Abstract
Analysis of proteins in biological samples opens up the possibility of discovering new markers of toxicity. The liver is one of the primary targets of drug-induced toxicity and it also secretes many plasma proteins, which can be measured clinically. Most of the plasma proteins secreted by the liver are secreted by hepatocytes, but there is little information regarding the protein profile secreted by these cells. The purpose of this study was to analyze the secreted proteome of primary rat hepatocytes in a collagen gel sandwich configuration by a Gel-LC-MS/MS procedure. We identified over 600 peptides corresponding to more than 200 proteins. The protein profile included over 50 plasma proteins secreted by the liver, suggesting that the cultured hepatocytes secrete many of the proteins that they produce in vivo. Our data also suggests that the hepatocytes are actively remodeling their environment, since we identified several structural extracellular matrix proteins as well as some proteins known to be secreted specifically during liver regeneration. We also identified two proteins, α1-antitrypsin and α2-macroglobulin, whose secretions appear to be down-regulated in cells exposed to aflatoxin B1. It was noted that a 15 nM dose of aflatoxin B1 led to substantially diminished levels of these proteins and that day 6 of incubation was the ideal timepoint for medium collection. These data suggest that proteins in the conditioned medium of hepatocyte sandwich culture might lead to discovery of biomarkers for drug chemical toxicity.
Keywords: hepatocytes, sandwich, proteome, proteomics, aflatoxin
Introduction
Analysis of proteins in biological samples has been used for decades for screening and diagnosing disease (1). For example, the measurement of enzymes in plasma, such as alanine aminotransferase (ALT) and aspartate aminotransferase (AST), has become a clinical standard for the detection of hepatic injury (2). In order to enrich biomarker databases, there have been significant efforts aimed at characterizing the “global” protein profile in plasma and tissue samples. This type of research is a promising field which could lead to the early diagnosis of diseases such as cancer, and it has already been shown that there are several differences in protein expression among cancerous and healthy tissues. Heat shock proteins, in particular, have been found to be differentially expressed in cancerous tissues such as liver tumors (3), tongue squamous cell carcinoma (4) and ductal carcinoma of the breast (5).
The liver has been a primary target for this type of analysis, since its protein expression has been shown to change after drug-induced toxicity. In order to establish a database of liver proteins, Fountoulakis, et al., analyzed the intracellular proteomics profile of the rat liver with two-dimensional electrophoresis and MALDI-MS1 resulting in the identification of 273 intracellular proteins (6). The effects of carbon tetrachloride on this profile were then investigated (7). Two stress proteins, catalase and uricase, were up-regulated, while α2-macroglobulin and senescence marker protein decreased (7). In a similar analysis, Ruepp, et al., (8) characterized the protein profile in the mouse liver after acetaminophen exposure and found a significant decrease in the heat shock proteins, within 15 min of exposure. The effects on protein expression of several other hepatotoxins was also investigated by Man, et al., (9), who found that dexamethasone and the steroid cyproterone acetate both upregulate haptoglobin, α1-antitrypsin and carboxylesterase in rat livers. Such analyses could therefore be used to determine whether toxins act through shared pathways.
Proteomics analysis of cultured hepatocytes appears especially promising since these cells secrete many plasma proteins, which can be potential biomarkers in clinical diagnostics. However, cultured hepatocytes rapidly lose liver-specific functions, limiting their use for studying drug metabolism, toxicity and protein expression. The dedifferentiation of hepatocytes in culture is thought to be partially due to lack of proper extracellular matrix which is known to regulate gene expression in the liver (10–12). It has been shown, however, that hepatocytes cultured between two layers of collagen, or a “collagen sandwich”, maintain drug metabolism (13–16), morphology (17–19) and bile secretion (20–22) for a week or longer.
In this study we have investigated the secreted protein profile of hepatocytes cultured in collagen sandwiches by analyzing proteins in conditioned medium. The advantage of analyzing secreted rather than intracellular proteins is that it allows for proteomic analysis without destroying the cultures. This type of approach could also be adapted to more complex cultures, such as bioreactors, where collecting the effluent medium is more convenient than lysing the cells. We have established a database of secreted proteins, from hepatocytes in culture, that includes a number of plasma proteins as well as extracellular matrix proteins. These data demonstrate that hepatocytes in this system secrete many of the proteins that they would excrete in vivo and that they also actively remodel their extracellular environment. In order to investigate whether secreted proteins can be used as markers of toxicity we exposed the hepatocytes to varying doses of the liver carcinogen aflatoxin B1 and found a dose-dependent decrease in the secretions of α1-antitrypsin and α2-macroglobulin. These results suggest that analysis of the secreted proteome can lead to a better understanding of the biology of cultured hepatocytes as well to the discovery of new biomarkers of toxicity.
Materials and Methods
Cell Culture
Twenty-four well plastic plates (Becton Dickinson, Franklin Lakes, NJ)) were coated with 110 µl/well of collagen solution. The collagen solution consisted of 2mg/mL bovine dermal collagen type I (Cohesion Technologies Inc., Palo Alto, CA) diluted with water. The solution also contained 10% (v/v) 10x phosphate buffered saline, which included 70 mM glucose and 0.44 M sodium bicarbonate. Hepatocytes were isolated from male Fisher 344 inbred rats (Taconic, Germantown, NY) with a protocol described in Powers, et al., (23) and were immediately plated on collagen-coated 24-well dishes. The cell viability, measured with trypan blue exclusion, was typically 87–91 %. The cells were incubated in Hepatocyte Growth Medium (24) with the following modifications: purified bovine serum albumin was omitted, the concentration of niacinamide was 2.5 mM, and the concentrations of ZnCl2, ZnSO4, CuSO4 and MnSO4 were 0.4 µM, 0.26 µM, 0.08 µM and 15 nM respectively. Furthermore, the medium was supplemented with 3 pM epidermal growth factor (EGF).
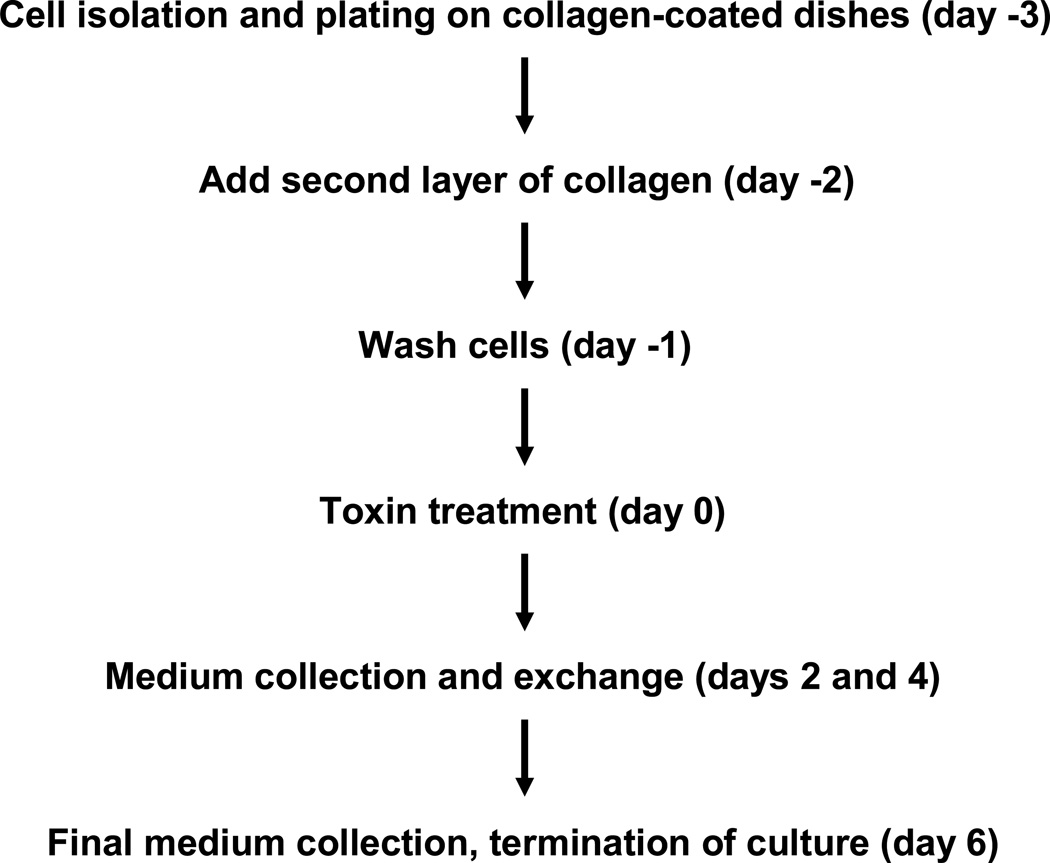
Cells were plated at a density of 100,000/well in a 24-well plate with 300 µL of medium and incubated at 37°C, 20% oxygen and 5% carbon dioxide. As Figure 1 shows, the medium was removed one day after plating and 55 µL of collagen solution was pipetted on top of cells to form the second layer of collagen. The collagen gelled within 1 h in the incubator and new medium was pipetted on the cells. On the following day the culture was washed four times with Hanks Buffer (Invitrogen, Carlsbad, CA) in order to remove contaminating proteins from the isolation procedure. In the control experiment without cells, collagen-coated plates were covered with 300 µL of medium, and underwent the same washing and medium exchange procedure as the plates with cells. Subsequently, the medium was changed every two days and samples for protein analysis were collected six days after washing.
Figure 1.
Protocol for preparation of collagen sandwiches and toxin treatment. For control cultures (no toxin treatment) there was a medium exchange on Day 0. For the “blank” cultures without cells, medium was pipetted on collagen-coated dishes on day −3 and the dishes were treated the same as the ones with cells.
Toxin treatment
Aflatoxin B1 was prepared fresh from powder (Sigma) before each experiment. The 1 mg powder in the sealed container was combined with 0.5 mL of DMSO using a syringe and needle. A sample of the solution was removed with the syringe, diluted 200fold with DMSO and its concentration was determined by UV absorption at 362 nm. The stock solution of aflatoxin was dissolved into DMSO at 1000 times the desired concentrations, and the final dilution was made into medium to ensure a DMSO concentration of 0.1 % (v/v). The control cells were treated with a 0.1% DMSO solution dissolved in medium. The cells were only exposed to toxin on Day 0 (Figure 1) and subsequent medium changes were carried out with toxin-free medium.
Albumin secretion
Albumin secretion was measured by enzyme-linked immunosorbent assay (ELISA) using sheep IgG fraction against rat albumin (ICN Pharmaceuticals, Costa Mesa, CA) and horseradish peroxidase-conjugated goat anti-rat IgG (Accurate Chemical, Westbury, NY). The absorbance was measured at 450 nm with a Spectramax 250 microplate spectrophotmeter (Molecular Devices Corp., Sunnyvale, CA). Medium samples were diluted 20–50-fold before measurement, and the exact concentrations were calculated after comparison with the absorbances of albumin standards (ICN Pharmaceuticals, Costa Mesa, CA).
Albumin removal
Two and a half milliliters of conditioned medium were concentrated to 100 µL using 3,000 Da cut-off microcon centrifugal filter units (Millipore, Billerica, MA). The immunoaffinity column was prepared by pipetting 200 µL of Protein-A agarose (Santa Cruz Biotechnology, Santa Cruz, CA) and 100 µL of rabbit anti-rat polyclonal antibody (Research Diagnostics Inc) into a 0.45 µm filter unit and mixing it for 2 h continuously. The column was washed 4 times with 500 µL of PBS to remove unbound antibody, and 200 µL of sample was added and incubated for 2 h while mixing continuously. The unbound sample was recovered by spinning and washing the column 2 times with 500 µL PBS. The unbound samples were pooled and concentrated to 30 µL using 3,000 Da cutoff microcon centrifugal filter units before being loaded onto the gel.
In-gel digestion
Gel electrophoresis was performed on a Protean III mini-gel apparatus (Biorad, Hercules, CA) using Tris-glycine, 8–16% gradient gel with 0.1% (w/v) SDS. Concentrated samples (30 µL) were mixed with 10 µL of 4X sample loading buffer (final buffer composition: 2% β-mercaptoethanol (v/v), 1% SDS (w/v), 12% glycerol (v/v), 50 mM Tris-HCl and a trace amount of bromophenol blue), boiled in water for 5 min, cooled and loaded directly into the gel. Gel electrophoresis was carried out under a constant current of 15 mA. The entire gel lane was cut into 30 equal sized gel slices, proteins were in-gel digested with trypsin and peptides were extracted as described by Shevchenko, et al., (25), with a few minor modifications as reported earlier (26). The extracted peptides were concentrated in a Speed-Vac to 10 µL, cleaned and desalted with C18 Zip-Tips (Millipore, Billerica, MA). The desalted peptides were dried completely in a Speed-Vac and redissolved in 0.6 µL of 0.1% TFA for LC-MS/MS analysis. For analysis of the hepatocyte medium, all 30 gel slices were analyzed individually. However, since the control medium had very low protein concentration, we consolidated the samples by combining peptides from consecutive slices after the extraction step, i.e., we combined peptides from slices 1 and 2, then slices 3 and 4, etc. resulting in 15 samples.
Silver staining and silver diamine staining
Fifty microliters of medium were concentrated and run on a 10% acrylamide one dimensional SDS-PAGE. Silver staining was performed as described by Shevchenko, et al, (25). Briefly, after electrophoresis the gel was fixed in 50% methanol and 5% acetic acid for 30 min and washed with 50% methanol for 10 min and then with water for 10 min. It was incubated for 1 min in 0.02% (w/v) sodium carbonate and washed twice with water for 1 min each time. The gel was incubated in chilled 0.1% silver nitrate solution for 20 min at 4 °C. It was then rinsed twice with water for 1 min each time and developed in 0.04% (w/v) formaldehyde in 2% (w/v) sodium thiosulfate solution with rocking. The staining was stopped with 5% acetic acid and then gels were stored in 1% acetic acid at 4 °C prior to in-gel digestion. For silver diamine staining the gel was fixed with three changes (1 h each) of 50% methanol containing 10% acetic acid, followed by 40% ethanol containing 5% acetic acid (fixative-2) for 1 h and oxidized with 0.7% periodate containing fixative-2 for 15 min. The gel was washed with three changes of water for 10 min each time. Afterwards, the gel was rocked in 150 mL silver diamine (5 mL 20% (w/v) silver nitrate solution, 28 mL 0.1M NaOH, 2mL of concentrated ammonium hydroxide and 115 mL deionized water) for 30 min and washed with water three times, 10 min each time. Development was done with developing solution (0.05% (w/v) formaldehyde and 0.005% (w/v) citric acid) and terminated with a 2.5% (v/v) acetic acid containing 12.5% (v/v) ethanol.
In-blot digestion
After gel electrophoresis, proteins were transferred to polyvinylidenedifluoride (PVDF) membrane (Millipore, Billerica, MA) at 30 volts and 4°C overnight. The membrane was stained with 0.1% (w/v) coomassie blue R-250. The protein that was down-regulated in aflatoxin treated cells, was excised from the control sample, destained, air-dried at room temperature and stored at −20°C until use. The membrane was wet with 1–2 µL of methanol and 100 µL of water. Then, 400 µL of methanol and 100 µL of dichloromethane were added stepwise while vortexing between steps. The protein was reduced with 100 µL of 0.5 M Tris, 10% acetonitrile, 5mM EDTA, 7mM dithiothreitol, pH 8.5 for 1h for 45°C. Free thiols were alkylated with 10 µL of 200mM iodoacetic acid in 0.5M NaOH. The membrane was blocked with 0.25% PVP-360 in 0.1% acetic acid for 20 min at room temperature and digested with trypsin (0.5µg in 100mM ammonium bicarbonate in 10% acetonitrile) at 37°C for 18 h. The supernatant was collected and washed with two changes of digestion buffer without trypsin, dried, and desalted with an in-house POROS-R2 (Applied Biosystem, Framingham, MA) capillary column.
Western blotting
After gel electrophoresis, proteins were transferred to a PVDF membrane at 30 volts for 2 h at room temperature. The membrane was blocked overnight in 5% (w/v) milk. Primary antibodies for α-l-antitrypsin and α2-macroglobulin, which were conjugated to horseradish peroxidase (Research Diagnostics Incorporated, Flanders, NJ) were diluted 1000 and 10,000-fold respectively. The membranes were incubated with antibodies for 1 h and were washed with a solution containing 0.5% (v/v) Tween and 0.1% (v/v) Triton-X 100. The membranes were incubated with a chemiluminescent substrate, Super Signal (Pierce Biotechnology, Rockford, IL) for 5 min. To visualize the proteins the membranes were exposed to x-ray film for 5–10 s.
Mass spectrometry
Chromatography for LC-MS/MS
Fused-silica capillary columns (75 µm i.d. × 360 µm o.d.; 14 cm length, tip 8 µm, New Objective, Worburn, MA) were packed in-house, with 5-µm C18 reverse-phase material (Protein&peptide C18; Vydac, Hesperia, CA) as described (27,28). The peptides were loaded onto the columns with a Rheodyne injector (0.5 µL internal loop) and separated with a 90 min linear gradient of 0 to 80% buffer B (0.08% TFA in 95% acetonitrile containing 5% methanol), at a flow rate of 120 nL/min, and then ramped back to the initial conditions [100% buffer A (0.1% TFA in 5% acetonitrile)] over an additional 60 min.
Tandem mass spectrometry
Full-scan MS spectra were acquired over the m/z range of 400–2000 using an Applied Biosystems (Framingham, MA) QSTAR XL quadrupole-time of flight (TOF) mass spectrometer equipped with a nanospray source (Proxeon Biosystems, Odense, Denmark). The electrospray interface design uses a micro-tee (Upchurch Scientific, Oak Harbor, WA) with a 1-inch piece of platinum rod, which was inserted into one stream of the micro-tee, to supply the electrical connection. The electrospray voltage was typically 2.3–2.7 kV. Data-dependent MS/MS analysis was performed on the three most intense peaks in each full-scan spectrum, using double and triple charge-states, with an exclusion time of 1 minute. Accumulation time and pulsar frequency were maintained at 3 s and 6.99 respectively; the mass tolerance was 50 mmu and the collision gas pressure setting was 6.
MALDI-TOF mass spectrometry
MALDI-TOF spectra were recorded on a PerSeptive Biosystems Voyager-DE STR in linear and reflector modes. MALDI matrix solution containing 10mg of nitrocellulose, 20 mg of α-cyanohydroxy cinnamic acid (CHCA) in 500µL of acetone and 500µL of 2-propanol as described elsewhere (29). Peptide mass fingerprinting searches were performed with Protein Prospector (http://prospector.ucsf.edu) or Mascot (http://www.matrixscience.com).
Data Processing and Analysis
Tandem MS spectra were searched against the National Center for Biological Information non-redundant (NCBInr) rat, bovine and complete mammalian proteome databases, using Agilent Spectrum Mill (30). This software includes a Data Extractor function that identifies good quality MS/MS spectrum for peptides by seeking CID fragment differences that correspond to known amino acids (sequence tag length >1) and thus functions as a filter to discard spectra that are unlikely to arise from peptides. By doing this, Spectrum Mill reduces the number of MS/MS spectra by about 35–40% and minimizes false positive identifications prior to searching the protein databases. To further minimize false-positives, the extracted MS/MS spectra were searched against the NCBInr databases for tryptic peptides only, with one allowed missed cleavage, in ‘identity mode’ to find unmodified peptides. This step was repeated in ‘homology-multi mode’ to search for mutations, post-translational modifications and chemical modifications. Search parameters were as follows: MS and MS/MS tolerance of 100 and 500 ppm, tryptic specificity allowing for up to one missed cleavage and fixed modification of carbamidomethylation of cysteine in identity mode and variable modification of oxidation of methionine (m), pyro-glutamic acid (q) and phosporylation of serine (s), threonine (t) and tyrosine (y) in homology-multi mode (mqsty). Spectrum Mill incorporates an algorithm that generates numerical scores for quality of identification for both peptides and proteins. The default thresholds considered to represent high-quality results are as follows: a.) in protein details mode; protein score >20, peptide score (scored percent intensity [SPI]) charge +1 (>9, >70%), peptide charge +2 (>9, >70%), peptide charge +3 (>9, >70%), peptide charge +4 (>9, >70%); b.) in peptide mode: SPI charge +1 (>13, >70%), peptide charge +2 (>13, >70%), peptide charge +3 (>13, >70%), peptide charge +4 (>13, >70%) . All autovalidations for our data were based on these thresholds. Additionally, spectra with lower scores (>9, >65%) were evaluated by visual examination and only good quality spectra were validated (acceptable signal-to-noise and the presence of at least three consecutive b or y ion fragments). The above parameters result in a protein being considered identified when either multiple spectra of moderate-to-good quality or at least 1 spectrum of excellent-to-good quality are obtained. Only proteins identified according to the above criteria were considered in developing the conclusions with respect to potential biomarkers. In cases of multiple protein database entries, based on the same set of peptides, only a single entry (highest molecular weight) was considered. A BLAST search was performed to confirm the identities for any single-peptide proteins. The proteins were further classified according to the Swiss-Prot and TrEMBL protein databases (us.expasy.org/sprot/).
Results
Albumin Depletion of Medium
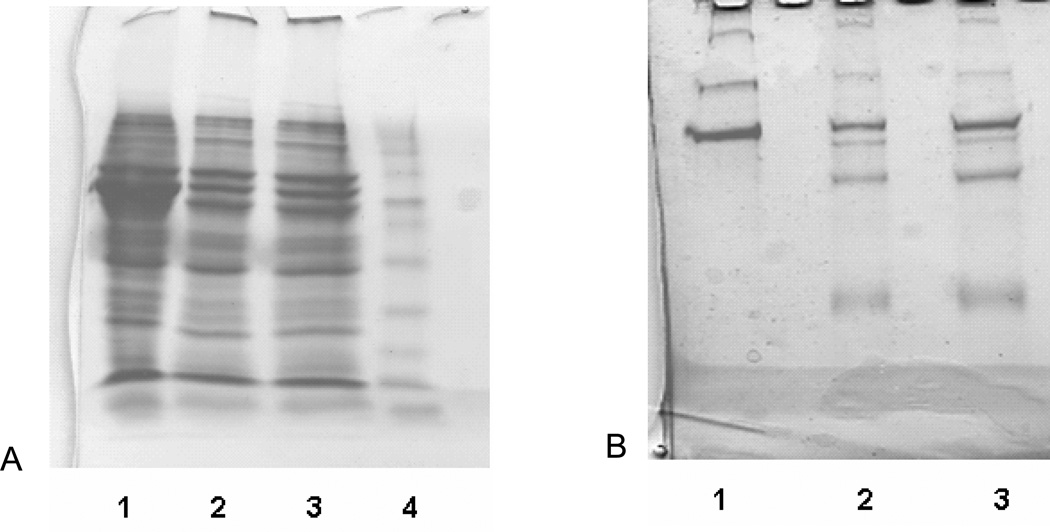
Lane 1 in Figures 2A and 2B correspond to 2.5 mL of hepatocyte and control medium respectively. Lanes 2 and 3 are duplicates of the medium depleted of albumin. Using the molecular weight markers it was estimated that most of the protein depletion is between 40 and 80 kDa. Using the ELISA albumin assay we estimated that this method we can remove up to 85% of the albumin (data not shown).
Figure 2.
2.5 ml of albumin depleted medium on an 8–16% gradient gel. (A) Hepatocyte medium, lane 1: before depletion, lanes 2 and 3: after depletion, lane 4: marker. (B) Control medium, lane 1: before depletion, lanes 2 and 3: after depletion.
Protein Profile
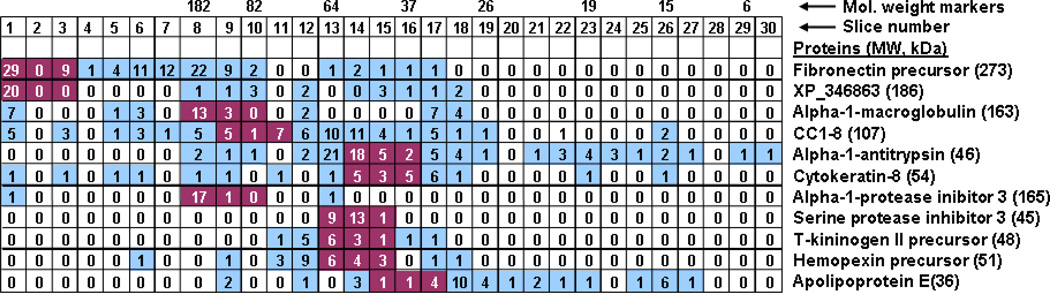
Proteins from hepatocyte medium were searched against the NCBInr rat proteome database. From the 30 slices we identified 604 peptides belonging to 204 proteins (the complete list of proteins is shown in the supplementary material.). Using the molecular weight markers we made a calibration curve to relate the gel slices with the molecular weights of proteins that they would be expected to contain (calibration curve not shown). Then, distribution of peptides for the proteins with the highest scores was mapped along the SDS-PAGE gel highlighting the regions where the peptides would be expected to be found based on the molecular weights of the proteins (Figure 3). These results show that many of the peptides are not found in the slices where they would be expected. This phenomenon is possibly due to proteolysis occurring over the course of the incubation period leading to proteins appearing at lower than expected molecular weight markers, and post-translational modifications (e.g. glycosylation) resulting in proteins with higher than expected molecular weights.
Figure 3.
Distribution of peptides across slices 1–30 for the proteins with the highest scores. Each square indicates the number of distinct peptides found in that slice. Based on the molecular weights of the proteins, the squares were shaded red if proteins would be expected to be found in that slice. The blue squares indicate the slices where peptides were detected, but where they were not expected to be found.
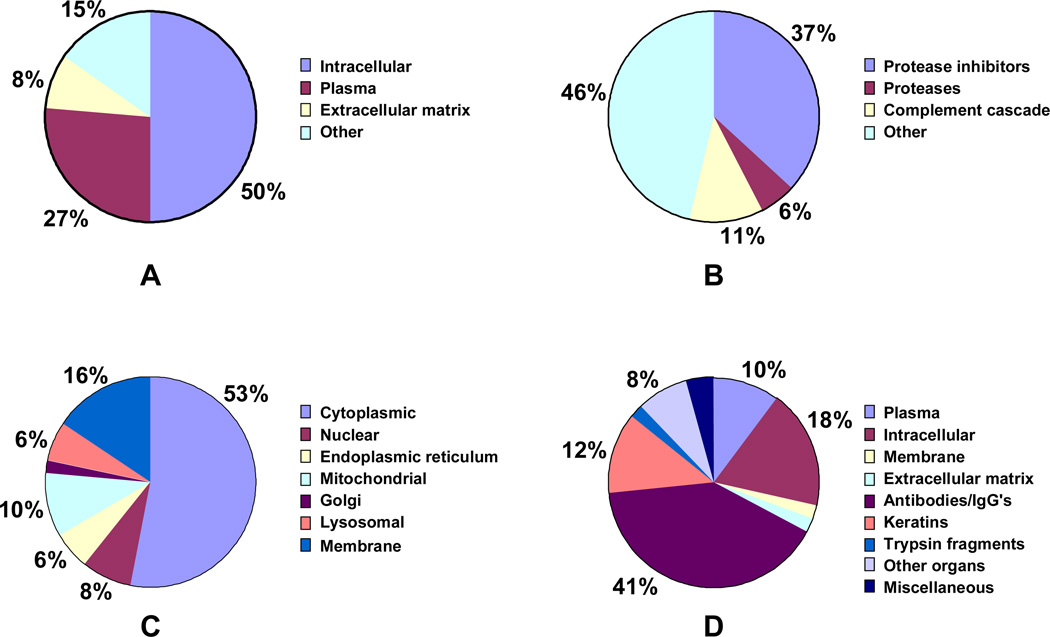
Of the 204 hepatocyte proteins, 50% were intracellular or membrane proteins, 26% were plasma proteins, 8% were extracellular matrix (ECM) proteins and 15% were miscellaneous (Figure 4A). Proteins were sorted according to their score, which was calculated with a Spectrum Mill algorithm based on the intensity and the signal-to-noise ratio of the peptide spectra. Proteins with scores higher than 20 were considered valid, and proteins with lesser scores were manually validated. Proteins with scores less than 20 were considered valid only if they were single-peptide proteins and their score was higher than 13. The protein with the highest score was fibronectin precursor (score of 551.57) but several plasma proteins were also present with high scores: α1-macroglobulin (271.85), α1-antitrypsin (212.75) and α1-proteinase inhibitor III (176.64). Of the 54 plasma proteins about 20 were protease inhibitors, and the rest included proteases, complement cascade proteins, metal-binding proteins and apolipoproteins (Figure 4B). Half of the proteins were either intracellular or membrane proteins, with proteins originating from various organelles including mitochondria, lysosomes, Golgi apparatus and the nucleus (Figure 4C). Proteins were classified as cytoplasmic if no specific organelle was listed or if they could be both intracellular and secreted. Furthermore, a number of proteins were found in both the cytoplasm and a specific organelle, in which case the protein was classified with the specific organelle. Several extracellular matrix proteins were also identified, including fibronectin, laminin and collagen type XVIII. Most of the miscellaneous proteins had unknown functions, while some had only been identified previously in other organs such as the central nervous system.
Figure 4.
Qualitative distribution of proteins in conditioned medium. (A) Total proteins in hepatocyte medium (B) Secreted proteins in hepatocyte medium (C) Intracellular proteins in hepatocyte medium (D) Proteins in blank medium.
The sample from the control medium, was analyzed using the complete mammalian database. The collagen was of bovine origin, but the bovine database is not well-annotated, so the complete mammalian database was searched. Of the 49 valid proteins (see supplementary material for a complete list of proteins) 20 were rabbit IgG’s probably due to the albumin depletion step, 10 were intracellular proteins or membrane proteins and there were several bovine proteins such as casein and albumin, most likely originating from the collagen matrix, which is also of bovine origin. In order to determine which proteins are common to the hepatocyte and the control medium, we analyzed both samples with the complete mammalian database and looked for common proteins (Table 1). There were 17 common proteins, including human keratin, human transferrin, rabbit IgG’s and bovine casein.
Table 1.
Common proteins between control and hepatocyte medium. The number of distinct peptides found per protein is shown for each sample in the corresponding columns
| Protein | Control | Hepatocyte | Mol. weight (kDa) | Species |
|---|---|---|---|---|
| Serum albumin | 22 | 174 | 69293.9 | Bovine |
| Keratin 1 | 136 | 65 | 66067 | Human |
| Transferrin | 25 | 51 | 77050.4 | Human |
| KRT10 protein | 72 | 42 | 58827.3 | Human |
| Alpha-S1 casein precursor | 28 | 25 | 24529.1 | Bovine |
| Cytokeratin 9 | 22 | 15 | 62129.7 | Human |
| Trypsin precursor | 17 | 13 | 24409.6 | Pig |
| Kappa-casein precursor | 10 | 12 | 17860.3 | Bovine |
| Serum albumin precursor | 5 | 9 | 68910.3 | Rabbit |
| Histidine-rich glycoprotein precursor | 3 | 7 | 58877.2 | Rabbit |
| Alpha-2-HS-glycoprotein precursor | 1 | 6 | 38387.1 | Rabbit |
| Ig gamma heavy chain constant region | 17 | 5 | 20576.3 | Rabbit |
| Alpha-S2 casein precursor | 4 | 4 | 26018.8 | Bovine |
| Ig mu heavy chain | 4 | 2 | 64455.2 | Rabbit |
| Immunoglobulin kappa chain | 9 | 1 | 25082.2 | Rabbit |
| Collagen alpha 1(I) chain precursor | 4 | 1 | 138763 | Dog |
| Hemoglobin beta chain | 1 | 1 | 16073.5 | Sheep |
| Bullous pemphigoid antigen 1 eA | 2 | 1 | 590881.8 | Human |
| Similar to rootletin | 1 | 1 | 234759 | Rat |
Analysis of conditioned medium from aflatoxin-treated cells
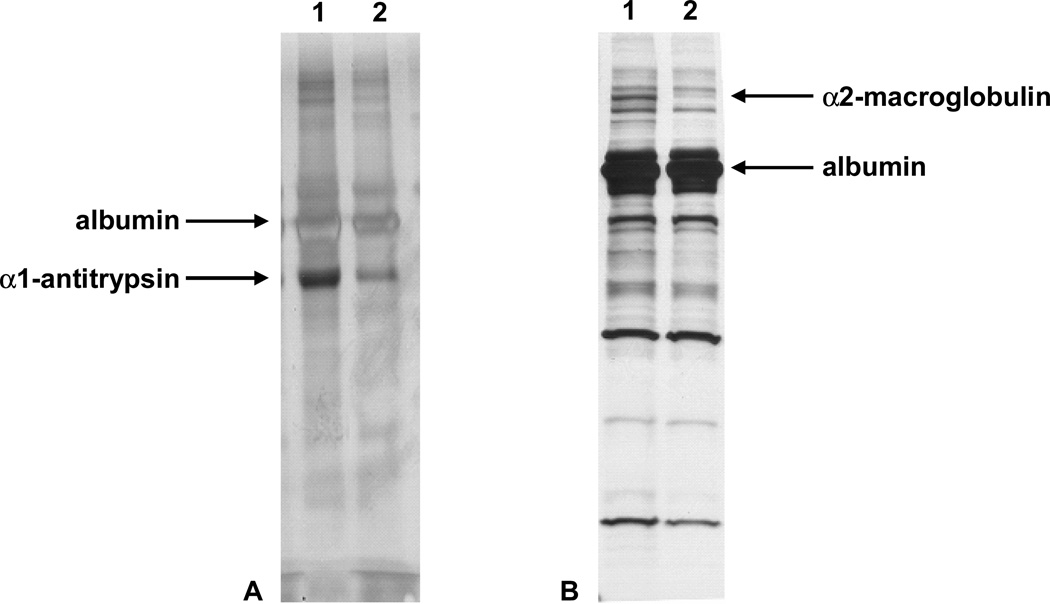
We noted a difference in the expression of two proteins in the media from control and aflatoxin-treated cells. Silver staining (Figure 5B) and silver diamine staining (Figure 5A) demonstrate diminished levels in a 55kDa protein (α1-antitrypsin) and a 180 kDa protein (α2-macroglobulin) respectively. It is interesting to note, that a decrease in α1-antitrypsin was only visible with silver diamine staining, not silver staining. Peptides were extracted with in-blot digestion (α1-antitrypsin) and in-gel digestion (α2-macroglobulin) for protein identification with mass spectrometry.
Figure 5.
SDS-PAGE gels of medium from control and aflatoxin-treated cells. Lane 1: 0.1% DMSO (vehicle), lane 2: 12 nM aflatoxin B1. (A) Silver diamine staining 12 days after toxin treatment. Both lanes were loaded with 100 µl of medium. (B) Silver staining 10 days after toxin treatment, 250 µl from control cells and 280 µl of toxin-treated cells.
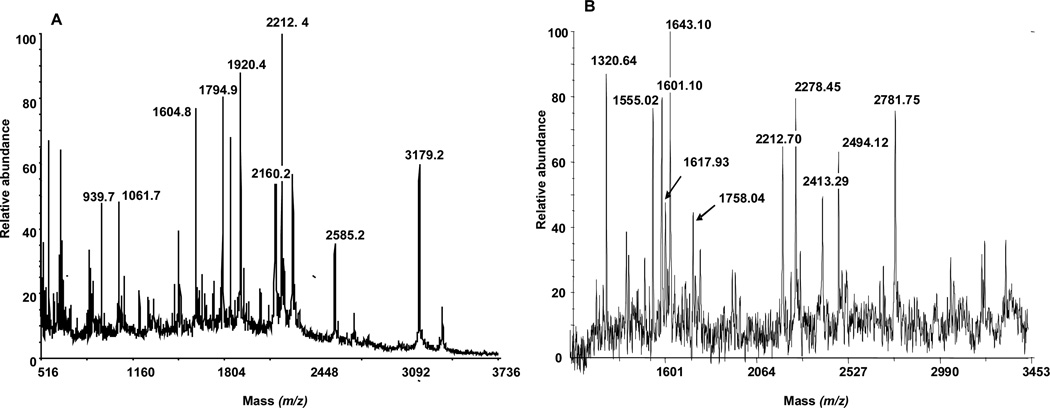
Figure 6 shows the MALDI-TOF spectra of the extracted peptides. The digestion of the 55 kDa protein resulted in peptides with the following m/z values: 939.7, 1061.7, 1604.8, 1794.9, 1920.4, 2160.2, 2585.2 and 3179.2 (Figure 6A). The digestion of the 180 kDa gave rise to peptides with m/z values of 1320.64, 1555.02, 1601.10, 1643.1, 1617.93, 1758.04, 2278.45, 2413.29, 2494.12 and 2781.75 (Figure 6B.) Using the Swiss Protein Database, we identified these proteins as α1-antitrypsin and α2-macroglobulin respectively. The identities of the proteins were verified with tandem mass spectrometry.
Figure 6.
MALDI-TOF spectra of proteins excised from coomassie blue stained blot (A) and silver stained gel (B) respectively.
(A) m/z values of 939.7, 1061.7, 1604.8, 1794.9, 1920.4, 2160.2, 2585.2 and 3179.2 corresponded to a1-antitrypsin.
(B) a2-macroglobulin was idenitified based on ions at m/z : 1320.64, 1555.02, 1601.10, 1643.1, 1617.93, 1758.04, 2278.45, 2413.29, 2494.12 and 2781.75.
Trypsin fragments with m/z values of 2212 and 2284, resulted from autodigestion, and served as internal calibrants.
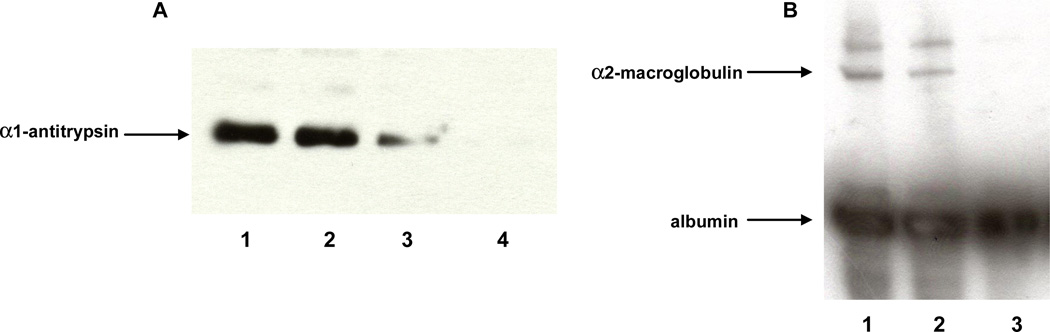
The dose-dependent down-regulation of the proteins was investigated with Western Blotting (Figure 7). Figures 7A and 7B show the Western blots of conditioned medium for α1-antitrypsin and α2-macroglobulin respectively six days after toxin-treatment. As the figures show, a 6 nMolar dose of aflatoxin does not change the secretion of these proteins significantly. A 15 nM dose decreases α1-antitrypsin at least 50%, and almost completely down-regulates α2-macroglobulin. A 30 nM dose decreases α1-antitrypsin to undetectable levels.
Figure 7.
(A) Western Blot of α1-antitrypsin 6 days after aflatoxin treatment. Lane 1: 0.1% DMSO (vehicle), lane 2: 6 nM aflatoxin B1, lane 3: 15 nM aflatoxin B1 Lane 4: 30 nM aflatoxin B1. (B) Western Blot of α2-macroglobulin and albumin 6 days after aflatoxin treatment. Lane 1: 0.1% DMSO (vehicle), lane 2: 6 nM aflatoxin B1, lane 3: 15 nM aflatoxin B1. The top two bands show the bottom band shows albumin, which we used to normalize gel loading.
Discussion
We have characterized a comprehensive proteomics profile of conditioned medium from rat hepatocytes in collagen sandwiches as well as control medium incubated over collagen alone. We were able to identify over 200 proteins in hepatocyte medium including plasma, extracellular, and intracellular proteins. The results show that hepatocytes secrete many plasma proteins in the collagen sandwich including proteases, protease inhibitors, proteins from the complement cascade and a number of transport plasma proteins such as selenoprotein P, Cc1–8 (iron-binding), ceruloplasmin precursor (copper-binding), and apolipoproteins. Our work clarified some of the mystery surrounding the survival of hepatocytes in collagen sandwiches. Beken, et al., (32) compared albumin secretion and the expression of glutathione S-transferases in collagen sandwiches made from rat tail collagen, commercially available collagen type I, and commercially available collagen supplemented with fibronectin, laminin and heparan sulfate proteoglycans, and found that after seven days of culture, there was no significant difference among the cultures. They hypothesized that hepatocytes cultured in collagen sandwiches secrete the necessary extra-cellular proteins and therefore do not need other types of matrix proteins for survival. Our data support this hypothesis and demonstrate that hepatocytes do secrete a number of matrix proteins including fibronectin precursor, laminin, collagen type XVIII and heparan sulfate proteoglycans. Initially, we hypothesized that the extracellular matrix proteins were contaminants from the collagen, but our results show that the only structural matrix protein in the control medium is a collagen alpha 1 precursor, which is the protein with which the plates were coated.
We also detected some proteins known to be produced by dedifferentiated cells during tissue repair and regeneration, which is consistent with earlier observations that hepatocyte isolation by collagenase-perfusion and addition of EGF to the medium can cause a down-regulation of transcription of hepatocyte-specific genes leading to a dedifferentiated phenotype mimicking tissue regeneration (39). One such protein was hepatocyte growth factor activator protein, which is usually produced as a zymogen and converted to its active form during tissue injury, in order to activate hepatocyte growth factor (HGF) (29). Another protein was connective tissue growth factor (CTGF), which can be produced by a number of cell types in the liver including fibroblasts, endothelial cells and stellate cells (30). CTGF is usually upregulated by TGF-β, but its production can also be stimulated by EGF (31), which is present in our medium. We also detected thrombospondin-1, which has been found near hepatocytes in congenital hepatic fibrosis tissue (32) and NG2, a chondroitin sulfate proteoglycan, which is usually associated with incompletely-differentiated or immature cells (33). NG2, originally though to be associated with precursor cells in the brain, is now known to be expressed in cartilage, muscle and bone (34), although to our knowledge it has not been isolated from liver. Another identified protein, reelin, has been regarded until recently as a protein involved in brain development, although it is now known to be also produced by the liver (35). In fact, it has been shown that reelin is produced by stellate cells during hepatic tissue repair, for example following CCl4 exposure (36).
The conditioned hepatocyte medium also contained two heparan sulfate proteoglycans (HSPG). The first HSPG was a precursor to agrin, a proteoglycan found in the basement membrane at neuromuscular junctions, which binds to dystroglycan, part of the dystrophin-glycoprotein complex (37). We were also able to identify dystroglycan, which was surprising, because the reported levels of both agrin and dystroglycan in the liver are very low (37). In fact, dystroglycan has only been detected in the liver from lysates of stellate cells, and its expression is usually up-regulated during liver fibrosis and regeneration (38). The second HSPG was syndecan-4 precursor, which is primarily expressed in the liver on the membranes of endothelial cells. Thus, these data suggest the presence of stellate and endothelial cells in the culture, although this is not surprising, since primary cell preparations frequently contain up to 5% of non-parenchymal cells. Stellate cells, in particular, are known to be involved in the remodeling of the extracellular matrix (39).
One application of proteome analysis is the identification of proteins that are down-regulated after exposure to a toxic compound. There are several challenges associated with characterizing the proteome of toxin-treated and control cells. First, the quantitation of proteins with mass spectrometry is very labor-intensive. The current approach for quantitative proteome analysis involves the preparation of isotope-coded affinity tags (ICAT) and differentially labeling the two samples, making it possible to use them as relative internal standards against each other (40). However, there are few positive controls for such experiments, since the up-regulation and down-regulation of proteins after toxin treatment has not been well-characterized. Another challenge is that the conditions of the toxin exposure (dose of toxin, time after exposure) need to be determined so that the dose is toxic but not lethal and the time interval allows for changes in protein expression. The toxicity of aflatoxin B1 in collagen sandwiches was previously investigated and the LD50 was found to be about 32 nM according to urea and albumin analysis (41). The doses for the aflatoxin treatment in this study were chosen accordingly, and it was found that α1-antitrypsin and α2-macroglobulin were not down-regulated at 6 nM, but they were undetectable at 30 nM. This result was found to be reproducible over approximately 10 experiments. It was also noted that day 6 after the toxin treatment was the earliest time-point when differences in α2-macroglobulin expression could be measured since this protein could not be detected in the medium of the control cells until that time. Interestingly, differences in the expression of αl-antitrypsin were evident just two days after toxin exposure. It was also observed that the toxin-treated cells did not resume production of these proteins even 20 days after exposure.
Our initial hypothesis was that exposure to aflatoxin B1 decreased the secretion of all proteins equally. Our Western blots, however, were normalized to secreted albumin by loading the samples such that the amount of albumin was the same in all lanes. These results suggest that α1-antitrypsin and α2-macroglobulin are specifically down-regulated after aflatoxin B1 exposure. Since these proteins are acute-phase proteins, one would expect that their expression would increase after toxic exposure; the opposite, however, is true. Interestingly, it has been found by others that the expression of α2-macroglobulin decreases after exposing rats to carbon tetrachloride (7), suggesting that a decrease in this protein could be a marker of toxicity in vivo. It is possible that one of the mechanisms through which aflatoxin B1 causes toxicity is through the down-regulation of these protease inhibitors by leading to uncontrolled breakdown of liver tissue. In fact, one of the risk factors for the development of hepatocellular carcinoma is a genetic deficiency in α1-antitrypsin (42). Thus, this study suggests that a proteomics approach can lead to the identification of new biomarkers and a better understanding of the mechanisms leading to hepatotoxicity.
Supplementary Material
Acknowledgements
This work was supported by the DuPont-MIT Alliance, a training grant from the National Institute of Environmental Health Sciences (T32-ES07020) and by the MIT Center for Environmental Health Sciences (NIEHS grant P30-ES02109)
Footnotes
Abbreviations: DMSO, dimethyl sulfoxide; CTGF, connective tissue growth factor; ECM, extracellular matrix; EGF, epidermal growth factor; ELISA, enzyme-linked immunosorbent assay; HSPG, heparan sulfate proteoglycan, MALDI, matrix-assisted laser desorption ionization; NCBInr , National Center for Biological Information non-redundant database; TGF-β, transforming growth factor beta; TOF, time-of-flight.
Supporting Information Available: Complete list of proteins in conditioned hepatocyte medium and “control” medium (incubated on collagen)
References
- 1.He QY, Chiu JF. Proteomics in biomarker discovery and drug development. J.Cell Biochem. 2003;89:868–886. doi: 10.1002/jcb.10576. [DOI] [PubMed] [Google Scholar]
- 2.Zimmerman HJ. Hepatotoxicity. New York: Lippincott Williams & Wilkins; 1999. [Google Scholar]
- 3.Kim J, Kim SH, Lee SU, Ha GH, Kang DG, Ha NY, Ahn JS, Cho HY, Kang SJ, Lee YJ, Hong SC, Ha WS, Bae JM, Lee CW, Kim JW. Proteome analysis of human liver tumor tissue by two-dimensional gel electrophoresis and matrix assisted laser desorption/ionization-mass spectrometry for identification of disease-related proteins. Electrophoresis. 2002;23:4142–4156. doi: 10.1002/elps.200290032. [DOI] [PubMed] [Google Scholar]
- 4.He QY, Chen J, Kung HF, Yuen AP, Chiu JF. Identification of tumor-associated proteins in oral tongue squamous cell carcinoma by proteomics. Proteomics. 2004;4:271–278. doi: 10.1002/pmic.200300550. [DOI] [PubMed] [Google Scholar]
- 5.Somiari RI, Sullivan A, Russell S, Somiari S, Hu H, Jordan R, George A, Katenhusen R, Buchowiecka A, Arciero C, Brzeski H, Hooke J, Shriver C. High-throughput proteomic analysis of human infiltrating ductal carcinoma of the breast. Proteomics. 2003;3:1863–1873. doi: 10.1002/pmic.200300560. [DOI] [PubMed] [Google Scholar]
- 6.Fountoulakis M, Suter L. Proteomic Analysis of Rat Liver. J. Chromatogr. B. 2002;782:197–218. doi: 10.1016/s1570-0232(02)00562-7. [DOI] [PubMed] [Google Scholar]
- 7.Fountoulakis M, de Vera MC, Crameri F, Boess F, Gasser R, Albertini S, Suter L. Modulation of gene and protein expression by carbon tetrachloride in the rat liver. Toxicol.Appl.Pharmacol. 2002;183:71–80. doi: 10.1006/taap.2002.9460. [DOI] [PubMed] [Google Scholar]
- 8.Ruepp SU, Tonge RP, Shaw J, Wallis N, Pognan F. Genomics and proteomics analysis of acetaminophen toxicity in mouse liver. Toxicol.Sci. 2002;65:135–150. doi: 10.1093/toxsci/65.1.135. [DOI] [PubMed] [Google Scholar]
- 9.Man WJ, White IR, Bryant D, Bugelski P, Camilleri P, Cutler P, Heald G, Lord PG, Wood J, Kramer K. Protein expression analysis of drug-mediated hepatotoxicity in the Sprague-Dawley rat. Proteomics. 2002;2:1577–1585. doi: 10.1002/1615-9861(200211)2:11<1577::AID-PROT1577>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 10.Ben Ze’ev A, Robinson GS, Bucher NL, Farmer SR. Cell-cell and cell-matrix interactions differentially regulate the expression of hepatic and cytoskeletal genes in primary cultures of rat hepatocytes. Proc.Natl.Acad.Sci.U.S.A. 1988;85:2161–2165. doi: 10.1073/pnas.85.7.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeCluyse EL, Bullock PL, Parkinson A. Strategies for restoration and maintenance of normal hepatic structure and function in long-term cultures of rat heaptocytes. Adv.Drug Del.Rev. 1996;22:133–186. [Google Scholar]
- 12.Gomez-Lechon MJ, Jover R, Donato T, Ponsoda X, Rodriguez C, Stenzel KG, Klocke R, Paul D, Guillen I, Bort R, Castell JV. Long-term expression of differentiated functions in hepatocytes cultured in three-dimensional collagen matrix. J.Cell Physiol. 1998;177:553–562. doi: 10.1002/(SICI)1097-4652(199812)177:4<553::AID-JCP6>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 13.Bader A, Fruhauf N, Zech K, Haverich A, Borlak JT. Development of a small-scale bioreactor for drug metabolism studies maintaining hepatospecific functions. Xenobiotica. 1998;28:815–825. doi: 10.1080/004982598239074. [DOI] [PubMed] [Google Scholar]
- 14.Kern A, Bader A, Pichlmayr R, Sewing KF. Drug metabolism in hepatocyte sandwich cultures of rats and humans. Biochem.Pharmacol. 1997;54:761–772. doi: 10.1016/s0006-2952(97)00204-9. [DOI] [PubMed] [Google Scholar]
- 15.Rotem A, Matthew HWT, Hsiao PH, Toner M, Tompkins RG, Yarmush ML. The Activity of Cytochrome P450IA1 in Stable Cultured Rat Hepatocytes. Toxicol.In Vitro. 1995;9:139–149. doi: 10.1016/0887-2333(94)00207-b. [DOI] [PubMed] [Google Scholar]
- 16.LeCluyse E, Madan A, Hamilton G, Carroll K, DeHaan R, Parkinson A. Expression and regulation of cytochrome P450 enzymes in primary cultures of human hepatocytes. J.Biochem.Mol.Toxicol. 2000;14:177–188. doi: 10.1002/(sici)1099-0461(2000)14:4<177::aid-jbt1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Moghe PV, Berthiaume F, Ezzell RM, Toner M, Tompkins RG, Yarmush ML. Culture matrix configuration and composition in the maintenance of hepatocyte polarity and function. Biomaterials. 1996;17:373–385. doi: 10.1016/0142-9612(96)85576-1. [DOI] [PubMed] [Google Scholar]
- 18.Ezzell RM, Toner M, Hendricks K, Dunn JCY, Tompkins RG, Yarmush ML. Effect of Collagen Gel Configuration on the Cytoskeleton in Cultured Rat Hepatocytes. Exp.Cell Res. 1993;208:442–452. doi: 10.1006/excr.1993.1266. [DOI] [PubMed] [Google Scholar]
- 19.Moghe PV, Ezzell RM, Toner M, Tompkins RG, Yarmush ML. Role of beta 1 integrin distribution in morphology and function of collagen-sandwiched hepatocytes. Tissue Eng. 1997;3:1–16. [Google Scholar]
- 20.Liu X, LeCluyse EL, Brouwer KR, Gan LS, Lemasters JJ, Stieger B, Meier PJ, Brouwer KL. Biliary excretion in primary rat hepatocytes cultured in a collagen-sandwich configuration. Am.J.Physiol. 1999;277:G12–G21. doi: 10.1152/ajpgi.1999.277.1.G12. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, LeCluyse EL, Brouwer KR, Gan LSL, Brouwer KLR. Biliary excretion of taurocholate (TC) in rat hepatocytes cultured in a collagen sandwich configuration (SC) Hepatology. 1996;24:973. [Google Scholar]
- 22.Liu X, Brouwer KL, Gan LS, Brouwer KR, Stieger B, Meier PJ, Audus KL, LeCluyse EL. Partial maintenance of taurocholate uptake by adult rat hepatocytes cultured in a collagen sandwich configuration. Pharm.Res. 1998;15:1533–1539. doi: 10.1023/a:1011994831139. [DOI] [PubMed] [Google Scholar]
- 23.Powers MJ, Janigian DM, Wack KE, Baker CS, Beer SD, Griffith LG. Functional behavior of primary rat liver cells in a three-dimensional perfused microarray bioreactor. Tissue Eng. 2002;8:499–513. doi: 10.1089/107632702760184745. [DOI] [PubMed] [Google Scholar]
- 24.Block GD, Locker J, Bowen WC, Petersen BE, Katyal S, Strom SC, Riley T, Howard TA, Michalopoulos GK. Population expansion, clonal growth, and specific differentiation patterns in primary cultures of hepatocytes induced by HGF/SF, EGF and TGF alpha in a chemically defined (HGM) medium. J.Cell Biol. 1996;132:1133–1149. doi: 10.1083/jcb.132.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal.Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 26.Nikov G, Bhat V, Wishnok JS, Tannenbaum SR. Analysis of nitrated proteins by nitrotyrosine-specific affinity probes and mass spectrometry. Anal.Biochem. 2003;320:214–222. doi: 10.1016/s0003-2697(03)00359-2. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh S, Jorgenson JW. Preparation and evaluation of slurry-packed liquid chromatography microcolumns with inner diameters from 12 to 33 microns. Anal.Chem. 1996;68:1212–1217. doi: 10.1021/ac950682m. [DOI] [PubMed] [Google Scholar]
- 28.Soglia JR, Turesky RJ, Paehler A, Vouros P. Quantification of the heterocyclic aromatic amine DNA adduct N-(deoxyguanosin-8-yl)-2-amino-3-methylimidazo[4,5-f]quinoline in livers of rats using capillary liquid chromatography/microelectrospray mass spectrometry: a dose-response study. Anal.Chem. 2001;73:2819–2827. doi: 10.1021/ac010218j. [DOI] [PubMed] [Google Scholar]
- 29.Fountoulakis M, Langen H. Identification of proteins by matrix-assisted laser desorption ionization-mass spectrometry following in-gel digestion in low-salt, nonvolatile buffer and simplified peptide recovery. Anal.Biochem. 1997;250:153–156. doi: 10.1006/abio.1997.2213. [DOI] [PubMed] [Google Scholar]
- 30.Liao H, Wu J, Kuhn E, Chin W, Chang B, Jones MD, O’Neil S, Clauser KR, Karl J, Hasler F, Roubenoff R, Zolg W, Guild BC. Use of mass spectrometry to identify protein biomarkers of disease severity in the synovial fluid and serum of patients with rheumatoid arthritis. Arthritis Rheum. 2004;50:3792–3803. doi: 10.1002/art.20720. [DOI] [PubMed] [Google Scholar]
- 31.Shimomura T, Denda K, Kitamura A, Kawaguchi T, Kito M, Kondo J, Kagaya S, Qin L, Takata H, Miyazawa K, Kitamura N. Hepatocyte growth factor activator inhibitor, a novel Kunitz-type serine protease inhibitor. J.Biol.Chem. 1997;272:6370–6376. doi: 10.1074/jbc.272.10.6370. [DOI] [PubMed] [Google Scholar]
- 32.El Youssef M, Mu Y, Huang L, Stellmach V, Crawford SE. Increased expression of transforming growth factor-beta1 and thrombospondin-1 in congenital hepatic fibrosis: possible role of the hepatic stellate cell. J.Pediatr.Gastroenterol.Nutr. 1999;28:386–392. doi: 10.1097/00005176-199904000-00008. [DOI] [PubMed] [Google Scholar]
- 33.Levine JM, Nishiyama A. The NG2 chondroitin sulfate proteoglycan: a multifunctional proteoglycan associated with immature cells. Perspect.Dev.Neurobiol. 1996;3:245–259. [PubMed] [Google Scholar]
- 34.Burg MA, Tillet E, Timpl R, Stallcup WB. Binding of the NG2 proteoglycan to type VI collagen and other extracellular matrix molecules. J.Biol.Chem. 1996;271:26110–26116. doi: 10.1074/jbc.271.42.26110. [DOI] [PubMed] [Google Scholar]
- 35.Smalheiser NR, Costa E, Guidotti A, Impagnatiello F, Auta J, Lacor P, Kriho V, Pappas GD. Expression of reelin in adult mammalian blood, liver, pituitary pars intermedia, and adrenal chromaffin cells. Proc.Natl.Acad.Sci.U.S.A. 2000;97:1281–1286. doi: 10.1073/pnas.97.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobold D, Grundmann A, Piscaglia F, Eisenbach C, Neubauer K, Steffgen J, Ramadori G, Knittel T. Expression of reelin in hepatic stellate cells and during hepatic tissue repair: a novel marker for the differentiation of HSC from other liver myofibroblasts. J.Hepatol. 2002;36:607–613. doi: 10.1016/s0168-8278(02)00050-8. [DOI] [PubMed] [Google Scholar]
- 37.Gesemann M, Brancaccio A, Schumacher B, Ruegg MA. Agrin is a high-affinity binding protein of dystroglycan in non-muscle tissue. J.Biol.Chem. 1998;273:600–605. doi: 10.1074/jbc.273.1.600. [DOI] [PubMed] [Google Scholar]
- 38.Bedossa P, Ferlicot S, Paradis V, Dargere D, Bonvoust F, Vidaud M. Dystroglycan expression in hepatic stellate cells: role in liver fibrosis. Lab Invest. 2002;82:1053–1061. doi: 10.1097/01.lab.0000024429.73158.de. [DOI] [PubMed] [Google Scholar]
- 39.Sato M, Suzuki S, Senoo H. Hepatic stellate cells: unique characteristics in cell biology and phenotype. Cell Struct.Funct. 2003;28:105–112. doi: 10.1247/csf.28.105. [DOI] [PubMed] [Google Scholar]
- 40.Gygi SP, Rist B, Griffin TJ, Eng J, Aebersold R. Proteome analysis of low-abundance proteins using multidimensional chromatography and isotope-coded affinity tags. J.Proteome Res. 2002;1:47–54. doi: 10.1021/pr015509n. [DOI] [PubMed] [Google Scholar]
- 41.Farkas D, Tannenbaum SR. Characterization of Chemically Induced Hepatotoxicity in Collagen Sandwiches of Rat Hepatocytes. Toxicol.Sci. 2005 doi: 10.1093/toxsci/kfi145. in press. [DOI] [PubMed] [Google Scholar]
- 42.Blum HE. Molecular targets for prevention of hepatocellular carcinoma. Dig.Dis. 2002;20:81–90. doi: 10.1159/000063163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.