Summary
Type-I interferons (IFN-I) are essential antiviral cytokines produced upon microbial infection. IFN-I elicits this activity through the upregulation of hundreds of IFN-I stimulated genes (ISGs). The full breadth of ISG induction demands activation of a number of cellular factors including the IκB kinase epsilon (IKKε). However, the mechanism of IKKε activation upon IFN receptor signaling has remained elusive. Here we show that TRIM6, a member of the E3-ubiquitin ligase tripartite motif (TRIM) family of proteins, interacts with IKKε and promotes induction of IKKε-dependent ISGs. TRIM6 and the E2-ubiquitin conjugase UbE2K cooperate in the synthesis of unanchored K48-linked poly-ubiquitin chains, which activate IKKε for subsequent STAT1 phosphorylation. Our work attributes a previously unrecognized activating role of K48-linked unanchored poly-ubiquitin chains in kinase activation and identifies the UbE2K-TRIM6-ubiquitin axis as critical for IFN signaling and antiviral response.
Introduction
Innate immune signaling pathways are activated when pathogen associated molecular patterns (PAMPs) contained in microbial products are recognized by host cell pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) and RIG-I-like receptors (RLRs) (Medzhitov et al., 1997; Meylan et al., 2006). Activation of these pathways ultimately results in production of type I interferons (IFN-I) and other cytokines essential for an effective anti-microbial response (Akira et al., 2006). Different adaptor proteins function to transmit downstream signals converging at the point of the IκB (IKK) and IKK-related kinases (Kawai et al., 2005; Yamamoto et al., 2003). The IKK-related kinases (TBK1 and IKKε) phosphorylate the transcription factors IRF3 and IRF7 required for type I IFN production (Hemmi et al., 2004; Sharma et al., 2003).
IFN-I receptor-mediated signaling triggers phosphorylation of signal transducers and activators of transcription 1 (STAT1) and STAT2. Together STAT1, STAT2, and a third transcription factor, IRF9, form the Interferon Stimulated Gene Factor 3 (ISGF3) complex, which is essential for induction of many IFN stimulated genes (ISGs) (Platanias, 2005). The tyrosine kinases JAK1 and TYK2 are critical for phosphorylation of tyrosine 701 (Y701) on STAT1 and its subsequent induction of a broad range of ISGs (Shuai et al., 1993). In addition to tyrosine phosphorylation, S708 has also been reported to modulate the transcriptional potential of STAT1 and is required for establishing an effective antiviral state (Tenoever et al., 2007). S708 is phosphorylated by IKKε during IFN signaling, but the mechanism by how IKKε becomes activated has remained elusive (Tenoever et al., 2007).
Ubiquitination of proteins is an important post-translational covalent modification process which has been thoroughly demonstrated to regulate signaling pathways in immune regulation and cytokine production (Jiang and Chen, 2012). Ubiquitin (Ub), a 76 amino acid protein, has seven lysines, each of which can be conjugated by another Ub to form a poly-Ub chain (Trempe, 2011). It is generally accepted that proteins covalently attached to poly-Ub chains linked through lysine 48 (K48) of Ub are targeted for degradation by the proteasome. Conversely, protein modification with K63-linked poly-Ub chains has non-proteolytic activating functions (Chen and Sun, 2009).
The TRIM family of proteins, which are characterized by the presence of a RING, B box and a coiled-coil domain, has been implicated in innate immune signaling pathways by acting as E3-Ub ligases (Rajsbaum et al., 2014). Recently, we have shown that an unprecedented large number of TRIMs positively regulates innate immune responses (Versteeg et al., 2013), however, only a limited number of these TRIMs that act as positive regulators have been characterized in detail. In this study we report that TRIM6 together with the E2 Ub conjugase UbE2K synthetizes unanchored K48-linked poly-Ub chains which activate IKKε to phosphorylate STAT1 S708, ultimately resulting in the induction of a subset of ISGs essential for the antiviral response in vitro and in vivo.
Results
TRIM6 plays a role in induction of IFNβ and ISGs but not of NF-κB dependent cytokines
We and others have recently reported that a large number of TRIMs are able to positively enhance innate immune signaling pathways (Versteeg et al., 2013; Uchil et al., 2013). TRIM6, which had thus far remained completely uncharacterized in immune regulation, was identified as a positive regulator of the type I IFN axis in our screen (Versteeg et al., 2013). We thus set out to investigate the role of TRIM6 in the activation of these innate immune pathways.
Firstly, the ability of TRIM6 to enhance RIG-I-mediated activation of IFN and NF-κB responsive promoters was investigated (Figure 1A, 1B and S1A). Exogenous TRIM6 expression enhanced the Sendai virus (SeV)- and the constitutively active RIG-I(2CARD)-dependent IFN response in a dose-dependent manner (Figure 1A). In contrast, RIG-I(2CARD)-dependent induction of the NF-κB promoter was minimally affected as compared to TRIM25 which has been previously reported to enhance RIG-I-dependent NF-κB activation (Gack et al., 2007) (Figure 1A). These results suggest that TRIM6 is a positive regulator of the IFNβ pathway acting downstream of MAVS/IPS-1 towards the TBK-1 and IKKε signaling axis (Figure S1A). Accordingly, TRIM6 also enhanced TBK-1- and IKKε-dependent IFN induction (Figure 1B).
Figure 1. TRIM6 enhances induction of IFNβ but not NF-kB-dependent cytokines.
(A-B) HEK-293T cells were transfected with the ISG54 IFN-stimulated responsive element (ISRE) or NF-kB reporter plasmid together with TRIM6 or TRIM25 and stimulating plasmids or SeV as indicated, followed by luciferase assay. Data are representative of 3 independent experiments and depicted is the mean ± SD (n=3). (C-E) hDCs transduced with lentiviruses expressing TRIM6 shRNAs were stimulated with LPS. At 2 h p.t. cells were harvested for RT-qPCR and plotted as percentage of control or fold induction over mock-induced samples. See also Figure S1.
To further validate these results, we performed loss of function experiments by silencing TRIM6 expression with short-hairpin lentiviral vector pools (shRNA) in primary human monocyte-derived dendritic cells (hDCs), from four different healthy human donors (Figure S1B). TRIM6 mRNA expression was consistently reduced over 80% in TRIM6-silenced (TRIM6si) hDCs (Figures 1C and S1C). At early time points after TLR4 activation by LPS (2 h p.t), IFNβ, ISG54, TNFα, IL-6 and IL-8 mRNA levels were induced about 10-80 fold in control cells compared to mock-treated samples (Figures 1D and S1D). Strikingly, while IFNβ and ISG54 mRNA levels were attenuated in TRIM6si hDCs as compared to control cells, IL-6 and IL-8 levels were not affected and in some cases even increased (Figures 1E and S1E). These data indicate that TRIM6 specifically acts as a positive regulator of the IFN system, yet does not play a role in promoting induction of pro-inflammatory cytokines.
Next, we set out to establish the importance of TRIM6 for mounting a cytokine response during virus infections. To this end, TRIM6 was silenced in lung epithelial A549 cells using short interfering RNA (siRNA), and subsequently analyzed for cytokine and ISG induction upon SeV or influenza A virus (IAV; PR8 strain) infection (Figure 2). TRIM6 was silenced by more than 75% at the mRNA (Figures 2A and S2A) and protein levels (Figure S2B). SeV and IAV infection strongly induced IFNβ induction -albeit with different kinetics, which was attenuated by TRIM6si without an effect on pro-inflammatory TNFα expression (Figure 2B). ISGs including RIG-I, OAS1, MxA and ISG54 were all reduced by TRIM6si (Figure 2C), whereas viral RNA replication was increased (Figure 2D). Together, these data demonstrate that TRIM6 is required for efficient IFN and ISG induction and that its depletion impairs the antiviral response as a result of attenuated IFN-I signaling, a phenotype reminiscent of that described for IKKε-deficient mice (Tenoever et al., 2007).
Figure 2. TRIM6 is required for efficient induction of IFNβ and ISGs.
(A-G) A549 cells were transfected with a control or TRIM6-specific siRNAs. (A-E) Forty h p.t., cells were mock-treated or infected with SeV (10 HAU/ml), IAV (PR8; 2 PFU/cell), (E) a recombinant NS1 RNA binding mutant virus (R38A/K41A), or (F-G) transfected with dsRNA poly (I:C). RNA levels were quantified by RT-qPCR. (E) At 8 h p.i. with IAV, cells were harvested for immunoblot (IB). (F) At 8 h p.t. with poly I:C, cells were harvested for RT-qPCR or (G) (4 h p.t.) EMSA analysis of ISGF3 binding to ISG promoters. (H) hDCs transduced with lentiviruses expressing TRIM6 shRNAs were infected with SeV or EMCV. Cells were harvested at 6 h p.i for RT-qPCR analysis or at 24 h p.i for virus titeration by RT-qPCR or plaque assay. Values are plotted as fold induction over mock samples. Data represent mean ± SD. (I) The effect of knockdown for all four donors combined was determined by two-way ANOVA. **p < 0.01; ***p < 0.001, N.S. (not significant). See also Figure S2
Immunoblot analyses showed that phosporylation of IRF3 and IKKε (a hallmark of activation) was reduced in TRIM6si cells upon infection with wild-type (WT) IAV or a recombinant virus (R38A/K41A) unable to effectively antagonize IFN induction (Figure 2E) (Talon et al., 2000). The effect of TRIM6 was not specific for RIG-I induced IFNβ induction, since TRIM6si also attenuated poly I:C-induced IFNβ and ISG54 mRNA expression (Figure 2F), as well as secreted IFNβ protein (Figure S2C) through MDA5. Consequently, the lower amounts of IFNβ production observed in TRIM6si cells attenuated downstream IFN signaling as indicated by a reduction of ISGF3 binding to the MxA, OAS1 and ISG15 promoters in an electrophoretic mobility shift assay (EMSA) (Figure 2G). These results indicate that TRIM6 is required for optimal signaling to induce IFNβ and ISGs during viral infection or dsRNA stimulation.
TRIM6 also contributes to the establishment of an efficient antiviral response against SeV (recognized by RIG-I) and Encephalomyocarditis virus (EMCV) (recognized by MDA5) (Figure 2H and I) in hDCs. At 6 h pi., IFNβ induction was significantly reduced in TRIM6si cells as compared to controls (Figure 2H, SeV p<0.001, EMCV p<0.003; Figure 2I), whereas the levels of IL6 were not significantly affected (Figures 2H-I). SeV replication (assessed by gRNA RT-qPCR) was significantly increased in 3 out of 4 donors (Figure 2I; p<0.001) whereas all 4 donors showed significant increase in EMCV viral titers (Figures 2H-I; p<0.001). These data demonstrate that TRIM6 is critical for efficient IFNβ production and establishment of an antiviral state in relevant primary human cells.
TRIM6 interacts through its C-terminal SPRY domain with IKKε
TRIM6 is required for optimal induction of IFNβ, but not NF-kB-dependent genes (Figures 1 and 2). We thus hypothesized that TRIM6 acts at the level of TBK-1, IKKε or IRF3 and investigated whether TRIM6 interacts with any of these three candidates. Co-immunoprecipitation (coIP) studies demonstrated that TRIM6 efficiently interacted with IKKε but not with the closely related kinase TBK-1 or the transcription factor IRF3 when co-expressed in HEK-293T cells (Figure 3A), suggesting that TRIM6 may be required for IKKε activation. These results were confirmed for endogenous TRIM6 and IKKε in primary hDCs upon LPS stimulation, SeV infection or IFNβ treatment (Figure 3B).
Figure 3. IKKε interacts with the C-terminal SPRY domain of TRIM6.
(A) Whole cell extracts (WCE) of HEK-293T cells co-transfected with HA-TRIM6 and either GFP-IRF3, FLAG-IKKε or TBK-1, were subjected to immunoprecipitation (IP) with anti-HA beads. (B) WCE of hDCs treated with SeV, LPS or IFNβ were subjected to IP with anti-IKKε antibody. (C) Localization of HA-TRIM6 and FLAG-IKKε in HeLa cells determined by confocal microscopy. (D) TRIM6 deletion mutants used for CoIP. (E) GST-pulldown of HEK-293T cells transfected with GST-TRIM deletion mutants and FLAG-IKKε. See also Figure S3.
Subsequently, confocal microscopy revealed that TRIM6 localizes in punctate cytoplasmic bodies as previously reported (Reymond et al., 2001), whereas IKKε exhibits a diffuse localization throughout the cytoplasm (Figure 3C). However, upon IKKε and TRIM6 co-expression, a substantial fraction of the total IKKε co-localized with TRIM6 and thus appears to be recruited to the cytoplasmic bodies where TRIM6 resides (Figure 3C). Endogenous IKKε and TRIM6 also co-localized in hDCs, and this co-localization increased upon IFNβ treatment and SeV infection, further substantiating the relevance of the IKKε-TRIM6 interaction in primary immune cells (Figure S3A).
To map the region of TRIM6 binding to IKKε, we generated deletion mutants of TRIM6 expressing the RING domain, Bbox domain, or the C-terminal SPRY domain (Figure 3D), and tested interaction with IKKε by coIP. The C-terminal SPRY domain of TRIM6 by itself specifically interacted with IKKε similar to full-length TRIM6, whereas a mutant lacking only the SPRY domain lost the ability to interact with IKKε (Figure 3E). Furthermore, the SPRY domain of TRIM6 interacted in vitro with baculovirus-produced recombinant IKKε protein (Figure S3B). Taken together these data indicate that TRIM6 interacts directly with IKKε through its SPRY domain.
TRIM6 is critical for the antiviral response mediated by the IFNβ signaling-IKKε axis
Although IKKε has been implicated in the signaling pathway to produce IFN-I, recent studies using IKKε-deficient mice demonstrated a predominant role for IKKε in the IFN signaling pathway by inducing a subset of IKKε–dependent ISGs (Figure 4A) (Perwitasari et al., 2011; Tenoever et al., 2007). Since TRIM6 interacted with IKKε, we sought to determine the role of TRIM6 in the IFN-I signaling pathway.
Figure 4. TRIM6 is critical for the antiviral response mediated by the IFNβ and IKKε axis.
(A) Schematics of the IFN mediated IKKε-dependent and IKKε-independent signaling for ISG induction. (B) WT, Ikbke-/- or Ddx58-/- MEFs were transfected with ISG54 luciferase reporter plasmid together with empty vector or TRIM6 plasmid. Twenty-four h p.t, cells were stimulated with IFNβ (100 U/ml) for 16 h, followed by luciferase assay. Data shown are representative of three independent experiments and depicted is the mean ± SD (n=3). (C,D,F,G,H) A549 cells were tranfected with TRIM6 siRNAs followed by stimulation with IFNβ (100U/ml). Cells were harvested for (C) RT-qPCR analysis of ISG mRNA, (D) IB or (F) EMSA analysis. (E) hDCs transduced with lentiviruses expressing TRIM6 shRNAs were stimulated with IFNβ (100U/ml) and subjected to IB. (G-H) A549 cells were transfected with TRIM6 siRNAs for 40 h and subsequently pre-treated with IFNβ (100U/ml) for 16 h. Cells were then infected with IAV expressing GFP (PR8-GFP) for 16 h. Cells were visualized by fluorescence microscopy (G) and quantified by FACS (H). *p < 0.05; **p < 0.01; ***p < 0.001, N.S. (not significant). See also Figure S4.
We first determined whether TRIM6 enhances the induction of ISGs upon IFN treatment and if IKKε is required for this effect (Figure 4A). To test this, TRIM6 was exogenously expressed in WT or Ikbke-/- (the gene encoding IKKε) murine embryonic fibroblasts (MEFs). TRIM6 overexpression effectively enhanced the IFNβ-induced ISG54 promoter activity in WT MEFs but not in Ikbke-/- cells (Figure 4B). This activity was not cell line-specific as TRIM6 was also able to enhance the IFNβ-induced ISG54 reporter activity in Ddx58-/- (the gene encoding RIG-I) MEFs (Figure 4B), which are only attenuated in their IFN induction, but not signaling. These results demonstrate that TRIM6 specifically requires IKKε to induce ISGs. In line with these results, TRIM6si attenuated IKKε-dependent ISG54 and OAS1 mRNA expression upon IFNβ treatment, whereas IKKε-independently regulated ISG15 and IRF7 mRNA expression was not affected (Figure 4C). Moreover, depletion of TRIM6 enhanced IFNγ-mediated expression of ISGs (Figure S4A), similar to the effects previously reported in Ikbke-/- MEFs (Ng et al., 2011).
As previously reported, IKKε underwent rapid activation by auto-phosphorylation on T501 upon IFNβ treatment of control A549 cells, which was impaired by TRIM6si (Figure 4D). In contrast, phosphorylation of STAT1 on Y701 and STAT2 on Y689 (which are IKKε- independent) were unaffected by TRIM6si (Figure 4D), indicating that TRIM6 is specifically required for optimal IKKε activation, but not for classic JAK1 and TYK2-mediated activation of STATs. As expected, TBK1 was not phosphorylated by IFNβ treatment and remained unaffected by TRIM6si (Figure S4B).
IKKε has been previously shown to phosphorylate S708 on STAT1, which is required for IKKε-dependent ISG induction (Figure 4A) (Perwitasari et al., 2011; Tenoever et al., 2007). Consistent with a role of TRIM6 on the IKKε-STAT1 arm of the IFN signaling pathway, IFN-induced S708 phosphorylation of STAT1 was impaired in TRIM6-silenced hDCs. In contrast, IKKε-independent STAT1-Y701 phosphorylation was not affected (Figure 4E). Moreover, IKKε auto-phosporylation on T501 was also impaired in TRIM6si hDCs (Figure 4E), which is in agreement with the results in A549 cells (Figure 4D). An IKKε-dependent role for TRIM6 was further substantiated by EMSA analysis, demonstrating that TRIM6si attenuated ISGF3 binding to the ‘IKKε-dependent’ OAS1 promoter (Figure 4F).
We next determined the requirement of TRIM6 for conferring biologically relevant IFN-mediated antiviral activity to virus infection. TRIM6si in non-stimulated A549 cells only slightly increased the replication IAV expressing GFP (Figure 4G-H). In contrast, TRIM6si attenuated antiviral activity of IFN pre-treatment, which resulted in a ∼ 3-fold increase in the fraction of virus-infected cells (∼ 35%, Figure 4G-H). Taken together, these results demonstrate that TRIM6 is required for optimal activation of the IKKε branch of the IFN-signaling pathway and is essential to establish an efficient antiviral state in cells.
IKKε interacts with K48-linked poly-ubiquitin chains in cell culture and in vivo
TRIM6, like other TRIM family members, contains a RING domain, which has been shown to confer E3-Ub ligase activity (Meroni and Diez-Roux, 2005). Since we have shown that TRIM6 is important for IKKε activity, we determined whether TRIM6 is involved in ubiquitination of IKKε, the type of Ub linkage it synthesizes, and if this modification is important for IKKε activity. To answer these questions, IKKε and different Ub lysine mutants were exogenously expressed in the presence or absence of TRIM6 and the levels of IKKε-associated Ub were assessed by CoIP. For these assays we used low amounts of IKKε to avoid over-activation of the cells.
IKKε pulled-down low levels of Ub in the absence of TRIM6 (Figure 5A, lane 4, IP). The amount of Ub that coIP with IKKε was greatly enhanced by TRIM6 (Figure 5A, compare IP lane 5 and 6 to lane 4), suggesting that TRIM6 synthesizes poly-Ub forms which either covalently or non-covalently interact with IKKε. These poly-Ub forms were predominantly K48-linked since coIP of a K48R Ub mutant with IKKε was reduced as compared to other lysine mutants or WT Ub (Figure 5A, compare IP lane 12 to other lanes). In fact, poly-Ub chains formed exclusively through K48-linkage (all other lysines mutated to arginine) coIP more efficiently with IKKε as compared to WT-Ub (Figure 5A, compare lane 14 to lane 6).
Figure 5. IKKε interacts with K48-linked poly-ubiquitin chains synthesized by TRIM6.
(A) HEK-293T cells were transfected with FLAG-IKKε together with HA-Ub WT (lanes 1-2, 4-6, 15) or different Ub mutants, and GST-TRIM6 (lane 6-14) or GST-RBCC (TRIM6 lacking SPRY domain: lane 15), or (B) FLAG-IKKε with HA-TRIM6 or TRIM6 C15A RING mutant. IP was performed with anti-FLAG beads. (C) Luciferase assay of HEK-293T cells transfected with ISG54 reporter together with TRIM6 WT or C15A mutant, and treated with IFNβ (100 U/ml) or (D) tranfected with TRIM6 siRNA followed by transfection with ISG54 reporter and TRIM6 plasmids and IFNβ treatment (100U/ml) 24 hr later. Depicted is the mean ± SD (n = 3). (E) hDCs transduced with lentiviruses expressing TRIM6 shRNAs were stimulated with IFNβ (100U/ml) and subjected to IKKε IP. (F) BALB/c mice were treated i.n. with 12,000 units of universal IFN. Lungs were collected and IKKε IP was performed. (G-H) Silencing of TRIM6 in vivo. BALB/c mice were treated i.n. with TRIM6-targeting or non-targeting PPMO at 24 and 48 hr prior to i.n. infection with 100 pfu IAV PR8. Lungs were collected for virus titer (G), depicted is the mean ± SD; n=3, or at 48 hrs pi. for IKKε IP (H). See also Figure S5.
Importantly, poly-Ub chains did not coIP with IKKε in the presence of a TRIM6 mutant lacking the C-terminal SPRY domain (Figure 5A, lane 15), which is required for TRIM6 binding to IKKε (see Figure 3E), indicating that formation of a TRIM6-IKKε complex is necessary for the IKKε-Ub interaction. Endogenous K48-linked Ub chains also coIP with IKKε, which was abolished in a TRIM6 E3 catalytic mutant (C15A; Figure 5B). In agreement, the same RING mutant failed to enhance the IFN signaling pathway (Figure 5C) or rescue ISRE reporter enhancement upon TRIM6si (Figure 5D). In line with these exogenous expression studies, association of endogenous IKKε with K48-linked poly-Ub chains increased upon IFNβ treatment in control hDCs, whereas it was reduced in TRIM6si cells (Figure 5E). Together, these results indicate that TRIM6 requires its E3 activity for IKKε-Ub association and enhancement of IFN signaling.
In an effort to ascertain that the TRIM6-IKKε-poly-Ub interaction was physiologically relevant and occurs in vivo, we investigated whether these three proteins interacted by coIP assays in the lungs of mice treated with universal type I IFN (Figure 5F). Intranasal (i.n.) IFN treatment resulted in rapid and transient STAT1 phosphorylation at Y701 (WCE, 45min-2hr), followed by STAT1 phosphorylation at S708 (WCE, 2-6hr). Consistent with the proposed role of IKKε in STAT1-S708 phosphorylation, activation of IKKε (T501 phosphorylation) correlated with STAT1-S708 phosphorylation. Consequently, coIP of poly-Ub chains and TRIM6 with IKKε also correlated with IKKε-T501 and STAT-S708 phosphorylation (Figure 5F).
To further demonstrate that TRIM6 is required for the formation of IKKε-Ub complexes and establishment of an efficient antiviral response in vivo, TRIM6 expression was silenced in the lungs of mice using peptide-conjugated phosphorodiamidate morpholino oligomers (PPMO) (Abes et al., 2006). In line with our results from hDCs and cell lines, the designed TRIM6 PPMOs efficiently and specifically downregulated TRIM6 protein levels in MEFs resulting in a reduction of ISG54 protein expression upon IFNβ treatement (Figure S5A). To test the effects of PPMO-mediated TRIM6si in vivo, we first determined the kinetics of IAV infection in vivo. Mice not treated with PPMO were infected i.n. with IAV for up to five days, and analyzed for TRIM6, IKKε and poly-Ub interaction in their lungs by coIP at different times p.i. (Figure S5B). Viral loads peaked at 3 d p.i. and remained high until 5 d p.i. (Figure S5C). Viral replication in the lungs of infected mice correlated with increased mRNA levels of M1 viral RNA, IFNβ, TRIM6 and the IFN-stimulated gene ADAR1 (Figure S5D). The protein levels of TRIM6, Ub and IKKε increased in the lungs of infected mice as compared with controls (Figure S5E, WCE). Furthermore, IKKε interaction with TRIM6 and Ub was detected as early as 1 d p.i. and increased in a time-dependent manner (Figure S5E, IP).
We next assessed the effects of PPMO-mediated TRIM6si on viral load, cytokine and ISG induction, and IKKε-Ub association at early time points p.i., before the peak-level of viral titer is typically observed in the lungs of this mouse model (Figure S5F). Treatment with PPMOs resulted in ∼50% reduction in TRIM6 mRNA and protein compared to non-targeting PPMO control in the lungs of infected and non-infected mice (Figure 5H, WCE and S5G). Reduction in TRIM6 expression in the lungs of mice correlated with increased viral load, as assessed by plaque assay (Figure 5G), RT-qPCR for viral M1 mRNA (Figure S5G), and immunoblot for viral nucleoprotein (NP) and NS1 protein (Figure 5H, WCE). The effects of TRIM6 silencing mirrored the phenotype previously observed in Ikbke-/- mice upon IAV infection (Tenoever et al., 2007). Namely, although the levels of IFNβ mRNA were higher in the lungs of TRIM6-silenced mice, induction of the IKKε-dependent ISGs ADAR1 and IFI44 was reduced as compared to controls (Figure S5G), consistent with defective IFN-I signaling. In contrast, the induction of pro-inflammatory cytokines TNFα and IL-6 as well as IL-12p40 (Figure S5G) was unaffected, indicating that TRIM6-silencing in vivo specifically affects the IFN-I-IKKε axis, consistent with our observations in human DCs.
Importantly, the formation of IKKε-Ub complexes and IKKε auto-phosphorylation were reduced in TRIM6si lungs as compared to controls (Figures 5H and S5H), indicating that TRIM6 is required for IKKε activation in vivo. In support of this, STAT1-S708 phosphorylation was also reduced in TRIM6si lungs (Figure 5H, WCE). In contrast, STAT1 induction (an IKKε-independent ISG) and STAT1-Y701 phosphorylation (which is IKKε-independent) were not affected in TRIM6si lungs (Figure 5H, WCE) indicating that TRIM6 specifically acts in the IKKε axis of the IFN-I signaling in vivo and confirms our results obtained in cell lines and human DCs.
Taken together, these data indicate that TRIM6 facilitates Ub chain synthesis required for efficient IKKε function in IFN-I signaling and demonstrate that TRIM6 is required for establishment of an efficient antiviral response in cultured cells and in vivo.
TRIM6 facilitates the synthesis of unanchored ubiquitin chains that interact with IKKε and are required for efficient type I IFN signaling
The previous analyses did not determine whether IKKε is covalently modified by Ub or whether it interacts non-covalently with ubiquitinated proteins or unanchored Ub chains. Furthermore, previous studies have also found Ub associated with IKKε (Friedman et al., 2008; Parvatiyar et al., 2010), but did not address whether these Ub chains were unanchored or covalently attached. To distinguish between these possibilities, a denaturing pull-down analysis of his-tagged Ub co-expressed with IKKε was performed (Figure S6A). In contrast to the results from the above-described coIP under native conditions, we did not detect ubiquitinated IKKε under denaturing pull-down conditions (Figure 6A), while ubiquitinated TRIM6 was readily detected, confirming the validity of the assay. Also, higher migrating –potentially ubiquitinated- forms of IKKε were not detected in the WCE (Figure 6A). Together, these data suggested that IKKε might interact non-covalently with TRIM6-synthesized unattached poly-Ub chains or other TRIM6-ubiquitinated cellular proteins.
Figure 6. IKKε interacts with unanchored polyubiquitin chains synthesized by TRIM6.
(A) HEK-293T cells, transfected with FLAG-IKKε plasmid together with empty vector or HA-TRIM6 and His-tagged Ub, were subjected to His-pulldown under denaturing conditions. (B) HEK-293T cells were transfected with empty vector or FLAG-IKKε together with HA-Ub, and GST-TRIM6 in the presence or absence of the isopeptidase T (IsoT) which cleaves only unanchored Ub. At 30 h p.t, IP with anti-FLAG antibody. FLAG-JAK1 was used as a control of covalently bound Ub which IsoT cannot cleave (right side). (C) ISG54 reporter assay of HEK-293T cells transfected with IKKε and TRIM6 in the presence or absence of IsoT, or (D) in the presence or absence of IsoT, OTU domain of the CCHFV or a mutant (C40A and H151A) lacking catalytic activity, followed by IFNβ treatment. Data shown is representative of 3 independent experiments (mean ± SD, n=3). (E-F) IKKε is recruited to “TRIM6 Ub-rich bodies”. (E) Confocal microscopy of HeLa cells transfected with HA-TRIM6 and FLAG-Ub, or (F) with HA-TRIM6, FLAG-IKKε and His-Ub. See also Figure S6.
As unanchored K63 Ub chains have recently been recognized as significant signaling mediators (Pertel et al., 2011; Xia et al., 2009; Zeng et al., 2010), we set out to determine whether the TRIM6-synthesized Ub moieties interacting with IKKε were in fact unanchored Ub chains. To this end we utilized co-expression of isopeptidase T/USP5 (IsoT), which has been well-established to specifically degrade unanchored Ub through recognition of its exposed C-terminal diglycine residues, while not deconjugating attached Ub chains (Figure S6B) (Reyes-Turcu et al., 2006; Zeng et al., 2010). Indeed, in agreement with our hypothesis, co-expression of IsoT decreased the amount of TRIM6-synthesized multimeric Ub interacting with IKKε in a dose-dependent manner (Figure 6B, left panel). Likewise, treatment of isolated IKKε-Ub complexes with recombinant IsoT in vitro also reduced the amount of poly-Ub chains that coIP with IKKε (Figure S6C). In contrast, IsoT expression did not have a major effect on multimeric Ub co- immunoprecipitated with JAK1, which was used as a control (Figure 6B, right panel). This suggests that JAK1-associated Ub chains are primarily covalently attached, whereas the poly-Ub chains associated with IKKε are unanchored and their synthesis is mediated by TRIM6.
Furthermore, IsoT co-expression reduced the TRIM6-dependent ISG54 reporter activation almost to the level of the control without TRIM6 (Figure 6C and 6D), suggesting that unanchored Ub chains are important for TRIM6-dependent IKKε enhancement. In agreement, cleavage of both anchored and unanchored poly-Ub chains by co-expression of the ovarian tumor (OTU) domain of Crimean Congo hemorrhagic fever virus (CCHFV) large (L) protein (Frias-Staheli et al., 2007), also negated TRIM6-mediated IKKε activation of the ISG54 promoter (Figure 6D). In contrast, a non-catalytic control mutant lacking this activity did not (Figure 6D).
We next determined whether the formation of the IKKε-Ub complex in vivo requires IFN signaling during infection and whether the Ub chains formed in vivo are indeed unanchored. To this end, WT and Ifnar1-/- mice were infected i.n. with IAV for up to five days, and analyzed for IKKε and poly-Ub interaction in their lungs by coIP at different times p.i.. In agreement with the results decribed above, K48-linked poly-Ub chains and TRIM6 coIP with IKKε and increased over time in the lungs of virus infected WT mice, but not Ifnar1-/- mice, indicating that the formation of this complex is IFN-I signaling dependent (Figure S6D). Importantly, the poly-Ub chains bound to IKKε in WT mice were unanchored since they were degraded by in vitro treatment with recombinant IsoT (Figure S6D; compare ‘day 3 WT’ in lane 4 with ‘day 3 WT + IsoT’ in lane 11).
Finally, to demonstrate that IKKε interacts directly with unanchored K48-linked Ub chains, a cell-free system with baculovirus-expressed recombinant IKKε purified from insect cells was established and used to determine IKKε binding to recombinant Ub chains in vitro (Figure S6E). IKKε interacted with K48-linked (lanes 9-10), but not K63-linked poly-Ub (lane 11) or K48-K63 mixed-linkage tetra-Ub (lane 12). Binding of IKKε to Ub required a minimum of four Ub units since di- or tri-Ub did not co-immunoprecipitate with IKKε under these conditions (Figure S6E).
Taken together, these results show that i) IKKε interacts directly with unanchored K48-linked poly-Ub chains, ii) TRIM6 enhances IKKε interaction with these unanchored K48-linked Ub chains, and iii) these Ub chains are important for TRIM6-IKKε-dependent ISG induction.
IKKε is recruited to TRIM6-ubiquitin-rich bodies
Next, it was addressed whether TRIM6 and IKKε co-localize with Ub in the cell by confocal microscopy. TRIM6 localized into defined cytoplasmic dots (Figure 6E), in agreement with previous findings ((Reymond et al., 2001) and Figure 3C). Exogenously expressed Ub alone exhibited weak staining distributed throughout the cell. This Ub staining increased when co-expressed with TRIM6 and extensively co-localized with the punctate cytoplasmic TRIM6 bodies (Figure 6E). Similarly, IKKε alone localized diffusely throughout the cytoplasm (Figures 6F and 3C), and re-localized into the same compartment as both TRIM6 and Ub upon co-expression of TRIM6 (Figure 6F).
Exogenous IsoT expression disrupted the TRIM6 punctate structures, without affecting TRIM6-IKKε co-localization (Figure S6F), as well as in vitro treatment with IsoT specifically degraded the unanchored poly-Ub chains associated with TRIM6, while not having an effect on covalently ubiquitinated TRIM25 used as a control (Figure S6G). Together these data suggest that unanchored Ub chains are essential for the formation of cytoplasmic “TRIM-Ub-rich bodies” but not for TRIM6-IKKε interaction. They indicate that i) TRIM6 enhances the synthesis of Ub forms in defined sub-cellular structures and ii) IKKε interacts with TRIM6 and requires TRIM6-mediated Ub synthesis for its activity.
TRIM6 and the E2 conjugase UbE2K cooperatively synthesize unanchored K48-linked ubiquitin chains required for IKKε activation
To demonstrate that TRIM6 can synthesize K48-Ub chains directly mediating IKKε activation, a TRIM6 in vitro ubiquitination assay was established. Ub conjugation requires the activity of E1 activating, E2 conjugating and E3 ligating enzymes. Since the E2 enzyme that couples TRIM6 to Ub conjugation had yet to be determined, first an in vitro screen with twenty-nine common E2-ligases was performed for their ability to synthesize poly-Ub chains in the presence of TRIM6.
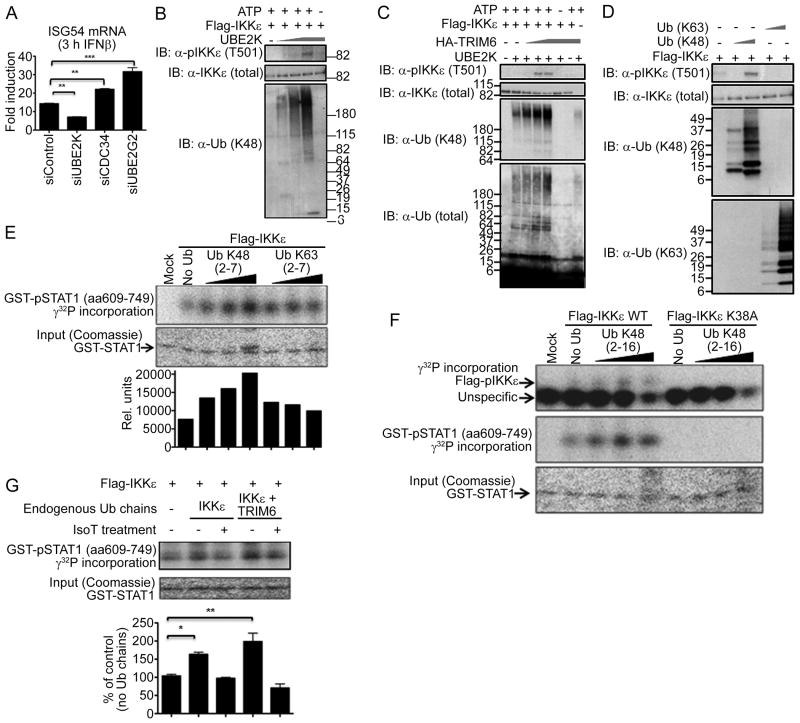
By these means we identified three E2 enzymes (UbE2K, CDC34, Ube2G2; data not shown) previously reported to facilitate K48-linked Ub chain synthesis (Chen and Pickart, 1990; Petroski and Deshaies, 2005; Shin et al., 2011), which were tested in knockdown experiments for their requirement in IFNβ-dependent ISG induction. Only UbE2K-silencing in A549 cells resulted in a decrease of ISG54 mRNA induction upon IFN stimulation as compared to control cells (Figure 7A). In contrast, silencing of CDC34 or UbE2G2 did not attenuate ISG54 mRNA induction, or even increased it (Figure 7A), despite similar knockdown (>80% data not shown).
Figure 7. The E2-ligase UbE2K and TRIM6 catalyze K48-linked ubiquitin chains that activate IKKε for STAT1 phosphorylation in vitro.
(A) Knockdown of endogenous E2 conjugases UbE2K, CDC34 and UbE2G2 in A549 cells. At 40 h p.t., cells were mock-treated or stimulated with IFNβ (500 U/ml) for 3 h and cells were harvested for RT-qPCR analysis. Values are depicted as fold induction over non-stimulated cells. (B) Purified FLAG-IKKε was incubated with recombinant E1, UbE2K (0.025-0.5 μM), mono-Ub and ATP. Activation of IKKε was assessed by phophorylation of T501. (C) Increasing amounts of TRIM6 were added to limiting amounts of UbE2K (0.025 μM). (D) Recombinant K48 or K63-linked poly-Ub chains (2-7 Ub molecules/chain ) were incubated with FLAG-IKKε and subjected to IB analysis. (E-F) In vitro STAT1 kinase assay. Recombinant K48 or K63-linked poly-Ub chains (2-7 Ub/chain; (E), or K48 2-16 Ub/chain (F)) were incubated with purified IKKε WT (E and F) or an IKKε kinase-dead mutant (K38A; (F)) STAT1 and γ32ATP. The reaction mixture was subjected to SDS-PAGE and visualized by autoradiography. The bands were quantified using ImageJ software. (G) Endogenous unanchored poly-Ub chains induce IKKε-mediated STAT1 phosphorylation. WCE of HEK-293T cells, transfected with FLAG-IKKε and HA-TRIM6 were subjected to FLAG-IP. Poly-Ub chains bound to IKKε were isolated by incubation at 75 °C for 5min, followed by treatment with IsoT. These Ub chains were used for IKKε-mediated STAT1 kinase assay. The bands were quantified using ImageJ software, normalized by the input (Coomassie), and expressed as percentage of control without Ub chains. The quantification data are from 2 independent experiments. See also Figure S7.
The UbE2K conjugase is known to elongate mono-Ub to produce K48-linked poly-Ub chains (Wilson et al., 2011), and has been reported to have the ability to synthesize unanchored poly-Ub chains in the absence of an E3 ligase in vitro (Chen and Pickart, 1990). Thus, as anticipated, K48-linked poly-Ub chains were produced in a UbE2K dose-dependent manner (Figure 7B). Under these conditions (at high concentrations of UbE2K and high amounts of synthesized K48 poly-Ub) IKKε auto-phosphorylation was detected on T501 (Fig 7B), demonstrating that UbE2K can mediate K48 Ub chain synthesis and IKKε activation. Moreover, K48-linked poly-Ub chain production and IKKε activation by the lowest concentration of UbE2K (0.025 μM) were enhanced by TRIM6 in a dose-dependent manner (Figure 7C), indicating that TRIM6 in combination with UbE2K acts as an E3 ligase driving the synthesis of K48 poly-ubiquitin. IKKε auto-phosphorylation was specific since T501 was not phosphorylated in the presence of an inactive IKKε kinase mutant (K38A; Figure S7A). As expected, poly-Ub synthesis was completely dependent on ATP and the presence of UbE2K, but not IKKε (Figure 7C). In addition, these poly-Ub chains were exclusively K48-linked since they were not produced from a K48R mono-Ub mutant substrate (Figure S7A). Conversely, chain synthesis of a Ub mutant with all lysines mutated except for K48 (K48only) was enhanced.
Next, IKKε was incubated in vitro with purified, recombinant poly-Ub chains after which IKKε activation was determined. Only a mixture of K48-linked poly-Ub chains (2-7 Ub molecules/chain) activated IKKε T501 auto-phosphorylation in a dose-dependent manner, whereas similar amounts of K63-linked chains did not (Figure 7D). This further demonstrates that specifically K48-linked poly-Ub chains -such as synthesized by UbE2K and TRIM6- activate IKKε.
We next confirmed that K48 poly-Ub-activated IKKε has indeed increased kinase activity towards its bona fide target STAT1. K48- but not K63-linked poly-Ub chains reproducibly increased purified STAT1 phosphorylation by IKKε in a dose-dependent manner (Figure 7E). Importantly, longer K48-linked poly-Ub chains (mixture of 2-16 Ubs/chain), which putatively more closely resemble the high molecular forms of Ub found in vivo (Figure 5F, S5E and S6D), induced even higher levels of STAT1 phosphorylation, as well as increased IKKε auto-phosphorylation (Figure 7F). Again, this was specific to IKKε as no phosphorylation was detected with the IKKε K38A mutant.
These results were substantiated by investigating whether isolated TRIM6-synthesized endogenous, unanchored poly-Ub chains could facilitate IKKε-dependent phosphorylation of STAT1 in vitro. To this end, endogenous Ub chains bound to IKKε were isolated as previously described for RIG-I (Zeng et al., 2010) (Figure S7B). In agreement with our other data, Ub chains isolated from IKKε alone, limitedly increased STAT1 phosphorylation (Figure 7G; compare lanes 1 and 2). Strikingly, endogenous Ub chains isolated from IKKε expressed in the presence of TRIM6, even further enhanced IKKε-dependent STAT1 phosphorylation within the limited experimental window (Figure 7G; compare lanes 2 and 4). Moreover, the poly-Ub-mediated increase in STAT1 phosphorylation was abrogated upon IsoT treatment, confirming that the enhancement of STAT1 phosphorylation depends on unanchored poly-Ub chains (Figure 7G; compare lanes 2 with 3, and 4 with 5).
Finally, to better understand the mechanism of IKKε activation by TRIM6 and poly-Ub chains, we mapped the region of IKKε required for interaction with TRIM6 and Ub by coIP using truncated forms of IKKε (Figure S7C). TRIM6 and poly-Ub chains interacted more efficiently with the kinase domain and IKKε mutants lacking their C-terminal CCDs (Figure S7D). However, TRIM6 interacted less efficiently with a shorter form of IKKε expressing both the kinase domain and the Ub-like domain (UBLD), suggesting that the UBLD could fold back on the kinase domain as previously reported (Ikeda et al., 2007), thereby interfering with TRIM6 binding to the IKKε kinase domain. Furthermore, residue S172 in the IKKε kinase domain was critical for interaction with TRIM6 and Ub (Figure S7D, last lane). In line with these observations, the IKKε S172A mutant did not activate the ISG54 promoter in a reporter assay (Figure S7E). In addition, TRIM6 did not enhance the IKKε S172A-dependent ISG54 reporter activity as compared to TRIM6 alone or WT-IKKε alone (Figure S7F).
Since unanchored K48 Ub chains induced transactivation of IKKε (Figure 7D), we hypothezised that TRIM6-synthesized poly-Ub chains are important for IKKε oligomerization and thus transactivation by binding to the kinase domain of IKKε. In line with this hypothesis, in vitro data indicated that IKKε runs as a dimer under native conditions (Figure S7G) and addition of K48-linked poly-Ub chains increased the oligomerization of IKKε with the strongest effect obtained in the presence of long K48-linked poly-Ub chains (Figure S7G). Thus, K48-linked poly-Ub chains promote oligomerization of IKKε suggesting that oligomerization is important for transactivation.
TRIM6 interacts with the JAK1 kinase
It remains to be determined how TRIM6 itself is regulated at the detailed molecular level. A possible mechanism could entail direct TRIM6 activation by the JAK1 and TYK2 kinases downstream of the IFN-I receptor. In support of this possibility, JAK1 interacted with TRIM6 and induced TRIM6 tyrosine phosphorylation (Figures S7H and S7I). Although the nature of this TRIM6 phosphorylated form remains to be determined, it is tempting to speculate that TRIM6 may be activated by JAK1 after IFN treatment.
Taken together, our results demonstrate that TRIM6 and UbE2K cooperate in the synthesis of K48-linked unanchored poly-Ub chains, which activate IKKε for STAT1 phosphorylation (Figure S7J). The IKKε-targeted phosphorylation site on STAT1 (S708) promotes antiviral activity through induction of a subset of ISGs. This phosphorylation is distinct from IKKε-independent STAT1-Y701 phosphorylation mediated by JAK1 and TYK2, which drives IKKε-independent ISG expression (Figure S7J).
Discussion
In this study we demonstrate that the Ub E3 ligase TRIM6 is required for efficient IFN-I signaling and establishment of an antiviral state by synthesis of K48-linked unanchored poly-Ub chains, which activate IKKε for STAT1 phosphorylation. Our findings indicate that these Ub chains play a role in both whole cell context and in vivo. This is supported by four lines of evidence: i) expression of IsoT, which specifically degrades unanchored Ub, reduced IFN-mediated ISG induction in cells, ii) knockdown of the E2-Ub ligase Ube2K, which is known to synthesize K48-linked poly-Ub chains (Chen and Pickart, 1990; Wilson et al., 2011), attenuated ISG expression upon IFN stimulation, iii) poly-Ub chains interact with IKKε in mouse lungs during viral infection in an IFN-I-dependent manner and iv) the formation of IKKε-Ub complexes is reduced in TRIM6si mouse lungs upon viral infection.
Importantly, TRIM6si not only impaired IFN signaling, but also impaired IFNβ production upon TLRs and RLRs stimulation in primary human DCs and human cell lines, although this was not the case in the lungs of IAV-infected mice. One possible explanation for this discrepancy could be differences in TRIM6 function in human and mouse. Alternatively, TRIM6 may have cell type specific effects or may act at multiple levels in signaling pathways of the IFN-I system. Moreover, increased IFN production in TRIM6si mice may be a reflection of increased viral replication as a result of diminished antiviral response. In addition, any defect on IFN production in TRIM6si cells in the lung may have been obscured by IFN produced from other cells in the lung that were not subject to TRIM6si, as for example, infiltrating cells.
Although it is clear that IKKε phosphorylates IRF3 and IRF7 for IFN induction (Sharma et al., 2003), production of IFN-I was not reduced in Ikbke-/- mice upon IAV infection (Tenoever et al., 2007). However, studies have suggested that TBK-1 and IKKε play a redundant role in IFN induction (Hemmi et al., 2004). Therefore it is still plausible that TRIM6 may also play a role by activating IKKε for IFNβ production in specific cell types, or under specific conditions. Alternatively, TRIM6 may have other signaling partners in addition to IKKε for IFN induction, as it has been shown for other TRIMs (Rajsbaum et al., 2014). The specific role of TRIM6 in the IFNβ induction pathway will need to be determined in future studies.
In our immunofluorescence experiments, TRIM6, IKKε and Ub co-localized in punctate cytoplasmic bodies. These structures, which have also been observed for other TRIMs, seem to be independent sub-cellular compartments since they do not co-localize with any markers for known cellular structures (Reymond et al., 2001). As we observed a dramatic increase in Ub staining in these TRIM6-containing bodies, we speculate that these structures may be “Ub factories” whereto other target proteins -such as IKKε- can be recruited during signaling. These structures required an intact TRIM6 coiled-coil region (data not shown, and (Reymond et al., 2001)), and formation of poly-Ub chains by TRIM6, suggesting that oligomerization is essential for their assembly. These sub-cellular structures may be an important compartmentalization mechanism in the cell.
Our data suggest that TRIM6 could be activated upon IFN stimulation by the JAK1 kinase by binding and phosphorylating TRIM6. However, TRIM6 activation may also involve ubiquitination by a yet unidentified E3-ligase since covalent K63-linked ubiquitination of both WT TRIM6 and its E3 ligase C15A mutant was detected.
In conclusion, our study identified TRIM6, UbE2K and K48-linked poly-Ub chains as novel players critical for the IKKε branch of the IFN-I signaling pathway and subsequent establishment of a protective antiviral response. Our study provides a framework for future work on the TRIM family of proteins, as well as the mechanism of kinase activation by unanchored K48-linked poly-Ub chains related to antiviral immunity. Wheras we report a unique function of unanchored K48-poly-Ub in kinase activation, it might be possible that these molecules also participate in other signaling process, as is the case for unanchored K63-poly-Ub (Pertel et al., 2011; Xia et al., 2009; Zeng et al., 2010). Free poly-Ub is therefore emerging as a secondary messenger that participates in regulation of signaling and further studies are needed to fully elucidate cellular processes that are controlled by the synthesis of unanchored Ub chains.
Experimental Procedures
In vitro ubiquitination assay and activation of IKKε
Ubiquitination assays in vitro were performed using the Ub conjugation initiation kit (Boston Biochem). In brief, a mix was prepared containing the E1 enzyme, Ub, and reaction buffer in the presence or absence of the E2 ligase UbE2K (Boston Biochem) (0.02-0.5mM) and the presence or absence of FLAG purified IKKε (0.1 μl) and HA-TRIM6 purified from HEK-293T cells with HA-beads (0.05-0.5 μl). The reaction was initiated with addition of 1mM Mg-ATP and stopped after 1h at 30°C by addition of Laemmli sample buffer (BioRad) containing β-Mercaproethanol and boiled for 5 min. Poly-Ub and IKKε auto-phosphorylation was detected by IB with α-pIKKε(T501) (Novus Biologicals).
Isolation and analysis of endogenous poly-ubiquitin chains
We followed a previously described protocol to isolate chains from RIG-I(2CARD) used by Zeng et.al. (Zeng et al., 2010). For detail information see Supplemental Information.
siRNA-mediated gene targeting
Transient gene targeting of endogenous TRIM6 in A549 or HEK-293T cells, seeded in 24-well plates (30,000 cell/well), was achieved by transfection of 10 picomol of non-targeting control or an siRNA specific for TRIM6 (see supplemntal information for sequences) with RNAiMAX (Invitrogen) according to the manufacturer's instructions.
TRIM6 gene targeting in human DCs with shRNA lenti-viral vectors
Peripheral blood mononuclear cells were isolated from buffy coats of healthy human donors by Ficoll density gradient centrifugation (Histopaque, Sigma Aldrich) as previously described (Fernandez-Sesma et al., 2006). For additional details see Supplemental Information. All human research protocols for this work have been reviewed and approved by the Institutional Review Board of the Mount Sinai School of Medicine.
In vitro oligomerization assay and native PAGE
Purified FLAG-IKKε protein was incubated with Ub chains in the presence or absence of 1mM of ATP and 2 mM MgCl2 in 50 mM Tris, 150 mM NaCl, 1 mM DTT buffer at 37°C for 15 minutes to allow formation of complexes. Reactions were stopped by addition of 1x Native PAGE sample buffer (Life) and subjected to native PAGE on a 3-12% Bis-Tris gel (Life), transferred on to PVDF membranes, fixed with 8% acetic acid in H2O and immunoblotting was performed using anti-FLAG antibodies.
TRIM6 gene targeting in vivo using PPMOs
The peptide-conjugated phosphorodiamidate morpholino oligomers (PPMO) were produced by previously published methods (Abes et al., 2006). All mouse experiments were carried out in accordance with institutional Animal Care and Use Committee (IACUC) guidelines, and have been approved by the IACUC of Icahn School of Medicine at Mount Sinai. Balb/c mice were anesthetized with ketamine, followed by i.n. administration with 100 μg (approximately 5 mg/kg) PPMO (in 40 μl of PBS). After 24 hr, PPMO treatment was repeated. After 48 h from the first PPMO administration, mice were infected i.n. with 1000 PFU of influenza virus (strain A/Puerto Rico/8/1934 H1N1 [PR8]). At 24 and 48 h.p.i. mice were euthanized and the lungs were collected and divided into 3 equivalent portions, followed by homogenization and subsequent plaque assay, real time PCR analysis and coIP studies. For more details see Supplemental Information.
Statistical Analysis
Statistical analysis was performed using Prism (Version 5.0, GraphPad Software, San Diego California USA). Student's paired t-test, or in defined cases two-way ANOVA with Bonferroni post-test were used. *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Material
Highlights.
TRIM6 plays a critical role in innate immune activation
TRIM6 and the E2 conjugase UbE2K synthesize unanchored K48-linked ubiquitin chains
Unanchored K48-linked ubiquitin chains activate the IKKε kinase
TRIM6 couples IFNαR engagement to activation of the IKKε kinase for ISG induction
Acknowledgments
We thank Richard Cadagan, Osman Lizardo and Dabeiba Bernal-Rubio for technical support. We thank Christopher F. Basler for kindly providing the IKKε deletion plasmids. Confocal microscopy was performed at the Mount Sinai Microscopy Shared Resource Facility. This work was supported in part by NIH R01AI080624 (B.R.T), P01AI090935, U19 AI083025, U01AI095611, U19AI106754 and HHSN272201000054C (A.G.-S.), NIH/NIAID 1P01AI90935, 1R01AI073450 and DARPA HR0011-11-C-0094 (A.F.-S) and by NIAID funded CEIRS (Center of Excellence in Influenza Researcha and Surveillance, contract # HHSN266200700010C) (A.G-S, and A-F-S).
Footnotes
Author contributions: R.R. and GA.V performed all aspects of this study. S.S, A.M.M, A.B.-V, C.M.-R, J.R.P, J.M., G.P., L.M., M.L.-R performed experiments. A.F.-S. and B.RT. provided reagents and advice. R.R., GA.V, and A.G.-S. organized this study and prepared the manuscript. H.M.M and DA.S designed and produced PPMOs. All authors discussed the results and commented on the manuscript. R.R. and GA.V. contributed equally to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abes S, Moulton HM, Clair P, Prevot P, Youngblood DS, Wu RP, Iversen PL, Lebleu B. Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J Control Release. 2006;116:304–313. doi: 10.1016/j.jconrel.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Bruni D, Sebastia J, Dunne S, Schroder M, Butler MP. A novel IRAK1-IKKepsilon signaling axis limits the activation of TAK1-IKKbeta downstream of TLR3. J Immunol. 2013;190:2844–2856. doi: 10.4049/jimmunol.1202042. [DOI] [PubMed] [Google Scholar]
- Chen Z, Pickart CM. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. The Journal of biological chemistry. 1990;265:21835–21842. [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Molecular cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Clark K, Peggie M, Plater L, Sorcek RJ, Young ER, Madwed JB, Hough J, McIver EG, Cohen P. Novel cross-talk within the IKK family controls innate immunity. The Biochemical journal. 2011;434:93–104. doi: 10.1042/BJ20101701. [DOI] [PubMed] [Google Scholar]
- Fernandez-Sesma A, Marukian S, Ebersole BJ, Kaminski D, Park MS, Yuen T, Sealfon SC, Garcia-Sastre A, Moran TM. Influenza virus evades innate and adaptive immunity via the NS1 protein. Journal of virology. 2006;80:6295–6304. doi: 10.1128/JVI.02381-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, et al. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell host & microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CS, O'Donnell MA, Legarda-Addison D, Ng A, Cardenas WB, Yount JS, Moran TM, Basler CF, Komuro A, Horvath CM, et al. The tumour suppressor CYLD is a negative regulator of RIG-I-mediated antiviral response. EMBO reports. 2008;9:930–936. doi: 10.1038/embor.2008.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, Sanjo H, Kawai T, Hoshino K, Takeda K, Akira S. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. The Journal of experimental medicine. 2004;199:1641–1650. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Hecker CM, Rozenknop A, Nordmeier RD, Rogov V, Hofmann K, Akira S, Dotsch V, Dikic I. Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes. The EMBO journal. 2007;26:3451–3462. doi: 10.1038/sj.emboj.7601773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Chen ZJ. The role of ubiquitylation in immune defence and pathogen evasion. Nature reviews. 2012;12:35–48. doi: 10.1038/nri3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature immunology. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- Ng SL, Friedman BA, Schmid S, Gertz J, Myers RM, Tenoever BR, Maniatis T. IkappaB kinase epsilon (IKK(epsilon)) regulates the balance between type I and type II interferon responses. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:21170–21175. doi: 10.1073/pnas.1119137109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar K, Barber GN, Harhaj EW. TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1-IKKi kinases. The Journal of biological chemistry. 2010;285:14999–15009. doi: 10.1074/jbc.M110.109819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perwitasari O, Cho H, Diamond MS, Gale M., Jr Inhibitor of kappaB kinase epsilon (IKK(epsilon)), STAT1, and IFIT2 proteins define novel innate immune effector pathway against West Nile virus infection. The Journal of biological chemistry. 2011;286:44412–44423. doi: 10.1074/jbc.M111.285205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nature reviews. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Rajsbaum R, Garcia-Sastre A, Versteeg GA. TRIMmunity: The Roles of the TRIM E3-Ubiquitin Ligase Family in Innate Antiviral Immunity. Journal of molecular biology. 2014;426:1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD. The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell. 2006;124:1197–1208. doi: 10.1016/j.cell.2006.02.038. [DOI] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. The EMBO journal. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science (New York, NY. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- Shin DY, Lee H, Park ES, Yoo YJ. Assembly of different length of polyubiquitins on the catalytic cysteine of E2 enzymes without E3 ligase; a novel application of non-reduced/reduced 2-dimensional electrophoresis. FEBS letters. 2011;585:3959–3963. doi: 10.1016/j.febslet.2011.11.002. [DOI] [PubMed] [Google Scholar]
- Shuai K, Ziemiecki A, Wilks AF, Harpur AG, Sadowski HB, Gilman MZ, Darnell JE. Polypeptide signalling to the nucleus through tyrosine phosphorylation of Jak and Stat proteins. Nature. 1993;366:580–583. doi: 10.1038/366580a0. [DOI] [PubMed] [Google Scholar]
- Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. Journal of virology. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenoever BR, Ng SL, Chua MA, McWhirter SM, Garcia-Sastre A, Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science (New York, NY. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- Trempe JF. Reading the ubiquitin postal code. Current opinion in structural biology. 2011;21:792–801. doi: 10.1016/j.sbi.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, Luban J, Mothes W. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. Journal of virology. 2013;87:257–272. doi: 10.1128/JVI.01804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, Garcia-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Chen T, Zhang J, Yang M, Li N, Xu X, Cao X. The E3 ubiquitin ligase Nrdp1 ‘preferentially’ promotes TLR-mediated production of type I interferon. Nature immunology. 2009;10:744–752. doi: 10.1038/ni.1742. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Edmondson SP, Flatt JW, Helms K, Twigg PD. The E2-25K ubiquitin-associated (UBA) domain aids in polyubiquitin chain synthesis and linkage specificity. Biochemical and biophysical research communications. 2011;405:662–666. doi: 10.1016/j.bbrc.2011.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science (New York, NY. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Wang L, Zhang M, Wang P, Yuan C, Qi J, Meng H, Gao C. Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I-mediated IFN-beta production and antiviral response by targeting NAP1. J Immunol. 2012;188:5311–5318. doi: 10.4049/jimmunol.1103506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.