Abstract
Small molecule control of intracellular protein levels allows temporal and dose-dependent regulation of protein function. Recently, we developed a method to degrade proteins fused to a mutant dehalogenase (HaloTag2) using small molecule hydrophobic tags (HyTs). Here, we introduce a complementary method to stabilize the same HaloTag2 fusion proteins, resulting in a unified system allowing bidirectional control of cellular protein levels in a temporal and dose-dependent manner. From a small molecule screen, we identified N-(3,5-dichloro-2-ethoxybenzyl)-2H-tetrazol-5-amine as a nanomolar HALoTag2 Stabilizer (HALTS1) that reduces the Hsp70:HaloTag2 interaction, thereby preventing HaloTag2 ubiquitination. Finally, we demonstrate the utility of the HyT/HALTS system in probing the physiological role of therapeutic targets by modulating HaloTag2-fused oncogenic H-Ras, which resulted in either the cessation (HyT) or acceleration (HALTS) of cellular transformation. In sum, we present a general platform to study protein function, whereby any protein of interest fused to HaloTag2 can be either degraded 10-fold or stabilized 5-fold using two corresponding compounds.
Keywords: Drug Target Validation, Hydrophobic Tag, Degron, Hsp70, Ubiquitin Proteasome System
Introduction
The use of small molecules to manipulate protein activity has proven helpful for delineating protein function and treating human disease1. Although great strides have been made in developing small molecule inhibitors for certain classes of proteins (e.g. kinases and G protein-coupled receptors), the great majority of proteins lack a potent small molecule inhibitor2. The cost and effort of developing potent small molecules often precludes many attractive drug targets from drug development programs. Therefore, a need exists for a streamlined method to establish whether a putative protein is critical for disease progression and thus could serve as a drug target.
An additional challenge in therapeutic development is that many drugs have off-target effects that can derail their further development. Thus, validating the on-target effects of new drugs in animal models is necessary. Although RNA interference3; 4 and genetic knockout have played a crucial role in drug target discovery and validation, there are many instances in which a small molecule-based target validation approach would provide utility5. For example, some of the more attractive features of small molecules include: high bioavailability, direct action on targeted proteins, precise dose dependence, rapid temporal control, and reversibility. Therefore, a small molecule approach would provide significant utility as an additional tool in drug target validation efforts.
Significant effort in the chemical biology community has focused on developing systems that induce the destabilization of a target fusion protein using small molecules (reviewed previously6). These generalized strategies should, in principal, allow researchers to assess the effects of small molecule inactivation of a target protein before a specific inhibitor has been developed. Recently, two methods were developed where the addition of a single small molecule is sufficient to induce the degradation of any target fused to a specific tag. Both techniques enable researchers to perform chemical knockdown of a protein of interest by fusing it to a single degradation-conferring protein. The first system fuses target proteins to FKBP12 containing a cryptic degron, which upon binding to a small molecule FK506 analog, leads to destabilization of the fusion protein7; 8. Recently, we developed a system whereby a small molecule is employed to append hydrophobic tag (HyT) compounds to the surface of the HaloTag2 protein9, thereby mimicking a misfolded protein state and inducing proteasome-mediated degradation of HaloTag2 fusion proteins10; 11. This system allows for degradation of approximately 90% of the fusion protein and has been shown to work in cell culture, zebrafish, and murine disease models.
Although the HaloTag2 system is an effective tool to degrade proteins of interest, it lacked a complementary method to evaluate the stabilization of target proteins. As evidenced by the recent development of sirtuin activators for diabetes and obesity12, protein kinase C activators for acute myeloid leukemia13, and p53 stabilizers for various cancers14, many studies require the ability to increase the activity of a protein of interest in disease models. In fact, a method to stabilize FKBP12 derivative fusion proteins with a FK506 analog has been shown to work in many systems, complementing the degradation system that employs the same small molecule but a different variant of FKBP128. However, an ideal system for the small molecule control of fusion proteins would employ one domain for both stabilization and destabilization so that the activity of a single fusion protein can be either increased or decreased via the use of two different small molecules.
Here, we describe a small molecule stabilizer of the HaloTag2 protein. This HaloTag2 Stabilizer (HALTS1) binds the HaloTag2 protein with nanomolar affinity, conferring a large thermodynamic stability to HaloTag2-fusion proteins. HALTS1-stabilized HaloTag2 shows drastically reduced interaction with heat shock protein Hsp70, resulting in reduced ubiquitination and reduced aggregation upon proteasome inhibition. Moreover, we demonstrate the functional utility of the combined HyT/HALTS system in probing the physiological role of putative drug targets by modulating the activity of an oncogenic HRas-HaloTag2 fusion protein in a focus formation assay. In summary, we have developed a system whereby any protein of interest fused to a single tag can be targeted with a small molecule to be degraded 10-fold or stabilized 5-fold in a reversible, dose-dependent manner, thereby providing new opportunities in drug target validation.
Results/Discussion
The stabilization of HaloTag2 with 22 point mutations provides the rational to screen for small molecule stabilizers of HaloTag2
HaloTag2 is a mutant bacterial dehalogenase9 with limited stability (t1/2 ~9h, Supplementary Information Figure S1) when expressed in eukaryotic cells. To increase its stability, 22 mutations have previously been introduced to create the more stable HaloTag7 protein for recombinant protein work15; 16. We hypothesized that these mutations would also provide increased stability in mammalian cells. In fact, when HEK293 FlpIn cells were used to insert HaloTag7- and HaloTag2-enhanced green fluorescent protein (EGFP) fusion proteins into identical genetic loci, we observed ten times lower levels of HaloTag2-EGFP, suggesting decreased intracellular stability for HaloTag2 (Figure 1a). Consistent with its decreased intracellular stability, we also observed aggregation of HaloTag2-EGFP upon treatment with a proteasome inhibitor, YU101, whereas no aggregation was observed with the more stable HaloTag7 (Figure 1b). These data suggest that HaloTag2 can render instability to fusion proteins and that it would be possible to stabilize HaloTag2 using a small molecule.
Figure 1.

HaloTag2 confers instability to EGFP, which can be reversed by HALoTag Stabilizer 1 (HALTS1) compounds. A) Flow cytometry analysis of steady state EGFP levels in HEK293 Flp-In cells expressing EGFP alone, EGFP fused to HaloTag2, or EGFP fused to HaloTag7. B) Treatment with 10 μM proteasome inhibitor, YU101, induces aggregation of EGFP HaloTag2 but not EGFP-HaloTag7. C) Structure of HALTS1 compounds, identified in a screen for compounds that reduce the rate of HyT13 induced HaloTag2-Luciferase degradation in cells. D) Structure of hydrophobic tags, HyT13 and HyT36, which have both previously been characterized 10; 11.
To find small molecule stabilizers of HaloTag2, we screened a 35,000 compound library of small molecules for compounds that prevent the degradation of HaloTag2-luciferase in response to the HaloTag2 destabilizer, HyT1310. The rationale for including HyT13 in the assay was to expedite the degradation of the HaloTag2 fusion protein, thereby enabling the cell-based screen to be completed in 24 hours. The top hits from the screen were largely able to prevent the HyT13 mediated degradation, and notably, a clear pharmacophore emerged among the top hits (Figure 1c). Three aminotetrazoles, containing mono- or dichloro-substituted benzyl amines were among the top four hits. After evaluating the efficacy and solubility of our top hits in follow-up assays, we decided to investigate further N-(3,5-dichloro-2-ethoxybenzyl)-2H-tetrazol-5-amine, hereafter referred to as HaloTag Stabilizer1 (HALTS1).
The bidirectional HyT/HALTS system allows temporal and dose-dependent control of HaloTag2 fusion protein levels in cells
To assess the kinetics of HaloTag2 stabilization, we treated HA-EGFP-HaloTag2 expressing cells with 1 μM HALTS1 for increasing lengths of time. Initial HALTS1-induced stabilization was observed by 6 hours and a new equilibrium was reached at 72 hours, at which point the fusion protein level had increased 4-fold (Figure 2a). Similarly, we treated the same cells with various concentrations of HALTS1 for 48 hours. Fusion protein stabilization was observed beginning at 50 nM with a maximum reached at 2.5 μM, at which point an almost 5-fold increase in protein level was observed (Figure 2b). From these cell culture studies, we estimated that the EC50 for HALTS1 is approximately 100 nM. No toxicity was observed with HALTS1, which was tested in an alamarBlue Cell Viability Assay with a 24 hour treatment of up to 200 μM HALTS1 (Supplementary Information, Figure S2). Given the ability of the HaloTag2 destabilizing ligand HyT36 to reduce HaloTag2 levels by 10-fold and the ability of HALTS1 to increase the protein levels ~5-fold, the total dynamic range for small molecule-control of HaloTag2 fusion proteins is expected to be about 50-fold. To obtain a more precise measure of the dynamic range, we analyzed EGFP-HaloTag2 expressing cells by flow cytometry after treatment with 10 μM HyT36 and 10 μM HALTS1. We confirmed that the dynamic range is 42-fold (Figure 2c). Additionally, after using HALTS1 to upregulate EGFPHaloTag2 levels 2.2-fold, HyT36 treatment was able to return levels to baseline within approximately 3 hours (Supplementary Information, Figure S3). As a secondary confirmation of the stabilization of HaloTag2 by HALTS1, we also showed that HALTS1-treated cells have higher EGFP-HaloTag2 levels in the presence of cycloheximide (Supplementary Information, Figure S4). These data demonstrate that HALTS1 is able, in a time- and dose-dependent manner, to stabilize HaloTag2 fusion proteins and thereby increase their levels in cells. Furthermore, the combination of HyTs and HALTS1 results in a HyT/HALTS system for bidirectional control of a single fusion protein.
Figure 2.
Characterization of HALTS1 activity in cells. A) HEK293 Flp-In cells expressing HA EGFP-HaloTag2 were treated for indicated times with 1 μM HALTS1. Western blot band intensity, via densitometry, is expressed as band intensity normalized to actin. B) The same cells were treated with the indicated concentrations of HALTS1 for 48 hours. The blots were probed with HA and actin antibodies. C) Cells were analyzed for whole cell EGFP content by flow cytometry after treatment with 10 μM HALTS1 and 10 μM HyT36 for 48 hours.
HALTS1 provides thermal stability to HaloTag7 by binding to it directly with a dissociation constant of 143 nM
To test our hypothesis that HALTS1 increases cellular HaloTag2 levels by binding to HaloTag2 directly and stabilizing it, we turned to differential scanning fluorimetry17. The technique involves incubating recombinant protein with the dye SYPRO Orange, which exhibits increased fluorescence upon interaction with hydrophobic residues that are exposed as proteins unfold. By measuring the amount of increased dye fluorescence as a product of increased solution temperature, the melting temperature of the protein can be determined. If HALTS1 binds directly to the target protein and stabilizes it, we would expect to observe an increase in the protein melting temperature, i.e. a thermal shift. Given our inability to obtain a baseline melting curve for the HaloTag2 protein in this in vitro binding assay, presumably due to its inherent instability, we instead used HaloTag7 because of its increased stability at the temperatures required for the study15. As shown in Figure 3, HaloTag7 exhibited a melting temperature of 59°C, and 333 μM HALTS1 increased its melting temperature to 74°C, resulting in a total 15°C thermal shift. In contrast, a simple chloroalkane, HyT32, binds HaloTag7 covalently but does not induce a detectable thermal shift (Supplementary Information, Figure S5; see also past work with hydrophobic tags11). We next used isothermal titration calorimetry (ITC) to determine the precise binding constant and physical parameters of this interaction. We found that HALTS1 binds HaloTag7 in vitro with a Kd of 143 nM, an enthalpy of −10.7 kcal/mol, and entropy of −4.8 cal/mol/deg. Since HaloTag2 protein is less stable than HaloTag7, we expect the HALTS1 interaction with HaloTag2 to have an even more robust cellular response than with HaloTag7. We further confirmed that HALTS1 binds to and stabilizes HaloTag2 by performing a cellular thermal shift assay as described recently by Molina and colleagues18 (Supplementary Information, Figure S6). In this cellular thermal shift assay, HALTS1 bound to HaloTag2 in cell lysate and stabilized it, increasing the temperature required for HaloTag2 to precipitate out of solution.
Figure 3.
HALTS1 binds to HaloTag7 directly and stabilizes it in vitro. Thermal shift assay using differential scanning fluorimetry to determine the melting temperature of recombinant HaloTag7 in the presence of 333 μM HALTS1 or vehicle.
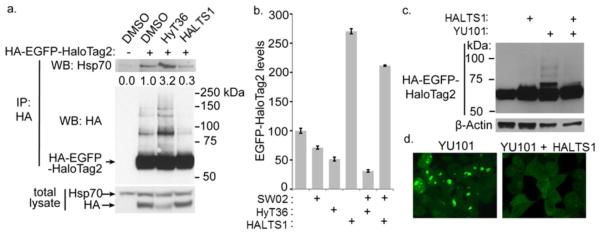
The HyTs/HALTS system modulates HaloTag2 fusion protein levels in cells through Hsp70 and the Ubiquitin-Proteasome System
We have previously shown that appending hydrophobic moieties to HaloTag2 leads to its degradation in cells by the proteasome10; 11. It is possible that HALTS1 prevents the intracellular turnover of HaloTag2 proteins by protecting HaloTag2 from the Ubiquitin-Proteasome System (UPS). Heat Shock Protein 70 (Hsp70) is a central chaperone involved in determining whether a protein is refolded or targeted for degradation by the UPS19. To study the potential interaction between Hsp70 and HaloTag2, we immunoprecipitated HA-EGFPHaloTag2 from cells treated for 2 hours with either HyT36 (10 μM) or HALTS1 (10 μM). Probing for Hsp70 in these immunoprecipitates confirmed that HyT36 leads to increased binding of Hsp70 to the fusion protein, whereas HALTS1 diminishes this interaction (Figure 4a). We did not detect any Hsp90 in the immunoprecipitates, suggesting that only the Hsp70 component of the chaperone system is involved in HaloTag2 stability surveillance. To confirm further that Hsp70 is responsible for the degradation of HaloTag2 fusion proteins, we treated cells with an Hsp70 activator, SW0220, with the expectation that the compound would enhance fusion protein degradation. We made three observations from these data: first, Hsp70 activation led to 30% degradation of the HaloTag2 fusion protein, demonstrating that Hsp70 is involved in the normal turnover of HaloTag2 proteins (Figure 4B). Second, whereas a low concentration of HyT36 led to a 50% degradation of HaloTag2 fusion protein, the combined effect of SW02 and HyT36 is additive, leading to 70% degradation. Hence, overactive Hsp70 enhances HyT36-mediated degradation. Third, even in SW02 treated cells, HALTS1 still exhibited a stabilizing effect on HaloTag2, further demonstrating that the small molecule acts upstream of Hsp70. Identical results were obtained when we increased Hsp70 levels using geldanamycin (Supplementary Information, Figure S7).
Figure 4.
Small molecule control of protein stability due to association with Hsp70 and engagement of UPS. A) HEK293 Flp-In cells expressing HA-EGFP-HaloTag2 were treated for 2 hours with 10 μM HyT36 or 10 μM HALTS1. The lysates were immunoprecipitated with anti-HA antibodies, and both the lysate and immunoprecipitate were probed with Hsp70 or HA antibodies. Western blot band intensity, via densitometry, is expressed as band intensity normalized to HA-EGFP-HaloTag2 + DMSO. B) The same cells as in (A) treated with 50 μM Hsp70 activator SW02, 10 μM HALTS1 or low 50 nM HyT36 for 24 hours were analyzed for EGFP fluorescence by flow cytometry. Data are represented as mean +/− SEM. C) Anti-HA Western blot of whole cell lysate from HA-EGFP-HaloTag2 expressing cells after being treated with vehicle, 10 μM HALTS1, 10 μM YU101 or both. The HA blot shows that HALTS1 treatment reduces the polyubiquitination of fusion protein. D) Treatment of the same cells with 10 μM YU101 over 16 hours induces visible aggregates, and the aggregation is blocked by inclusion of 10 μM HALTS1.
Interestingly, in the immunoprecipitates shown in Figure 4A, we noticed higher molecular weight HA-EGFP-HaloTag2 bands that appeared to correlate with increased (in the case of HyT36 treatment) or decreased (in the case of HALTS1) ubiquitination. To confirm that HALTS1 prevents HaloTag2 ubiquitination, we used the proteasome-specific inhibitor YU10121 and observed the appearance of additional higher molecular weight bands, characteristic of polyubiquitinated proteins. Inclusion of HALTS1 in the media completely blocked this ubiquitination (Figure 4c). Further, the aggregation of HaloTag2 fusion proteins in cells upon proteasome inhibition can be completely prevented by HALTS1 (Figure 4d). These results demonstrate that in cells, the thermodynamic stabilization of HaloTag2 protein by HALTS1 prevents the recruitment of Hsp70 and thus protects the protein from the UPS.
HALTS1 binding to the active site of HaloTag2 induces conformational changes that may be involved in stabilization
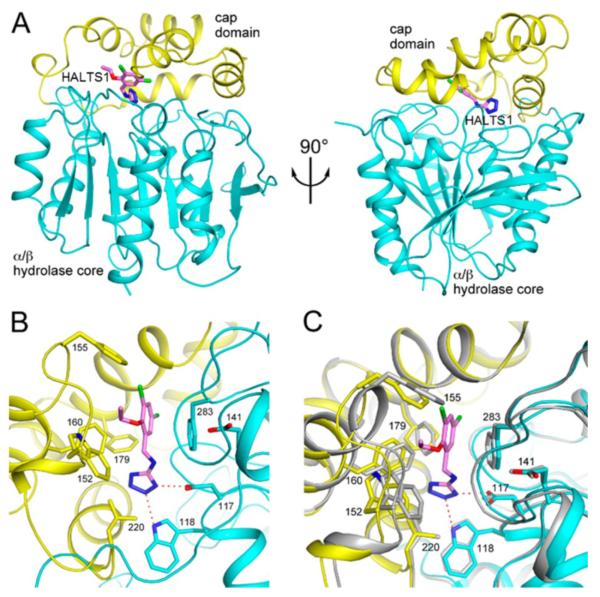
We next determined the crystal structure of HaloTag2 alone and in complex with HALTS1 at 2.3 Å resolution (Table S1) to provide direct, molecular evidence for its mechanism of action. We found that the compound is bound in the active site of the enzyme, in a deep tunnel between the α/β hydrolase core and the helical cap domain (Fig. 5a). In fact, the compound is completely shielded from the solvent and is not visible from the surface of the protein. The tetrazole ring of HALTS1 is bound deepest in the tunnel, and also has hydrogen-bonding interactions with the side chains of residues Asp117 and Trp118 (Fig. 5b). The phenyl ring of the compound has π-stacking interactions with Phe283, which is a histidine residue in the wild-type haloalkane dehalogenase. The cap domain contributes several hydrophobic, especially aromatic, side chains to the binding site.
Figure 5.
Crystal structure of HaloTag2 in complex with HALTS1. (A). Schematic drawing, in two views, of the overall structure of HaloTag2 (in cyan and yellow for the α/β hydrolase core and cap domain) in complex with HALTS1 (in pink). (B). The binding site for HALTS1 in HaloTag2. Selected side chains in the binding site are shown. Hydrogen-bonding interactions are indicated with dashed lines in red. (C). Overlay of the structures of the HaloTag2-HALTS1 complex (in color) and HaloTag2 alone (in gray). Produced with the program PyMOL (www.pymol.org). See also Table S1.
Compared to the structure of HaloTag2 alone, conformational changes in the main chain and side chain of many residues in the cap domain are clearly visible in the HALTS1:HaloTag2 complex (Fig. 5c). Smaller conformational changes in several of the loops in the α/β hydrolase core that form the binding tunnel are also observed. These conformational changes upon HALTS1 binding, as well as the fact that the HALTS1 fills an internal cavity of HaloTag2, may explain the observed thermal stabilization of the structure. The conformation of free HaloTag2 is more similar to that of the wild-type Rhodococcus haloalkane dehalogenase22.
Use of the HyT/HALTS system to study the physiological effects of either decreasing or increasing in activity of therapeutically relevant proteins
Given the robust destabilization and stabilization of HaloTag2 fusion proteins by HyT36 and HALTS1, respectively, we sought to demonstrate the functional utility of modulating levels of HaloTag2-fusion proteins and used oncogenic Ras as a model. NIH-3T3 cells expressing HRasG12V undergo cellular transformation and as a result, these cells begin to grow as multilayered foci. We infected NIH-3T3 cells with the HaloTag2-HRasG12V construct at low multiplicity of infection (MOI) so that the cells form only limited foci. The levels of HaloTag2-HRasG12V in these cells can be readily increased by HALTS1 and decreased by HyT36 (Figure 6A). Correspondingly, the elevated levels of oncogenic Ras lead to qualitative appearance of multilayered foci, whereas HyT36 treatment leads to disappearance of foci and reversion to an untransformed phenotype (Figure 6B). These results demonstrate that modulating HaloTag2 fusion protein levels with HyT36 or HALTS1 can lead to robust phenotypic consequences.
Figure 6.
The severity of cellular transformation with oncogenic HaloTag2-HRas can be modulated with HALTS1 and HyT36. A) NIH-3T3 cells expressing HA–HaloTag2–HRasG12V were treated with vehicle, 1 μM HALTS1 or 1 μM HyT36 for 4 days. The lysates were prepared for immunoblotting, and the blots were probed with HA-specific and β-actin–specific antibodies. B) The same cells were sparsely plated and treated with 1 μM HALTS1 or 1 μM HyT36 for 4 days. Representative images were taken. C) A protein of interest can be fused to HaloTag2 and either destabilized with HyT36 or stabilized with HALTS1 in a reversible and dose-dependent manner.
Conclusions
Functional validation of putative disease genes remains a significant challenge. RNAi technology provides great utility, but systemic delivery of RNAi species has been difficult to achieve. Transgenic animals with inducible shRNA or inducible genetic knockouts (e.g. Cre-LOX) can overcome many of the hurdles associated with delivery, but the timescale of protein knockdown and the catalytic nature of RNAi can make functional assignment difficult. Furthermore, adaptations associated with long-term genetic ablation may confound the true effect of gene disruption. Small molecule approaches, with a different profile of advantages and disadvantages, may therefore provide significant utility as an additional tool in drug target validation.
Given the impracticality of developing a potent and specific inhibitor/activator pair for every protein of interest, there exists an unmet need for a general small molecule-based method to modulate target proteins. We have recently demonstrated that a number of proteins of interest fused to HaloTag2 can be readily degraded by destabilizing the HaloTag2 portion of the protein with HyT molecules. We believe the ability to stabilize HaloTag2 fusion proteins with HALTS1 will greatly expand the utility of the HaloTag2 system. Hence, HALTS1 complements our previous HyT molecules, resulting in a system to manipulate protein levels in a rapid and dose dependent manner and thereby study physiological effects inaccessible to traditional approaches (Figure 6C). Thus, to functionally characterize a protein of interest, one need only generate a single HaloTag2 fusion protein, making the system particularly useful in studying transgenic animal models. For instance, in a single animal model one can test the functional consequences of inactivating and overexpressing a protein of interest. Additionally, this system can theoretically be combined synergistically with others, e.g. the FKBP12-based destabilization method mentioned earlier, to conduct more intricate experiments. For example, one could control multiple proteins in a signaling pathway in a temporal and dose-dependent manner.
In our previous studies we have demonstrated that the proteasome is responsible for degrading HaloTag2 proteins upon hydrophobic tagging. Here we extend the dissection of molecular mechanisms dictating small molecule control over protein stability. Hydrophobic tags, which reduce the thermal stability of HaloTag2, lead to increased binding by the quality control chaperone Hsp70. As refolding of the HaloTag2 protein fails, the HaloTag2 protein is ubiquitinated, presumably by the E3 ubiquitin ligase CHIP or Parkin, which are known to associate with Hsp70. Increased ubiquitination leads to enhanced binding to the proteasome and ultimately degradation of the fusion protein. HALTS1, on the other hand, stabilizes the HaloTag2 protein, rendering it less likely to interact with Hsp70, and as a result, HaloTag2 is protected from degradation via the ubiquitin-proteasome system. These observations could inform future studies on how therapeutic inhibitors that also stabilize or destabilize target proteins exert their effect. For instance, anti-androgen bicalutamide and anti-estrogen fulvestrant both lead to the degradation of their respective nuclear hormone receptors via the UPS23; 24. However, the molecular mechanisms leading to this degradation are unclear. Similar to our results, irreversible Her2 inhibitor, Canertinib, has been shown to promote the degradation of Her2 protein via increasing the binding of Her2 to Hsp7025. Given the direct role for Hsp70 in modulating HaloTag2 and Her2 stability in response to small molecules, we propose that Hsp70 client proteins could be particularly attractive targets for a hydrophobic tagging approach.
The control over HaloTag2 stability with HyT and HALTS1 molecules may allow for future modifications that add additional customization to assay design. Akin to reports with FKBP12, one could employ mutagenesis to skew the dynamic range of HaloTag2 stabilities with the two molecules. For instance, mutagenesis could yield a HaloTag2 mutant that would exhibit an even larger dynamic range. We envision that in addition to the HaloTag2 version reported here, one could design additional HaloTag2 variants for specific purposes.
In summary, we present here a system that allows researchers to modulate the levels of a protein of interest with a small molecule to either increase (HALTS1) or decrease (HyT) the protein’s abundance in cells. The result is a general method to probe the physiological role of any target protein using small molecules.
Methods
Cell Culture and Materials
HA-EGFP-HaloTag2, HA-EGFP-HaloTag7 and HA-EGFP were stably incorporated into FRT recombination-based HEK 293 Flp-In cells according to manufacturer’s protocol (Invitrogen). All cells were grown at 37 °C in DMEM and supplemented with 10% FBS, 100 U/ml carbenicillin and 100 μg/ml streptomycin. HRASG12V was obtained from Addgene plasmid 9051 (provided by R. Weinberg) and cloned into a retroviral pEYK3.1 vector by excising EGFP. Retrovirus was generated in GP2-293 cells (Clontech) with a pVSV-G vesicular stomatitis virus retroviral vector and a corresponding pEYK plasmid, and NIH-3T3 cells were infected. Rabbit HA-specific (C29F4) and Hsp70 (D69) antibodies were purchased from Cell Signaling, and β-actin–specific antibody was purchased from Sigma (clone AC-74). HALTS1 was purchased from ChemBridge (9074451), YU101, HyT13 and HyT36 have been synthetized in our laboratory, and geldanamycin was purchased from InvivoGen. HyT36, HyT13, HALTS1, YU101, SW02 and geldanamycin were stored and aliquoted in DMSO as 1000x stock solutions.
Immunoblotting
The indicated cells were washed twice with cold PBS, and the cells were lysed in lysis buffer (1× PBS, 1% NP-40, 1 mM EDTA, 40 mM HEPES) with protease inhibitors. The lysates were cleared by centrifugation at 10,000g for 5 min. The total protein concentration was determined by Bradford assay, and 50 μg of protein was loaded onto an 8% Bis-Tris gel. To solubilize polyubiquitinated and aggregated proteins upon proteasome inhibition, samples were lysed with an SDS lysis buffer (1× PBS, 1% NP-40, 1% SDS, 1% sodium deoxycholate, 1 mM EDTA, 40 mM HEPES) with protease inhibitors and sonicated. The blots were processed by standard procedures with the indicated antibodies, and the band intensities were quantified by ImageJ (US National Institutes of Health).
HaloTag2 Aggregation Assays
HEK293 KG, KGH2, or KGH7 cells were seeded overnight on coverslips. They were then treated for 16 hours with media containing 10 μM HALTS1, 10 μM YU101, both, or vehicle alone (0.2% DMSO). Cells were then fixed with 4% paraformaldehyde, mounted on slides, and visualized for EGFP fluorescence.
Small Molecule HaloTag2 Stabilization Screen
The screen was carried out at the Yale Center for Molecular Discovery. HEK293 cell line stably expressing HaloTag2-luciferase was plated in 384-well plates at 10K cell/well. Twenty four hours later, a 15K ChemBridge DIVERSet or 20K Maybridge Diversity compound libraries were pinned onto plates at a final concentration of 10 μM. One hour later, 10 μM HyT13 was added. Luciferase assays with Steady Glo (Promega) were performed 24 hours later. For each plate, a control set of DMSO and HyT13 alone was included. The fraction of HaloTag2-luciferase remaining for each well was calculated as a ratio of luciferase activity for each well and the difference between DMSO and HyT13 treated wells. This ratio is plotted in Figure 1C as a percentage for the top 30 compounds preventing degradation.
Recombinant Protein
HaloTag7 was amplified from pFN18A vector (Promega) and cloned into the pET-28a(+)vector (Invitrogen). The resulting plasmid was then transformed into BL21 (DE3) PLysS chemically competent E. coli (Invitrogen). Two liters of these bacteria were induced in exponential phase with IPTG (1 mM, 16 hr) at room temperature. The bacteria were resuspended in equilibration buffer used with the HisPur™ Cobalt Purification Kit (Thermo Scientific), spiked with 0.1% NP-40 and protease inhibitors. The bacteria were lysed with Branson Sonifier 450, and cellular debris was removed by centrifugation at 10,000g for 10 min. The recombinant protein was purified using the HisPur™ Cobalt Purification Kit (Thermo Scientific), then dialyzed 1:100 against PBS (Gibco) twice. Elutions and dialyzed proteins were assayed for purity using a Coomassie stain, and protein levels were quantified using the Bio-Rad Protein Assay.
Thermal Shift Assays
Purified HaloTag7 protein was diluted in PBS to either 30 μM or 10 μM, and SYPRO Orange Protein Gel Stain stock (Sigma Aldrich) was diluted 1:1000 into the protein solution. A 1 μL volume of various compound concentrations was then placed into each well of a Low 96-well Clear Multiplate PCR Plate (Bio-Rad), 30 μL of the protein-SYPRO Orange mixture was placed into each well, and the plates were covered with Microseal ‘B’ Film (Bio-Rad). Real-time thermal cycling was then performed using the CFX96 Real-Time System (Bio-Rad). The reaction mixture was first equilibrated at 25 °C for 5 minutes. Thereafter, the temperature of the solution was increased by 1 °C and incubated for 5 sec repeatedly to generate a melting curve from 25-100 °C. Protein thermal stability was then quantified in terms of Hex fluorescence at each temperature of the melting curve. Raw data files were exported from the CFX software and formatted in Microsoft Office Excel. Data were trimmed to include the lower limit and upper limit values flanking the melting temperature and Tm values were calculated as the inflection point of the resulting sigmoidal curve using PRISM™ (GraphPad Software, Inc.).
Isothermal Titration Calorimetry
Recombinant HaloTag7 was dialyzed 1:3000 into buffer containing 50 mM Tris (pH = 7.4) and 200 mM NaCl. HaloTag7 was then diluted with buffer, and DMSO was added to 2%. HALTS1, dissolved in DMSO, was added to buffer to a final concentration of 2% DMSO and 100 μM. Both solutions were filtered and degassed before the assay. An ITC200 calorimeter (MicroCal) was used to titrate compound into a sample well containing 200 μL of protein. Data was analyzed using Origin software.
Immunoprecipitation
Indicated cells were grown in 10-cm plates to 80% confluency and treated for 2 hours with vehicle, 10 μM HyT36 or 10 μM HALTS1. The cells were washed with cold PBS and lysed in HENG buffer (50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 20 mM Na2MoO4, 2 mM EDTA, 5% glycerol, 0.5% Triton X-100). The lysates were cleared by centrifugation at 10,000g for 5 min. Five hundred micrograms of protein was immunoprecipitated with 30 μL of anti-HA Affinity Gel (Sigma) at 4°C for 3 hours. The beads were washed 3 times with HENG buffer, and the protein complexes were released by boiling in sample buffer.
Focus Formation Assay
One hundred thousand NIH-3T3 cells infected with HA–HaloTag2–Hras1G12V at MOI=0.1 were plated onto 10-cm cell culture plates in 10% FBS with DMEM. The next day, the medium was replaced with DMEM containing 1% FBS and the cells were administered either vehicle, 1 μM HyT13 or 1 μM HALTS1. On day 4, the plates were photographed and the cells were harvested for immunoblotting.
Supplementary Material
Acknowledgements
We thank J. Everett, M. Maglaqui, E. Kohan, and H. Wang for their contributions to the NESG sample production and crystallization pipeline. We wish to acknowledge financial support from the US NIH (R01AI084140) and the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG) Program. Protein crystallography was supported as a Community Outreach Project of the NIH Protein Structure Initiative (grant U54 GM094597).
Footnotes
Supporting Information This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Crews CM. Targeting the undruggable proteome: the small molecules of my dreams. Chem Biol. 2010;17:551–5. doi: 10.1016/j.chembiol.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verdine GL, Walensky LD. The challenge of drugging undruggable targets in cancer: lessons learned from targeting BCL-2 family members. Clin Cancer Res. 2007;13:7264–70. doi: 10.1158/1078-0432.CCR-07-2184. [DOI] [PubMed] [Google Scholar]
- 3.Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 4.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 5.Sioud M. Promises and challenges in developing RNAi as a research tool and therapy. Methods Mol Biol. 2011;703:173–87. doi: 10.1007/978-1-59745-248-9_12. [DOI] [PubMed] [Google Scholar]
- 6.Banaszynski LA, Wandless TJ. Conditional control of protein function. Chem Biol. 2006;13:11–21. doi: 10.1016/j.chembiol.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 7.Bonger KM, Chen LC, Liu CW, Wandless TJ. Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nat Chem Biol. 2011;7:531–7. doi: 10.1038/nchembio.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, Wandless TJ. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006;126:995–1004. doi: 10.1016/j.cell.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Los GV, Encell LP, McDougall MG, Hartzell DD, Karassina N, Zimprich C, Wood MG, Learish R, Ohana RF, Urh M, Simpson D, Mendez J, Zimmerman K, Otto P, Vidugiris G, Zhu J, Darzins A, Klaubert DH, Bulleit RF, Wood KV. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008;3:373–82. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 10.Neklesa TK, Tae HS, Schneekloth AR, Stulberg MJ, Corson TW, Sundberg TB, Raina K, Holley SA, Crews CM. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat Chem Biol. 2011;7:538–43. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tae HS, Sundberg TB, Neklesa TK, Noblin DJ, Gustafson JL, Roth AG, Raina K, Crews CM. Identification of hydrophobic tags for the degradation of stabilized proteins. Chembiochem. 2012;13:538–41. doi: 10.1002/cbic.201100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–58. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Hampson P, Chahal H, Khanim F, Hayden R, Mulder A, Assi LK, Bunce CM, Lord JM. PEP005, a selective small-molecule activator of protein kinase C, has potent antileukemic activity mediated via the delta isoform of PKC. Blood. 2005;106:1362–8. doi: 10.1182/blood-2004-10-4117. [DOI] [PubMed] [Google Scholar]
- 14.Basse N, Kaar JL, Settanni G, Joerger AC, Rutherford TJ, Fersht AR. Toward the Rational Design of p53-Stabilizing Drugs: Probing the Surface of the Oncogenic Y220C Mutant. Chem Biol. 2010;17:46–56. doi: 10.1016/j.chembiol.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Ohana RF, Encell LP, Zhao K, Simpson D, Slater MR, Urh M, Wood KV. HaloTag7: A genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification. Protein Expression and Purification. 2009;68:110–120. doi: 10.1016/j.pep.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Encell LP, Friedman Ohana R, Zimmerman K, Otto P, Vidugiris G, Wood MG, Los GV, McDougall MG, Zimprich C, Karassina N, Learish RD, Hurst R, Hartnett J, Wheeler S, Stecha P, English J, Zhao K, Mendez J, Benink HA, Murphy N, Daniels DL, Slater MR, Urh M, Darzins A, Klaubert DH, Bulleit RF, Wood KV. Development of a dehalogenase-based protein fusion tag capable of rapid, selective and covalent attachment to customizable ligands. Curr Chem Genomics. 2012;6:55–71. doi: 10.2174/1875397301206010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 18.Molina DM, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–7. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 19.Mandal AK, Gibney PA, Nillegoda NB, Theodoraki MA, Caplan AJ, Morano KA. Hsp110 chaperones control client fate determination in the hsp70-Hsp90 chaperone system. Molecular biology of the cell. 2010;21:1439–1448. doi: 10.1091/mbc.E09-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans CG, Wisen S, Gestwicki JE. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1-42) aggregation in vitro. J Biol Chem. 2006;281:33182–91. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 21.Elofsson M, Splittgerber U, Myung J, Mohan R, Crews CM. Towards subunit-specific proteasome inhibitors: synthesis and evaluation of peptide α’, β’- epoxyketones. Chemistry & Biology. 1999;6:811–822. doi: 10.1016/s1074-5521(99)80128-8. [DOI] [PubMed] [Google Scholar]
- 22.Newman J, Peat TS, Richard R, Kan L, Swanson PE, Affholter JA, Holmes IH, Schindler JF, Unkefer CJ, Terwilliger TC. Haloalkane dehalogenases: Structure of a Rhodococcus enzyme. Biochemistry. 1999;38:16105–16114. doi: 10.1021/bi9913855. [DOI] [PubMed] [Google Scholar]
- 23.Waller A, Sharrard R, Berthon P, Maitland N. Androgen receptor localisation and turnover in human prostate epithelium treated with the antiandrogen, casodex. Journal of Molecular Endocrinology. 2000;24:339–351. doi: 10.1677/jme.0.0240339. [DOI] [PubMed] [Google Scholar]
- 24.Long X, Nephew KP. Fulvestrant (ICI 182,780)-dependent interacting proteins mediate immobilization and degradation of estrogen receptor-alpha. J Biol Chem. 2006;281:9607–15. doi: 10.1074/jbc.M510809200. [DOI] [PubMed] [Google Scholar]
- 25.Citri A, Alroy I, Lavi S, Rubin C, Xu W, Grammatikakis N, Patterson C, Neckers L, Fry DW, Yarden Y. Drug-induced ubiquitylation and degradation of ErbB receptor tyrosine kinases: implications for cancer therapy. EMBO J. 2002;21:2407–17. doi: 10.1093/emboj/21.10.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.