Abstract
Disruption of microRNA (miRNA) expression patterns is now being recognized as a hallmark of human cancer. The causes of these altered profiles are diverse, and, among them, we found the existence of defects in the miRNA processing machinery. However, little is known about how these alterations affect the biology of the underlying tumors. Herein, we show that colorectal cancer cells with an impairment in DICER1, a major miRNA biogenesis gene, undergo enrichment of tumor stemness features and an epithelial-to-mesenchymal transition. These phenotypes are associated with the downregulation of miRNAs, such as miR-34a, miR-126 and those of the miR-200 family, that target critical coding genes in these pathways. Most importantly, DICER1 impairment also induces the acquisition of a greater capacity for tumor initiation and metastasis, two properties associated with cancer stem cells.
Keywords: DICER1, miRNA, colon cancer, stemness, metastasis
Introduction
MicroRNAs (miRNAs) belong to a conserved group of small non-coding RNAs (18–24 nucleotides) that are transcribed as long miRNA precursors and processed into mature miRNAs by several enzymatic and biochemical steps that require the activity of DICER1.1, 2 Sequence-specific interactions of miRNAs with the 3′-untranslated region of target mRNAs inhibit translation or induce degradation of these mRNAs.1, 2 The fact that one miRNA can control the expression of multiple gene targets makes miRNAs important regulators of physiological and pathological processes, including cancer.3, 4 In this regard, miRNA signatures relate to the tumoral differentiation stage of the tumor5 and are indicative of a patient's clinical outcome.3, 6 Importantly, an miRNA expression profile of human tumors has emerged that is characterized by a defect in miRNA production and global miRNA downregulation.5, 6, 7 Mechanisms that explain this miRNA deregulation in cancer include the failure of miRNA post-transcriptional control,8 miRNA transcriptional silencing by CpG island promoter hypermethylation9, 10 oncogene-mediated transcriptional repression11 and mutational impairment of the miRNA processing machinery.12, 13
The role of DICER1 in tumorigenesis is attracting increasing interest in the field. Although full deletion of Dicer has deleterious effects on tumor growth in some cancer mouse models,14 it is currently believed that Dicer may be a haploinsufficient tumor suppressor that facilitates mouse and human cancer development.12, 14, 15 In this context, the hemizygous loss of Dicer has been observed in various tumor types,14 and low levels of Dicer expression are known to predict poor survival of patients with chronic lymphocytic leukemia, breast, lung, ovarian, nasopharyngeal, colorectal and bladder cancer.16, 17, 18, 19, 20, 21, 22, 23 Furthermore, low levels of Dicer expression occur in colorectal cancer liver metastasis24 and are associated with an enhanced epithelial-to-mesenchymal transition (EMT) and metastasis in breast cancer cells.25, 26
Loss of Dicer function also severely affects stem cell proliferation and differentiation during development,27, 28, 29 so one way that DICER1 defects might contribute to human tumors is by paving the way for generating cancer stem cells that are important for sustaining tumor growth.30 Herein, we show that colorectal cancer cells impaired for DICER1 activity develop an enhanced stemness phenotype in association with a higher metastatic potential.
Results
Impaired DICER1 function causes upregulation of colon cancer stem cell markers
To evaluate the effect of an impaired function of DICER1 in the stemness of colorectal tumors, we used human colon cancer cells hypomorphic for the enzyme activity,31 because Dicer −/− vertebrate cells are known to be inviable.28 The colorectal cancer cell lines HCT-116, DLD-1 and RKO were disrupted at exon 5 of DICER1, interrupting a well-conserved segment of the N-terminal helicase domain, by using an adeno-associated targeting vector to originate HCT-116-Dicerex5-, DLD-1-Dicerex5- and RKO-Dicerex5-defective cell lines that have a reduced production of mature miRNAs.31
We first studied whether the generation of DICER1-impaired cells created any shift in the pattern of expression of colorectal cancer stem cell markers such as CD133, CD166 and CD44.32, 33 Quantification by flow cytometry of the percentage of cells expressing each marker showed that if there were no substantial differences in CD133 and CD166 between parental and hypomorphic cells (data not shown), Dicerex5 colon cells become enriched in a population with a high level of CD44 expression (Figure 1a). Immunofluorescence (Figure 1b), western blot (Figure 1c) and quantitative reverse transcriptase PCR (qRT–PCR) (Figure 1d) confirmed the overexpression of CD44 in HCT-116-Dicerex5, DLD-1-Dicerex5 and RKO-Dicerex5 cells relative to the parental lines. For the DICER1-impaired HCT-116 cells, which showed the greatest increase in the number of cells with enhanced CD44 expression, we also observed increased expression of standard stem cell markers, such as Sox2, Sox9 and Nanog (Supplementary Figure S1), and the well-characterized colorectal cancer stem cell marker Lgr5 (Supplementary Figure S1).34, 35, 36
Figure 1.
CD44 expression is induced in colon cancer cells with impaired Dicer function. (a) Representative results of CD44+ cell quantification by flow cytometry in parental and Dicerex5 cells. Percentage of CD44+ cells and CD44high cells are indicated in each panel. Blue numbers indicate the mean intensity of fluorescence of CD44+ cells, as quantified by Flow-Jo software. (b) Representative immunofluorescence images of CD44-expressing cells in parental and Dicerex5 cells. Nuclei were stained with DAPI. Scale bars, 30 μm. (c) Levels of CD44 protein was analyzed by western blot in whole-cell protein extracts of parental and Dicerex5 cells. Nucleolin expression was used as loading control. (d) Results (mean±s.e.) of the CD44 mRNA quantification in parental and Dicerex5 cells. Results were calculated as mRNA levels in Dicerex5 cells relative to their respective parental cells. ‘*' indicates significant differences in CD44 expression between parental and Dicer mutant cells (t-test; P<0.05).
DICER1 defects are linked to the generation of subpopulations of colorectal cancer cells with enhanced stem cell features
We next investigated the molecular features of the population of Dicerex5 cells with the highest CD44 expression identified by flow cytometry (Figure 1a). Although most (98.1%) of the parental HCT-116 cells expressed high levels of the epithelial marker EpCAM (Figure 2a),37 we identified in HCT-116-Dicerex5 cells two subpopulations: CD44high/EpCAMlow and CD44high/EpCAMhigh cells (Figure 2b).
Figure 2.
The generation of a population of CD44high/EpCAMlow cells with enhanced expression of stem cell markers is linked to impaired Dicer function in HCT-116 cells. (a) Quantification of the percentage of EpCAM+ cells in HCT-116 parental and Dicerex5 cells by flow cytometry. The percentage of EpCAM+, EpCAMhigh (high) and EpCAMlow cells (low) are indicated in each panel. (b) Representative results of flow cytometry assays, showing the distribution of the CD44+/EpCAM+ cells in HCT-116 parental and Dicerex5 cells. Limit lines were adjusted to allow the quantification of the percentage of cells expressing high levels of CD44 and low levels of EpCAM proteins. Numbers inside the panels indicate the percentage of each cell population in this assay. Dashed square mark out CD44low/EpCAMhigh and CD44high/EpCAMhigh cells, and circles indicate CD44high/EpCAMlow cells. (c) CD44 and EpCAM protein levels in CD44low/EpCAMhigh and CD44high/EpCAMlow cells were analyzed by western blot assay. Nucleolin and β-actin expression were used as loading control. (d) Representative immunoflurescence images showing the expression of CD44 and EpCAM in HCT-116 parental and CD44low/EpCAMhigh and CD44high/EpCAMlow subsets, isolated from HCT-116-Dicerex5 cells. Nuclei were stained with DAPI. Scale bars, 30 μm. (e) Quantification of CD44, LGR5 and SOX2 mRNA by qRT–PCR in the indicated HCT-116-Dicerex5 cell subpopulations. The results show the mean±s.e. of mRNA levels of CD44high/EpCAMhigh and CD44high/EpCAMlow cells relative to CD44low/EpCAMhigh cells (2–3 samples for each population were analyzed). ‘*' indicates significant differences between both cell populations (t-test; P<0.05).
To further characterize these subpopulations of CD44high cells, which only appeared with DICER1 impairment, we isolated these fractions by sorter-fluorescence-activated cell sorting (FACS) and compared them with the CD44low/EpCAMhigh cells. All subpopulations of HCT-116-Dicerex5 cells exhibited upregulated CD44 expression compared with that in the parental cells, as quantified by qRT–PCR (1.75±0.1-fold increase in CD44low/EpCAMhigh cells, 3.61±0.39-fold increase in CD44high/EpCAMhigh cells and 4.68±0.65-fold increase in CD44high/EpCAMlow cells). Furthermore, western blot (Figure 2c) and immunofluorescence assays (Figure 2d) confirmed that CD44high/EpCAMlow cells had higher levels of CD44 and lower levels of EpCAM than CD44low/EpCAMhigh cells. In addition, the expression of Sox2 significantly increased in both populations of CD44high cells, although only the CD44high/EpCAMlow cells showed an upregulation in the expression of Lgr5 compared with CD44low/EpCAMhigh cells (Figure 2e). These results indicate that the CD44high cell subset, which exhibit enhanced stem cell features, is formed by a heterogeneous population of cells, distinguishable by EpCAM and Lgr5 expression.
When CD44low/EpCAMhigh cells were regrown in culture, they were able to reconstitute the cell heterogeneity of the original HCT-116-Dicerex5 cells (Supplementary Figure S2). In contrast, fewer than 20% of CD44low/EpCAMhigh cells were recovered from the CD44high/EpCAMlow population after 14 days of culture (Supplementary Figure S2). Given that both cell population have the same genetic mutation (Supplementary Figure S2), our data indicate that the two subsets of cells show a degree of plasticity, and CD44high/EpCAMlow cells may be dynamically generated from CD44low/EpCAMhigh cells, rapidly reconstituting HCT-116-Dicerex5 cell heterogeneity.
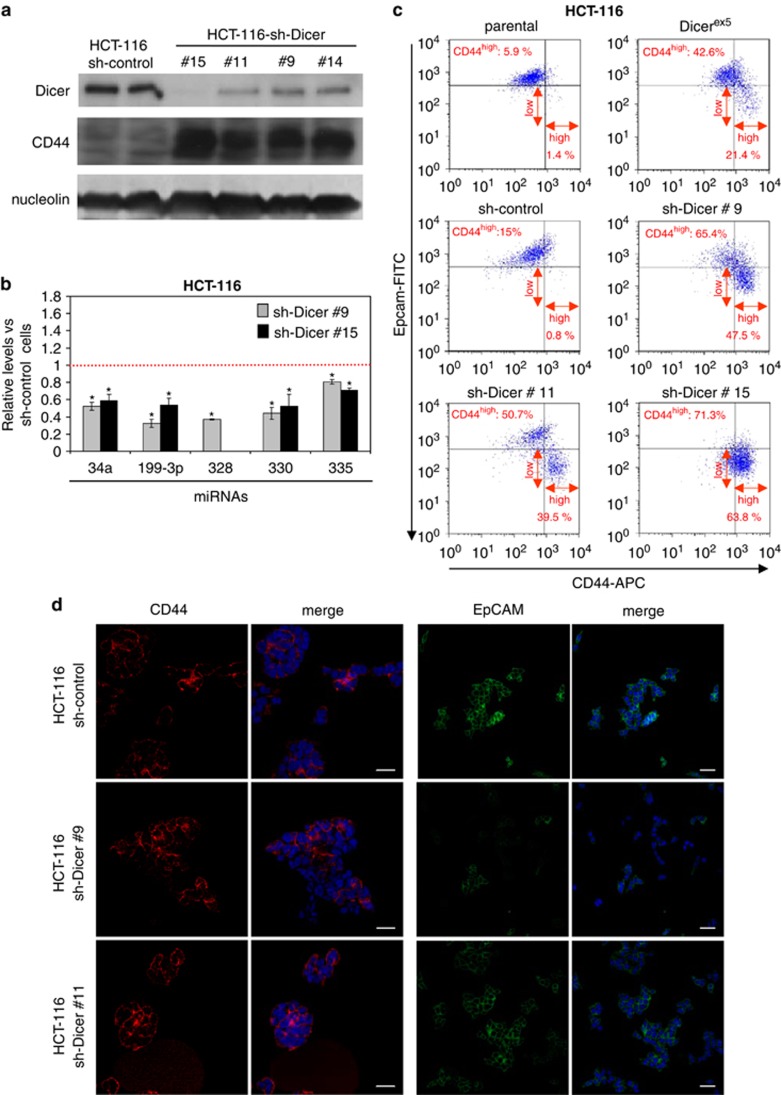
We also confirmed that the generation of the CD44high cells and, specifically of CD44high/EpCAMlow subpopulation, is directly dependent on the impaired function of Dicer by knocking down DICER1 expression using short hairpin RNA (shRNAs) in HCT-116 parental cells (Figure 3). Using four shRNA constructs, we induced a large decrease in DICER1 protein expression (Figure 3a) that was associated with a significant reduction in the expression levels of mature miRNAs (Figure 3b). As we have described above for the genetically impaired Dicerex5 cells, shRNA-mediated depletion of DICER1 induced the upregulation of CD44 expression and downregulation of EpCAM expression in HCT-116 cells (Figures 3c and d). Most importantly, a subpopulation of CD44high/EpCAMlow cells emerged specifically within the HCT-116 shRNA-Dicer cells. These were more abundant in cells with weaker Dicer expression (Figure 3c). We were also able to re-introduce the wild-type DICER1 in HCT-116-Dicerex5 cells. We observed that this DICER1 stably transfected clone diminishes the CD44high cell subset and its two subpopulations, CD44high/EpCAMlow and CD44high/EpCAMhigh (Supplementary Figure S3).
Figure 3.
DICER1 knockdown induces the generation of CD44high/EpCAMlow subset of cells in HCT-116. (a) Dicer and CD44 protein levels were determined by western blot analysis in whole-cell protein extracts of sh-control cells and of different clones of sh-DICER1 cells. Nucleolin protein was used as loading control. (b) Quantification by qRT–PCR of mature miRNAs involved in CD44 and the Wnt/β-catenin pathway regulation. Results represent the levels (mean±s.e.) of the indicated miRNAs in two different clones of sh-DICER1 cells relative to sh-control cells. ‘*' indicates significant differences between the compared cells (t-test; P<0.05). (c) Representative results of flow cytometry assays showing the generation of the CD44high/EpCAMlow subpopulation in sh-DICER1 cells. The percentage of this cell subpopulation is indicated in red for each analyzed cell line. (d) Representative images showing CD44 and EpCAM expressing cells in sh-control and sh-DICER1 cells, as analyzed by immunofluorescence assays. Nuclei were stained with DAPI. Scale bars, 30 μm.
Global decrease of miRNA expression in DICER1-impaired cells mediates cancer stemness
Once we had shown that the impaired function of DICER1 enhanced the stem cell features of colon cancer, in particular generating a population of CD44high cells, we sought to identify a mechanism that might explain this phenotype. One possibility is that, within the spectrum of reduced production of mature miRNAs in Dicerex5 cells,31 miRNAs targeting key coding genes in stemness features could be critically affected.
In this regard, different miRNAs targeting and downregulating CD44 have been identified in human cells, such as miR-34a,38 miR-199-3p,39 miR-328,40 miR-33041 and miR-520.42 Herein, we found that expression levels for the five miRNAs were significantly diminished in HCT-116-Dicerex5, DLD-1-Dicerex5 and RKO-Dicerex5 in comparison with the corresponding parental cell lines (Figure 4a). Downregulation of most of these miRNAs was also observed in DICER1 shRNA-depleted HCT-116 cells (Figure 3b). Stable transfection of DICER1 in HCT-116-Dicerex5 increased miRNA expression (Supplementary Figure S3). As CD44high/EpCAMhigh and CD44high/EpCAMlow cells showed a similar upregulation of CD44 expression, we next compared the levels of this subset of miRNAs between CD44high/EpCAMlow and CD44low/EpCAMhigh cells. Importantly, the studied miRNAs were also significantly decreased in the subpopulation of CD44high/EpCAMlow cells compared with the CD44low/EpCAMhigh set (Figure 4b). These differences were not associated with a higher downregulation of the DICER1 protein in CD44high/EpCAMlow cells (Supplementary Figure S4). To show that the observed effects were not restricted to microsatellite unstable colorectal cancer cells such as HCT-116, we depleted DICER1 expression by the shRNA approach in the microsatellite stable SW480 colorectal cancer cell line. Following efficient DICER1 depletion validated by western blot, we have observed that DICER1 shRNA-mediated downregulation is associated with a diminished expression of the studied mature miRNAs and an upregulation of the stem cell markers CD44, Sox 9 and Sox2 (Supplementary Figure S5). Thus, it resembles the data derived from the DICER1 genetic disruption in the microsatellite unstable cells. Therefore, we conclude that DICER1 impairment causes the downregulation of miRNAs that target CD44 expression. This could help explain the emergence of this cancer stem cell marker in the molecular context under consideration here.
Figure 4.
Reduction of multiple miRNAs that target CD44 and the Wnt/β-catenin pathway mediates the upregulation of CD44 expression in cells with impaired Dicer function. (a) Quantification of mature CD44-targeting miRNAs by qRT–PCR. Results (mean±s.e. from three independent assays) show the levels of the indicated miRNAs in Dicerex5 cells relative to their respective parental cells. (b) Results (mean±s.e. from three independent assays) indicate the levels of the indicated miRNAs in CD44high/EpCAMlow cells relative to CD44low/EpCAMhigh cells. (c) Representative images showing the cellular localization of β-catenin in HCT-116 and DLD-1 parental and Dicerex5 cells (upper panel), and in sh-control and Dicer knockdown cells (lower panel). Cells exhibiting nuclear location of β-catenin are indicated with white arrowhead. Scale bars, 30 μm. (d) Quantification of AXIN2, CCDN1 and OLFM4 mRNAs in the indicated cells. Mean of mRNA levels±s.e. in each cell population is represented as relative value to their respective parental cells or to CD44low/EpCAMhigh cells. OLFM4 expression was not detected (nd) in HCT-116 cells. (e) Results (mean±s.e.) indicate the levels of indicated miRNA that regulate the Wnt/β-catenin pathway in Dicerex5 cells relative to their respective parental cells. (f) Results show the levels (mean±s.e. from 2–3 independent assays) of the indicated miRNAs in CD44high/EpCAMlow cells relative to CD44low/EpCAMhigh cells. ‘*' indicates significant differences between compared cell populations (t-test; P<0.05).
The Wnt/β-catenin signaling is another miRNA-targeted pathway that could contribute to the upregulation of CD44 in DICER1 hypomorphic cells. The expression of CD44 is transcriptionally induced by β-catenin/T-cell factor (TCF) in response to the activation of the pathway.43 Although two of the studied colon cancer cell lines, HCT-116 and DLD-1, carry a heterozygous mutation in the CTNNB1 and APC locus, respectively, leading to an activated β-catenin signaling,44, 45, 46 this pathway can be further upregulated by the alteration of miRNA homeostasis.47 In this regard, we found that the frequency of cells with β-catenin labeling within the nucleus increased in Dicerex5 cell lines and HCT-116-Dicer knockdown cells compared with parental and HCT-116-shRNA-control cells, respectively (Figure 4c). To further determine the induction of β-catenin activity upon Dicer impairment, we used the 7TGC lentivirus in which seven binding sites of Tcf/Lef proteins drive the expression of the EGFP gene reporter, whereas transduced cells are detected by SV40 promoter-driven mCherry expression (Supplementary Figure S6). We found a significant increase of enhanced green fluorescent protein (EGFP+)/mCherry+ cells in HCT-116 Dicer mutant cells compared with parental cells, as determined by direct fluorescence detection by microscopy and flow cytometry quantification (Supplementary Figure 6). In contrast to the low percentage of HCT-116 parental cells exhibiting activated β-catenin, the 66.4% of DLD-1 parental cells showed a strong β-catenin activation, and not so significant increase in the percentage of EGFP+/mCherry+ cells was detected in DLD-1-Dicerex5 cells (Supplementary Figure S6). Parental RKO or RKO-Dicer mutant cells did not show detectable β-catenin expression by western blot, and β-catenin activation was not detected in these cells (Supplementary Figure S6). In agreement with the HCT-116 and DLD-1 data, we observed an upregulation of β-catenin target genes, such as AXIN2, CCND1 and OLFM4 (Figure 4d), along with the aforementioned LGR5 (Supplementary Figure S1). Therefore, we analyzed the levels of previously reported miRNAs that regulate the Wnt/β-catenin pathway in Dicerex5 cells. Levels of miR-15, miR-16-1, miR-25 and miR-335, which directly or indirectly target different modulators of the Wnt/β-catenin pathway,48, 49, 50 as well as the previously mentioned miR-34a, which also negatively regulates the Wnt/β-catenin pathway,51 were significantly reduced in Dicerex5 cells relative to parental cells (Figure 4e). Consistent with these findings, miRNAs targeting the Wnt/β-catenin pathway were even more downregulated in CD44high/EpCAMlow cells than in CD44low/EpCAMhigh cells (Figure 4f). Thus, the impaired expression of miRNAs targeting the Wnt/β-catenin pathway is another mechanism that helps explain the enhancement of CD44 expression in DICER1-compromised cells.
DICER1 impairment increases tumor initiation potential
Once we had demonstrated that DICER1 impairment was associated with the acquisition of cancer stem cell features, we next considered whether this stemness phenotype was associated with particular tumor biology properties. The most obvious approach was to assess its impact on tumor growth. We found that HCT-116-Dicerex5, DLD-1-Dicerex5 and RKO-Dicerex5 showed a reduced proliferation rate, measured by the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay, in comparison with the parental colorectal cancer cell lines (Figure 5a and Supplementary Figure S7). DICER1 shRNA-depleted HCT-116 cells also grew slower than shRNA-scrambled cells (Supplementary Figure S7). The same phenomenon was observed in the selected subpopulations: CD44high/EpCAMlow cells proliferated significantly more slowly than CD44low/EpCAMhigh and CD44high/EpCAMhigh cells (Figure 5b). Subcutaneous injection of HCT-116-, HCT-116-Dicerex5- and HCT-116-shRNA DICER1-depleted cells in immunodeficient mice demonstrated that the tumors generated from the DICER1-impaired cells grew more slowly than those generated from the same amount of HCT-116 parental or shRNA-control cells (Figures 5c and e and Supplementary Figure S7). In the isolated subpopulations, tumors generated from CD44high/EpCAMlow cells grew more slowly than CD44low/EpCAMhigh cells or CD44high/EpCAMhigh cells (Figures 5d and e).
Figure 5.
Dicer-impaired cells show a reduced cell proliferation and enhanced tumor-initiating ability. (a, b) Cell proliferation was compared (a) between HCT-116 parental and Dicerex5 cells, and (b) between CD44low/EpCAMhigh, CD44high/EpCAMhigh and CD44high/EpCAMlow cells by MTT assays. Results represent absorbance (mean±s.e.) of each time point tested. (c, d) Results (mean±s.e. of tumor volume) represent tumor growth kinetics after injection of c HCT-116 parental and Dicerex5 cells (n=10 mice), or (d) CD44low/EpCAMhigh, CD44high/EpCAMhigh and CD44high/EpCAMlow cells (n=10 mice) in immunodeficient mice. ‘*' indicates significant differences between the examined cells (t-test; P<0.05). (e) Images showing tumors generated after injection of the indicated cells at the time of mice kill. (f) Serial dilutions of the indicated cells were injected in immunodeficient mice (10 mice injected for each cell dilution). Days to tumor detection was recorded for each cell dilution and cell population. Results shown on the right column represent the frequency of TICs in each cell population and their confidence intervals, as calculated by using ELDA software.
The slow tumor growth observed in DICER1-deficient cells could be a consequence of the impaired proliferation of these cells and might not directly reflect their ability to initiate tumor growth. To determine the frequency of tumor-initiating cells in HCT-116 with proficient (parental and shRNA-control cells) and deficient DICER1 function (Dicerex5- and shRNA-depleted DICER1 cells), we injected serial dilutions of these cells into immunodeficient mice and monitored the generation and growth of tumors over 5 weeks. We observed that, although the initial detection of tumors was frequently delayed in mice injected with Dicerex5- or shRNA-depleted DICER1 cells, these cells featured a higher frequency of tumor-initiating cells compared with the parental or shRNA-control samples (Figure 5f and Supplementary Figure S7). Interestingly, while the isolated CD44low/EpCAMhigh subpopulation presented a similar tumor-initiating cell frequency than heterogeneous HCT-116-Dicerex5 cells, both populations of CD44high cells (CD44high/EpCAMhigh and CD44high/EpCAMlow cells) showed a significant enrichment in tumor-initiating cells (Figure 5f). We conclude that DICER1 defects are associated with an enhanced tumor-initiating ability that is related to the gain of cancer stem cells, identified by a high expression of the CD44 marker.
DICER1 impairment induces an EMT transition and enhances metastatic potential
In addition to an increase in the frequency of tumor-initiating cells, DICER1 impairment could also be related to a higher risk of cancer progression and dissemination. Data from previous studies suggest that a low level of Dicer expression is associated with a greater predisposition to develop metastasis,25, 52, 53 although, on the other hand, epithelial cancer stem cells exhibit some features of mesenchymal cells, and the EMT transition is associated with acquisition of stemness and distal metastasis.54, 55, 56, 57, 58 Thus, we wondered whether DICER1 hypomorphic cells, with enhanced cancer stem cell features, also show an acquisition of mesenchymal features and are prone to metastasize.
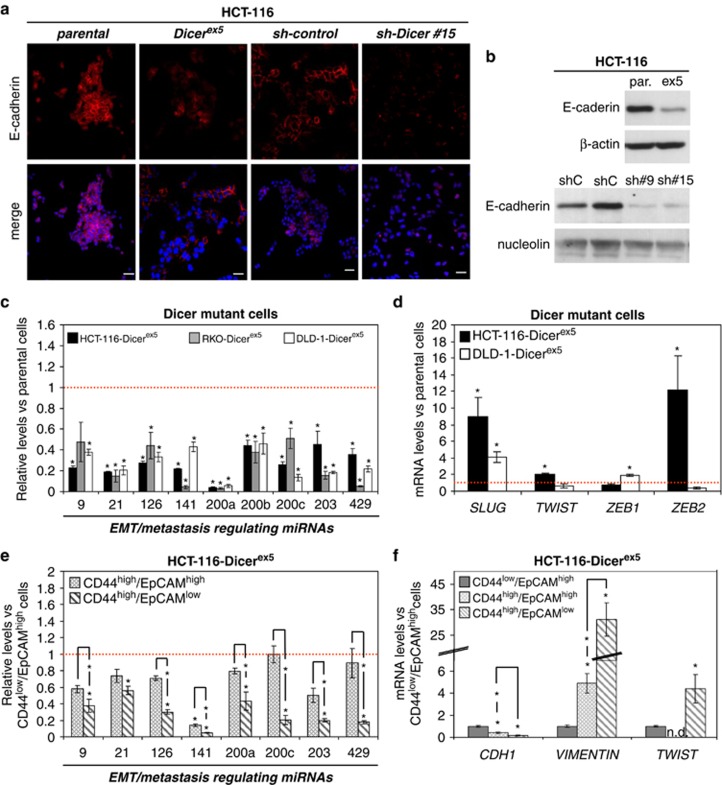
With respect to the EMT transition, we observed that both Dicerex5 and HCT-116 shRNA-depleted DICER1 cells, relative to parental and shRNA control cells, experienced a significant reduction in E-cadherin expression (Figures 6a and b and Supplementary Figure S8) and an upregulation of the mesenchymal marker Vimentin (Supplementary Figure S8). Thus, in addition to exhibiting a cancer stem cell pattern, DICER1-impaired cells undergo an EMT transition that could also be explained by a defect in the production of the miRNAs controlling these features. We found in HCT-116-Dicerex5, DLD-1-Dicerex5 and RKO-Dicerex5 cells that all the EMT-regulating miRNAs examined were significantly downregulated (Figure 6c), including miR-200 family members (that is, miR-200a, miR-200b, miR-200c, miR-141 and miR-429), which directly target the expression of Zeb1 and Zeb2.59, 60, 61 A similar downregulation of miR-200 members was also observed in DICER1 shRNA-depleted cells (Supplementary Figure S8). This was not just an in vitro observation. We analyzed the DICER1 expression levels in the primary colorectal tumors (n=140) available from The Cancer Genome Atlas. Herein, we observed that cases with lower expression of DICER1 presented a significant reduced expression of the epithelial marker E-cadherin (analysis of variance (ANOVA)-test, P=0.01) and the EMT regulator miR-200c (ANOVA test, P=0.05) (Supplementary Figure S9). Thus, it resembles the data derived from the DICER1 genetic disruption in the cell lines, where these targets were also downregulated.
Figure 6.
EMT is induced in cells with impaired Dicer function, coinciding with a global decrease of EMT and metastasis suppressor miRNAs. (a) Representative images of E-cadherin immunodetection in HCT-116 parental and Dicerex5 cells, as well as in sh-control and sh-DICER1 cells. Nuclei were stained with DAPI. Scale bars, 30 μm. (b) The levels of E-cadherin protein were determined by western blot assays in the indicated cell lines. Nucleolin and β-actin were used as loading control. (c) Quantification by qRT–PCR of mature miRNAs that regulate EMT program and metastasis in the indicated cells. Results represent mean±s.e. of miRNA levels in Dicerex5 cells relative to their parental cells. (d) The expression of EMT-inducer transcription factors was quantified by qRT–PCR. Results (mean±s.e.) were expressed as mRNA levels in Dicerex5 cells relative to parental cells. (e) Levels (mean±s.e.) of the indicated miRNAs in CD44high/EpCAMhigh and CD44high/EPCAMlow cells relative to CD44low/EpCAMhigh cells, as quantified by qRT–PCR. (f) The expression of Twist (EMT-inducer transcription factor), as well as CDH1 (E-cadherin) and VIMENTIN was quantified by qRT–PCR in the indicated cells. Results (mean±s.e.) indicate the mRNA levels in CD44high/EpCAMhigh and CD44high/EpCAMlow cells relative to CD44low/EpCAMhigh cells. ‘*' indicates significant differences between the compared cells (t-test; P<0.05).
Following the downregulation of these miRNAs in the colorectal cancer cell lines, we found upregulation of most EMT-inducing transcriptional factors, such as the Zeb, Slug and Twist proteins, in Dicerex5 cells (Figure 6d). It is of particular note that all the EMT-regulating miRNAs analyzed were further reduced in CD44high/EpCAMlow cells than in CD44low/EpCAMhigh cells (Figure 6e), and in a higher magnitude than in the other identified subpopulation with enhanced stem cell features (CD44high/EpCAMhigh) (Figure 6e). A further increase of Twist and Vimentin expression, and a decrease in E-cadherin expression were also consistently observed in the CD44high/EpCAMlow subpopulation, whereas the subset of CD44high/EpCAMhigh cells showed a lower induction of these EMT markers (Figure 6f). Thus, the EMT process may be further induced in the subpopulation of Dicerex5 cells with the highest level of expression of cancer stem cell markers and reduced expression of EpCAM. The enhanced cancer progression of DICER1-deficient cells is also highlighted by the observation that other miRNAs inhibiting metastasis formation and angiogenesis, such as miR-126 (Figure 6c),62, 63, 64, 65 miR-34a (Figure 4a) and miR-335 (Figure 4e),38, 66, 67 were also downregulated in Dicerex5 cells.
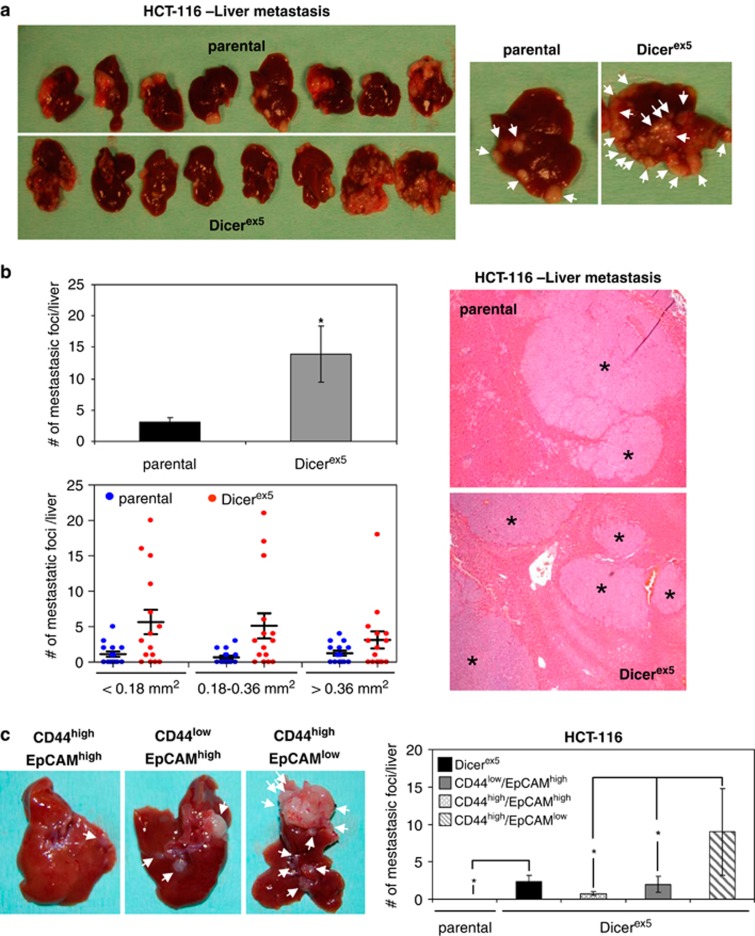
Finally, as with the growth and tumor-initiating experiments, we also conducted our study of metastasis formation in DICER1-impaired cells in an in vivo context. We injected equal amounts (2 × 106 cells) of HCT-116 and HCT-116-Dicerex5 cells into the spleens of immunodeficient mice. To check the dissemination ability of these cells in the absence of tumor growth, spleens were resected 48 h after tumor cell injection, and the generation of liver metastasis was quantified 7 weeks later. Following this procedure, HCT-116 parental and Dicerex5 cells were able to generate multiple metastatic lesions in the liver. Macroscopical analysis of the liver revealed that Dicerex5 cells induced smaller metastatic lesions than those cells seen in the parental cells (Figure 7a), which is entirely consistent with the slow tumor growth associated with DICER1 impairment described above (Figure 5c). However, and most importantly, when serial sections were microscopically analyzed, we found that Dicerex5 cells had developed significantly more metastatic foci in the liver than was the case with the parental cells (Figure 7b). Next, we compared the capacity of isolated subpopulations of HCT-116-Dicerex5 cells to initiate liver metastasis by injecting a more reduced amount of cells (3 × 105). HCT-116-Dicerex5 cells generated significantly more metastatic lesions than HCT-116 parental cells (Figure 7c). Importantly, we found that CD44high/EpCAMlow cells developed a significantly higher number of metastatic lesions than the other subpopulations of HCT-116 Dicer mutant cells (Figure 7c). These results indicate that DICER1 defects in colon cancer enhance the production of metastasis-initiating cells, which are characterized by a high expression of CD44, a reduced expression of EpCAM and the enhancement of EMT markers.
Figure 7.
Dicer-impaired function in colon cancer cells promotes metastasis, associated to CD44high/EpCAMlow cells. (a) Images show macroscopic metastatic lesions (indicated by white arrows on the right panel) developed in the liver of immunodeficient mice after injecting 2 × 106 of HCT-116 parental and Dicerex5 cells in the spleen. (b) Representative images of liver section from mice injected with parental and Dicerex5 cells after hematoxylin and eosin staining (right panel). ‘*' indicates the metastatic foci identified in these samples. The quantification of metastatic foci detected per liver (mean± s.e.) is showed in the upper left panel, and the number of these metatastic foci categorized according to size is represented in the lower left panel. ‘*' indicates significant differences between parental and Dicer mutant cells (t-test; P<0.05). (c) Comparison of the metastasis-initiating capacity between the separate HCT-116-Dicerex5 cells. Images show macroscopic metastasis developed in the liver of mice after intra-spleen injection of the CD44low/EpCAMhigh-, CD44high/EPCAMhigh- and CD44high/EpCAMlow-isolated cells. White arrows indicate metastatic lesions (left panel). Results on the right panel show the number of metastatic lesions developed per liver (t-test; P<0.05).
Discussion
A current hypothesis in cancer cell biology suggests that tumor growth is sustained by a specific population of tumor cells, the cancer stem cells, which exhibit features reminiscent of adult stem cells, such as the ability to self-renew and differentiate.30 Cancer stem cells have been identified in most solid tumors by using different stem cell surface markers, including CD133, CD44, EpCAM, CD24 and Lgr5, or the expression of ALDH1.30 Moreover, cancer stem cells and adult stem cells resemble their gene expression profiles to a degree.68, 69 The activation of this stem cell signature in human epithelial cancers is correlated with recurrent disease, metastasis and poor prognosis.70, 71 Cancer stem cells are important in clinical cancer because they are more resistant to antitumor therapies than most other types of tumor cell.55, 72, 73, 74, 75, 76, 77 In addition, cancer stem cells have some features that make them likely candidates for metastasis-initiating activities, such as the expression of EMT markers, which is associated with the migration of tumor cells and the ability to initiate tumor growth in distal tissues.54, 55, 56, 57, 58 An increasing body of evidence indicates that the establishment and maintenance of cancer stem cell features can be orchestrated by genetic and epigenetic mechanisms.78 Our results indicate that defects in one of the main enzymes involved in the generation of miRNAs, DICER1, contribute to the generation of cancer stem cells.
Several studies have demonstrated the role of specific miRNAs regulating self-renewal/differentiation of tumor cells, EMT and metastasis78, 79 in recent years. In addition, downregulation of Dicer in cancer cells promotes metastasis.26, 53 However, data regarding the correlation of altered Dicer expression and colon cancer progression are scarce.22, 24, 80 In this study, we have demonstrated that DICER1 hypomorphic colorectal cells show significantly stronger expression not only of CD44, but also of other cancer stem cell markers such as Sox9, Sox2, Lgr5 and Nanog. In addition, Dicerex5 cells generated not only a cellular population that show high levels of CD44 and an enhanced tumor-initiating capability, but also a specific subpopulation of CD44high/EpCAMlow cells endowed with metastasis-initiating activity. These subpopulations of cells also appeared in DICER1 shRNA-depleted cells, and their abundances were decreased after restoring the DICER1 function, indicating that the promotion of stemness and metastastic features depends on the impairment of Dicer function. The subset of colorectal cancer cells considered here is highly reminiscent of the one observed in melanoma: cells with reduced proliferation rate, enhanced cancer stem cell features and EMT traits.81, 82 Overall, high levels of expression of EpCAM have been found in cells with higher proliferation rates,83, 84 whereas diminished levels of EpCAM are associated with cell invasion85 and dissemination.86 These observations are in agreement with our data, as the CD44high/EpCAMlow cells, with lower proliferation rate, showed the strongest EMT induction and metastasis generating activity. From a mechanistic standpoint, the appearance of stemness, loss of epithelial characteristics and gain of mesenchymal features, and increased metastatic potential in DICER1-impaired cells are associated with the defective production of the miRNAs that regulate these pathways, such as miR-199-3p,39 miR-328,40 miR-330,41 miR-520,42 miR-126,62, 63, 64, 65 miR-34a, miR-335,38, 66, 67 miR-15, miR-16-1, miR-25 and miR-200 family members,59, 60, 61 which were further decreased in the subset of CD44high/EpCAMlow cell population.
One critical finding of this study is that DICER1-deficient colorectal cancer cells, although not having an obvious proliferative advantage, have an enhanced ability to initiate tumors and metastasis. In this regard, it has been demonstrated that the overexpression of CD44 promotes metastasis in prostate and colon cancer cells,38, 87, 88 and, independently, that a consequence of the deregulation of miRNAs promoting cancer stem cell features is the induction of metastasis.78 Our study shows that the two processes are linked: DICER1 hypomorphic-mediated loss of miRNAs induces cancer stem cell properties associated with enhanced tumor dissemination.
Materials and methods
Cell culture
Parental HCT-116, RKO and DLD-1 colorectal cancer cell lines and the isogenic Dicerex5 cells were a generous gift from Dr Bert Vogelstein (Johns Hopkins Kimmel Comprehensive Cancer Center, Baltimore, MD, USA). All cells were grown in DMEM (GIBCO Life Technologies, Auckland, New Zeland) supplemented with 10% fetal bovine serum (FBS; Life Technologies, Foster City, CA, USA) and 1% penicillin/streptomycin (PAA Laboratories, Pasching, Austria) in a humidified 37 °C, 5% CO2 incubator.
DicerEx5Genomic PCR
Genomic DNA was extracted and PCR was performed to amplify across the Dicer-exon5 insertion point, as previously described.31
Dicer shRNA and cell transductions
Parental HCT-116 and SW480 cells were transfected with Human 29mer shRNA (pRS HuSH) constructs against DICER1 or shRNA-control (scramble) constructs (OriGene Technologies, Inc, Rockville, MD, USA). Stable knockdown clones were selected in the presence of 0.5 μg/ml puromycin and then propagated in the presence of 0.3 μg/ml puromycin. Knockdown efficiency was evaluated 48h post transfection by western blot using anti-Dicer antibodies. For the lentiviral transduction, lentivirus was produced in 293T using standard protocols, and psPAX2 and pMD2.G as packaging vectors. TCF/LEF reporter driving expression of EGFP (7TGC) was used to compare the activity of Wnt/β-catenin signaling in HCT-116, DLD-1 and RKO parental and Dicerex5 cells. EGFP and mCherry expression was evaluated by flow cytometry analysis or direct fluorescence detection by confocal microscope. Construction of DICER1-Flag-pLPC retroviral expression vector: The DICER1-Flag-pLPC retroviral expression vector was constructed by subcloning the DICER1-Flag frame, using Hind III/EcoRI restriction sites, from the pCMV-Tag4B-DICER1 construct into an empty pLPC vector.
Immunofluorescence assays
Cells were cultured directly on coverslips, washed with PBS and fixed in 4% paraformaldehyde. They were permeabilized with 0.1% Triton X-100 in PBS, blocked with 5% bovine serum albumin (BSA) in PBS for 1 h at room temperature (RT) and incubated with primary antibodies in 2% BSA/PBS overnight at 4 °C. Cells were washed with PBS containing 0.1% Tween-20 (PBS-T) and incubated with Alexa Fluor 488- and/or 568-conjugated antibodies (1:1000) in 2% BSA/PBS-T for 1 h at RT. DNA was counterstained with 4′, 6-diamidino-2-phenylindole (DAPI). Cells were visualized using a Leica TCS SP5 confocal microscope (Leica Microsystems, Heidelberg GMbH, Germany). The following antibodies were used: anti-CD44 (1:250; Cell Signaling, Danvers, MA, USA), anti-E-cadherin (1:100; Abcam, Cambridge, UK), anti-β catenin (1:100; Abcam); and anti-Vimentin (1:100; Millipore, Billerica, MA, USA). For EpCAM staining, a direct immunofluorescence protocol was followed, incubating cells directly with a fluorescent primary antibody anti-Epcam-FITC (1:5; Becton Dickinson, San Jose, CA, USA) for 2 h at RT.
Western blot analysis
Whole-cell protein extracts were resolved in 6–15% Bis-Tris SDS–polyacrylamide gel for electrophoresis (Bio-Rad, München, Germany) and transferred onto a nitrocellulose membrane (Amersham, Buckinghamshire, UK). After transfer, proteins were incubated with the following antibodies: anti-CD44 (1:1000; Cell Signaling); anti-Dicer (1:1000; Abcam), anti-E-cadherin (1:500; Abcam), anti-β catenin (1:500; Millipore), anti-Epcam (1:1000; Abcam), anti-nucleolin (1:1000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), anti-tubulin (1:5000; Sigma, St Louis, MO, USA) and anti-β actin (1:20 000; Sigma). Antibody binding was detected with a secondary antibody coupled to horseradish peroxidase using enhanced chemiluminescence.
qRT–PCR assays
Total RNA was isolated using TRIzol reagent (Invitrogen, Frederick, MD, USA) according to the manufacturer's instructions. RNA samples were DNase I-treated (Turbo DNA-free, Ambion, Austin, TX, USA), and reverse-transcribed using oligo(dT)-primer and the ThermoScript RT–PCR System (Invitrogen). Real-time PCRs were performed using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), and ACTIN, GUSB and HPRT1 expression was used for housekeeping purposes. Primer sequences are available in Supplementary Table S1. A TaqMan miRNA Reverse Transcription Kit (Applied Biosystems) was used in conjunction with TaqMan miRNA assays to quantify the levels of mature miRNAs, as previously described.89
Flow cytometry analysis and sorting
Cell were harvested by trypsin treatment and suspensions were prepared shortly before analysis. For flow cytometry analysis, cells (2 × 105 cells/100 μl buffer, 5% FBS in PBS) were stained with anti-EpCAM-FITC (Becton Dickinson), anti-CD133/1-PE (Miltenyi Biotec, Bergisch Gladbach, Germany), anti-CD166-PE (R&D Systems, Minneapolis, MN, USA) and/or monoclonal anti-CD44-APC (BD Pharmingen, San Jose, CA, USA) for 30 min at 4 °C. Cells were washed twice with washing buffer (0.5% BSA/2 mM EDTA in PBS) and were finally diluted in 300 μl analysis buffer (2.5% FBS/2 mM EDTA in PBS). For FACS-sorting, 106 cells were incubated in 700 μl blocking buffer in the presence of anti-CD44-APC and anti-EpCAM-FITC, and cells were finally resuspended in 700 μl analysis buffer. The dead cells and debris were excluded after propidium iodide staining (Sigma). Analyses of sorting experiments were performed with a BD Biosciences (San Jose, CA, USA) FACSaria apparatus.
MTT assay
Cell viability was measured daily for 7 days using the MTT assay. Briefly, 5 × 102 cells were seeded per well in replicates on 96-well plates. Cell proliferation was calculated by measuring absorbance (mean±s.e.) at 595 nm every 24 h.
In vivo tumor and metastasis development assays
Parental or Dicerex5 cells (106 cells in 100 μl of DMEM), or CD44low/Epcamhigh-and CD44high/Epcamlow-isolated cells (4000 cells in 100 μl of DMEM) were mixed with BD matrigel (BD Biosciences) at 1:1 ratio and then subcutaneously injected into the dorsal side of each flank of 6-week-old male athymic nude mice (Harlan Laboratories, Indianapolis, IN, USA). Tumor development was monitored on average of every 2 days for 1–3 months. Mice were killed when their tumors reached a critical size. To carry out limited dilution assays, 102, 103, 104 and 105 HCT-116 parental and Dicerex5 cells or sh-DICER1 and sh-control or CD44low/EpCAMhigh, CD44high/EpCAMhigh and CD44high/EpCAMlow cells were subcutaneously injected in each dorsal flank of nude mice. The presence and size of tumors were monitored every 2 days for 4–5 weeks. Tumor-initiating cells frequency was calculated using the ELDA program (http://bioinf.wehi.edu.au/software/elda). To test the ability of HCT-116 parental and Dicerex5 cells to develop metastasis, 2 × 106 or 3 × 105 of HCT-116 parental and Dicerex5 cells or 3 × 105 CD44low/EpCAMhigh, CD44high/EpCAMhigh and CD44high/EpCAMlow cells were injected into the spleens of anesthetized mice. Spleens were resected 48 h after injection, and hepatic metastases were examined 5–7 weeks later. Metastatic foci were counted under a bright-field microscope (Zeiss AX10 Scope.A1) at × 20 magnification. Metastatic lesions in the liver were categorized into three groups according to the area of the parenchyma covered (<0.18; 0.18–0.36; and >0.36 mm2), and the number of foci in each category was recorded. Animal housing and handling, and all procedures involving mice were approved by the Bellvitge Biomedical Research Institute (IDIBELL) Ethics Committee.
DICER1 expression analysis in primary colorectal tumor samples
Gene (UNC-AgilentG4502A-07-3) and miRNA (BCGSC-IlluminaGA-miRNASeq) expression data for primary colorectal adenocarcinoma samples (n=140) was obtained from The Cancer Genome Atlas data portal (https://tcga-data.nci.nih.gov/tcga). Samples were grouped according to their DICER1 expression levels. Therefore, the samples were split in octiles, and the groups with highest and lowest DICER1 expression were used for subsequent comparisons. Significant differences in candidate gene expression between the groups were assessed using the ANOVA test.
Acknowledgments
We thank Dr Bert Vogelstein (Johns Hopkins Kimmel Comprehensive Cancer Center) for the generous gift of the parental and Dicerex5 isogenic cell lines; R Alvarez, J Comas and E Castaño (Universitat de Barcelona-SCT) for technical support with flow cytometry; and the staff of the IDIBELL animal facility for mouse care. This work was supported by grants from the Spanish Ministry of Science and Innovation (SAF2008-01173 and SAF2011–22803), the European Research Council Advanced Grant EPINORC no. 268626, the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreements no. HEALTH-F2-2011-259015—COLTHERES project and HEALTH-F5-2010-258236—SYSCOL project, Cellex Foundation, Botin Foundation, and the Health and Science Departments of the Catalan Government (Generalitat de Catalunya). MSI was an ESTOOLS Postdoctoral Research Fellow. VdS-D was funded by a Spanish Ministry of Science and Innovation fellowship. ME is an ICREA Research Professor.
The authors declare no conflicts of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, et al. Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 2007;67:2456–2468. doi: 10.1158/0008-5472.CAN-06-2698. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2222. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- Lopez-Serra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31:1609–1622. doi: 10.1038/onc.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L, et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med. 2012;366:234–242. doi: 10.1056/NEJMoa1102903. [DOI] [PubMed] [Google Scholar]
- Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnaw SM, Rakha EA, Abdel-Fatah TM, Nolan CC, Hodi Z, Macmillan DR, et al. Loss of Dicer expression is associated with breast cancer progression and recurrence. Breast Cancer Res Treat. 2012;135:403–413. doi: 10.1007/s10549-012-2169-3. [DOI] [PubMed] [Google Scholar]
- Shu GS, Yang ZL, Liu DC. Immunohistochemical study of Dicer and Drosha expression in the benign and malignant lesions of gallbladder and their clinicopathological significances. Pathol Res Pract. 2012;208:392–397. doi: 10.1016/j.prp.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Wu D, Tao J, Xu B, Li P, Lu Q, Zhang W. Downregulation of Dicer, a component of the microRNA machinery, in bladder cancer. Mol Med Report. 2012;5:695–699. doi: 10.3892/mmr.2011.711. [DOI] [PubMed] [Google Scholar]
- Zhu DX, Fan L, Lu RN, Fang C, Shen WY, Zou ZJ, et al. Downregulated Dicer expression predicts poor prognosis in chronic lymphocytic leukemia. Cancer Sci. 2012;103:875–881. doi: 10.1111/j.1349-7006.2012.02234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faggad A, Kasajima A, Weichert W, Stenzinger A, Elwali NE, Dietel M, et al. Down-regulation of the microRNA processing enzyme Dicer is a prognostic factor in human colorectal cancer. Histopathology. 2012;61:552–561. doi: 10.1111/j.1365-2559.2011.04110.x. [DOI] [PubMed] [Google Scholar]
- Guo X, Liao Q, Chen P, Li X, Xiong W, Ma J, et al. The microRNA-processing enzymes: Drosha and Dicer can predict prognosis of nasopharyngeal carcinoma. J Cancer Res Clin Oncol. 2012;138:49–56. doi: 10.1007/s00432-011-1058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann J, Wang CJ, Gnosa S, Wallin A, Hinselwood D, Sun XF, et al. Dicer and miRNA in relation to clinicopathological variables in colorectal cancer patients. BMC Cancer. 2011;11:345. doi: 10.1186/1471-2407-11-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101:673–683. doi: 10.1038/sj.bjc.6605193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Tang F, Kaneda M, O'Carroll D, Hajkova P, Barton SC, Sun YA, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH. van de Wetering M, Begthel H, van den Born M et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Kemper K, Prasetyanti PR, De Lau W, Rodermond H, Clevers H, Medema JP. Monoclonal antibodies against Lgr5 identify human colorectal cancer stem cells. Stem Cells. 2012;30:2378–2386. doi: 10.1002/stem.1233. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JC, Park JK, Jiang J, Kim JH, Nagorney DM, Roberts LR, et al. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2010;403:120–125. doi: 10.1016/j.bbrc.2010.10.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CH, Lee DY, Deng Z, Jeyapalan Z, Lee SC, Kahai S, et al. MicroRNA miR-328 regulates zonation morphogenesis by targeting CD44 expression. PLoS One. 2008;3:e2420. doi: 10.1371/journal.pone.0002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyapalan Z, Deng Z, Shatseva T, Fang L, He C, Yang BB. Expression of CD44 3'-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011;39:3026–3041. doi: 10.1093/nar/gkq1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R, et al. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;154:515–523. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Ilyas M, Tomlinson IP, Rowan A, Pignatelli M, Bodmer WF. Beta-catenin mutations in cell lines established from human colorectal cancers. Proc Natl Acad Sci USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 1998;58:1130–1134. [PubMed] [Google Scholar]
- Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther. 2010;10:219–222. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- Negrini M, Calin GA. Breast cancer metastasis: a microRNA story. Breast Cancer Res. 2008;10:203. doi: 10.1186/bcr1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton R, Chatterjee SS, Simundza J, Cowin P, Dasgupta R. A systematic screen for micro-RNAs regulating the canonical Wnt pathway. PLoS One. 2011;6:e26257. doi: 10.1371/journal.pone.0026257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NH, Kim HS, Kim NG, Lee I, Choi HS, Li XY, et al. p53 and MicroRNA-34 Are Suppressors of Canonical Wnt Signaling. Sci Signal. 2011;4:ra71. doi: 10.1126/scisignal.2001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Dai Y, Zhang L, JiangC LiZ, Yang J, et al. miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis. 2012;34:415–425. doi: 10.1093/carcin/bgs329. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14:43–47. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou A, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci USA. 2010;107:18115–18120. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang R, Law WL, Chu AC, Poon JT, Lam CS, Chow AK, et al. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell. 2010;6:603–615. doi: 10.1016/j.stem.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Chen X, Yu Y, Su L, Yu B, Li J, et al. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Hamada S, Satoh K, Fujibuchi W, Hirota M, Kanno A, Unno J, et al. MiR-126 acts as a tumor suppressor in pancreatic cancer cells via the regulation of ADAM9. Mol Cancer Res. 2012;10:3–10. doi: 10.1158/1541-7786.MCR-11-0272. [DOI] [PubMed] [Google Scholar]
- Sasahira T, Kurihara M, Bhawal UK, Ueda N, Shimomoto T, Yamamoto K, et al. Downregulation of miR-126 induces angiogenesis and lymphangiogenesis by activation of VEGF-A in oral cancer. Br J Cancer. 2012;107:700–706. doi: 10.1038/bjc.2012.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tome M, Lopez-Romero P, Albo C, Sepulveda JC, Fernandez-Gutierrez B, Dopazo A, et al. miR-335 orchestrates cell proliferation, migration and differentiation in human mesenchymal stem cells. Cell Death Differ. 2011;18:985–995. doi: 10.1038/cdd.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png KJ, Yoshida M, Zhang XH, Shu W, Lee H, Rimner A, et al. MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev. 2011;25:226–231. doi: 10.1101/gad.1974211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B, Driessens G, Goossens S, Youssef KK, Kuchnio A, Caauwe A, et al. A vascular niche and a VEGF-Nrp1 loop regulate the initiation and stemness of skin tumours. Nature. 2011;478:399–403. doi: 10.1038/nature10525. [DOI] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Eramo A, Ricci-Vitiani L, Zeuner A, Pallini R, Lotti F, Sette G, et al. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 2006;13:1238–1241. doi: 10.1038/sj.cdd.4401872. [DOI] [PubMed] [Google Scholar]
- Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Dylla SJ, Beviglia L, Park IK, Chartier C, Raval J, Ngan L, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One. 2008;3:e2428. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749–1758. doi: 10.1038/sj.onc.1210811. [DOI] [PubMed] [Google Scholar]
- Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren G, et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69:1951–1957. doi: 10.1158/0008-5472.CAN-08-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz P, Iliou MS, Esteller M. Epigenetic alterations involved in cancer stem cell reprogramming. Mol Oncol. 2012;6:620–636. doi: 10.1016/j.molonc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Li Y, Fan H, Liu Z, Pestell RG. MiRNAs regulate stem cell self-renewal and differentiation. Front Genet. 2012;3:191. doi: 10.3389/fgene.2012.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber C, Horst D, Hlubek F, Kirchner T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur J Cancer. 2011;47:1414–1419. doi: 10.1016/j.ejca.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- Ghislin S, Deshayes F, Middendorp S, Boggetto N, Alcaide-Loridan C. PHF19 and Akt control the switch between proliferative and invasive states in melanoma. Cell Cycle. 2012;11:1634–1645. doi: 10.4161/cc.20095. [DOI] [PubMed] [Google Scholar]
- Munz M, Kieu C, Mack B, Schmitt B, Zeidler R, Gires O. The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene. 2004;23:5748–5758. doi: 10.1038/sj.onc.1207610. [DOI] [PubMed] [Google Scholar]
- Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- Gosens MJ, van Kempen LC, van de Velde CJ, van Krieken JH, Nagtegaal ID. Loss of membranous Ep-CAM in budding colorectal carcinoma cells. Mod Pathol. 2007;20:221–232. doi: 10.1038/modpathol.3800733. [DOI] [PubMed] [Google Scholar]
- Gorges TM, Tinhofer I, Drosch M, Rose L, Zollner TM, Krahn T, et al. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer. 2012;12:178. doi: 10.1186/1471-2407-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YJ, Lai HM, Chang YW, Chen GY, Lee JL. Direct reprogramming of stem cell properties in colon cancer cells by CD44. Embo J. 2011;30:3186–3199. doi: 10.1038/emboj.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Park YS, Kim HJ, Kin CH, Lim SW, Huh JW, et al. CD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasion. Int J Oncol. 2012;41:211–218. doi: 10.3892/ijo.2012.1453. [DOI] [PubMed] [Google Scholar]
- Iliou MS, Lujambio A, Portela A, Brustle O, Koch P, Andersson-Vincent PH, et al. Bivalent histone modifications in stem cells poise miRNA loci for CpG island hypermethylation in human cancer. Epigenetics. 2011;6:1344–1353. doi: 10.4161/epi.6.11.18021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.