Abstract
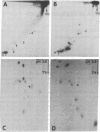
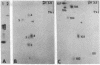
tRNAMetf binds to 23S rRNA of Escherichia coli, forming a complex with a melting temperature of 75 degrees (in 0.6 M NaCl). The regions within the RNAs that bind to each other have been isolated and their nucleotide sequences have been determined. The interacting region in tRNAMetf is 17 nucleotides long, extending from G5 in the acceptor stem to D21 (D = 5.6-dihydrouridine) in the D loop. The sequence in 23S rRNA is complementary to that sequence except for an extra Up in the middle and allowing a Gp.D base pair. We propose that association of these two sequences may play a role in initiation of protein synthesis by tRNAMetf. In addition, part of this sequence in 23S rRNA may also stabilize tRNA binding to the ribosome during elongation of nascent polypeptides.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Branlant C., Widada J. S., Krol A., Ebel J. P. Studies on the primary structure of the ribosomal 23S RNA of Escherichia coli: II. A characterisation and an alignment of 24 sections spanning the entire molecule and its application to the localisation of specific fragments. Nucleic Acids Res. 1977 Dec;4(12):4323–4345. doi: 10.1093/nar/4.12.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Cole P. E., Hilbers C. W., Shulman R. G. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J Mol Biol. 1974 Jul 25;87(1):63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E., Dingman C. W., Peacock A. C. Electrophoretic characterization of bacterial polyribosomes in agarose-acrylamide composite gels. J Mol Biol. 1969 Apr 14;41(1):139–147. doi: 10.1016/0022-2836(69)90131-4. [DOI] [PubMed] [Google Scholar]
- Dube S. K., Marcker K. A., Clark B. F., Cory S. The nucleotide sequence of N-formyl-methionyl-transfer RNA. Products of complete digestion with ribonuclease T-1 and pancreatic ribonuclease and derivation of their sequences. Eur J Biochem. 1969 Mar;8(2):244–255. doi: 10.1111/j.1432-1033.1969.tb00521.x. [DOI] [PubMed] [Google Scholar]
- Dube S. K., Marcker K. A. The nucleotide sequence of N-formyl-methionyl-transfer RNA. Partial digestion with pancreatic and T-1 ribonuclease and derivation of the total primary structure. Eur J Biochem. 1969 Mar;8(2):256–262. doi: 10.1111/j.1432-1033.1969.tb00522.x. [DOI] [PubMed] [Google Scholar]
- Dube S. K. Recognition of tRNA by the ribosome. A possible role of 5 S RNA. FEBS Lett. 1973 Oct 1;36(1):39–42. doi: 10.1016/0014-5793(73)80332-1. [DOI] [PubMed] [Google Scholar]
- Erdmann V. A. Structure and function of 5S and 5.8 S RNA. Prog Nucleic Acid Res Mol Biol. 1976;18:45–90. [PubMed] [Google Scholar]
- Herr W., Noller H. F. A fragment of 23S RNA containing a nucleotide sequence complementary to a region of 5S RNA. FEBS Lett. 1975 May 1;53(2):248–252. doi: 10.1016/0014-5793(75)80030-5. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Dahlberg J. E. Small ribonucleic acids of Escherichia coli. I. Characterization by polyacrylamide gel electrophoresis and fingerprint analysis. J Biol Chem. 1973 Jul 25;248(14):5024–5032. [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Klug A. Crystallographic refinement of yeast phenylalanine transfer RNA at 2-5A resolution. J Mol Biol. 1976 Dec 25;108(4):619–649. doi: 10.1016/s0022-2836(76)80109-x. [DOI] [PubMed] [Google Scholar]
- Kurland C. G. Structure and function of the bacterial ribosome. Annu Rev Biochem. 1977;46:173–200. doi: 10.1146/annurev.bi.46.070177.001133. [DOI] [PubMed] [Google Scholar]
- Lund E., Dahlberg J. E. Spacer transfer RNAs in ribosomal RNA transcripts of E. coli: processing of 30S ribosomal RNA in vitro. Cell. 1977 Jun;11(2):247–262. doi: 10.1016/0092-8674(77)90042-3. [DOI] [PubMed] [Google Scholar]
- Nishimura S. Minor components in transfer RNA: their characterization, location, and function. Prog Nucleic Acid Res Mol Biol. 1972;12:49–85. [PubMed] [Google Scholar]
- Nomura M., Lowry C. V., Guthrie C. The initiation of protein synthesis: joining of the 50S ribosomal subunit to the initiation complex. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1487–1493. doi: 10.1073/pnas.58.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Resolution of multiple ribonucleic acid species by polyacrylamide gel electrophoresis. Biochemistry. 1967 Jun;6(6):1818–1827. doi: 10.1021/bi00858a033. [DOI] [PubMed] [Google Scholar]
- Rich A., RajBhandary U. L. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Sanger F., Brownlee G. G., Barrell B. G. A two-dimensional fractionation procedure for radioactive nucleotides. J Mol Biol. 1965 Sep;13(2):373–398. doi: 10.1016/s0022-2836(65)80104-8. [DOI] [PubMed] [Google Scholar]