Abstract
Digitonin extracts of mitochondria from cardiolipin-containing (wild type) and cardiolipin-lacking (crd1Δ mutant) Saccharomyces cerevisiae subjected to colorless native polyacrylamide gel electrophoresis in the presence of 0.003% digitonin displayed a supercomplex composed of homodimers of complexes III and IV in the former case but only the individual homodimers in the latter case. To avoid treatment with any detergent or dye, we compared organization of the respiratory chain in intact mitochondria from wild type and cardiolipin-lacking cells by using a functional analysis developed previously for the study of the organization of the respiratory chain of S. cerevisiae (Boumans, H., Grivell, L. A., and Berden, J. A. (1998) J. Biol. Chem. 273, 4872–4877). Dependence of the kinetics of NADH oxidation via complexes III, IV, and cytochrome c on the concentration of the complex III-specific inhibitor antimycin A was studied. A linear relationship between respiratory activity and saturation of complex III with antimycin A was obtained for wild type mitochondria consistent with single functional unit kinetics of the respiratory chain. Under the same conditions, cardiolipin-lacking mitochondria displayed a hyperbolic relationship indicating cytochrome c pool behavior. No release of cytochrome c from cardiolipin-lacking mitochondria or mitoplasts under our standard experimental conditions was detected. Identical cytochrome c pool behavior was observed for both wild type and cardiolipin-lacking mitochondria in the presence of a chaotropic agent, which disrupts the interaction between respiratory complexes. The results demonstrate that cardiolipin is essential for association of complexes III and IV into a supercomplex in intact yeast mitochondria.
Cardiolipin, a phospholipid exclusively found in bacterial membranes and the inner membrane of mitochondria, supports structural integrity and modulates activity of many multimeric complexes of these energy-transducing membranes (1–4). In particular, considerable evidence implicates the general requirement of cardiolipin for the activities and structure of cytochrome bc1 complex (complex III)1 (5, 6) and cytochrome c oxidase (complex IV) (7). It was demonstrated by blue native (BN)-PAGE that complexes III and IV form a supercomplex in yeast and mammalian mitochondria composed of a heterodimer of individual homodimers of complex III and complex IV that remains intact after detergent solubilization (8, 9). Using BN-PAGE analysis of digitonin extracts from Saccharomyces cerevisiae mitochondria, we previously found that, in contrast to wild type yeast, complexes III and IV do not associate to form a supercomplex in mitochondrial extracts from cardiolipin-lacking yeast mutants (crd1Δ). Expression in the crd1Δ mutant of a low copy plasmid carrying a “tet-off ”-regulated CRD1 gene (that encodes cardiolipin synthase) allowed us to demonstrate that the level of the supercomplex in the digitonin extracts was dependent on the cardiolipin content of the mitochondria. We suggested a specific role for cardiolipin in supporting a critical interaction between complexes III and IV in vivo that cannot be substituted by the elevated levels of phosphatidylglycerol in the mutant (10).
Although BN-PAGE analysis was developed to study native assembly of mitochondrial complexes (11), the possible dissociation of loosely bound subunits of the supercomplex caused by the anionic dye Coomassie Blue (Serva Blue G) in the absence of cardiolipin was suggested (12). Colorless native (CN)-PAGE might be a preferred method to identify membrane protein complexes in cardiolipin-lacking mutants, because low levels of nonionic detergent normally replace the anionic dye in the gel (13). In a comparative study, no significant amount of the supercomplex was detected in the mutant lacking cardiolipin when using BN-PAGE, whereas mainly supercomplex was found in the same mutant when CN-PAGE was employed in the absence of detergent (12). It was concluded that the anionic dye induced dissociation of the loosely associated complexes III and IV in the absence of cardiolipin and that the supercomplex still existed in the mutant mitochondria, although cardiolipin was essential for its stabilization. On the other hand, because no detergent or anionic dye was introduced during gel separation, the possibility of aggregation between detergent-solubilized membrane components resulting in loss of components was not excluded.
In this study, we employed CN-PAGE at concentrations of digitonin in the gel that were much lower than that used to extract the supercomplex. Next, we investigated the cardiolipin-dependent association of complexes III and IV into a supercomplex in intact yeast mitochondria. To avoid treatment of mitochondria with any detergent or dye, we compared the organization of the respiratory chain in wild type and cardiolipin-lacking mitochondria using a functional, kinetic analysis developed previously for the study of the organization of the respiratory chain of S. cerevisiae (14). In that study, titration with the complex III-specific inhibitor antimycin A was used to distinguish the organization of the respiratory chain into a single functional unit from independent localization of the redox components of the respiratory chain in the lipid bilayer connected by randomly diffusing small carriers, Coenzyme Q (CoQ) and cytochrome c, which behave in the membrane as homogeneous diffusible pools. It was found that the S. cerevisiae respiratory chain was titrated as a single functional unit (14), which is in good agreement with a supercomplex model in wild type mitochondria. Using the same kinetic analysis, we report here that the cytochrome c in the respiratory chain in intact mitochondria lacking cardiolipin displays pool behavior. This result is consistent with our previous observation based on the molecular organization of solubilized components of the respiratory chain (10) and demonstrates for the first time that cardiolipin plays an essential role in supercomplex formation in intact mitochondria, rather than merely providing added stability to the supercomplex.
EXPERIMENTAL PROCEDURES
Yeast Strain and Growth Media
Wild type yeast strain DL1 and its CRD1 null derivatives (crd1Δ) lacking cardiolipin, YZD5 (15), were grown at 30 °C in YPEG growth medium consisting of 1% Bacto yeast extract, 2% Bacto peptone, and 1% ethanol (v/v) and 3% glycerol as a carbon source.
Mitochondrial Isolation
Mitochondria were isolated as previously described (16) with the following modifications. Cells were grown in YPEG media to the mid-exponential phase of growth, harvested at 1500 × g (GSA rotor, Sorval) for 5 min at 4 °C, washed with cold water, and resuspended in 100 mM Tris-SO4 (pH 9.4) at a ratio of 2 ml/g wet weight cells. Thereafter, the cells were incubated with 10 mM dithiothreitol at 30 °C for 15 min with gentle shaking, centrifuged at 2000 × g (Sorval, SS-34 rotor here and later, if not specified) for 5 min, washed with 2 ml of 1 M cold sorbitol/1 g of cells, and digested with Zymolase-20T (25 mg/g of wet weight cells) in buffer containing 1 M sorbitol, 20 mM potassium Pi buffer (pH 7.2) at 37 °C for ~30–40 min. Spheroplasts were washed with 1 M cold sorbitol and lysed at 4 °C by Dounce homogenization in 0.5 M sorbitol, 10 mM Tris-HCl (pH 7.5), 0.02% bovine serum albumin supplemented with 1/1000 volume of Protease Inhibitor Mixture Set IV (Calbiochem®). After homogenization, the samples were centrifuged at 1500 × g for 5 min, and then the supernatant was transferred to a new tube and centrifuged again at 1500 × g for 5 min. The supernatant was once again centrifuged at 12,000 × g for 10 min. Pellets were washed with 20 ml of SE buffer (250 mM sucrose, 10 mM MOPS, 1 mM EDTA (pH 7.2)) and centrifuged at 1500 × g for 5 min. Supernatant from the last centrifugation was centrifuged at 12,000 × g for 10 min at 4 °C. Mitochondria (pellets) were suspended in SE buffer and frozen in 0.1-ml aliquots at −80 °C. Protein concentration was measured using the Bradford method (Bio-Rad) with bovine serum albumin as the standard.
Mitoplast Preparation
Isolated mitochondria in SE buffer were diluted 15-fold with hypotonic buffer (20 mM HEPES-KOH (pH 7.4)). After incubating on ice for 30 min, the mitoplasts were centrifuged at 14,000 × g for 10 min at 4 °C and redissolved in SE buffer (17, 18). Changes in the ratio of mitochondrial outer membrane porin (voltage-dependent anion channel or VDAC) to subunit 2 of complex IV were estimated by Western blotting with antibody to these proteins (Molecular Probes) and were used as an indicator of mitoplast formation.
CN-PAGE
Mitochondria (75 μg of protein) were solubilized in 15 μl of buffer containing 1% digitonin (w/v recrystallized digitonin), 50 mM potassium acetate, 30 mM HEPES-KOH (pH 7.4), 10% glycerol, 0.1 mg/ml α2-macroglobulin, 0.1% (v/v) protease inhibitor mixture set IV for 30 min on ice. After solubilization, the samples were centrifuged at 125,000 × g for 30 min (Beckman TL-100 ultracentrifuge, TLA100.3 rotor). The supernatant (15 μl) was transferred to a new tube, supplemented with 1.5 μl of sample buffer containing 500 mM aminocaproic acid, and subjected to electrophoresis in a 4–8% polyacrylamide gradient gel at 80 V for 1 h and then for 4 h at 200 V at 4 °C, cathode buffer (50 mM Tricine, 15 mM BisTris (pH 7.0)) and anode buffer (50 mM BisTris, pH 7.0). Low concentrations of the mild detergent digitonin (0.003 or 0.01%, w/v) were added into the gel to achieve separation of the protein complexes in the gel. After CN-PAGE, the gel was soaked in Western blotting buffer (20% methanol (v/v), 0.02% SDS, 20 mM Tris-HCl (pH 8.3), 0.15 M glycine) with gentle shaking for 60 min. Proteins were transferred to nitrocellulose sheets using a semidry apparatus (MilliBlot™-Graphite Electroblotter I, Millipore Corporation) at 40 V for 1.5 h at 4 °C. Cathode buffer was used as the buffer during transfer. Proteins were then specifically located using monoclonal antibody (Molecular Probes) specific for subunit 3 (Cox3p) of complex IV or polyclonal antibody (Gottfried Schatz, Biozentrum, Basel, Switzerland) specific for the cytochrome b component (Cobp) of complex III. Final detection was with secondary antibodies linked to horseradish peroxidase using SuperSignal® West Pico chemiluminescent substrate (Pierce) and a Bio-Rad Fluor-S™Max MultiImager for quantification. High molecular mass native markers from Amersham Biosciences were used as standards and included thyroglobulin (669 kDa) and ferritin (440 kDa).
Cytochrome b Concentration in Mitochondria
The cytochrome b concentration was determined spectroscopically (Hitachi U-3000) from the absorption difference spectra. Either solid dithionite or 10 μM ferricyanide was added to either reduce or oxidize the cytochrome, respectively, and the visible difference spectrum (reduced versus oxidized) was determined. An absorbance coefficient of 28.0 for the maximum (at 563 nm) minus the minimum (at 577 nm) difference was used to determine cytochrome b concentration (19).
Electron Transfer Activity of Complex III
Q2H2 (reduced CoQ2): cytochrome c oxidoreductase activity of isolated mitochondria (0.02–0.1 mg/ml) was determined at 30 °C by measuring the reduction of 18 μM yeast ferricytochrome c at 550 nm by 25 μM Q2H2 in 20 mM potassium Pi (pH 7.4), 2 mM EDTA, and 0.5 mM KCN in the presence of different concentrations of antimycin A (14) using a Hitachi U-3000 spectrophotometer. An extinction coefficient for reduced cytochrome c at 550 nm of 18.5 mM−1 cm−1 was used to calculate the reductase activity (20).
Respiratory Activity of Isolated Mitochondria
Oxygen consumption rates were measured at 30 °C with a Clark oxygen electrode. Rates were determined from the slope of a plot of O2 concentration versus time. Mitochondria (0.03–0.06 mg/ml) were incubated in buffer containing 0.6 M sorbitol, 25 mM potassium Pi (pH 7.0), 1 mM EDTA, and 1 mM MgCl2 in a final volume of 3 ml. After 2 min of preincubation with antimycin A (dissolved in methanol), 0.5 mM NADH was added to the reaction chamber (14). Control samples had an equal volume of methanol. Titrations with antimycin A were carried out at least three times.
Definition and Kinetics of Pool Function of Cytochrome c
As established by Boumans et al. (14), the kinetic behavior of the respiratory chain of yeast is consistent with an ordered molecular assembly, which means that the number of subsequent acceptors a carrier, such as CoQ or cytochrome c, can reach is limited to 1. As organization of the system deviates from an ordered array, the number of subsequent acceptors increases from 1, dependent on the degree of pool function of a freely diffusing carrier. When cytochrome c is present as a pool, respiratory activity (Vo) can be described as a function of the maximum rate of reduction (Vred) of cytochrome c and the maximum rate of oxidation (Vox) of cytochrome c by equation 1 (14) previously derived in the study of another mobile carrier, CoQ (21).
| (Eq. 1) |
| (Eq. 2) |
| (Eq. 3) |
A putative pool behavior of specifically cytochrome c can be studied by titration of the respiratory activity of yeast mitochondria with NADH as a substrate by the complex III-specific inhibitor antimycin A (14), which targets the quinone reduction site of the complex and binds in a 1:1 ratio (22). In this case, antimycin A titrates the reduction rate of cytochrome c (Vred). The respiratory chain of S. cerevisiae, unlike that of mammals, does not contain complex I (23). Instead, inner membrane-bound NADH dehydrogenases Ndi1p (internal, faces the matrix) and Nde1p (external, faces the intermembrane space) are present, which transfer the electrons without translocation of protons across the membrane (24). When NADH is used as a substrate in isolated S. cerevisiae mitochondria, it reduces CoQ via external NADH dehydrogenase and provides the highest reduction rate for cytochrome c, which was previously established (14) to be a necessary prerequisite for measuring cytochrome c pool function. The titrated respiratory activity, Vx, can be described by equation 2, where Vx is the activity at a given antimycin A concentration and x is the fraction of complex III inhibited at a given antimycin A concentration (14). Equation 3 is derived from equation 2 by dividing both sides by Vo (initial respiratory activity without antimycin A) and was used to fit the data using the Kaleida Graph program. The y-axis on graphs (Fig. 3) represents the relative NADH oxidase activity (Vx/Vo). The x-axis represents the fractional saturation by antimycin A. Saturation with antimycin A was determined in the fitting procedure of the experimental data, with equation 3 as the concentration of antimycin A at which the curve crosses the x-scale (Vx/Vo = 0; complete inhibition) and was set to 1. Vred/Vo, the maximum rate of reduction (which is titrated with antimycin A) relative to the Vo, is referred to as Vtitr in Table I. The values of this coefficient for curves represented in Fig. 3 were obtained in the fitting procedure of the experimental data using equation 3 in the Kaleida Graph program. If the value for Vtitr is close to 1, the above hyperbolic relationship (equation 3) approximates a straight line relationship between relative NADH oxidase activity and relative saturation by antimycin A. This would be consistent with complex III, cytochrome c, and complex IV functioning as a single unit. The deviation of Vtitr from 1 reflects the degree of pool function of cytochrome c and represents its ability to provide interconnection between individual complexes III and IV randomly embedded in the membrane (for details, see Ref. 14).
Fig. 3. Titration of NADH oxidation with antimycin A.
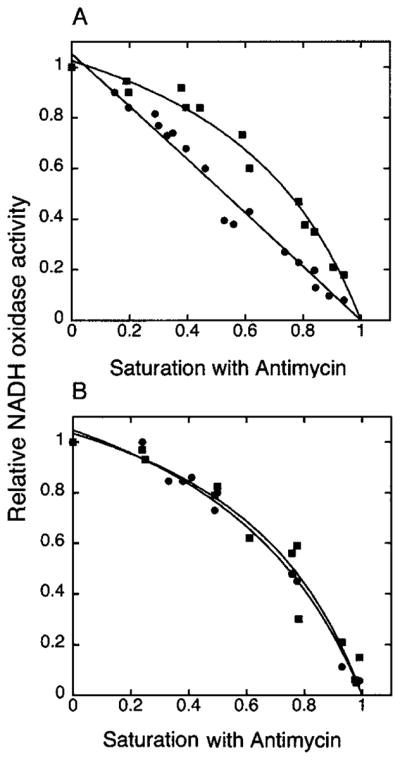
Respiration of isolated mitochondria from wild type (DL1, CRD1) (A) and cardiolipin-lacking cells (YZD5, crd1Δ) (B) was measured in standard buffer (0.6 M sorbitol, 25 mM potassium Pi (pH 7.0), 1 mM EDTA, 1 mM MgCl2, 0.5 mM NADH) (●) and in the standard buffer supplemented with 50 mM trichloroacetic acid (pH 7.0) (■) in the presence of different concentrations of antimycin A. Respiratory activities of both types of mitochondria ranged from 320 to 350 nmol of O2/mg/min for the preparations used in this figure.
Table I. Reaction rates of NADH oxidation titrated with antimycin A.
The deviation of Vtitr from 1 reflects the degree of pool function of cytochrome c (14).
| Buffera plus | Vtitr (CRD1) | Vtitr (crd1Δ) |
|---|---|---|
| 25 mM KPi | 1.07 ± 0.08 | 2.8 ± 0.3 |
| 25 mM KPi/50 mM TCAb | 3.0 ± 0.2 | 3.1 ± 0.4 |
Buffer was 0.6 M sorbitol, 1 mM EDTA, 1 mM MgCl2, pH adjusted to 7.0.
pH adjusted to 7.0.
Cytochrome c Release from Mitochondria or Mitoplast
Mitochondria (5 mg/ml protein) or mitoplasts (5 mg/ml protein) isolated from both wild type and crd1Δ cells after exponential growth at 30 °C in YPEG were resuspended in mitochondrial respiratory buffer containing either 0.6 M sorbitol, 25 mM potassium Pi (pH 7.0), 1 mM EDTA, 1 mM MgCl2, or the above buffer plus 300 mM KCl (pH 7.0). The samples were supplemented with 0.5 mM NADH and the same concentration of methanol as described under “Experimental Procedures” to mimic the exact conditions of the oxygen consumption measurement. After incubation under the same condition as in the respiration experiment for the indicated time, the samples were centrifuged at 10,000 × g for 5 min. Supernatants were centrifuged once more to avoid mitochondrial contamination and concentrated by 10% trichloroacetic acid precipitation in the presence of 0.5% (w/v) bovine serum albumin. The resulting supernatants (released cytochrome c) and pellets (mitochondrial-bound cytochrome c) were analyzed by SDS-PAGE and Western blotting (15) with a monoclonal anticytochrome c antibody (BD Biosciences) for the presence of cytochrome c. The same amount of total pellet protein in sufficient amounts for detection was loaded in each lane, whereas all the protein recovered from the supernatant was loaded in each lane.
RESULTS
Lack of Supercomplex Formation between Complexes III and IV as Detected by CN-PAGE
The anionic dye Coomassie Blue used in BN-PAGE analysis may result in the dissociation of weakly bound complexes (12). Therefore, a gentler technique of CN-PAGE in which the anionic dye is replaced by low levels of nonionic detergent was employed to verify the results obtained by BN-PAGE (10). To minimize nonspecific membrane protein aggregation in the absence of the anionic dye, low concentrations of mild detergent digitonin (0.01 or 0.003% w/v), which were much lower then those used for extraction (1%), were added during CN-PAGE. In CN-PAGE, the direction and mobility of protein complexes are determined by the intrinsic charge of the complexes and their apparent molecular size, whereas in BN-PAGE, the presence of the protein-bound anionic dye is assumed to normalize all protein components to the same charge to mass ratio, so that electrophoretic mobility within the gel is dependent only on apparent molecular size.
Consistent with the results using BN-PAGE analysis (10, 12), digitonin extracts of mitochondria isolated from CRD1 cells and subjected to CN-PAGE (Fig. 1) showed similar supercomplex formation as detected by antibodies specific for a component of complex IV (Cox3p) and a component of complex III (Cobp) (Fig. 1, lanes 1 and 3). Digitonin extracts of mitochondria isolated from crd1Δ cells showed primarily bands in which mobilities were consistent with a homodimer of complex III (~600 kDa) (Fig. 1, lane 4) and a homodimer of complex IV (~400 kDa, Fig. 1, lane 2). No supercomplex was observed using CN-PAGE analysis of mitochondria lacking cardiolipin (Fig. 1, lanes 2 and 4), but trace amounts of intermediate molecular species containing both Cox3p and Cobp were present. In the experiment presented in Fig. 1, 0.003% (w/v) digitonin was introduced into the gel for CN-PAGE. A gel with 0.01% (w/v) digitonin displayed the same protein pattern (data not shown).
Fig. 1. Lack of supercomplex formation in crd1Δ cells as detected by CN-PAGE.
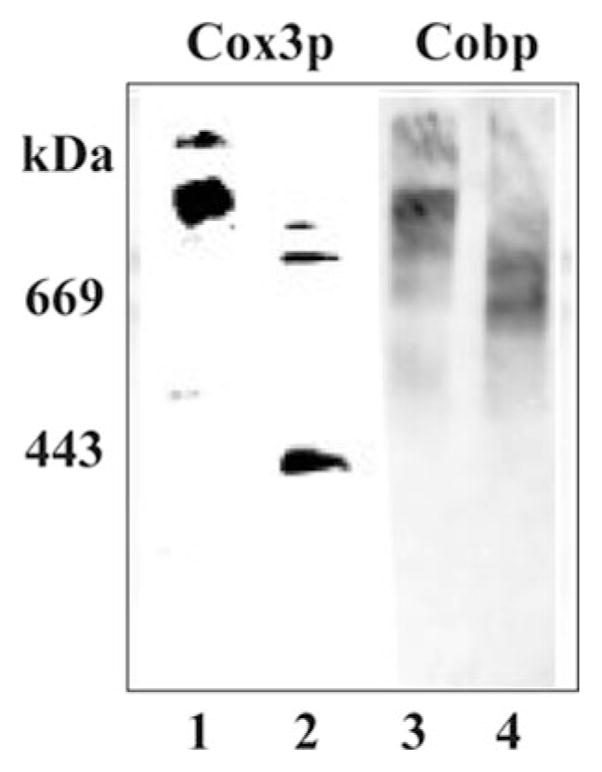
DL1 (CRD1, lanes 1 and 3) and YZD5 (crd1Δ, lanes 2 and 4) were grown in YPEG at 30 °C and harvested in the exponential phase of growth. Mitochondria were isolated, solubilized by digitonin, and displayed (75 μg of protein/lane) by CN-PAGE, as described under “Experimental Procedures.” Digitonin (0.003%) was added to the gel. The specificity of the antibodies used (Cox3p, lanes 1 and 2; Cobp, lanes 3 and 4) in each blot is indicated. The mobility of standard proteins of the indicated weight average Mr is on the left.
Titration of Q2H2:Cytochrome c Oxidoreductase Activity in Mitochondria with Antimycin A
To establish the lack of supercomplex formation between complexes III and IV in cardiolipin-lacking (crd1Δ) intact mitochondria, as suggested by CN-PAGE, we used an independent kinetic approach that avoids membrane solubilization by digitonin and the introduction of charged dyes. We first compared the sensitivity of complex III to antimycin A in wild type and cardiolipin-deficient mitochondria. Antimycin A is a specific inhibitor of complex III, which binds to the complex in a 1:1 ratio (22). Because it was shown previously for purified complex III that delipidation can change the affinity of the complex for antimycin A (25), we tested whether antimycin A has the same inhibitory effect on complex III in mitochondria from wild type (CRD1) and crd1Δ strains.
The concentration of cytochrome b was measured by absorption difference spectra as described under “Experimental Procedures.” Both CRD1 and crd1Δ mitochondria contained cytochrome b at a concentration of ~0.2 nmol/mg protein. Mitochondria from both strains had a similar Q2H2:cytochrome c oxidoreductase activity (500–600 nmoles/min/mg). Under the conditions of our experiments, a linear relationship between antimycin A concentration and electron transfer activity was observed in both wild type, as reported by Ref. 14, and cardiolipin-deficient mitochondria. In both cases, the relative inhibition of complex III activity by antimycin A can be viewed directly as the relative saturation of the complex with the antimycin A, suggesting that lack of cardiolipin did not alter the sensitivity of complex III to antimycin A (Fig. 2).
Fig. 2. Titration of the Q2H2:cytochrome c oxidoreductase (complex III) activity with antimycin A in mitochondria from wild type and crd1Δ cells.
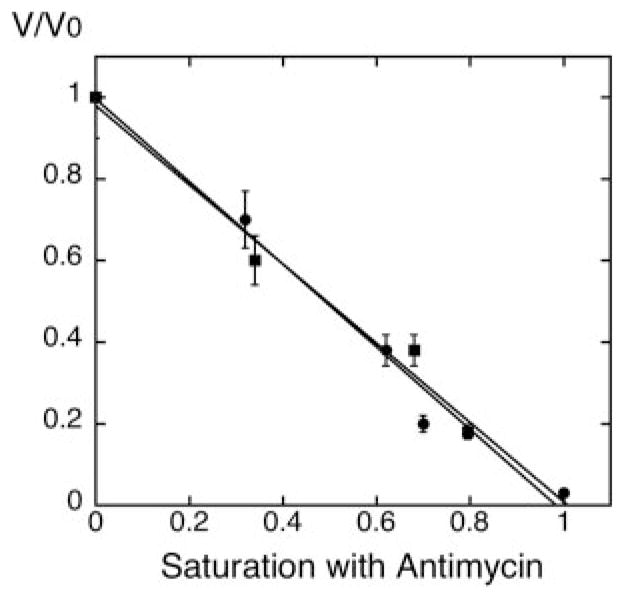
Mitochondria were isolated from both wild type (DL1, CRD1, ●) and cardiolipin-lacking cells (YZD5, crd1Δ, ■) grown at 30 °C in YPEG to exponential phase. Q2H2:cytrochrome c oxidoreductase activity of isolated mitochondria was determined with different concentrations of antimycin A, as described under “Experimental Procedures.” V/Vo represents the complex III activity at each antimycin A concentration relative to the activity with no antimycin A. The x-axis represents the fractional saturation by antimycin A with complete inhibition set to 1.
Pool Versus Non-Pool Behavior of Cytochrome c
The kinetic characterization of the electron carrier cytochrome c between complexes III and IV should provide evidence of whether mitochondria lacking cardiolipin contain a physiologically functional supercomplex or individual complexes. Mitochondria from both CRD1 and crd1Δ cells were analyzed for oxygen consumption using NADH as the substrate. If, as in wild type cells, complexes III and IV are associated into a supercomplex with channeling of cytochrome c, a linear relationship (Vtitr approaching 1) between the saturation of complex III with antimycin A and inhibition of the relative respiratory activity should be observed, because inhibition of complex III would reflect the inhibition of the whole supercomplex. If complexes III and IV exist as individual complexes, then cytochrome c would mediate electron transfer between them as a freely diffusible carrier producing interconnection between non-inhibited complexes III and complexes IV. In this case, inhibition of any one complex III assembly would not proportionately inhibit reduction of cytochrome c molecules or complex IV assemblies, and a hyperbolic inhibition curve (Vtitr > 1) should be observed (for details of the method, see “Experimental Procedures” and Ref. 14).
When using standard buffer with 25 mM Pi, a nearly linear relationship between relative respiratory activity and saturation with antimycin A was observed for wild type mitochondria (Fig. 3A, ●), confirming previous results and the single functional unit model of the electron transport chain of S. cerevisiae (14). This result is consistent with a cytochrome c molecule shuttling an electron from complex III to complex IV as a tightly associated supercomplex (Vtitr of 1.07) (Table I). Under the same conditions, mitochondria from crd1Δ cells displayed a hyperbolic inhibition curve (Fig. 3B, ●), indicating cytochrome c behaves as a freely diffusible carrier with Vtitr approaching 3 (Table I). When standard buffer was supplemented with a chaotropic agent (50 mM trichloroacetic acid (pH 7.0)) to dissociate the respiratory supercomplex (14), hyperbolic inhibition curves were obtained in both CRD1 (Fig. 3A, ■, and Table I, Vtitr 3.0) and crd1Δ mitochondria (Fig. 3B, ■, and Table I, with Vtitr also near 3). Data from Fig. 3 and the derived constants in Table I demonstrate the high level of dissociation of the functional supercomplex composed of complexes III and IV and cytochrome c in crd1Δ mitochondria, which is comparable with the level of their dissociation in the presence of a chaotropic agent.
Lack of Release of Cytochrome c from Mitochondria and Mitoplasts
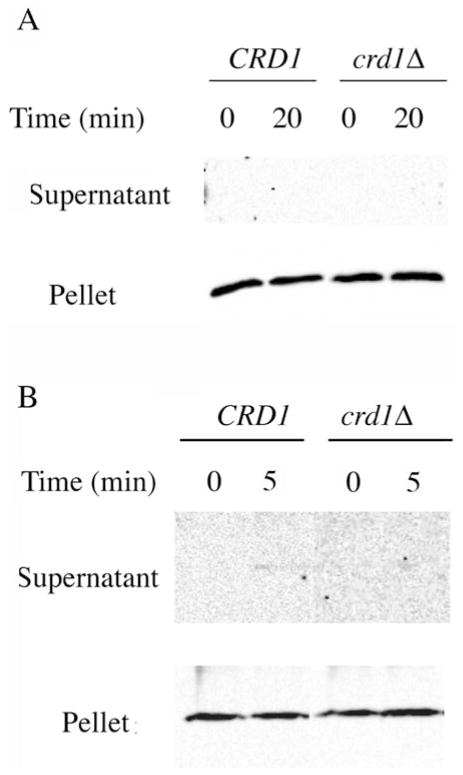
According to previous experiments (14), cytochrome c in wild type mitochondria exhibited pool behavior under two conditions, namely, upon the treatment of mitochondria with a chaotropic agent or with solutions of high ionic strength. In the first case, dissociation of the supercomplex composed of complexes III and IV was suggested. In the second case, release of cytochrome c from the outer surface of the inner mitochondrial membrane into the intermembrane space was suggested. Thus, in our experiments, the hyperbolic curve (Fig. 3B, ●) between relative NADH oxidase activity and saturation of complex III with antimycin A in cardiolipin-lacking mitochondria can be explained in two ways, dissociation of complexes III and IV (as in the case of the chaotropic agent) or release of cytochrome c into the intermembrane space under experimental conditions (as in the case of high ionic strength). No release of cytochrome c from either wild type or cardiolipinlacking mitochondria was observed even after a 20-min treatment under the same condition as used in the respiration measurement (Fig. 4A). High salt (300 mM KCl) released cytochrome c from crd1Δ mitochondria after 10 min of treatment and after 15 min for wild type mitochondria (data not shown). These results are identical to those previously reported (26) but where the low salt conditions employed were considerably lower (5 mM Tris-HCl, ionic strength of 5 mM) than those used here (25 mM potassium Pi, ionic strength of 50 mM). Therefore, under the conditions of our kinetic analysis, cytochrome c is still tightly associated with mitochondria.
Fig. 4. Analysis of cytochrome c release from mitochondria and mitoplasts of wild type and crd1Δ cells.
Mitochondria (A) and mitoplasts (B) isolated from both wild type and crd1Δ cells were incubated in the respiration buffer (0.6 M sorbitol, 25 mM potassium Pi, 1 mM EDTA, 1 mM MgCl2 (pH 7.0), 0.5 mM NADH and methanol (4 μl/3 ml)) for the indicated time. Samples were processed as described under “Experimental Procedures.” SDS-PAGE and Western blotting were used to detect the presence of cytochrome c in the supernatants (released cytochrome c) and pellets (mitochondrial- or mitoplast-bound cytochrome c).
To exclude the possibility that the absence of cytochrome c release from mitochondria is a result of the impermeability of the outer membrane in the absence of high salt, we repeated the same experiments using mitoplasts from wild type and cardiolipin-lacking mitochondria. In mitochondria treated with hypotonic buffer on ice for 30 min (see “Experimental Procedures”), 30% of the initial mitochondrial outer membrane protein porin remained in the mitoplast prepared from wild type and cardiolipin-lacking cells, whereas >70% of the initial mitochondrial inner membrane protein subunit 2 of complex IV was still present in the mitoplasts, indicating that the outer membrane was disrupted to allow detection of the release of cytochrome c from the inner membrane. Under respiration conditions, no release of cytochrome c from the mitochondrial inner membrane was detected with either wild type or crd1Δ mitoplasts (Fig. 4B). Release of cytochrome c was detected only under high ionic strength conditions (data not shown), which further confirmed that the pool behavior of cytochrome c detected in cardiolipin-deficient mitochondria, as shown in Fig. 3B, (●), was not because of the release of cytochrome c from the mitochondrial inner membrane but rather the dissociation of complexes III and IV in the absence of cardiolipin.
DISCUSSION
Based on BN-PAGE analysis of complexes III and IV from S. cerevisiae, we previously concluded that these two complexes do not form a supercomplex in detergent-solubilized extracts of mitochondria devoid of cardiolipin (10) as they do in extracts of mitochondria from wild type cells (9). In a subsequent report (12), it was concluded that cardiolipin stabilizes supercomplex formation in wild type cells but that the supercomplex was still the predominant form observed in cardiolipin-lacking mitochondria based on analysis of detergent-solubilized complexes using CN-PAGE. The authors rationalized that the presence of anionic dye Coomassie Blue in the BN-PAGE system acted as an ionic detergent that selectively dissociated a more weakly associated supercomplex lacking cardiolipin but not a more stable complex containing cardiolipin. However, we noted that the CN-PAGE system employed completely lacked digitonin or other detergents in the gel. From cells grown on ethanol/glycerol, the ratio of supercomplex displayed on the gels relative to complex V (ATP synthase that was unaffected by the lack of cardiolipin) was significantly lower even for the cardiolipin-containing samples, suggesting that complexes III and IV (either as a supercomplex or individually) were poorly represented and therefore were possibly lost because of aggregation in the absence of detergent or anionic dye.
To resolve these differences in the behavior of complexes III and IV in native gel electrophoresis, we carried out CN-PAGE analysis in the presence of low levels of digitonin in the gel to prevent possible aggregation, in contrast to the above report (12). We rationalized that, if the anionic dye in the BN-PAGE system (and not the 1% digitonin used for solubilization) dissociated the supercomplex, then inclusion of 0.003% digitonin in the gel would prevent nonspecific aggregation of proteins but not specific weak interactions between complexes III and IV. Inclusion of low amounts of digitonin resulted in comparable recovery of complexes III and IV from wild type and cardiolipin-lacking mitochondria but predominately supercomplex in the former case and primarily homodimers of individual complexes III and IV in the latter case, as we previously reported using BN-PAGE (10).
The contradictory results from different native electrophoresis methods (Refs. 10, 12, 27, and this study) lead us to apply an independent functional analysis of the organization of the respiratory chain of S. cerevisiae in intact mitochondria. Kinetic analysis was successfully used to discriminate between two models of organization of redox complexes of the respiratory chain, random organization of individual respiratory enzymes, or their assembly into supercomplexes (Ref. 28 and references therein). Kinetic analysis of cytochrome c behavior (14) in the study of the organization of the respiratory chain of S. cerevisiae is also in good agreement with previous BN-PAGE results (8, 10), which showed the organization of complexes III and IV mostly in a supercomplex in wild type mitochondria.
Our kinetic data from this study confirmed that cytochrome c does not behave as a pool in wild type mitochondria. However, consistent with BN-PAGE and CN-PAGE, the respiratory chain in intact cardiolipin-lacking mitochondria exhibits, with respect to cytochrome c, pool behavior. As mentioned above (see “Results”), two factors might contribute to our observed pool behavior of cytochrome c in cardiolipin-deficient mitochondria. First, complexes III and IV may not be associated with each other in the absence of cardiolipin. Second, complexes III and IV could still be associated with each other, but the pool behavior would be caused by the release of cytochrome c to the intermembrane space in the absence of cardiolipin. However, it was previously demonstrated that cytochrome c remained tightly bound to the inner mitochondrial membrane of wild type and cardiolipin-lacking (but enriched in phosphatidylglycerol) mitochondria from S. cerevisiae after permeabilization under low ionic strength conditions (26). We established that there was no release of cytochrome c from the cardiolipin-lacking mitochondria or mitoplasts under our experimental conditions, which employed higher ionic strength conditions than used previously (26). This experimental evidence rules out reduced affinity of cytochrome c for the outer surface of the inner mitochondrial membrane as the basis for pool behavior in the absence of cardiolipin. The possibility that complexes III and IV still associate with each other in the absence of cardiolipin but that cytochrome c, although bound to the membrane, loses its ability to donate electrons directly from one partner to another within the supercomplex is less probable. In any case, lack of cardiolipin results in the lack of the assembly of a functional supercomplex with channeling of cytochrome c from complex III to IV within the supercomplex, implying structural changes in supercomplex organization in the absence of cardiolipin. Taken together with our structural data from BN-PAGE and CN-PAGE, the results demonstrate significant dissociation of the supercomplex composed of complexes III and IV in intact mitochondria lacking cardiolipin.
The state of the respiratory chain of yeast mitochondria organized into a functional and structural unit or “respirasome” (29) differs from organization of the respiratory chain of higher eukaryotes in which CoQ and cytochrome c exhibit pool behavior and BN-PAGE and CN-PAGE reveal heterogeneity in distribution of the individual redox complexes between monomeric, dimeric, and supercomplex organization (30). A shift in the state of redox enzymes of S. cerevisiae toward a supercomplex organization might provide a protective mechanism against significant changes in the respiratory capacity of the chain under conditions of glucose repression, when a strong decrease in the level of redox enzymes in the membrane takes place (14). In contrast to higher eukaryotes, where complexes III and IV are organized into a supercomplex through additional interactions with the large complex I, the yeast supercomplex is formed only through interactions between complexes III and IV (9). This might suggest a more specific role for cardiolipin in formation of supercomplexes in yeast mitochondria.
Acknowledgments
We thank Dr. A. Tsai for advice in the experiments with antimycin A, Dr. V. Shinkarev for critical comments on the interpretation of kinetic data, and Dr. R. Kulmacz for making available equipment for measurement of mitochondria oxygen consumption.
Footnotes
This work was supported in part by National Institutes of Health Grant GM56389 (to W. D.).
The abbreviations used are: complex III, cytochrome bc1 complex; complex IV, cytochrome c oxidase; BN, blue native; CN, colorless native; CoQ, coenzyme Q; Q2H2, reduced CoQ2; YPEG, yeast extract, peptone, ethanol, glycerol; MOPS, 4-morpholinepropanesulfonic acid; Cox3p, subunit 3 of complex IV; Cobp, cytochrome b component of complex III.
References
- 1.Schlame M, Rua D, Greenberg ML. Prog Lipid Res. 2000;39:257–288. doi: 10.1016/s0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- 2.Palsdottir H, Hunte C. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Dowhan W, Mileykovskaya E, Bogdanov M. Biochim Biophys Acta. 2004;1666:19–39. doi: 10.1016/j.bbamem.2004.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mileykovskaya E, Zhang M, Dowhan W. Biochemistry (Mosc) 2005;70:154–158. doi: 10.1007/s10541-005-0095-2. [DOI] [PubMed] [Google Scholar]
- 5.Gomez B, Jr, Robinson NC. Biochemistry. 1999;38:9031–9038. doi: 10.1021/bi990603r. [DOI] [PubMed] [Google Scholar]
- 6.Lange C, Nett JH, Trumpower BL, Hunte C. EMBO J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sedlak E, Robinson NC. Biochemistry. 1999;38:14966–14972. doi: 10.1021/bi9914053. [DOI] [PubMed] [Google Scholar]
- 8.Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. J Biol Chem. 2000;275:18093–18098. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- 9.Schagger H, Pfeiffer K. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Mileykovskaya E, Dowhan W. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 11.Schagger H, von Jagow G. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 12.Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 13.Krause F, Reifschneider NH, Vocke D, Seelert H, Rexroth S, Dencher NA. J Biol Chem. 2004;279:48369–48375. doi: 10.1074/jbc.M406085200. [DOI] [PubMed] [Google Scholar]
- 14.Boumans H, Grivell LA, Berden JA. J Biol Chem. 1998;273:4872–4877. doi: 10.1074/jbc.273.9.4872. [DOI] [PubMed] [Google Scholar]
- 15.Zhang M, Su X, Mileykovskaya E, Amoscato AA, Dowhan W. J Biol Chem. 2003;278:35204–35210. doi: 10.1074/jbc.M306729200. [DOI] [PubMed] [Google Scholar]
- 16.Glick BS, Pon LA. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 17.Glick BS, Brandt A, Cunningham K, Muller S, Hallberg RL, Schatz G. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- 18.Glick BS. Methods Enzymol. 1995;260:224–231. doi: 10.1016/0076-6879(95)60140-6. [DOI] [PubMed] [Google Scholar]
- 19.Berden JA, Slater EC. Biochim Biophys Acta. 1970;216:237–249. doi: 10.1016/0005-2728(70)90215-x. [DOI] [PubMed] [Google Scholar]
- 20.Kubota T, Yoshikawa S, Matsubara H. J Biochem (Tokyo) 1992;111:91–98. doi: 10.1093/oxfordjournals.jbchem.a123725. [DOI] [PubMed] [Google Scholar]
- 21.Kroger A, Klingenberg M. Eur J Biochem. 1973;39:313–323. doi: 10.1111/j.1432-1033.1973.tb03129.x. [DOI] [PubMed] [Google Scholar]
- 22.Tsai AL, Palmer G. Biochim Biophys Acta. 1982;681:484–495. doi: 10.1016/0005-2728(82)90191-8. [DOI] [PubMed] [Google Scholar]
- 23.de Vries S, Marres CA. Biochim Biophys Acta. 1987;895:205–239. doi: 10.1016/s0304-4173(87)80003-4. [DOI] [PubMed] [Google Scholar]
- 24.Marres CA, de Vries S, Grivell LA. Eur J Biochem. 1991;195:857–862. doi: 10.1111/j.1432-1033.1991.tb15775.x. [DOI] [PubMed] [Google Scholar]
- 25.Tsai AL, Palmer G. Biochim Biophys Acta. 1986;852:100–105. doi: 10.1016/0005-2728(86)90061-7. [DOI] [PubMed] [Google Scholar]
- 26.Iverson SL, Enoksson M, Gogvadze V, Ott M, Orrenius S. J Biol Chem. 2004;279:1100–1107. doi: 10.1074/jbc.M305020200. [DOI] [PubMed] [Google Scholar]
- 27.Schagger H. Biochim Biophys Acta. 2002;1555:154–159. doi: 10.1016/s0005-2728(02)00271-2. [DOI] [PubMed] [Google Scholar]
- 28.Genova ML, Bianchi C, Lenaz G. Ital J Biochem. 2003;52:58–61. [PubMed] [Google Scholar]
- 29.Chance B, Williams GR. Nature. 1955;176:250–254. doi: 10.1038/176250a0. [DOI] [PubMed] [Google Scholar]
- 30.Schagger H, Pfeiffer K. J Biol Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]