cleft (Fig. 3 b). Although the overall conformation of the Ag-binding cleft of MR1 was not appreciably altered in accommodating Ac-6-FP when compared with 6-FP (root mean square deviation (RMSD) of 0.23 Å between MR1-6-FP and MR1–Ac-6-FP over Cα atoms within the α1 and α2 domains), the presence of the additional moiety did cause conformational changes within the vicinity of the MR1-binding pocket. Namely, to accommodate the acetyl group, Trp69 was repositioned slightly (0.9 Å displacement of Nε1), Tyr152 was moved away (1.1 Å displacement of OH), and the buried residue, Gln153, was repositioned (2.6 Å displacement of Cδ) such that its side chain was orientated away from the binding pocket (Fig. 3, c and d). This reorientation of Gln153 caused an altered conformation of Glu149 (2.2 Å displacement of Cδ; Fig. 3 c), a surface-exposed MR1 residue that contacts the MAIT TCR (discussed below). Thus, subtle plasticity within the MR1-Ag–binding cleft allowed for ready accommodation of the acetyl group, with these additional contacts correlating with 8°C increase in the stability of MR1–Ac-6-FP in comparison to MR1-6-FP (Fig. 1 c). Moreover, these conformational changes affected the conformation of buried residues within the MR1-binding pocket that were transmitted to the MAIT TCR-MR1 contact zone.
The TRBV6-1+ MAIT TCR docked onto MR1-6-FP and MR1–Ac-6-FP (RMSD 0.25 Å over all Cα atoms and buried surface area [BSA] of 1,100 and 1,210 Å2, respectively) in a similar manner. Namely, the MAIT TCR docked orthogonally and centrally over the MR1-Ag–binding cleft, where Tyr95α of the TRAJ33 chain contacted the α1 and α2 helices of MR1 (Fig. 3 d), whereas the germline-encoded TRBV6-1 region contacted the α1 helix of MR1 only (not depicted). Interestingly, although the CDR3β loop was of identical sequence within these ternary complexes, their respective conformations were different, and this was attributable to Ac-6-FP induced conformational changes (Fig. 3 c). The CDR3β loop, which made an extensive interface with MR1–Ac-6-FP (BSA 24%), bridged the α1 and α2 helices of MR1 and was directly positioned over the Ag-binding pocket (Fig. 3 c). The Ac-6-FP–induced reorientation of Glu149 caused a marked remolding of a major region of the CDR3β loop so as to prevent the electrostatic repulsion of the remodeled Glu149 with Glu99β, which swung down (6.3 Å displacement of Cδ) to slot in between Trp69 and Tyr152 of MR1 (Fig. 3 c). Notably, the remodeling of Glu99β also resulted in a subtle, albeit important conformational change within the CDR3α loop. Namely, Glu99β “pulled” Tyr95α toward the ligand, enabling it, together with Glu99β from the CDR3β loop backbone, to form direct interactions with the nonstimulatory ligand (Fig. 3, c and d). These direct contacts with the Ac-6-FP are in contrast to the TRBV6-1+ MAIT TCR-MR1-6-FP ternary complex in which no direct contacts were observed between 6-FP and the TCR (Fig. 3 d). Thus, an MR1-restricted ligand can cause conformational changes to the MR1-binding cleft that are transmitted to the MAIT TCR, which subsequently leads to the hypervariable CDR3β loop making direct contacts with the Ag itself.
Impact of CDR3β variability on MAIT TCR recognition
Given that the CDR3β loop played a prominent and central role in interacting with MR1, and the conformation of the CDR3β loop could be altered indirectly in an Ag-dependent manner, we asked whether CDR3β loop hypervariability could impact on MAIT TCR-MR1-Ag recognition. To establish this, we selected six TRBV6-1+ MAIT TCRs, termed B-B10, B-F3-C1, B-G8, B-C10, C-C10, and C-A11, from published sequence data (Tilloy et al., 1999) and the original WT TCR (Tilloy et al., 1999; clone A-F7; described previously [Reantragoon et al., 2012]). Although exhibiting some minor differences in the N-region usage at the TRAV1-2-TRAJ33 junctional boundaries, the TCRs principally differed in their CDR3β composition (Table 1). These 7 MAIT TCRs were chosen because they had limited sequence identity within the CDR3β loops and differed in sequence length (11–13 aa), thereby permitting a broad perspective on CDR3β hypervariability on MAIT TCR recognition.
Table 1.
MAIT TCR usage and sequences
| TCR (name) | TRAJ | CDR3α | TRBJ | TRBV | CDR1β | CDR2β | CDR3β |
| WT (A-F7) | 33 | 89AVKDSNYQLI98 | 2-2 | 6-1 | 27MNHNS31 | 49SASEGT54 | 92ASSVWTGEGSGELF105 |
| B-B10 | 33 | 89AGMDSNYQLI98 | 1-5 | 6-1 | 27MNHNS31 | 49SASEGT54 | 92ASTLGQEGQPQH103 |
| B-F3-C1 | 33 | 89ASIDSNYQLI98 | 2-2 | 6-1 | 27MNHNS31 | 49SASEGT54 | 92ASSETDPNTGELF104 |
| B-G8 | 33 | 89AAMDSNYQLI98 | 2-2 | 6-1 | 27MNHNS31 | 49SASEGT54 | 92ASSGGDSGELF102 |
| B-C10 | 33 | 89AAEDSNYQLI98 | 2-6 | 6-1 | 27MNHNS31 | 49SASEGT54 | 92ASSYEVSGANVLT104 |
| C-C10 | 33 | 89AVVDSNYQLI98 | 2-3 | 6-1 | 27MNHNS31 | 49SASEGT54 | 92ASSPPGGTDTQ*Y103 |
| C-A11 | 33 | 89AVRDSNYQLI98 | 1-3 | 6-1 | 27MNHNS31 | 49SASEGT54 | 92ASSAAVEGGNTIY104 |
| #1 | 12 | 89AVMDSSYKLI98 | 2-1 | 6-4 | 27MRHNA31 | 49SNTAGT54 | 92ASSRGDYNEQF102 |
| #2 | 12 | 89AVMDSSYKLI98 | 2-1 | 6-4 | 27MRHNA31 | 49SNTAGT54 | 92ASSASGGAYNEQF104 |
| #3 | 12 | 89AVMDSSYKLI98 | 2-3 | 6-4 | 27MRHNA31 | 49SNTAGT54 | 92ASSPGTSGGADTQY105 |
| #4 | 20 | 89AVRDGDYKLS98 | 2-2 | 6-4 | 27MRHNA31 | 49SNTAGT54 | 92ASSAGASTGELF103 |
| #5 | 33 | 89AVSDSNYQLI98 | 2-2 | 6-4 | 27MRHNA31 | 49SNTAGT54 | 92ASSDSGGGTGELF104 |
| #6 | 33 | 89AVMDSNYQLI98 | 2-1 | 6-4 | 27MRHNA31 | 49SNTAGT54 | 92ASSGGTNNEQF102 |
| C-F7 | 33 | 89AFMDSNYQLI98 | 2-1 | 20 | 27DFQATT32 | 50SNEGSKA56 | 95SARTSGDFGEQF107 |
J segments as part of the CDR regions are shown in italics; N regions (known for sequences reported here (MAIT #1-5) or previously (MAIT #6; Reantragoon et al., 2012) are shown in bold. Sequence numbering is indicated. *, Q in TRBJ2-3 but R in Tilloy et al. (1999).
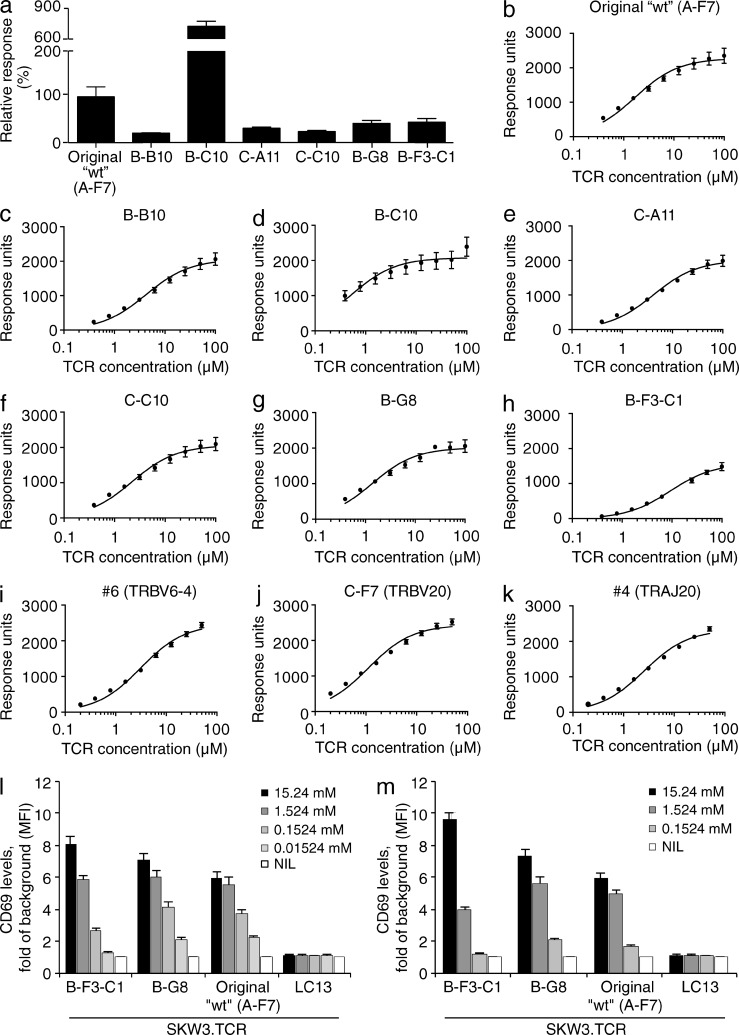
The seven TRBV6-1+ MAIT TCRs were expressed, refolded and, as judged by chromatographic profiles, assessed to be fully folded. Initially, we undertook surface plasmon resonance (SPR)–based studies to measure the binding affinity against MR1–5-OP-RU and MR1–Ac-6-FP (Table 2 and Fig. 4, a–k). The A-F7 WT Kdeq for MR1–5-OP-RU was ∼2.0 µM, consistent with previous measurements (Patel et al., 2013). The Kdeq values for the remaining six TRBV6-1+ TCRs toward MR1–5-OP-RU were mostly similar (Table 2), thereby providing insight into why MR1 tetramers of this ligand effectively stain all MAIT cells (Reantragoon et al., 2013; Corbett et al., 2014). Nevertheless, some MAIT TCRs (e.g., B-F3-C1) bound more weakly (approximate Kdeq = 9.1 µM) in comparison to the A-F7 WT value, whereas the B-C10 TCR showed moderately improved binding (approximate Kdeq = 0.5 µM) to the MR1–5-OP-RU complex (Table 2). Interestingly, great variation in relative binding affinities was observed against the MR1–Ac-6-FP complex (Fig. 4 a and Table 2). For example, the B-C10 TCR showed a marked preference for MR1–Ac-6-FP (approximate Kdeq = 35.4 µM) in comparison to the other TRBV6-1+ TCRs (Kdeq > 200 µM). Accordingly, the CDR3β loop can modulate the affinity toward MR1-restricted ligands. A MAIT TCR with a high affinity for MR1–Ac-6-FP seems rare, however, as we did not observe MR1–Ac-6-FP tetramer+ PBMCs (Fig. 1 b).
Table 2.
Summary of SPR affinities and MAIT cell activation status
| MAIT TCR | TRAJ | TRBV | Kdeq (μM), mean duplicate MR1–5-OP-RU | Activation by MR1–5-OP-RU | Kdeq (μM), mean duplicate Ac-6-FP |
| Original WT (A-F7) | 33 | 6-1 | 2.0 ± 0.6 | + | >200 |
| B-B10 | 33 | 6-1 | 4.2 ± 0. 6 | ND | >200 |
| B-C10 | 33 | 6-1 | 0.5 ± 0.1 | ND | 35.4 ± 3.0 |
| C-A11 | 33 | 6-1 | 4.2 ± 0. 5 | ND | >200 |
| C-C10 | 33 | 6-1 | 2.2 ± 0.3 | ND | >200 |
| B-G8 | 33 | 6-1 | 1.4 ± 0.2 | + | >200 |
| B-F3-C1 | 33 | 6-1 | 9.1 ± 1.0 | + | >200 |
| #1 | 12 | 6-4 | 1.5 ± 0.1 | + | >200 |
| #2 | 12 | 6-4 | 2.2 ± 0.2 | + | >200 |
| #3 | 12 | 6-4 | 1.9 ± 0.2 | + | >200 |
| #1 Y95F | 12 | 6-4 | 35 ± 2 | - | >200 |
| #2 Y95F | 12 | 6-4 | 20 ± 1 | - | >200 |
| #3 Y95F | 12 | 6-4 | 33 ± 2 | - | >200 |
| #4 | 20 | 6-4 | 2.4 ± 0.4 | + | >200 |
| #6 | 33 | 6-4 | 2.7 ± 0.7 | +a | N/D |
| C-F7 | 33 | 20 | 1.0 ± 0.3 | +a | N/D |
5-OP-RU presented by C1R.MR1 incubated with MAIT cell–activating bacteria (Reantragoon et al., 2012) or rRL-6-CH2OH (Patel et al., 2013).
Figure 4.
MAIT TCR binding affinity and activation. (a) Relative binding of MAIT TCRs original WT (clone A-F7), B-B10, B-C10, C-A11, C-C10, B-G8, and B-F3-C1 against MR1–Ac-6-FP determined by SPR at 100 µM TCR. (b–k) Equilibrium binding curves for MAIT TCRs. (b) original WT (TRBV6-1, clone A-F7), (c) B-B10, (d) B-C10, (e) C-A11, (f) C-C10, (g) B-G8, (h) B-F3-C1, (i) #6 (TRBV6-4/TRAJ33), (j) C-F7 (TRBV20), and (k) #4 (TRAJ20) against MR1–5-OP-RU determined by SPR. Data are representative of the mean response for each TCR concentration and SEM in duplicate (n = 2). Dose response to synthetic rRL-6-CH2OH (l) or 5-A-RU (m) preincubated with C1R.MR1 cells by TRAV1-2-TRAJ33-TRBV6-1 MAIT TCR+ SKW3.B-F3-C1, SKW3.B-G8, and SKW3.original WT (A-F7) or control TCR+ SKW3.LC13 cells. Data shows mean ± SEM fold of background MFI of CD69 expression for gated SKW3.TCR cells from 1 experiment (triplicate samples). These experiments were performed twice, yielding similar results.
To establish whether the differences in affinity for MR1–5-OP-RU between the B-F3-C1 MAIT TCR and a high-affinity MAIT TCR (B-G8) impacted on functional outcome, we transduced the B-F3-C1 and B-G8 MAIT TCRs into the SKW3 T cell line (SKW3.B-F3-C1 and SKW3.B-G8). We then assessed their ability to be activated by synthetic rRL-6-CH2OH and 5-A-RU in the presence of C1R.MR1 cells and compared activation to the previously transduced SKW3.MAIT TCR (WT, clone A-F7; Fig. 4, l and m). Notably, the SKW3.B-F3-C1 showed reduced activation by synthetic rRL-6-CH2OH and 5-A-RU when these ligands were not used at saturating concentrations, thereby providing further supporting evidence for the CDR3β loop fine-tuning MAIT TCR reactivity.
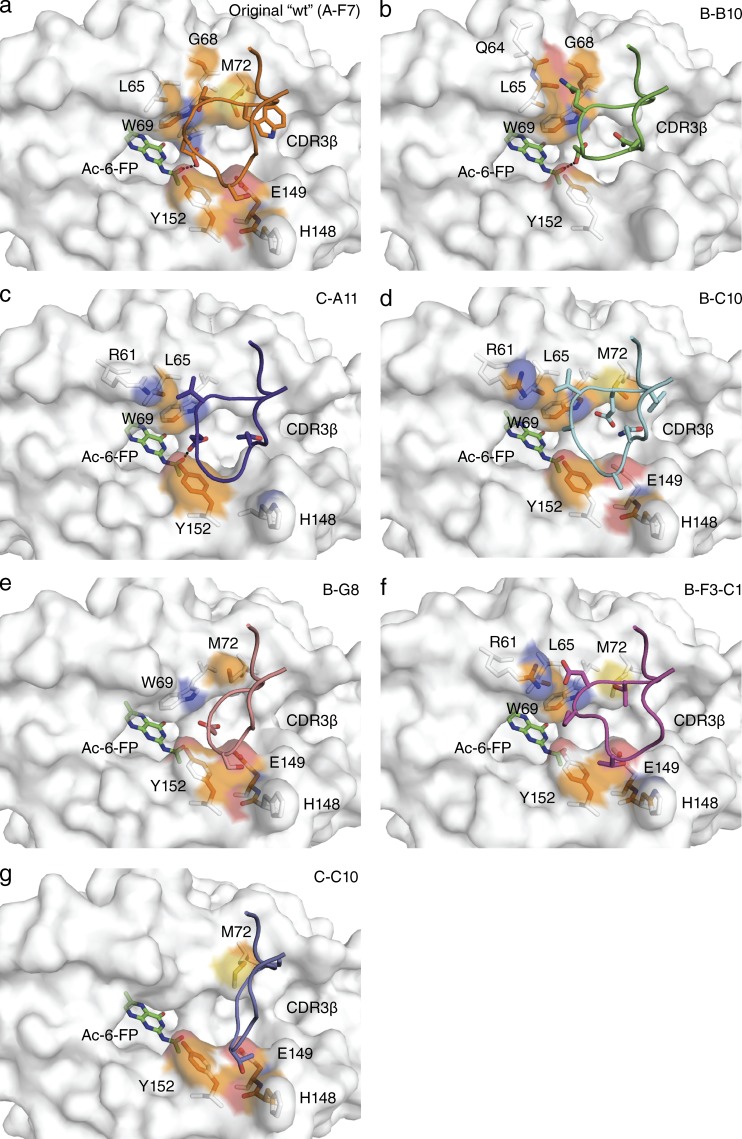
Next, we sought a structural basis for the observed effects of CDR3β impacting on MAIT TCR-MR1-Ag recognition. To establish this, we determined the structures of the B-B10, B-F3-C1, B-G8, B-C10, C-C10, and C-A11 MAIT TCRs in complex with MR1–Ac-6-FP and compared them to the original WT (clone A-F7) MAIT TCR MR1–Ac-6-FP ternary complex (Table S1 and Fig. 5, a–g). All seven TRBV6-1+ MAIT TCRs sat similarly onto MR1–Ac-6-FP, thereby indicating that the variations within the CDR3β loop do not impact on MAIT TCR-MR1 docking. Moreover, with the exception of the CDR3β loop-mediated contacts, the majority of the MAIT TCR-MR1 interatomic contacts did not alter appreciably, thereby indicating that variations within the CDR3β loop are not transferrable to the invariant MAIT TCR α chain (with the exception of Tyr95α, discussed below). While the CDR3β loops were similarly disposed atop MR1–Ac-6-FP, the extent of contacts varied between the seven TRBV6-1+ MAIT TCRs (BSA values ranging from 15 to 24%), with the CDR3β loop of the original WT (clone A-F7) TCR contributing greatest to the interface and the C-C10 and B-G8 TCRs contributing the least (Figs. 5, a–g). The CDR3β loops that are relatively rich in glycine residues result in a reduced number of CDR3β-MR1 contacts, while the B-C10 and the A-F7 WT MAIT TCR possess an aromatic residue (Tyr and Trp, respectively, Figs. 5, a and c) that made conserved interactions with MR1. Further, although there is little/no sequence identity between the CDR3β loops, a negatively charged residue (Asp/Glu) within this loop (Table 1) converged to a similar position at the interface in four of the seven MAIT TCRs, with the Asp/Glu residue neighboring Tyr95α from the CDR3α loop.
Figure 5.
Differential CDR3β usage and MAIT TCR-MR1-Ag recognition. Structural representation of the MR1 surface in white with Ac-6-FP bound, shown in green. MAIT TCR CDR3β loops for the 7 structures are indicated for (a) original WT (TRBV6-1, clone A-F7) in orange, (b) B-B10 in green, (c) C-A11 in purple, (d) B-C10 in cyan, (e) B-G8 in salmon, (f) B-F3-C1 in magenta, and (g) C-C10 in slate. Contacting residues between MR1 and the CDR3β loops are shown in stick representation with the contact surface on MR1 shown according to the MR1 element type: carbon, orange; nitrogen, blue; oxygen, red; and sulfur, yellow. CDR boundaries are listed in Table 1.
In all the MAIT TCR-MR1-Ac-6-FP ternary complexes, the additional acetyl moiety caused Gln153 to swing away, which subsequently displaced Tyr152, which caused an altered position of Tyr95α from the CDR3α loop such that it now directly contacts the Ac-6-FP antigen (Fig. 3 d and not depicted). Thus, the ability of Ac-6-FP to contact the MAIT TCR appears to be an intrinsic feature of the ligand itself and independent of CDR3β sequence variation. Interestingly, the CDR3β loop was also observed to directly contact the Ag, with the acidic residue in the original WT (A-F7), B-B10 and C-A11 MAIT TCRs (Figs. 5, a–c) directly interacting with the acetyl moiety of Ac-6-FP. However, direct CDR3β–Ag interactions do not necessarily correlate with improved binding affinities for this ligand, as the B-C10 MAIT TCR was observed to have the highest affinity for the MR1-Ac-6-FP complex, indicating that other factors, such as CDR3β flexibility can play an important role in the energetics of the MAIT TCR-MR1-Ag interaction.
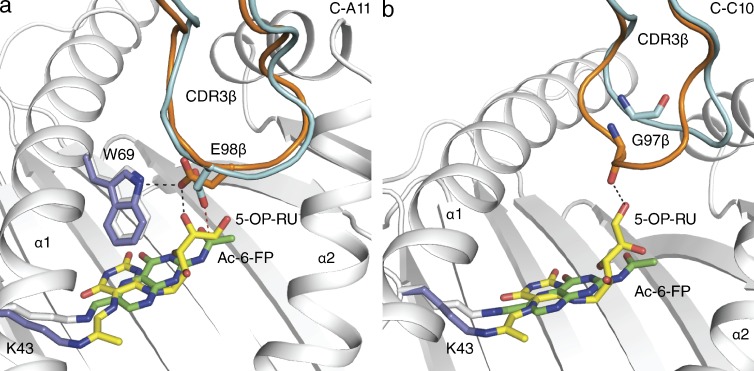
As the CDR3β loop can differently interact with MR1–Ac-6-FP, we next asked whether this loop would show altered flexibility in binding a different ligand, namely MR1–5-OP-RU. Accordingly, we determined the structures of the B-B10, B-F3-C1, C-C10, and C-A11 MAIT TCRs in complex with MR1–5-OP-RU (Table S2). The CDR3β loops of the B-B10 and B-F3-C1 TCRs did not alter significantly between the MR1–Ac-6-FP and MR1–5-OP-RU complexes, regardless of whether the CDR3β loop directly contacted the Ag or not (unpublished data). However, the presence of the additional ribityl moiety caused moderate and marked conformational changes within the CDR3β loop of the C-A11 and C-C10 MAIT TCRs, respectively (Fig. 6, a and b). Regarding the C-A11 MAIT TCR, the ribityl moiety, whereas maintaining the hydrogen bond with Glu98β nevertheless caused a reorientation of the Glu98β side chain to form a direct H-bond to Trp69 of MR1 (Fig. 6 a). The repositioning of Glu98β (2.0 Å displacement of Oε2) subsequently resulted in a remodeling of the main chain of the CDR3β loop (RMSD 0.43 Å over Cα atoms). Interestingly, although the CDR3β loop of the C-C10 TCR did not directly contact Ac-6-FP, within the C-C10 TCR-MR1–5-OP-RU ternary complex, this loop was drawn toward the ligand (6.5 Å displacement of Gly97 O), implying a greater degree of flexibility in the C-C10 CDR3β loop (Fig. 6 b). The movement of the CDR3β loop enabled its main chain to directly contact the ribityl group of 5-OP-RU through Gly97, which itself exhibited some flexibility in that it reoriented its aliphatic moiety (not shown) to enable favorable contacts with the CDR3β loop (Fig. 6 b). Thus, our data demonstrate that the flexible CDR3β loop of the MAIT TCR can fine-tune its interactions and responsiveness to MR1 in an Ag-dependent manner.
Figure 6.
Impact of different Ags on MAIT TCR-MR1-Ag recognition. Structural differences between the ternary complexes of MR1–Ac-6-FP-MAIT TCR (MR1: white, Ac-6-FP: green, MAIT TCR: cyan) and MR1–5-OP-RU-MAIT TCR (MR1: slate, 5-OP-RU: yellow, MAIT TCR: orange). Remodeling of the (a) C-A11 and (b) C-C10 CDR3β loop between Ac-6-FP and 5-OP-RU complexes, with hydrogen bonds indicated in black dashed lines and vdw contacts in red dashed lines. CDR boundaries are listed in Table 1.
Differential Vβ usage
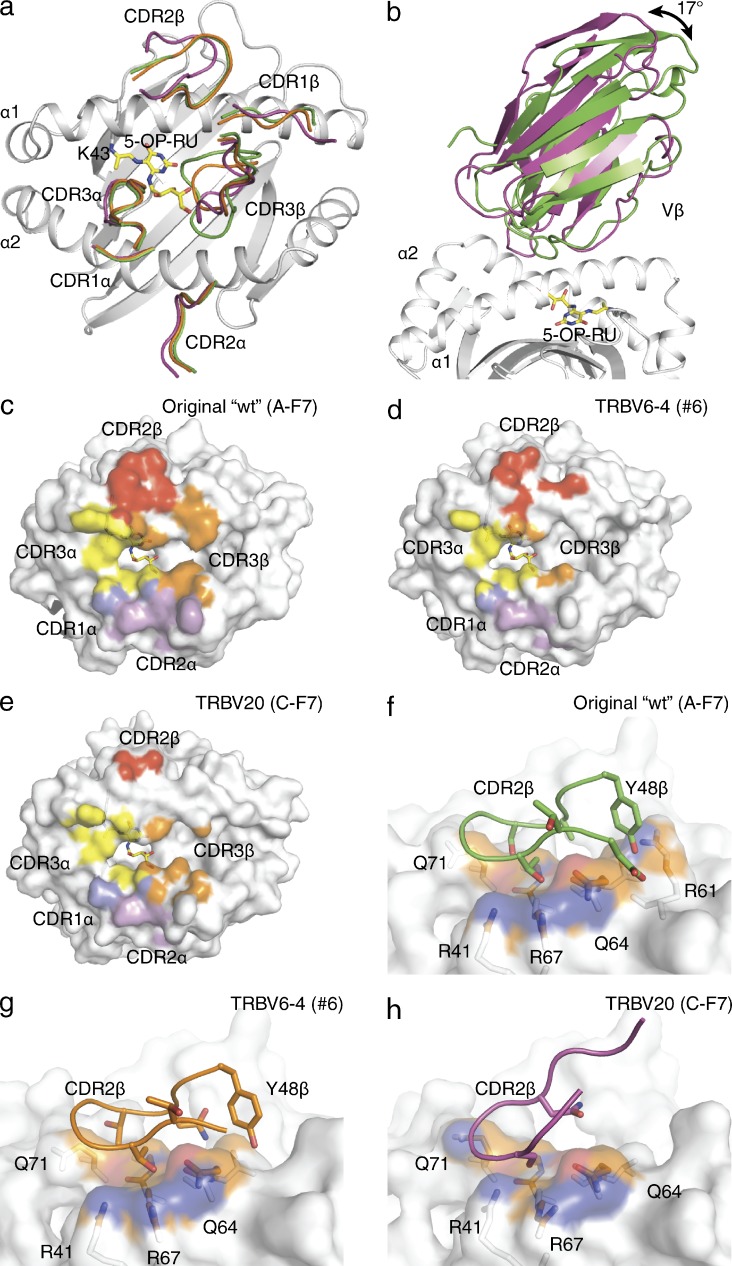
In addition to the varied CDR3β usage, the MAIT TCR repertoire is characterized by differential TRBV usage, including the predominant utilization of the TRBV6-1, TRBV6-4, and TRBV20 genes in humans (Reantragoon et al., 2013). Accordingly, we aimed to understand the molecular basis for how the differential TRBV usage can be accommodated while maintaining MR1-Ag recognition. We determined the structures of a TRBV6-4 (clone described previously; Reantragoon et al., 2012; MAIT TCR #6) and a TRBV20 TCR derived from published sequence data (clone C-F7; Tilloy et al., 1999) and described previously (Reantragoon et al., 2012) in complex with MR1–5-OP-RU (Table S3) and compared them to the TRBV6-1 ternary complex (Fig. 7, a–h).
Figure 7.
TRBV usage and MAIT TCR-MR1-Ag recognition. Comparison of the MR1–5-OP-RU- MAIT TCR C-F7 (TRBV20), original WT (TRBV6-1, clone A-F7) and #6 (TRBV6-4) complex variable domains and CDRβ loops in magenta, green and cyan, respectively. (a) Overlay of the CDR loops from the three complexes on the MR1–5-OP-RU groove. (b) Comparison of relative angles for the Vβ-domains of original WT (TRBV6-1, clone A-F7) and C-F7 (TRBV20) MAIT TCR structures. Footprints of the MAIT TCRs on the MR-5-OP-RU surface for (c) original WT (TRBV6-1, clone A-F7), (d) #6 (TRBV6-4), and (e) C-F7 (TRBV20). Contacting residues between MR1 and the (f) original WT (TRBV6-1, clone A-F7), (g) #6 (TRBV6-4), and (h) C-F7 (TRBV20) CDR2β loops shown in stick representation with the contact surface on MR1 colored according to the MR1 element type: carbon, orange; nitrogen, blue; oxygen, red. CDR boundaries are listed in Table 1.
The TRBV6-1 and TRBV6-4 TCRs docked similarly onto MR1–5-OP-RU (RMSD of 0.51 Å over all Cα atoms between the TCR Vα and MR1 α1 and α2 domains; Fig. 7 a). The TRBV6-1 and TRBV6-4 chains share 83% sequence identity, with some of these differences located within the CDRβ loops and at the MAIT TCR-MR1 interface (Table 2). However, the CDR1β loop does not make appreciable contacts with MR1 in either ternary complex, and thus the Asn28β → Arg28β substitution in TRBV6-4 does not impact on MR1-Ag recognition (Fig. 7, c and d). The TRBV6-4 CDR2β loop shows a more prominent contribution to the BSA at the MR1 interface than TRBV6-1 (BSA of 46 and 26%, respectively), whereas the framework regions retain similar contributions between the two structures (BSA of 19% for both complexes; Fig. 7, c and d). In contrast, the CDR3β loop showed a larger contribution to BSA for TRBV6-1 than TRBV6-4 (53 to 29%, respectively). The affinities of the TRBV6-1 and TRBV6-4 TCRs toward MR1–5-OP-RU were similar (approximate Kdeq 2.0 µM and 2.7 µM, respectively; Table 2). Thus, the TRBV6-4 TCR compensates for the lower contribution of the CDR3β loop to the interface BSA by increasing the relative contribution of CDR2β, to achieve a similar docking mode; nevertheless, the overall BSA was lower for TRBV6-4 than TRBV6-1 (1050 Å2 and 1160 Å2, respectively).
Although the overall docking mode between the TRBV6-1 and TRBV20+ TCRs onto MR1–5-OP-RU was similar (RMSD 0.79 Å over all Cα atoms between the TCR Vα and MR1 α1 and α2 domains), greater variation in the interatomic contacts were observed due to the lower sequence identity between the respective TCRβ chains (35% sequence identity between Vβ domains; Table 1 and Fig. 7, c and e). Whereas the TCR α-chain of the TRBV20+ MAIT #4 TCR was situated very similarly atop MR1 when compared with the TRBV6-1+ original WT (A-F7) MAIT TCR, the TRBV20 β-chain was juxtaposed differently to that of the TRBV6-1 β-chain, resulting in a 17-degree rotation away from the α1 helix of MR1 (Fig. 7 b). This difference can be attributed to differing Vα-Vβ packing as well as differing Vβ-MR1 contacts. The BSA contribution of the TRBV20 CDR1β loop was marginally higher than the TRBV6-1 complex (4 and 1%, respectively; Fig. 7 e). Conversely, TRBV20 framework regions had reduced contributions compared with that of the TRBV6-1 and TRBV6-4 TCRs (BSA of 4, 9 and 8%, respectively; Fig. 7 e). This was principally due to the TRBV20 lacking Tyr48β (equivalent residue Thr49β; Fig. 7, f and g), which had the effect of lifting the β-chain away from the MR1 interface. Although the CDR2β loop was devoid of any large residues, thereby reducing the extent of MR1-mediated contacts via this loop, the relative contribution of the CDR2β was similar to TRBV6-1 (10 and 13%, respectively). In addition the relative contribution of the CDR3β to the BSA was similar between TRBV20 and TRBV6-1 (22% and 25%, respectively). Despite this, the overall contact area was decreased in TRBV20 (1070 Å2). Accordingly, the TRBV20 chain appeared nonideally disposed to interact with MR1, akin to TRBV6-4. However, in the TRBV20 ternary complex the CDR3β loop played a major role in interacting the MR1–5-OP-RU akin to TRBV6-1 (BSA of 22 and 25%, respectively), thereby structurally compensating for the reduced interactions from the germline encoded regions of the TCR β-chain. Despite the differences in contact area between the complexes the Kdeq, determined by SPR (Table 2), for MR1–5-OP-RU was comparable for each TCR (∼2.0, 2.7, and 1.0 µM for TRBV6-1 [original WT, clone A-F7], TRBV6-4 [#6], and TRBV20 [#4], respectively). This indicates how the MAIT TCR can readily tolerate some variations within the germline-encoded regions of the TCR β-chain.
Atypical TRAJ gene usage
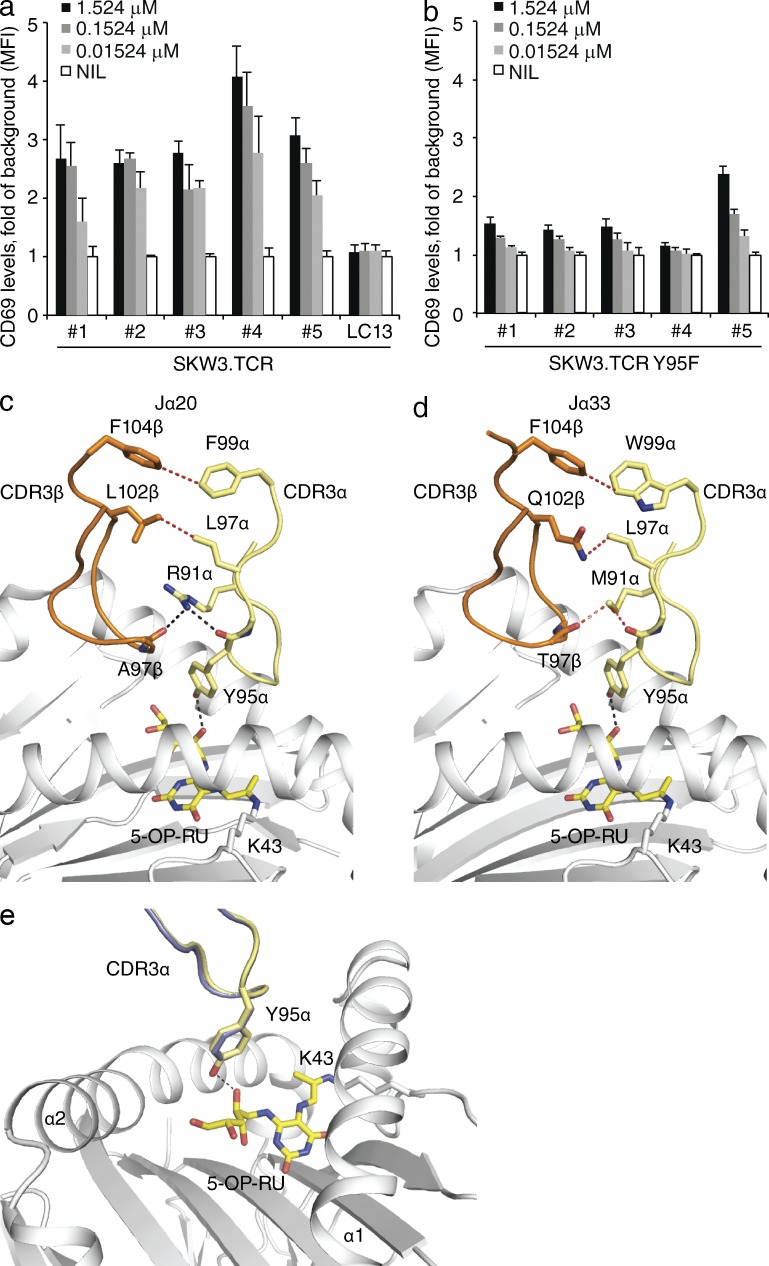
Although the invariant MAIT TCR α-chain is characterized by TRAJ33 gene usage, we recently demonstrated that the TCR α-chain could also consist of TRAV1-2 joined with the TRAJ20 or TRAJ12 gene segments (Reantragoon et al., 2013). Although the sequences differ between the TRAJ33, TRAJ12 and TRAJ20 regions, Tyr95α was conserved (Table 1), thereby suggesting a conserved functional role in mediating contacts with the Ag. To formally establish this, we transduced genes encoding wild-type or mutant Y95F TRAJ20+ (#4) and three TRAJ12+, TRBV6-4+ MAIT TCRs (#1–3) differing in CDR3β into SKW3 cells, and compared their activation to that of TRAJ33+ MAIT TCR-transduced SKW3 cells (all these TCRs were obtained from single cell sorted PBMCs reactive with MR1-Ag tetramer (Reantragoon et al., 2013; Fig. 8 a). SKW3 cell lines expressing TRAJ33+ (#5), TRAJ20+ (#4) and TRAJ12+ (#1–3) MAIT TCRs responded equivalently to the activating ligand containing synthetic rRL-6CH2OH in the presence of C1R.MR1 cells, and moreover, activation of all the mutant Y95F SKW3 cell lines (Fig. 8 b) was impaired, thereby indicating a commonality in function between the TRAJ12, TRAJ20, and TRAJ33 MAIT TCRs.
Figure 8.
TRAJ usage and MAIT TCR recognition. (a) Dose response to synthetic rRL-6-CH2OH preincubated with C1R.MR1 cells by WT (a) or mutant Y95F (b) TRAV1-2-TRBV6-4 TRAJ12+, TRAJ20+, and TRA33+ SKW3.MAIT. Data shows mean ± SEM fold of background MFI of CD69 expression for gated SKW3.TCR cells from one experiment (triplicate samples). These experiments were performed twice yielding similar results. Comparison of the MAIT TCR TRBV6-4 (c) TRAJ20 (#4) and (d) TRAJ33 (#6) MR1–5-OP-RU structures with MR1, 5-OP-RU, CDR3α, and CDR3β loops shown in white, yellow, pale yellow, and orange, respectively. (e) Comparison of the positioning of CDR3α loops and Tyr95α for TRAJ20 (#4) and TRAJ33 (#6) in slate and pale yellow, respectively. Contacts between CDR3α, CDR3β, and 5-OP-RU are shown with dashed lines in black for hydrogen bonds and red for vdw contacts. CDR boundaries are listed in Table 1.
To directly investigate this, we determined the structure of a TRAJ20+ MAIT TCR in complex with MR1–5-OP-RU (Table S3). The TRAJ20+ MAIT TCR-MR1-5-OP-RU complex was very similar to the TRAJ33+ MAIT TCR-MR1–5-OP-RU complexes with the general positions of the interactions being the same, thereby indicating that divergent TRAJ gene usage does not disrupt the consensus MAIT TCR-MR1 docking mode. Moreover, whereas the sequence differences between TRAJ20 and TRAJ33 gene usage resulted in an altered interaction network within the TCR α/β-chain junctional interface (Fig. 8 c), Tyr95α adopted a conserved binding mode with the ribityl tail of the 5-OP-RU ligand (Fig. 8 d). In addition SPR experiments revealed that the dissociation constant for TRAJ20 TCR was comparable to TRAJ33 TCR (∼2.4 and 2.7 µM, respectively; Table 2). Thus, TRAJ20 and TRAJ12 gene heterogeneity is readily accommodated within the MAIT TCR–MR1 binding axis.
DISCUSSION
MAIT cells, type I NKT cells, and germline-encoded mycolyl lipid-reactive (GEM) cells are T cells that express distinct semiinvariant TCRs, which are restricted to MR1, CD1d, and CD1b, respectively (Rossjohn et al., 2012; Van Rhijn et al., 2013). Here, the TCR α-chain is generally fixed, whereas greater repertoire diversity is found within the TCR β-chain. Within the type I NKT cell axis, it is established that the semiinvariant TCR acts like a pattern recognition receptor (Scott-Browne et al., 2007). Although the NKT TCR-CD1d docking mode is conserved, variations in TCR β-chain usage can impact on the functional hierarchy of ligand recognition, in a mechanism that involves the CDR2β and CDR3β loops functionally collaborating to enable recognition (Mallevaey et al., 2009). Moreover, NKT TCR β-chains appear to have a penchant for recognizing particular ligands, with the TCR β-chain transmitting conformational changes to the invariant TCR α-chain (Wun et al., 2011). Notably, NKT TCR autoreactivity is mediated via its CDR3β loop (Matulis et al., 2010), which directly contacts CD1d, with the extent of autoreactivity being independent on the interactions with the CD1d-restricted Ag (Pellicci et al., 2011). Further, within the type I NKT system, variations within the TRAV and TRAJ usage is observed, with such differential TCR α-chain usage correlating with differential Ag specificity within the consensus-docking framework (Uldrich et al., 2011). Thus, although we have a growing understanding of type I NKT TCR recognition of CD1d, it is unclear whether these emerging generalities would be mirrored by MAIT TCR-MR1 recognition, a vitamin B metabolite sensing system. Our data, presented here, provides fundamental insight into the similarities and dissimilarities underpinning NKT TCR and MAIT TCR recognition.
We formally establish that, despite the varied TRAJ, TRBV, and CDR3β usage, the MAIT TCR-MR1-Ag docking mode is conserved, analogous to type I NKT TCRs recognizing CD1d–Ag complexes (Rossjohn et al., 2012). We show how differing TRAJ gene usage converges to similar MAIT TCR-MR1-Ag recognition, with the key CDR3α-encoded residue, Tyr95α, making conserved interactions with the vitamin B metabolite (Reantragoon et al., 2012). Markedly dissimilar to NKT TCR recognition, however, is that the nongermline-encoded CDR3β loop of the MAIT TCR can directly contact the Ag, with the nature of the CDR3β contacts being dynamically and directly modulated by the Ag itself. Moreover, the extent of CDR3β contacts mediated by the MAIT TCR is, on average, much greater than that for NKT TCR recognition, where for the most part the CDR3β loop plays a more peripheral role (Rossjohn et al., 2012). It also appears that, for the ligands tested, the CDR3β loop of the MAIT TCR impacts on MAIT recognition independently of the proximal CDR1β and CDR2β loops. Further, while the CDR3β loop of the NKT TCR can have a huge influence on CD1d autoreactivity (Gapin et al., 2013), the impact of the MAIT TCR CDR3β loop appears to play a more subtle fine-tuning role, with the range of affinity values being much more clustered when compared with the effect the NKT TCR’s CDR3β loop can have.
In describing a new ligand that competitively inhibits MAIT TCR recognition, Ac-6-FP, we demonstrate that the MR1-binding pocket has sufficient plasticity to accommodate this ligand, and simultaneously show how the ligand itself can markedly affect MR1 stability, which results in greater MR1 cell-surface up-regulation and more potent inhibition. Interestingly, the Ac-6-FP ligand can transmit its effects on MAIT TCR recognition indirectly, by altering the conformation of buried MR1 residues that subsequently affect surface-exposed MR1 residues that directly contact the TCR. In this regard, this is analogous to how buried polymorphic MHC residues can impact on TCR recognition (Archbold et al., 2009; Gras et al., 2012) and some CD1d-restricted ligands can impact on F′-pocket dynamics and hence TCR recognition (McCarthy et al., 2007; Li et al., 2010; Wun et al., 2011). This raises the possibility that weak agonists could selectively be stimulatory toward a subset of MAIT cells, with a key-determining factor being the CDR3β loop. Accordingly, MAIT TCR heterogeneity, while converging toward a consensus footprint on MR1, can nevertheless fine-tune MAIT TCR recognition.
In MHC-restricted immunity, altered peptide ligands (APLs) can exert markedly differing biological outcomes. Nevertheless, the TCR-MHC-APL docking modes of APL ranging from potent agonists to strong antagonists are very similar (Ding et al., 1999; Degano et al., 2000), thereby indicating that subtle differences at the TCR-pMHC interfaces, and ensuing biophysical parameters of the TCR-pMHC interaction, play a major role in determining the antigenicity of a given peptide (Huppa et al., 2010; Dushek and van der Merwe, 2014). The studies reported here are analogous to that of MHC-restricted APLs, in that the MAIT TCR-MR1-Ag docking mode between stimulatory and nonstimulatory MAIT cell antigens are very similar, yet markedly differing biological outcomes are elicited. The affinity of one MAIT TCR toward Ac-6-FP (approximate Kdeq 34 µM) was shown to be appreciably higher than the remainder of the MAIT TCRs examined (Kdeq > 200 µM). Consistent with this, the MR1–Ac-6-FP tetramers have shown that the majority of MAIT cells do not bind with sufficiently high affinity to enable MAIT cell staining. Nevertheless, we are able to observe the MAIT TCR–MR1–Ac-6-FP interaction structurally, in a similar manner to which low-affinity TCR–pMHC interactions have been observed (Yin et al., 2012). Interestingly, the affinity of the nonstimulatory Tyr95αPhe MAIT TCR mutant toward MR1–5-OP-RU was ∼30 µM, thereby indicating that the affinity threshold required for MAIT cell activation is below this value and that an ∼15-fold reduction in affinity can equate to a complete loss of activation. Moreover, a key feature between the stimulatory and nonstimulatory MAIT Ags is the direct contact between Tyr95α of the MAIT TCR and the MAIT Ag. Thus, our findings simultaneously provide insight into the factors that govern MAIT cell antigenicity.
MATERIALS AND METHODS
C1R.MR1, Jurkat.MAIT, and SKW3.MAIT cell lines.
C1R APCs expressing MR1 (CIR.MR1) have previously been described (Reantragoon et al., 2012). For the generation of Jurkat.MAIT and SKW3.MAIT cells, full-length TCRα and TCRβ genes were cloned into a self-cleaving 2A peptide-based (MSCV)-IRES-GFP (pMIG) vector (Szymczak et al., 2004) and stably transduced into TCR-deficient Jurkat clone 76 cells (Jurkat76; Heemskerk et al., 2003), Jurkat RT3-T3.5 cells (Jurkat; American Type Culture Collection no. T1B-153), or SKW3 cells (DSMZ accession code 53) using the retroviral Murine Stem Cell Virus (MSCV) expression system. Transduced cell lines were FACS sorted based on fluorescence of GFP and flow cytometric staining for CD3, which co-expresses on the cell surface with TCR.
Activation of Jurkat.MAIT and SKW.MAIT cells.
Jurkat or SKW3 cells (105) transduced with genes encoding MAIT TCRα- and TCRβ-chain genes were tested for activation by co-incubation with compounds (synthetic rRL-6-CH2OH or 5-A-RU, synthesized as described previously; Corbett et al., 2014) and C1R.MR1 cells (105) for 16 h. For inhibition, 6-FP, Ac-6-FP, or control pterins (Schircks Laboratories) were added to C1R.MR1 cells for 1 h before addition of Jurkat.MAIT cells and 0.02 µM synthetic rRL-6-CH2OH. Cells were subsequently stained with PE-conjugated anti-CD3 (BD), and APC-conjugated anti-CD69 (BD) mAbs for 30 min on ice, before analysis of CD69 surface expression by flow cytometry. Activation of Jurkat.MAIT or SKW.MAIT cells was measured by an increase in surface CD69 expression.
Jurkat76 cells (105) transduced with genes encoding the WT original (clone A-F7) MAIT TCR α- and β-chain genes were co-incubated with compounds (6-FP, Ac-6-FP; 5-OP-RU; Schircks Laboratories) and C1R.MR1 cells (105) in 200 µl media for 21 h. IL-2 production was measured as a mean of Jurkat76.MAIT activation in ELISA (BD OptEIA kit) using 100 µl of supernatant, frozen/thawed to kill cells. In brief, IL-2 was assayed with biotinylated anti–IL-2 mAb and o-Phenylenediamine dihydrochloride (OPD; Sigma-Aldrich) substrate conversion by HRP-Streptavidin detected at 492 nm emission.
Detection of up-regulation of MR1 on the cell surface.
C1R.MR1 cells (105) were incubated for 2, 4, or 24 h with compounds. After this, cells were washed and stained with MR1-specific mAb 26.5 on ice for 30 min, followed by PE-conjugated anti–mouse-IgG on ice for 30 min before analysis. For the comparison of C1R and C1R.MR1 expression levels over time, cells (2 × 105) were incubated for 1, 2, 4, 12, or 24 h with 10 µM 6-FP or Ac-6-FP, or no treatment (media alone). Cells were then washed and stained with biotin-conjugated 26.5 mAb, followed by PE-conjugated streptavidin.
Generation of soluble MR1-ligand, MR1 K43A (empty), C-terminal cysteine-tagged-MR1/ligand, MR1 tetramers, and soluble MAIT TCRs.
Genes were expressed in E. coli inclusion bodies, refolded and purified as described previously (Patel et al., 2013). Tetramers were generated as described previously (Corbett et al., 2014) and prepared simultaneously.
Tetramer staining of PBMCs.
PBMCs were isolated from whole blood of healthy donors (authorized by the Australian Red Blood Cross Service Material Supply Agreement with the University of Melbourne) as described previously (Reantragoon et al., 2013). Approximately 1.2 × 106 human PBMCs were stained with live/dead discriminator (Live/Dead Fixable Aqua Dead Cell Stain kit; Invitrogen) PE-labeled, human tetrameric, MR1–5-OP-RU, MR1-6-FP or MR1–Ac-6-FP at 5 µg/ml, anti-CD3-Alexa Fluor 700 (BD), and anti-CD161-PE-Cy7 (BioLegend) mAb for 30 min on ice in the dark. Cells were then washed three times with wash buffer (2% fetal calf serum in PBS) and resuspended in fixing solution (2.1% glucose and 1% paraformaldehyde in PBS). Data were acquired using an LSR Fortessa (BD) and analyzed using FlowJo software.
Thermostability of soluble MR1 by fluorescence-based thermal shift assay.
Soluble MR1-6-FP, MR1–Ac-6-FP, MR1–5-OP-RU and MR1-K43A (empty) were purified by gel filtration before the assay. To generate MR1-K43A-5-OP-RU (loaded), MR1-K43A was incubated in TBS with a 10 M excess of 5-OP-RU, prepared as described previously (Corbett et al., 2014), for 4 h at room temperature in the dark. To separate 5-OP-RU–loaded MR1-K43A from unloaded MR1-K43A, complex of soluble MAIT TCR-MR1-K43A-5-OP-RU was purified by gel filtration. MR1-K43A-5-OP-RU was subsequently separated from MAIT TCR through the addition of excess 12H8 mAb specific for TCR (∼1.1:1.0 molar ratio 12H8 mAb to MAIT TCR in MR1-TCR complex), followed by gel filtration whereby MAIT TCR complexed with 12H8 mAb shifted to earlier elution volume as compared with MR1-K43A-5-OP-RU.
For the fluorescence-based thermal shift assay, triplicate samples consisting of 5 µg of MR1 and 2.5 µg of SYPRO Orange (Sigma-Aldrich) in 20 µl TBS were prepared on ice. Samples were heated from 25 to 95°C at a rate of 1°C/min using a Stratagene Mx3005P Real Time PCR machine. Thermal protein unfolding caused unquenching of SYPRO Orange fluorescent signal, resulting in an increase of fluorescence signal (excitation at 492 nm, emission 610 nm). Emission signal at 610 nm was plotted against temperature and half maximum melt point (Tm50), at which 50% of the protein is unfolded, was determined upon base line correction, normalization, and nonlinear curve fit using the software GraphPad Prism. The experiment was performed three times.
Surface plasmon resonance.
All surface plasmon resonance (SPR) experiments were conducted in duplicate at 25°C on a BIAcore 3000 instrument using HBS buffer: 10 mM HEPES-HCl (pH 7.4), 150 mM NaCl, and 0.005% surfactant P20 supplied by the manufacturer (GE Healthcare). Biotinylated C-terminal cysteine-tagged-MR1-Ag (generated as described previously e.g., (Patel et al., 2013)) were immobilized on a SA-Chip with a surface density of ∼3000-4000 response units (RU). Various concentrations of MAIT TCRs (between 0.2 and 50 µM) were injected over the captured MR1-Ag at 10 µl/min. The final response was calculated by subtracting the response of a biotin-labeled flow cell alone from the TCR-MR1-Ag complex. The equilibrium data were analyzed using GraphPad Prism to determine affinity constants as the result of two independent experiments.
Crystallization, structure determination and analysis.
Crystals of the soluble MAIT TCR-MR1-Ag complexes were obtained using the hanging drop vapor diffusion method. The MR1-β2M-5-OP-RU, MR1-β2M-Ac-6-FP and MAIT TCRs were concentrated to between 2–4 mg/ml, mixed in a 1:1 molar ratio, then 0.5 µl added to 0.5 µl of a precipitant solution consisting 0.2 M sodium acetate, 0.1 M bis-tris propane with a pH ranging between 6 and 6.5, and varying concentrations of PEG 3350 between 8–14% wt/vol depending on the complex. Crystals were observed after incubation at 20°C for 24 h in dark conditions. Cryoprotected before diffraction experiments by soaking in the crystallization condition modified with between 10–15% vol/vol glycerol before cooling to 100K. Diffraction images were collected at the Australian Synchrotron MX2 beamline, diffracting in either a C2, P31 (MAIT TCR TRBV20 only) or P41212 (MAIT TCR TRAJ20 only) spacegroups. The data were processed using either XDS (Kabsch, 2010) or programs from the CCP4 Suite (Winn et al., 2011). The phase problem was solved by molecular replacement using PHASER (McCoy, 2007), using MR1 ternary complex (Protein Data Bank accession no. 4L4T; Patel et al., 2013) with CDR loops and ligands removed and using the Rfree reflection set from the model. The initial solution was refined in Phenix using simulated annealing refinement, with all subsequent refinement steps performed using BUSTER 2.10. Restraints for 5-OP-RU and Ac-6-FP were generated using the Grade Web Server, with model building performed in COOT using MolProbity for validation (Emsley and Cowtan, 2004). All molecular graphics were made with PyMOL. The buried surface area was calculated with Areaimol (Winn et al., 2011) and RMSD values were calculated using the align function in PyMOL. Within the C2 crystal form there are two complexes per asymmetric unit, with these two complexes being very similar to each other. In relation to the Ag-binding pocket of the structures reported here and previously (Patel et al., 2013), minor perturbations around the side chains of E149 and Q153 were observed, but this lies within coordinate error. Accordingly, unless explicitly stated, only one complex per asymmetric unit will be discussed in the manuscript.
Accession information.
The coordinates have been deposited in the Protein Data Bank under the following accession codes: 4PJ5, 4PJ7, 4PJ8, 4PJ9, 4PJA, 4PJB, 4PJC, 4PJD, 4PJE, 4PJF, 4PJG, 4PJH, 4PJI, and 4PJX.
Online supplemental material.
Table S1 shows data collection and refinement statistics for the ternary complexes MR1-Ac-6-FP-MAIT TCR. Table S2 shows data collection and refinement statistics for the ternary complexes MR1-5-OP-RU-MAIT TCR. Table S3 shows data collection and refinement statistics for the ternary complexes MR1-5-OP-RU-MAIT TCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20140484/DC1.
Supplementary Material
Acknowledgments
We thank the Australian synchrotron for assistance with data collection, and the Australian Phenomics Network Histopathology Service for use of facilities.
This research was supported by the National Health and Medical Research Council of Australia (NHMRC) and the Australian Research Council. We thank Ted Hansen for the 26.5 Mab. R. Reantragoon was supported by the Faculty of Medicine, Chulalongkorn University and Chulalongkorn Hospital, Thai Red Cross Society scholarships. N.A. Gherardin was supported by a Leukemia Foundation postgraduate scholarship. D.P. Fairlie and D.I. Godfrey were supported by NHMRC Senior Principal Research Fellowships; O. Patel was supported by an ARC Future Fellowship; and J. Rossjohn was supported by an NHMRC Australia Fellowship.
The authors declare that they have no competing financial interests.
Footnotes
Abbreviations used:
- 5-A-RU
- 5-amino-6-D-ribitylaminouracil
- 5-OP-RU
- 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil
- 6-FP
- 6-formylpterin
- Ac-6-FP
- acetyl-6-FP
- Ag
- antigen
- APL
- altered peptide ligand
- CDR
- complementarity-determining region
- MAIT cell
- mucosal associated invariant T cell
- MR1
- MHC related protein-1
- rRL-6-CH2OH
- reduced 6-hydroxymethyl-8-D-ribityllumazine
- vdw
- van der Waals
References
- Archbold, J.K., Macdonald W.A., Gras S., Ely L.K., Miles J.J., Bell M.J., Brennan R.M., Beddoe T., Wilce M.C., Clements C.S., et al. 2009. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. J. Exp. Med. 206:209–219 10.1084/jem.20082136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkinshaw, R.W., Kjer-Nielsen L., Eckle S.B., McCluskey J., and Rossjohn J.. 2014. MAITs, MR1 and vitamin B metabolites. Curr. Opin. Immunol. 26:7–13 10.1016/j.coi.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Borg, N.A., Wun K.S., Kjer-Nielsen L., Wilce M.C., Pellicci D.G., Koh R., Besra G.S., Bharadwaj M., Godfrey D.I., McCluskey J., and Rossjohn J.. 2007. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 448:44–49 10.1038/nature05907 [DOI] [PubMed] [Google Scholar]
- Chua, W.-J., Kim S., Myers N., Huang S., Yu L., Fremont D.H., Diamond M.S., and Hansen T.H.. 2011. Endogenous MHC-related protein 1 is transiently expressed on the plasma membrane in a conformation that activates mucosal-associated invariant T cells. J. Immunol. 186:4744–4750 10.4049/jimmunol.1003254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, W.J., Truscott S.M., Eickhoff C.S., Blazevic A., Hoft D.F., and Hansen T.H.. 2012. Polyclonal mucosa-associated invariant T cells have unique innate functions in bacterial infection. Infect. Immun. 80:3256–3267 10.1128/IAI.00279-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett, A.J., Eckle S.B., Birkinshaw R.W., Liu L., Patel O., Mahony J., Chen Z., Reantragoon R., Meehan B., Cao H., et al. 2014. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature. 509:361–365 10.1038/nature13160 [DOI] [PubMed] [Google Scholar]
- Croxford, J.L., Miyake S., Huang Y.-Y., Shimamura M., and Yamamura T.. 2006. Invariant V(alpha)19i T cells regulate autoimmune inflammation. Nat. Immunol. 7:987–994 10.1038/ni1370 [DOI] [PubMed] [Google Scholar]
- Degano, M., Garcia K.C., Apostolopoulos V., Rudolph M.G., Teyton L., and Wilson I.A.. 2000. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity. 12:251–261 10.1016/S1074-7613(00)80178-8 [DOI] [PubMed] [Google Scholar]
- Ding, Y.-H., Baker B.M., Garboczi D.N., Biddison W.E., and Wiley D.C.. 1999. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 11:45–56 10.1016/S1074-7613(00)80080-1 [DOI] [PubMed] [Google Scholar]
- Dushek, O., and van der Merwe P.A.. 2014. An induced rebinding model of antigen discrimination. Trends Immunol. 35:153–158 10.1016/j.it.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusseaux, M., Martin E., Serriari N., Péguillet I., Premel V., Louis D., Milder M., Le Bourhis L., Soudais C., Treiner E., and Lantz O.. 2011. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161hi IL-17-secreting T cells. Blood. 117:1250–1259 10.1182/blood-2010-08-303339 [DOI] [PubMed] [Google Scholar]
- Eckle, S.B.G., Turner S.J., Rossjohn J., and McCluskey J.. 2013. Predisposed αβ T cell antigen receptor recognition of MHC and MHC-I like molecules? Curr. Opin. Immunol. 25:653–659 10.1016/j.coi.2013.07.010 [DOI] [PubMed] [Google Scholar]
- Emsley, P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Gapin, L., Godfrey D.I., and Rossjohn J.. 2013. Natural Killer T cell obsession with self-antigens. Curr. Opin. Immunol. 25:168–173 10.1016/j.coi.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey, D.I., Rossjohn J., and McCluskey J.. 2010. Fighting infection with your MAITs. Nat. Immunol. 11:693–695 10.1038/ni0810-693 [DOI] [PubMed] [Google Scholar]
- Gold, M.C., and Lewinsohn D.M.. 2013. Co-dependents: MR1-restricted MAIT cells and their antimicrobial function. Nat. Rev. Microbiol. 11:14–19 10.1038/nrmicro2918 [DOI] [PubMed] [Google Scholar]
- Gold, M.C., Cerri S., Smyk-Pearson S., Cansler M.E., Vogt T.M., Delepine J., Winata E., Swarbrick G.M., Chua W.J., Yu Y.Y., et al. 2010. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. 8:e1000407 10.1371/journal.pbio.1000407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gras, S., Burrows S.R., Turner S.J., Sewell A.K., McCluskey J., and Rossjohn J.. 2012. A structural voyage toward an understanding of the MHC-I-restricted immune response: lessons learned and much to be learned. Immunol. Rev. 250:61–81 10.1111/j.1600-065X.2012.01159.x [DOI] [PubMed] [Google Scholar]
- Hashimoto, K., Hirai M., and Kurosawa Y.. 1995. A gene outside the human MHC related to classical HLA class I genes. Science. 269:693–695 10.1126/science.7624800 [DOI] [PubMed] [Google Scholar]
- Heemskerk, M.H.M., Hoogeboom M., de Paus R.A., Kester M.G.D., van der Hoorn M.A.W.G., Goulmy E., Willemze R., and Falkenburg J.H.F.. 2003. Redirection of antileukemic reactivity of peripheral T lymphocytes using gene transfer of minor histocompatibility antigen HA-2-specific T-cell receptor complexes expressing a conserved alpha joining region. Blood. 102:3530–3540 10.1182/blood-2003-05-1524 [DOI] [PubMed] [Google Scholar]
- Huang, S., Gilfillan S., Cella M., Miley M.J., Lantz O., Lybarger L., Fremont D.H., and Hansen T.H.. 2005. Evidence for MR1 antigen presentation to mucosal-associated invariant T cells. J. Biol. Chem. 280:21183–21193 10.1074/jbc.M501087200 [DOI] [PubMed] [Google Scholar]
- Huang, S., Gilfillan S., Kim S., Thompson B., Wang X., Sant A.J., Fremont D.H., Lantz O., and Hansen T.H.. 2008. MR1 uses an endocytic pathway to activate mucosal-associated invariant T cells. J. Exp. Med. 205:1201–1211 10.1084/jem.20072579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S., Martin E., Kim S., Yu L., Soudais C., Fremont D.H., Lantz O., and Hansen T.H.. 2009. MR1 antigen presentation to mucosal-associated invariant T cells was highly conserved in evolution. Proc. Natl. Acad. Sci. USA. 106:8290–8295 10.1073/pnas.0903196106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa, J.B., Axmann M., Mörtelmaier M.A., Lillemeier B.F., Newell E.W., Brameshuber M., Klein L.O., Schütz G.J., and Davis M.M.. 2010. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature. 463:963–967 10.1038/nature08746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illés, Z., Shimamura M., Newcombe J., Oka N., and Yamamura T.. 2004. Accumulation of Valpha7.2-Jalpha33 invariant T cells in human autoimmune inflammatory lesions in the nervous system. Int. Immunol. 16:223–230 10.1093/intimm/dxh018 [DOI] [PubMed] [Google Scholar]
- Kabsch, W.2010. Xds. Acta Crystallogr. D Biol. Crystallogr. 66:125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawachi, I., Maldonado J., Strader C., and Gilfillan S.. 2006. MR1-restricted V alpha 19i mucosal-associated invariant T cells are innate T cells in the gut lamina propria that provide a rapid and diverse cytokine response. J. Immunol. 176:1618–1627 10.4049/jimmunol.176.3.1618 [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen, L., Patel O., Corbett A.J., Le Nours J., Meehan B., Liu L., Bhati M., Chen Z., Kostenko L., Reantragoon R., et al. 2012. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 491:717–723 [DOI] [PubMed] [Google Scholar]
- Le Bourhis, L., Martin E., Péguillet I., Guihot A., Froux N., Coré M., Lévy E., Dusseaux M., Meyssonnier V., Premel V., et al. 2010. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11:701–708 10.1038/ni.1890 [DOI] [PubMed] [Google Scholar]
- Li, Y., Girardi E., Wang J., Yu E.D., Painter G.F., Kronenberg M., and Zajonc D.M.. 2010. The Vα14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J. Exp. Med. 207:2383–2393 10.1084/jem.20101335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sagaseta, J., Dulberger C.L., Crooks J.E., Parks C.D., Luoma A.M., McFedries A., Van Rhijn I., Saghatelian A., and Adams E.J.. 2013. The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proc. Natl. Acad. Sci. USA. 110:E1771–E1778 10.1073/pnas.1222678110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallevaey, T., Scott-Browne J.P., Matsuda J.L., Young M.H., Pellicci D.G., Patel O., Thakur M., Kjer-Nielsen L., Richardson S.K., Cerundolo V., et al. 2009. T cell receptor CDR2 beta and CDR3 beta loops collaborate functionally to shape the iNKT cell repertoire. Immunity. 31:60–71 10.1016/j.immuni.2009.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, J.L., Naidenko O.V., Gapin L., Nakayama T., Taniguchi M., Wang C.-R., Koezuka Y., and Kronenberg M.. 2000. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J. Exp. Med. 192:741–754 10.1084/jem.192.5.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulis, G., Sanderson J.P., Lissin N.M., Asparuhova M.B., Bommineni G.R., Schümperli D., Schmidt R.R., Villiger P.M., Jakobsen B.K., and Gadola S.D.. 2010. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol. 8:e1000402 10.1371/journal.pbio.1000402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, C., Shepherd D., Fleire S., Stronge V.S., Koch M., Illarionov P.A., Bossi G., Salio M., Denkberg G., Reddington F., et al. 2007. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J. Exp. Med. 204:1131–1144 10.1084/jem.20062342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy, A.J.2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63:32–41 10.1107/S0907444906045975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierovics, A., Yankelevich W.J., and Cowley S.C.. 2013. MAIT cells are critical for optimal mucosal immune responses during in vivo pulmonary bacterial infection. Proc. Natl. Acad. Sci. USA. 110:E3119–E3128 10.1073/pnas.1302799110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki, Y., Miyake S., Chiba A., Lantz O., and Yamamura T.. 2011. Mucosal-associated invariant T cells regulate Th1 response in multiple sclerosis. Int. Immunol. 23:529–535 10.1093/intimm/dxr047 [DOI] [PubMed] [Google Scholar]
- Patel, O., Pellicci D.G., Uldrich A.P., Sullivan L.C., Bhati M., McKnight M., Richardson S.K., Howell A.R., Mallevaey T., Zhang J., et al. 2011. Vβ2 natural killer T cell antigen receptor-mediated recognition of CD1d-glycolipid antigen. Proc. Natl. Acad. Sci. USA. 108:19007–19012 10.1073/pnas.1109066108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, O., Kjer-Nielsen L., Le Nours J., Eckle S.B., Birkinshaw R., Beddoe T., Corbett A.J., Liu L., Miles J.J., Meehan B., et al. 2013. Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nat. Commun. 4:2142 10.1038/ncomms3142 [DOI] [PubMed] [Google Scholar]
- Pellicci, D.G., Patel O., Kjer-Nielsen L., Pang S.S., Sullivan L.C., Kyparissoudis K., Brooks A.G., Reid H.H., Gras S., Lucet I.S., et al. 2009. Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 31:47–59 10.1016/j.immuni.2009.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicci, D.G., Clarke A.J., Patel O., Mallevaey T., Beddoe T., Le Nours J., Uldrich A.P., McCluskey J., Besra G.S., Porcelli S.A., et al. 2011. Recognition of β-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat. Immunol. 12:827–833 10.1038/ni.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfalvi, A., Gomori E., Magyarlaki T., Pal J., Banati M., Javorhazy A., Szekeres-Bartho J., Szereday L., and Illes Z.. 2008. Invariant Valpha7.2-Jalpha33 TCR is expressed in human kidney and brain tumors indicating infiltration by mucosal-associated invariant T (MAIT) cells. Int. Immunol. 20:1517–1525 10.1093/intimm/dxn111 [DOI] [PubMed] [Google Scholar]
- Porcelli, S., Yockey C.E., Brenner M.B., and Balk S.P.. 1993. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- alpha/beta T cells demonstrates preferential use of several V beta genes and an invariant TCR alpha chain. J. Exp. Med. 178:1–16 10.1084/jem.178.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon, R., Kjer-Nielsen L., Patel O., Chen Z., Illing P.T., Bhati M., Kostenko L., Bharadwaj M., Meehan B., Hansen T.H., et al. 2012. Structural insight into MR1-mediated recognition of the mucosal associated invariant T cell receptor. J. Exp. Med. 209:761–774 10.1084/jem.20112095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reantragoon, R., Corbett A.J., Sakala I.G., Gherardin N.A., Furness J.B., Chen Z., Eckle S.B., Uldrich A.P., Birkinshaw R.W., Patel O., et al. 2013. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 210:2305–2320 10.1084/jem.20130958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegert, P., Wanner V., and Bahram S.. 1998. Genomics, isoforms, expression, and phylogeny of the MHC class I-related MR1 gene. J. Immunol. 161:4066–4077 [PubMed] [Google Scholar]
- Rossjohn, J., Pellicci D.G., Patel O., Gapin L., and Godfrey D.I.. 2012. Recognition of CD1d-restricted antigens by natural killer T cells. Nat. Rev. Immunol. 12:845–857 10.1038/nri3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne, J.P., Matsuda J.L., Mallevaey T., White J., Borg N.A., McCluskey J., Rossjohn J., Kappler J., Marrack P., and Gapin L.. 2007. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat. Immunol. 8:1105–1113 10.1038/ni1510 [DOI] [PubMed] [Google Scholar]
- Serriari, N.E., Eoche M., Lamotte L., Lion J., Fumery M., Marcelo P., Chatelain D., Barre A., Nguyen-Khac E., Lantz O., et al. 2014. Innate mucosal-associated invariant T (MAIT) cells are activated in inflammatory bowel diseases. Clin. Exp. Immunol. 176:266–274 10.1111/cei.12277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak, A.L., Workman C.J., Wang Y., Vignali K.M., Dilioglou S., Vanin E.F., and Vignali D.A.. 2004. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 22:589–594 10.1038/nbt957 [DOI] [PubMed] [Google Scholar]
- Tang, X.-Z., Jo J., Tan A.T., Sandalova E., Chia A., Tan K.C., Lee K.H., Gehring A.J., De Libero G., and Bertoletti A.. 2013. IL-7 licenses activation of human liver intrasinusoidal mucosal-associated invariant T cells. J. Immunol. 190:3142–3152 10.4049/jimmunol.1203218 [DOI] [PubMed] [Google Scholar]
- Tilloy, F., Treiner E., Park S.-H., Garcia C., Lemonnier F., de la Salle H., Bendelac A., Bonneville M., and Lantz O.. 1999. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J. Exp. Med. 189:1907–1921 10.1084/jem.189.12.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiner, E., Duban L., Bahram S., Radosavljevic M., Wanner V., Tilloy F., Affaticati P., Gilfillan S., and Lantz O.. 2003. Selection of evolutionarily conserved mucosal-associated invariant T cells by MR1. Nature. 422:164–169 10.1038/nature01433 [DOI] [PubMed] [Google Scholar]
- Tsukamoto, K., Deakin J.E., Graves J.A., and Hashimoto K.. 2013. Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics. 65:115–124 10.1007/s00251-012-0666-5 [DOI] [PubMed] [Google Scholar]
- Uldrich, A.P., Patel O., Cameron G., Pellicci D.G., Day E.B., Sullivan L.C., Kyparissoudis K., Kjer-Nielsen L., Vivian J.P., Cao B., et al. 2011. A semi-invariant Vα10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties. Nat. Immunol. 12:616–623 10.1038/ni.2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rhijn, I., Kasmar A., de Jong A., Gras S., Bhati M., Doorenspleet M.E., de Vries N., Godfrey D.I., Altman J.D., de Jager W., et al. 2013. A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat. Immunol. 14:706–713 10.1038/ni.2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn, M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67:235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wun, K.S., Cameron G., Patel O., Pang S.S., Pellicci D.G., Sullivan L.C., Keshipeddy S., Young M.H., Uldrich A.P., Thakur M.S., et al. 2011. A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity. 34:327–339 10.1016/j.immuni.2011.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., Li Y., and Mariuzza R.A.. 2012. Structural basis for self-recognition by autoimmune T-cell receptors. Immunol. Rev. 250:32–48 10.1111/imr.12002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.