SUMMARY
Redox-mediated posttranslational modifications represent a molecular switch that controls major mechanisms of cell function. Nitric oxide (NO) can mediate redox reactions via S-nitrosylation, representing transfer of an NO group to a critical protein thiol. NO is known to modulate neurogenesis and neuronal survival in various brain regions in disparate neurodegenerative conditions. However, a unifying molecular mechanism linking these phenomena remains unknown. Here we report that S-nitrosylation of myocyte enhancer factor 2 (MEF2) transcription factors acts as a redox switch to inhibit both neurogenesis and neuronal survival. Structure-based analysis reveals that MEF2 dimerization creates a pocket, facilitating S-nitrosylation at an evolutionally conserved cysteine residue in the DNA binding domain. S-Nitrosylation disrupts MEF2-DNA binding and transcriptional activity, leading to impaired neurogenesis and survival in vitro and in vivo. Our data define a novel molecular switch whereby redox-mediated posttranslational modification controls both neurogenesis and neurodegeneration via a single transcriptional signaling cascade.
INTRODUCTION
Redox modification of cysteine, reminiscent of phosphorylation and ubiquitination of critical amino acids, regulates protein activity (Hess et al., 2005). One type of redox modification involves reaction of nitric oxide (NO) with cysteine thiol. In this manner, NO functions as a signaling molecule influencing multiple targets. In brain, NO regulates physiologic functions including neural development, neurotransmitter release, and synaptic plasticity, but excess NO promotes neuronal damage in a variety of neurodegenerative disorders (Nakmura et al., 2013). Previously, we and our colleagues demonstrated that NO reacts with critical cysteine thiols, termed S-nitrosylation, on various proteins to modulate their activity (Hess et al., 2005). Recently, we found that S-nitrosylation of the transcription factor myocyte enhancer factor 2C (MEF2C) occurs in cell-based Parkinson’s disease (PD) models, resulting in inhibition of transcriptional activity and resultant cell death via a PGC1α-mediated pathway (Ryan et al., 2013). Heretofore, however, it remained unknown if S-nitrosylation of a common target could molecularly ‘switch’ off both neurogenesis and neuronal survival. Here, we demonstrate that S-nitrosylation of MEF2A or MEF2C at an inhibitory ‘cysteine switch’ abrogates DNA binding. This redox reaction shuts off two distinct transcriptional cascades involved in neuronal survival and neurogenesis.
MEF2 integrates diverse signals and plays roles in the immune, muscular, and nervous systems. In brain, we and others have shown that various MEF2 isoforms modulate neurogenesis and neuronal survival (Flavell et al., 2006; Li et al., 2008; Mao et al., 1999; Okamoto et al., 2000; Okamoto et al., 2002; Shalizi et al., 2006). Here, we used cerebral ischemia models to study the influence of NO on MEF2 in neuronal cell death, as both NO and MEF2 have been previously linked to such damage (Huang et al., 1994; Okamoto et al., 2002). Additionally, as a tractable paradigm to study dysfunctional neurogenesis in the adult nervous system, we used Alzheimer’s disease (AD) models because NO and MEF2 have both been implicated in the generation of new neurons (Packer et al., 2003; Li et al., 2008).
RESULTS
S-Nitrosylation of MEF2 Transcription Factors
To confirm that the consensus motif for nitrosylation on MEF2 (Stamler et al., 1997) can indeed be S-nitrosylated (to form SNO-MEF2), we initially expressed Myc-tagged MEF2C in human embryonic kidney (HEK) 293 cells, and exposed the transfected cells to the physiological NO donors, S-nitrosocysteine (SNOC) or nitroso-S-glutathione (GSNO). Using the biotin switch assay in which biotin is selectively substituted for the NO moiety of SNO-cysteine residues (Jaffrey et al., 2001), we found that these NO donors induced S-nitrosylation of MEF2C (Figure S1A).
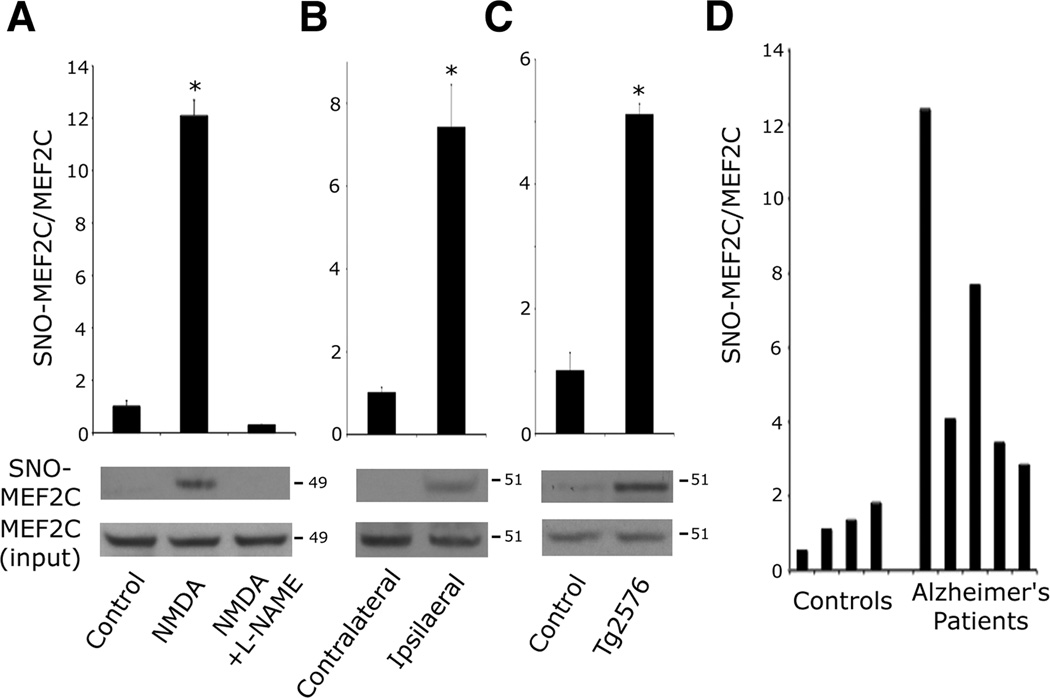
Next, we asked if endogenous SNO-MEF2 is increased in neurodegenerative conditions. We primarily focused on MEF2C because this isoform is known to predominate in cerebrocortical neurons, although our findings pertain to all isoforms (Leifer et al., 1993; Li et al., 2008). Since neuronal cell death triggered by excessive activation of the N-methyl-d-aspartate (NMDA)-type glutamate receptor is known to be mediated, at least in part, by generation of endogenous NO (Dawson et al., 1991), and MEF2C activity has been shown to ameliorate such damage (Okamoto et al., 2002), we exposed cultured rat cerebrocortical neurons to 50 µM NMDA and monitored SNO-MEF2C formation by biotin switch assay. We observed SNO-MEF2C within 30 min of NMDA application, an effect blocked by co-treatment with the NO synthase (NOS) inhibitor l-NG -nitroarginine methyl ester (l-NAME; Figures 1A and S1B). Since apoptosis does not occur in this paradigm until the next day, these results imply that MEF2C is S-nitrosylated at an early stage of cell-death signaling (Bonfoco et al., 1995; Cho et al., 2009). NO-induced neurotoxicity has also been implicated in in vivo studies, including focal cerebral ischemia (stroke) (Hara and Snyder, 2007; Huang et al., 1994). Robust NO production has been observed during the first few hours of cerebral ischemia in rodent middle cerebral artery (MCA) occlusion models (Kader et al., 1993; Zhang et al., 1995). Accordingly, we occluded the right MCA of mice for 1 h and observed S-nitrosylation of MEF2C in the ischemic but not the contralateral cerebral cortex (Figures 1B and S1C), showing that MEF2C is S-nitrosylated in vivo during ischemia.
Figure 1. MEF2 is S-Nitrosylated In Vitro and In Vivo during Neurodegeneration.
(A) Excitotoxic NMDA (50 µM) was administered to cortical neurons in the presence or absence of NOS inhibitor l-NAME (500 µM). After 30 min, endogenous SNO-MEF2C was detected by biotin switch. SNO proteins were precipitated from neuronal lysates, and SNO-MEF2C detected with anti-MEF2C-specific antibody. Quantitative densitometry shown above immunoblots. Values are mean + SEM (n = 3 independent experiments, *p < 0.0001 by ANOVA with post-hoc Scheffé's test).
(B) SNO-MEF2C in contralateral or ipsilateral cortex 1 h after MCA occlusion. Values are mean + SEM (n = 3 animals, *p = 0.001 by t test).
(C) SNO-MEF2C in brains from WT littermates and Tg2576 AD mice. Values are mean + SEM (n = 4 animals in each group, *p = 0.001 by t test).
(D) SNO-MEF2C in brain tissues from human control or AD patients by biotin switch.
See also Figure S1.
In addition to promoting acute neuronal injury in conditions such as stroke, excessive NO generation has been observed in chronic neurodegenerative diseases, including human AD brains (Fernandez-Vizarra et al., 2004; Vodovotz et al., 1996). By biotin switch assay, we examined brains of Tg2576 mice, an AD model overexpressing amyloid precursor protein (APP) with the Swedish mutation, and in postmortem human AD brains. We found increased SNO-MEF2C levels in both mouse (Figures 1C and S1D) and human AD brains but not in controls (Figures 1D and S1E). Taken together, these findings suggest that MEF2C is S-nitrosylated in vivo both in acute neurodegenerative conditions such as stroke and in chronic neurodegenerative conditions such as AD.
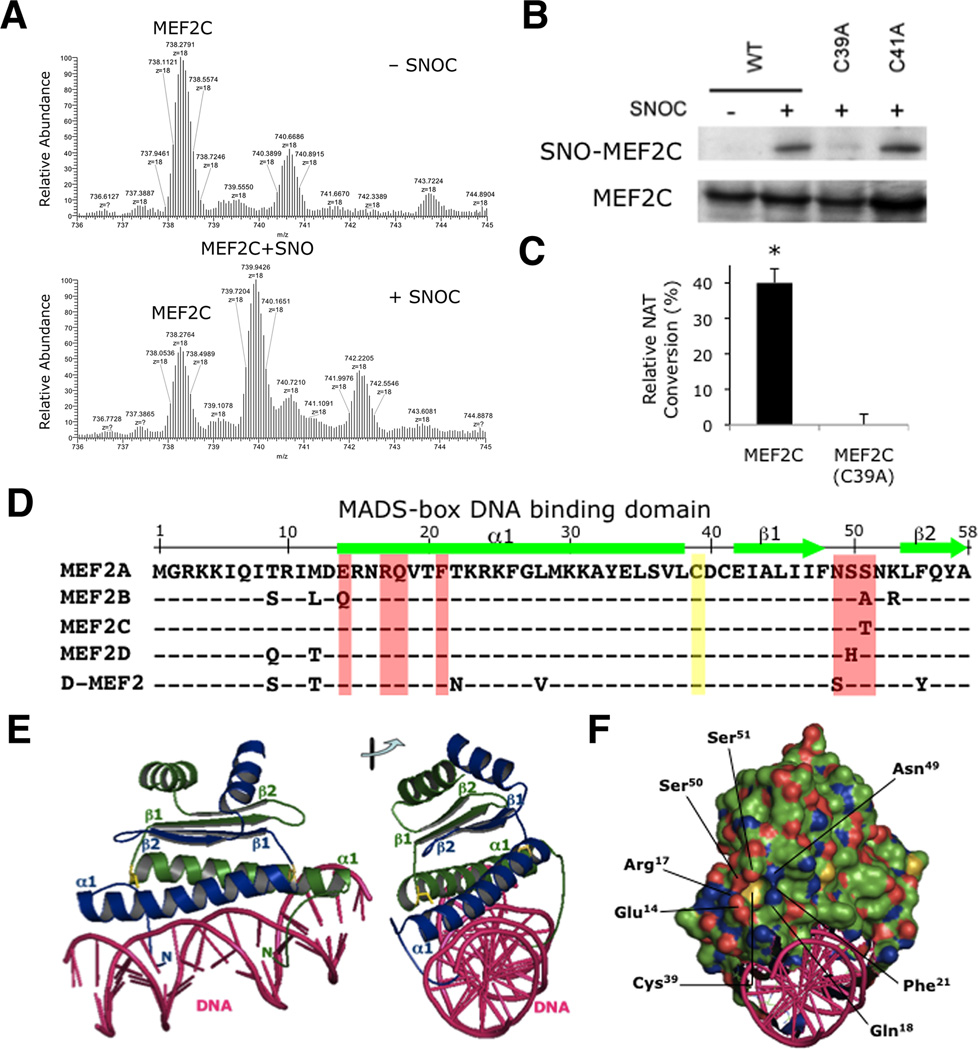
To determine which cysteine residue(s) is S-nitrosylated on MEF2C and other MEF2 isoforms, we first performed a top-down analysis of the protein using an LTQ (linear trap quadrupole)-Orbitrap-XL mass spectrometry with electron transfer dissociation (ETD). Exposure to the physiological NO donor SNOC shifted the MEF2C spectral cluster reflecting S-nitrosothiol formation on a single cysteine residue (739.9426 m/z), while expected clusters of MEF2C with two S-nitrosothiols (741.4986 m/z) or more were not observed. These results indicate the presence of one S-nitrosylation site on MEF2C (Figure 2A). To identify that site, we mutated each Cys residue in MEF2C to Ala, and then transfected expression vectors encoding these mutated forms into HEK293 cells. We exposed the cells to SNOC and analyzed S-nitrosylated MEF2C by the biotin switch method. Mutation of Cys39 (C39A) abrogated formation of SNO-MEF2C, whereas other mutations such as MEF2C(C41A) did not (Figures 2B and S2A, densitometric quantification). To confirm S-nitrosylation of Cys39, we employed a direct biochemical analysis for S-nitrosothiols, as described previously, by monitoring conversion of 2,3-diaminonaphthalene (DAN) to 2,3-naphthotriazole (NAT) from immunoprecipitates of HEK293 cells transfected with wild-type (WT) or mutant MEF2C(C39A) (Wink et al., 1999). In this assay, the C39A mutation abolished SNO-MEF2C formation (Figure 2C), supporting the notion that Cys39 is the primary site of S-nitrosylation.
Figure 2. S-Nitrosylation of MEF2 at Cys39 in the MADS-Box DNA Binding Domain.
(A) Recombinant MEF2C protein was infused into a mass spectrometer (LTQ-Orbitrap-XL with ETD) to obtain molecular ion spectra in the presence or absence of NO donor, 50 µM SNOC (n = 2 independent experiments).
(B) HEK293T cells transfected with WT myc-tagged MEF2C (WT), MEF2C(C39A) or MEF2C(C41A) were incubated with SNOC. SNO-MEF2C was determined by biotin switch. SNO proteins were precipitated and SNO-MEF2C detected with anti-myc antibody (n = 3 independent experiments; quantification shown in Figure S2A).
(C) HEK293 cells stably expressing nNOS were transfected with V5-tagged MEF2C (WT) or V5-tagged MEF2C(C39A), and nNOS activated with Ca2+ ionophore A23187. V5-tagged MEF2C proteins were immunoprecipitated with anti-V5 antibody and subjected to DAN assay. Values are mean + SEM (n = 3 independent experiments, *p < 0.05 by t test).
(D) Sequence alignment of MADS-box DNA binding domains of MEF2 (human isoforms MEF2A-D; Drosophila D-MEF2). α-Helix (bars) and β-strands (arrows). Cys39 S-nitrosylation site (yellow). Residues involved in pocket formation (red).
(E) Cys39 (yellow) is symmetrically located at the hinge between α1-helix and β1-strands in the MEF2 (monomer A in blue, monomer B in green)-DNA (pink) complex (PDB ID: 1EGW). Right: viewed at 90°.
(F) Electrostatic view of MEF2 (negative charge in red, positive charge in blue). Cys39 is present at the bottom of the pocket created by Glu14, Arg17, Ser50 Ser51, Asn49, Phe21, and Gln18.
(G) Cys39 of MEF2 monomer A (blue) is surrounded by residues (Glu14 Arg17, Ser50, Ser51, Asn49, Phe21, and Gln18; red) of monomer B (green). Right: complex viewed at 90°.
See also Figure S2.
S-Nitrosylation of MEF2 Inhibits DNA Binding and Transcriptional Activity
Next, we investigated the effect of SNO-MEF2 on transcriptional activity. We noted that Cys39 is conserved in the N-terminal MADS domain among all MEF2 family members, and other MEF2 isoforms can also be S-nitrosylated at this position (Figures 2D, S2B, and S2C). Since MEF2 dimerizes and binds to DNA through the MADS domain, we studied the effect that SNO-MEF2 might exert on dimerization and DNA binding. The structure of the MEF2/DNA complex has been analyzed by NMR spectroscopy and X-ray crystallography (Han et al., 2005; Han et al., 2003; Santelli and Richmond, 2000). Our analysis of these structures revealed that Cys39 is located at a hinge region where the α-helix stretch (α1) turns into a β-sheet (β1) (Figures 2D and 2E). Intriguingly, the electrostatic view demonstrates that Cys39 is at the bottom of a pocket created by seven amino acids: Glu14, Arg17, Gln18, Phe21, Asn49, Ser50, and Ser51 (Figure 2F). These residues are highly conserved throughout the entire MEF2 family (red in Figure 2D). Additionally, these same residues are often part of an amino-acid motif facilitating S-nitrosylation of a critical Cys residue (Greco et al., 2006; Hao et al., 2006), suggesting that the pocket may form such a nitrosylation motif.
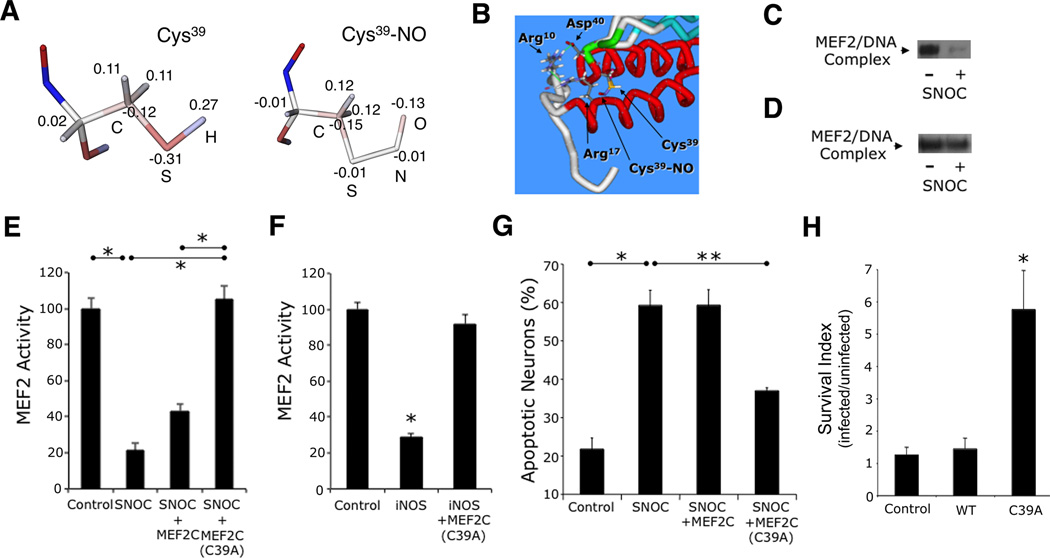
To determine whether S-nitrosylation alters MEF2 transcriptional activity, we first used a molecular dynamics approach to analyze the interaction patterns of Cys39 and SNO-Cys39 residues. Using the crystal structure of the solved MEF2A/DNA complex (Santelli and Richmond, 2000), we modified the Cys39 residues to SNO-Cys in both polypeptide chains of the dimer in silico. In order to determine the geometry and electrical properties of the NO-substituted side chain of Cys39, we then performed quantum mechanical calculations. The SNO-Cys dipeptide (CH3CO-NH-CH(CH2SNO)CO-NH-CH3) was used as a model molecule for deriving essential force-field parameters necessary for molecular mechanics modeling. The geometry of the SNO-Cys dipeptide was optimized using the quantum mechanical GAMESS program (Schmidt et al., 1993), and the charges on the dipeptide atoms were determined to reproduce the quantum mechanically-derived electrostatic potential around the entire dipeptide using the RESP method (Bayly et al., 1993) and R.E.D.III program (Grivel et al.). We compared the resultant atomic charges of the SNO-Cys dipeptide sidechain with the charge distribution of the unmodified Cys residue, for which molecular mechanical parameters are available in the AMBER force field (Cornell et al., 1995). Substitution of the H atom with the NO group in Cys39 replaced the positively-charged single atom with two negatively-charged atoms, making the entire side chain electronegative (Figure 3A). This modification should, in theory, substantially affect the interaction of Cys39, located at the hinge region of the DNA binding domain (Figures 2D and 2E), with the surrounding residues essential for MEF2/DNA interaction.
Figure 3. S-Nitrosylation of MEF2 Impairs its DNA Binding, Transcriptional, and Pro-Survival Activities.
(A) Computational structure analysis of S-nitrosylated Cys39. Atomic charge distribution for normal Cys and Cys-NO residues obtained from quantum mechanical calculations (see text). Electrostatic potentials: positive charge (blue) and negative charge (red). Values show excess atomic charge.
(B) Optimization of molecular mechanical geometry to show possible orientation of the Cys-NO sidechain in MEF2. Key residues of dimer interface are highlighted.
(C) EMSA of MEF2/DNA-binding activity. Nuclear extracts from cortical neurons were exposed to 50 µM SNOC (n = 3 independent experiments). MEF2/DNA-binding complex (arrow).
(D) EMSA of mutant MEF2/DNA-binding activity. In vitro translated MEF2C(C39A) was exposed to SNOC (n = 3 independent experiments).
(E) Effect of NO on MEF2 reporter gene activity. Cortical neurons were transfected with empty vector (pcDNA 3.1) or expression vectors for MEF2C or MEF2C(C39A), MEF2 luciferase reporter construct, and renilla luciferase vector to control for transfection efficiency. Luciferase activity was measured 4 h after 50 µM SNOC exposure. Bars represent mean + SEM (n = 12 cultures derived from 3 independent platings, *p < 0.0001 by ANOVA with post-hoc Scheffé's test).
(F) Effect of iNOS transfection on endogenous MEF2 reporter gene activity. Cortical neurons were transfected with empty vector (pcDNA 3.1) or expression vectors for iNOS with or without MEF2C(C39A), and MEF2 luciferase reporter construct plus renilla luciferase; 24 h after transfection, luciferase activity was measured. Bars represent mean + SEM (n = 12 cultures derived from 3 independent platings, *p < 0.0001 by ANOVA with post-hoc Scheffé's test).
(G) Effect of MEF2 S-nitrosylation on neuronal apoptosis. Cortical neurons were transfected with control vector, MEF2C, or MEF2C(C39A) plus enhanced green fluorescent protein (EGFP) vector to identify transfected neurons. Cultures were then exposed to 50 µM SNOC and analyzed 8 h later. Hoechst DNA dye was used to assess condensed, apoptotic nuclei. Bars represent mean + SEM (n = 9 cultures derived from 3 independent platings, *p < 0.01, **p < 0.05 by ANOVA with post-hoc Scheffé's test). (H) Stereotactic injection of S-nitrosylation-resistant MEF2C(C39A) mutant (LV-C39A) but not WT MEF2C into striatum improved neuronal survival in stroke penumbra. Values are mean + SEM of survival index, representing the inverse of the apoptotic ratio for infected vs. uninfected neurons for each construct (n ≥ 4 animals for each condition, *p < 0.05 by ANOVA with post-hoc Scheffé's test).
See also Figure S3.
We found that the solved atomic structure shows the Cys39 region has a network of interacting residues that controls orientation of the N-terminal polypeptide arm (composed of residues 1–17), which is involved in DNA binding (Figure 3B). One important interaction is a salt bridge between the Arg10 and Asp40 residues located in different chains of the MEF2 dimer. Another important interaction is between Arg10 and Arg17 within a single polypeptide chain. The importance of this second interaction is experimentally supported since mutation of Arg17 to Val has been shown to decrease MEF2-DNA binding activity (Molkentin et al., 1996). We thus obtained a plausible conformation of SNO-Cys39 by molecular mechanical optimization using the AMBER program and its parm99 force field (Figure 3B) (Case et al., 2005; Cornell et al., 1995). This type of -SNO orientation could potentially perturb interactions between Arg17 and Arg10, thus affecting the strength of the salt bridge between Arg10 and Asp40. This perturbation, in turn, could dislocate the position of the MEF2 N-terminal arm from the vicinity of DNA, thus reducing DNA binding.
To test empirically whether S-nitrosylation of MEF2 impairs DNA binding, as predicted by the atomic structure and quantum mechanical considerations, we exposed nuclear extracts of cortical neurons to NO donor and analyzed DNA binding by electrophoretic mobility shift assay (EMSA) (Krainc et al., 1998; Leifer et al., 1993). We found that exposure to SNOC decreased MEF2 binding to DNA, with binding specificity documented by supershift (Figures 3C and S3A). To determine the contribution of SNO-Cys39 to this effect, we mutated Cys39 to Ala, translated the mutant protein MEF2C(C39A) in vitro, and assayed DNA binding by EMSA (Huang et al., 2000). We observed that mutant MEF2C(C39A), unlike WT, bound to DNA irrespective of SNOC exposure (Figure 3D). The fact that SNOC had little if any effect on MEF2C(C39A)-DNA binding is consistent with the notion that the action of NO on MEF2C-DNA binding involved S-nitrosylation at Cys39.
To analyze whether this effect of NO leads to a decrease in MEF2 transcriptional activity, we performed reporter gene assays in cortical neurons. We transfected neurons with a MEF2-dependent luciferase reporter and exposed the cells to SNOC. SNOC significantly attenuated MEF2 activity compared to control (Figures 3E and S3B). Under these conditions, we observed no cell death in our cortical cultures (Okamoto et al., 2002); hence, toxicity cannot account for the decrease in luciferase reporter activity. Transfection with S-nitrosylation-resistant MEF2C(C39A) completely abrogated the decrease in transcriptional activity engendered by SNOC, consistent with the notion that formation of SNO-Cys39 suppresses MEF2 activity. We also observed a decrement in MEF2 transcriptional activity in neurons transfected with inducible NOS; this suppressive effect was blocked by co-transfection with MEF2C(C39A) (Figures 3F and S3C). Since aberrant NOS expression has been associated with neurodegenerative conditions such as AD (Fernandez-Vizarra et al., 2004; Vodovotz et al., 1996), we postulated that impaired MEF2 transcriptional activity resulting from S-nitrosylation could contribute to the neurodegenerative process.
Formation of SNO-MEF2C Contributes to Neuronal Cell Death both In Vitro and In Vivo
We next investigated the potential link between SNO-MEF2 and neurodegeneration. We had previously shown that MEF2C is not only the predominant MEF2 isoform in the cerebrocortex but that conditional knock out of this isoform selectively in the brain results in decreased neuronal cell counts and synapses in vivo (Leifer et al., 1993; Li et al., 2008). If inactivation of MEF2C activity via S-nitrosylation contributes to neurotoxicity, expression of S-nitrosylation-resistant MEF2C(C39A) should rescue neurons from NO-mediated cell death. To test this hypothesis, we expressed MEF2C(C39A) in cortical neurons in culture, and then exposed the cells to SNOC. MEF2C(C39A) significantly ameliorated NO-induced cell death, as assessed by monitoring apoptotic nuclei (Figure 3G) and cleaved caspase-3 (Figure S3D). In contrast, this protective effect was not observed following transfection with WT MEF2C, which is sensitive to S-nitrosylation. These results provide evidence that the known antiapoptotic function of MEF2 (Mao et al., 1999; Okamoto et al., 2000; Okamoto et al., 2002) is abrogated by S-nitrosylation.
To investigate the involvement of SNO-MEF2 in neuronal cell death in vivo, we used the MCA stroke model since we had observed an increase in SNO-MEF2C in this mouse model (see Figure 1B, above). We stereotactically injected lentiviral expression vectors for GFP alone (LV-GFP), WT MEF2C with GFP (LV-WT), or mutant MEF2(C39A) with GFP (LV-C39A) into the striatum, representing the penumbral area where neuronal apoptosis is known to occur after MCA ischemia. We then induced MCA occlusion and examined apoptotic neurons in infected vs. uninfected neurons for each construct 14 h post reperfusion. We observed a significant difference in the survival index with the various constructs, with mutant MEF2(C39A) affording neuroprotection (Figures 3H and S3E). These results support the notion that SNO-MEF2 contributes to neuronal cell death in vivo since the non-nitrosylatable MEF2C construct, unlike control vector or WT MEF2C, enhanced neuronal survival after stroke.
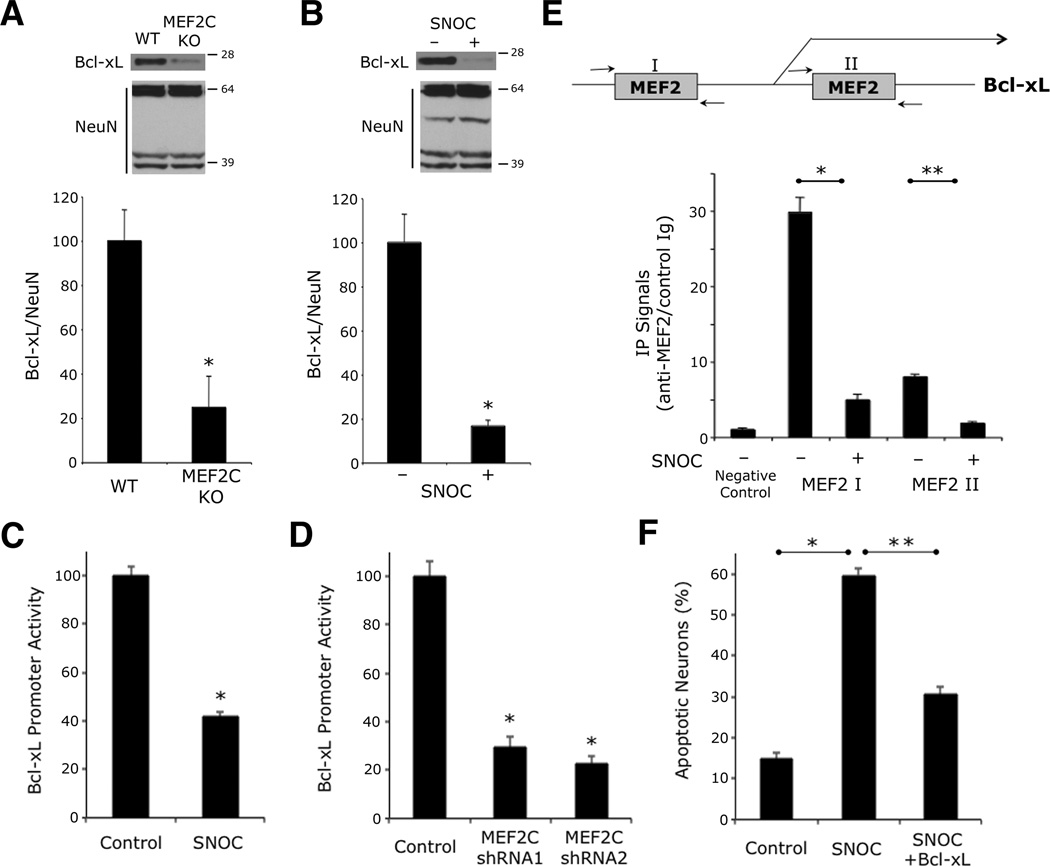
Next, we studied the mechanism whereby SNO-MEF2 promotes neuronal cell death. Initially, we performed a systematic analysis of the promoter regions of known cell death/survival genes to look for potential MEF2 binding sites and found multiple such sites on the published Bcl-xL promoter. Mice that are null for the Bcl-xL gene manifest massive neuronal death. Interestingly, we also found that Bcl-xL levels are decreased in the cortex of conditional MEF2C knockout mice (Figures 4A and S4A) (Li et al., 2008). Hence, we tested if S-nitrosylation of MEF2 would decrease Bcl-xL promoter activity and neuronal expression of Bcl-xL prior to neuronal cell death, and found that this was indeed the case (Figures 4B, 4C, and S4B). Additionally, MEF2C knockdown by RNA interference (RNAi) (Figure S4C) resulted in a decrease in Bcl-xL promoter activity (Figure 4D), consistent with the notion that NO exposure exerted a similar effect via formation of SNO-MEF2C. As further evidence, we found in chromatin immunoprecipitation (ChIP) experiments using anti-MEF2 antibody, that NO exposure diminished the association of MEF2 to its cognate sites on the Bcl-xL promoter (Figure 4E). Moreover, transfection with Bcl-xL ameliorated NO-induced neuronal cell death (Figure 4F and S4D). Taken together, our findings suggest that MEF2 transcriptionally activates Bcl-xL to afford neuroprotection, whereas S-nitrosylation of MEF2 limits Bcl-xL expression and thus contributes to NO-induced neuronal cell death.
Figure 4. SNO-MEF2—Induced Suppression of Bcl-xL Contributes to NO-Induced Neuronal Cell Death.
(A and B) Western blot analysis of Bcl-xL expression. Cell lysates were collected from cerebrocortex of CamKII-Cre/MEF2C conditional knockout mouse brain (A) and from cultured rat cerebrocortical neurons exposed to 50 µM SNOC (B). NeuN insured equal loading (NeuN antibody is known to recognize multiple bands). Quantitative densitometry shown below immunoblots. Values are mean + SEM (n = 3 for each panel, *p < 0.05 by t test).
(C) NO exposure significantly reduces Bcl-xL promoter activity. Cortical neurons were transfected with a luciferase reporter gene controlled by the Bcl-xL promoter. SNOC (50 µM) was added 4 h prior to luciferase assay. Bars represent mean + SEM (n = 12 cultures from 3 platings, *p < 0.0001 by Mann-Whitney test).
(D) Reporter assays using Bcl-xL luciferase construct cotransfected with non-targeting shRNA (control) or two shRNAs against MEF2C. Bars represent mean + SEM (n = 12 cultures derived from 3 platings, * p < 0.0001 by ANOVA with post-hoc Dunnett’s test).
(E) NO decreases association of endogenous MEF2 with endogenous Bcl-xL promoter in cortical neurons. Chromatin immunoprecipitation (ChIP) assay was performed for two MEF2 binding sites in the Bcl-xL promoter region. ‘Negative Control’ value indicates virtually no binding 2 kb upstream of the two MEF2 consensus sites. Bars represent mean + SEM (n = 3 independent experiments, *p < 0.0001, **p < 0.05 by ANOVA with post-hoc Scheffé's test).
(F) Amelioration of NO-induced cell death in neurons expressing Bcl-xL. Neurons transfected with control vector (pCMV-Sport) or Bcl-xL were exposed to 50 µM SNOC. Bars represent mean + SEM (n = 11 cultures from 3 platings, *p < 0.0001, **p < 0.05 by Kruskal-Wallis test).
SNO-MEF2 Disrupts Adult Neurogenesis both In Vitro and In Vivo
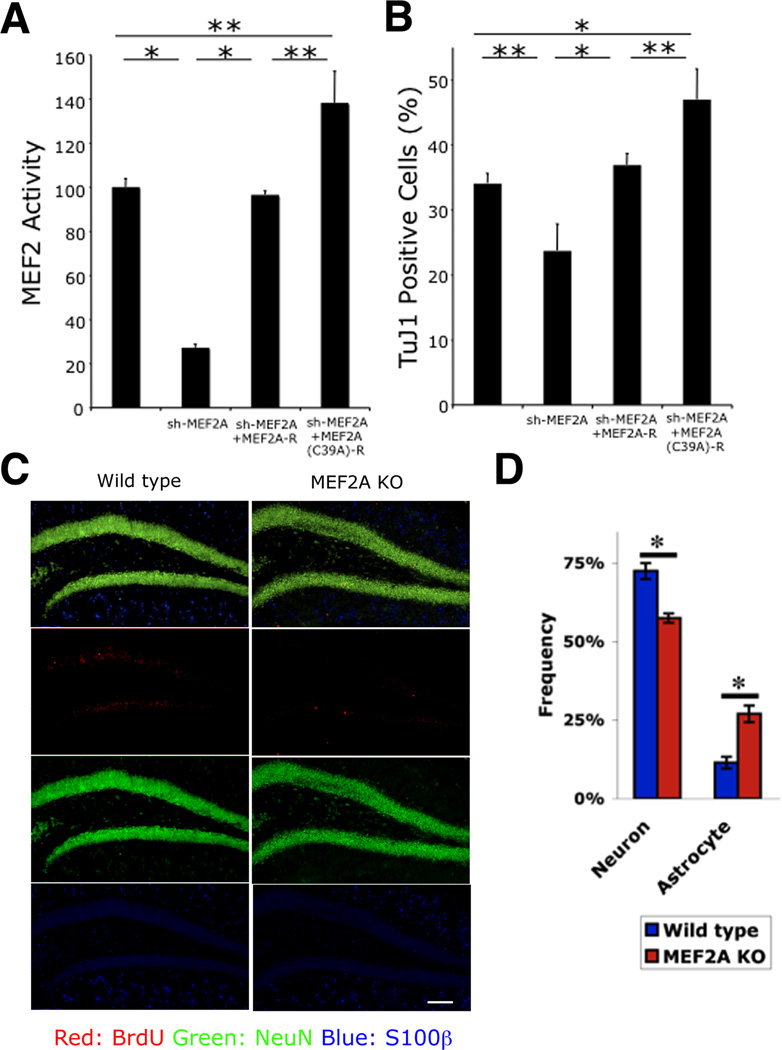
In several neurodegenerative conditions including AD, in addition to neuronal cell injury and death, adult neurogenesis is also affected (Ohab et al., 2006; Winner et al., 2011). Neurogenesis normally persists in the adult hippocampus and contributes to specific types of learning and memory (Ma et al., 2009; Zhao et al., 2008). Recent studies in animal models have suggested that NO negatively regulates adult neurogenesis in the hippocampus, but the mechanism for this effect has remained elusive (Moreno-Lopez et al., 2004; Packer et al., 2003). To investigate the potential effects of NO-related redox regulation of MEF2 on adult neurogenesis, we performed experiments to determine the expression and involvement of MEF2 family members in this process both in vitro and in vivo. Initially, we found that MEF2A was the predominant isoform during neurogenesis in vitro of adult rat hippocampal neural progenitor/stem cells (NSCs) (Figure S5A), so we studied MEF2A as a representative MEF2 isoform. To begin to elucidate the function of MEF2A in adult neurogenesis, we used an RNAi approach. A short hairpin RNA (shRNA) targeted specifically to MEF2A (sh-MEF2A) efficiently knocked down MEF2A expression (Figure S5B). The sh-MEF2A also reduced MEF2-dependent luciferase reporter gene activity in NSCs (Figure 5A). Co-expression of an RNAi-resistant form of MEF2A (MEF2A-R) rescued this decrement in MEF2 activity (Figures S5B and 5A). To investigate whether MEF2A regulates neurogenesis in NSCs, we transfected sh-MEF2A, allowed neurogenesis to proceed for two days (Gage et al., 1995), and then analyzed the number of cells expressing the early neuronal marker TuJ1. Knockdown of MEF2A decreased the generation of TuJ1+ cells (Figure 5B) without inducing cell death at this stage of development (Figure S5C). In contrast, MEF2A-R abrogated the decrement in neurogenesis induced by MEF2A knockdown (Figure 5B), making it unlikely that an off-target effect of RNAi could account for the defect in neurogenesis.
Figure 5. MEF2A is Essential for Adult Hippocampal Neurogenesis In Vitro and In Vivo.
(A) Effect of non-nitrosylatable MEF2 on transcriptional activity during neurogenesis. NSCs were transfected in vitro with MEF2 luciferase reporter, and MEF2A shRNA expression vector, together with expression plasmids encoding shRNA-resistant MEF2A (MEF2A-R) or shRNA and S-nitrosylation—resistant MEF2A [MEF2A(C39A)-R]. MEF2 transcriptional activity was significantly decreased after MEF2A knockdown; MEF2 activity was restored to control levels by MEF2A-R expression and above control levels by MEF2A(C39A)-R. Data are mean + SEM (n = 15 cultures derived from 3 platings, *p < 0.001, **p < 0.01 by ANOVA with post-hoc Scheffé's test).
(B) Effect of non-nitrosylatable MEF2A on neurogenesis. NSCs were transfected as indicated and induced towards neural lineage for 2 d. TuJ1+ cells decreased after MEF2A knockdown. Neurogenesis recovered to control levels with MEF2A-R expression and above control levels with MEF2A(C39A)-R. Data are mean + SEM (n = 15 cultures from 3 platings, *p < 0.001, **p < 0.01 by ANOVA with post-hoc Scheffé's test).
(C and D) Decreased neurogenesis in MEF2A KO mice in vivo. Control and MEF2A KO mice were injected with BrdU and sacrificed 4 week later. Coronal sections of dentate gyrus were immunostained for BrdU (red, newly-proliferating cells), NeuN (green, neuron), and S100β (blue, astrocyte) (C), and stereologically quantified (D). Data are mean + SEM (n = 5 animals for each group, *p < 0.05 by t test). Scale bar, 100 µm.
See also Figure S5.
We next utilized MEF2A knockout mice in order to validate the role of this MEF2 isoform in adult hippocampal neurogenesis in vivo. 5-Bromodeoxyuridine (BrdU) was injected into the mice to label hippocampal NSCs. Four weeks later, we examined differentiation of the labeled NSCs by double staining brain sections for BrdU plus NeuN, a mature neuronal marker, or S100β, an astrocytic marker (Figure 5C). We observed a significant decrease in NeuN/BrdU double-positive cells and a significant increase of S100β/BrdU double-labeled cells (Figure 5D). Together, these data indicate that MEF2A plays an important role in adult hippocampal neurogenesis, both in vitro and in vivo, akin to the role of this family of transcription factors we previously demonstrated during embryonic neurogenesis in the developing cerebrocortex (Li et al., 2008).
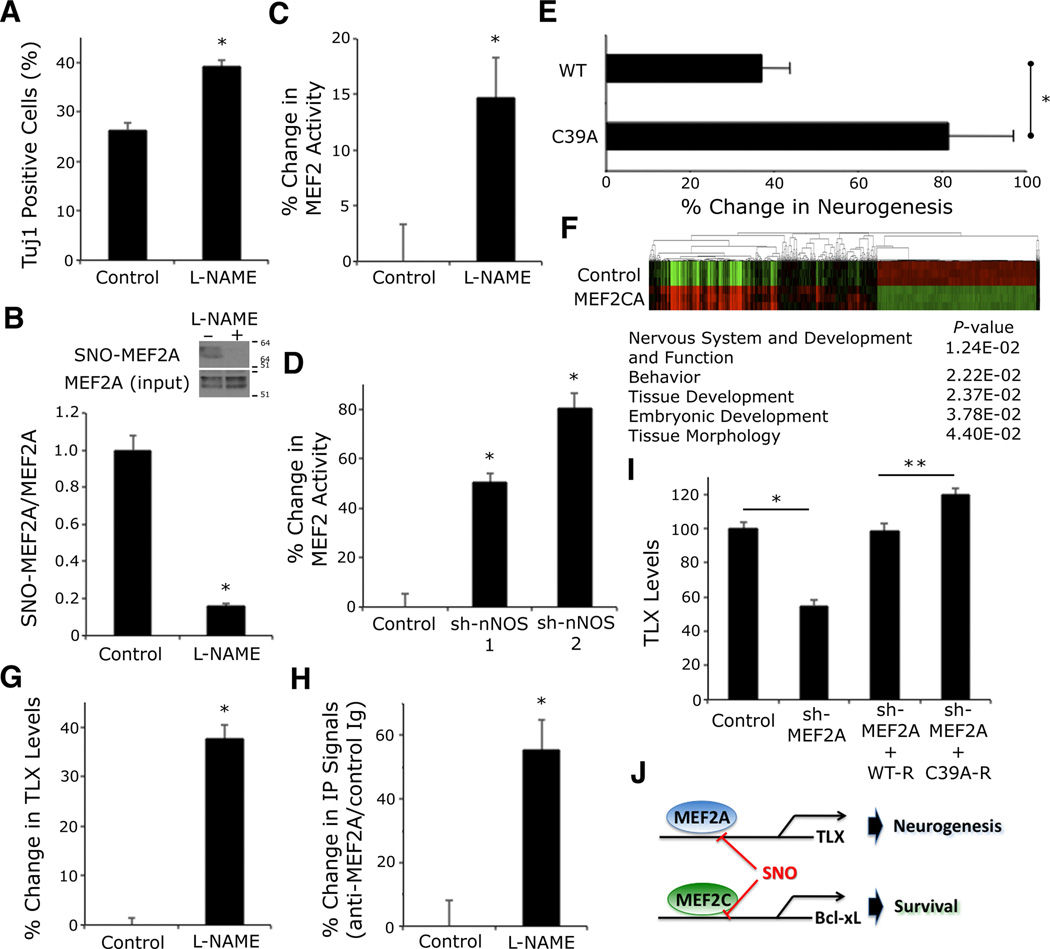
Concerning the mechanism of the effect of endogenous NO on adult hippocampal neurogenesis, we observed that the NOS inhibitor l-NAME significantly increased the generation of new neurons in cultures of NSCs (Figure 6A). We next asked whether formation of SNO-MEF2A mediated, at least in part, the inhibitory effect of NO on adult neurogenesis. First, we demonstrated the existence of SNO-MEF2A in cultures of NSCs under resting conditions (Figures 6B and S6A). Second, we found that l-NAME or two short hairpin RNAs against neuronal NOS (sh-nNOS-1 and −2, Figure S6B) increased MEF2 activity (Figures 6C and 6D ).
Figure 6. S-Nitrosylation of MEF2A Regulates TLX Expression.
(A) l-NAME (200 µM) increased TuJ1+ cells differentiating from NSCs within 2 d of neural induction in vitro. Data are mean + SEM (n > 80 cells per condition from 3 platings, *p < 0.0001 by t test).
(B) Western blot shows l-NAME inhibited S-nitrosylation of MEF2A in NSCs. Biotin switch assay of cell lysates detected SNO-MEF2A (MEF2A manifests multiple bands on immunoblots due to phosphorylated and sumoylated forms (Flavell et al., 2006; Shalizi et al., 2006)). Quantitative densitometry above immunoblots. Values are mean + SEM (n = 3 independent experiments, *p = 0.001 by t test).
(C) l-NAME increased MEF2 transcriptional activity in NSCs monitored by luciferase reporter assay. Data are mean + SEM (n = 16 cultures derived from 4 platings, *p < 0.05 by Mann Whitney test).
(D) nNOS RNA-interference augmented MEF2 transcriptional activity. NSCs were transfected with MEF2 luciferase reporter plus control (non-targeting shRNA) or two independent nNOS shRNA expression plasmids. Data are mean + SEM (n = 12 cultures from 3 platings, *p < 0.0001 by ANOVA with post-hoc Dunnett’s test).
(E) LV-GFP, LV-WT MEF2, or LV-C39A was stereotactically injected into the dentate gyrus of Tg2576 AD mice. Neurogenesis was stereologically assessed by comparison of GFP, NeuN, and EdU triple-positive cells for LV-WT or LT-C39A as a percentage of LV-GFP. Bars represent mean + SEM (n ≥ 4 animals for each condition, *p < 0.05 by t test).
(F) Heatmap of MEF2-dependent gene expression with gene ontology analysis. Total RNA from human NSCs transfected with constitutively-active MEF2 (MEF2CA) vs. vector control (ptd-Tomato) was used for global expression profiling (n = 3 independent platings and transfections).
(G) l-NAME enhanced TLX expression. NSCs were treated with l-NAME, and TLX expression analyzed by quantitative immunofluorescence using deconvolution microscopy. Data are mean + SEM (n = 100 cells per condition from 3 platings, *p < 0.0001 by Mann Whitney test).
(H) ChIP analysis of TLX promoter in NSCs treated with l-NAME. MEF2A immunoprecipitation was followed by real-time PCR for TLX promoter. Data are fold-change over control Ig immunoprecipitation value (mean + SEM; n = 5 experiments, *p < 0.05 by t test).
(I) SNO-MEF2 decreased TLX levels. NSCs were transfected with MEF2A shRNA expression vector plus sh-resistant (R) WT or MEF2(C39A) mutant expression vector, and TLX expression analyzed by quantitative immunofluorescence. Data are mean + SEM (n > 70 cells per condition from 3 platings, *p < 0.0001, **p < 0.01 by Kruskal-Wallis test).
To examine the functional consequences of S-nitrosylation of MEF2A at the Cys39 residue on adult neurogenesis, we generated the non-nitrosylatable mutant MEF2A(C39A). Similar to our findings with MEF2C(C39A), the DNA binding activity of this mutation was not affected by NO. Accordingly, an RNAi-resistant form of MEF2A(C39A), designated MEF2A(C39A)-R, resulted in increased MEF2 transcriptional activity compared to MEF2A-R in NSCs (Figure 5A). In parallel, MEF2A(C39A)-R generated more neurons from NSCs than MEF2A-R (Figure 5B). These results support the notion that S-nitrosylation of MEF2A has a negative impact on neurogenesis from NSCs.
Since we had shown that SNO-MEF2 levels in the brain were elevated in advanced cases of human AD and in AD model mice (Figure 1C), and since neurogenesis is impaired under these conditions (Winner et al., 2011), we next investigated the potential role of SNO-MEF2 in disrupting neurogenesis in vivo. For this purpose, we labeled adult neural progenitors with EdU in ~6 month-old Tg2576 AD mice, which are known to manifest impaired neurogenesis (Winner et al., 2011). We then stereotactically injected lentiviral vectors expressing GFP alone (LV-GFP) into the left dentate gyrus as a control, and WT MEF2 with GFP (LV-WT) or mutant MEF2(C39A) with GFP (LV-C39A) into the right dentate gyrus to determine if the non-nitrosylatable mutant vs. WT MEF2 would rescue neurogenesis in this AD mouse model. The infected progenitors were allowed to differentiate into neurons for 4 wk. Although lentivirus can infect many cell types, we concentrated here on the effect of MEF2 on NSCs by monitoring cells that were also co-labeled with EdU (indicating that the cells had divided) and also NeuN (indicating that they had subsequently become mature neurons). We then stereologically assessed EdU/GFP/NeuN-positive cells and found that mutant MEF2(C39A) significantly increased neurogenesis compared to WT (Figure 6E), consistent with the notion that SNO-MEF2 inhibits neurogenesis in AD brain.
Next, to investigate the molecular pathway whereby MEF2 transcriptional activity regulates neurogenesis, we performed a microarray on mRNA isolated from NSCs expressing a constitutively active form of MEF2 (MEF2CA), which enhances overall MEF2 activity (Molkentin et al., 1996; Okamoto et al., 2002). Heatmap and gene ontology analysis of the MEF2-regulated transcripts demonstrated that the most significant enrichment of genes occurred in pathways involved in nervous system development and function (Figure 6F and Table S1). Of particular interest, one of these genes was nuclear receptor tailless (TLX). TLX had been shown previously to be required for neurogenesis in the adult hippocampus (Zhang et al., 2008). Moreover, we found an evolutionarily conserved MEF2-binding site in the promoter of TLX (Sharov et al., 2006). When we exposed NSCs in culture to NOS inhibitor l-NAME, we observed an increase in TLX levels (Figure 6G). Similarly, we found an increase in TLX in the dentate gyrus of l-NAME-treated mice in vivo (Figure S6C). Importantly, endogenous MEF2A occupied the TLX promoter, and NOS inhibition enhanced this association (Figures 6H and S6D). In contrast, RNAi knockdown of MEF2A resulted in diminished levels of TLX, while an RNAi-resistant form of the MEF2(C39A) mutant increased TLX compared to RNAi-resistant WT (Figure 6I). These data are consistent with the notion that MEF2A is a transcriptional regulator of TLX expression and, via S-nitrosylation of MEF2A, NO may influence TLX levels.
Conservation of SNO-Cys39 in MEF2 proteins across species and kingdoms
Our structure-based molecular and biochemical studies define a novel mechanism whereby MEF2 possesses a pocket surrounding Cys39 in the MADS DNA-binding domain, and S-nitrosylation of this Cys residue disrupts DNA binding activity and hence MEF2 transcriptional activity. Interestingly, the MADS domain is highly conserved in MEF2 proteins across both the plant and animal kingdoms (Figures S2B and S2C). In particular, Cys39 is perfectly conserved in all MEF2 sequences. Similarly, Arg17, Gln18, Phe21, and Asn/Ser49, which form the pocket for S-nitrosylation of Cys39, are highly conserved across species. Intriguingly, in parallel with MEF2 regulation of neurogenesis in animals, similar MADS family genes in plants control floral development (Ng and Yanofsky, 2001), which is also negatively regulated by NO (He et al., 2004). In fact, Cys39 of SEP3, an Arabidopsis MADS family protein, can also be S-nitrosylated (Figure S6E), tempting one to speculate that the mechanism described here may be conserved in many plants and animals.
DISCUSSION
Our findings show that redox-modification by NO reacting with a critical cysteine residue of the MEF2 family of transcription factors acts as a molecular switch to control neurogenesis and cell death in the mammalian brain. We found that MEF2A enhances adult hippocampal neurogenesis in vitro and in vivo, and S-nitrosylation of MEF2A inhibits this effect, at least in part via inhibition of the TLX cascade (Figure 6J). S-Nitrosylation of MEF2A may thus prevent premature differentiation of adult NSCs, while maintaining rests of these cells under physiological conditions. In contrast, we demonstrate in mature neurons that S-nitrosylation of another MEF2 isoform, MEF2C, contributes to NO-induced cell death in cerebrocortical neurons during excitotoxic/ischemic insult and in AD model mice, at least in part by suppressing the antiapoptotic Bcl-xL pathway (Figure 6J). Interestingly, this pathway is distinct from the antiapoptotic MEF2C-PGC1α cascade we recently reported to be affected by S-nitrosylation in human induced pluripotent stem cell (iPSC)-derived dopaminergic neurons as models of PD (Ryan et al., 2013). Taken together, our findings suggest that distinct MEF2-initiated transcriptional events are both cell type- and context-dependent. Moreover, multiple isoforms of MEF2 may well participate in this redox-switching mechanism.
Generation of excessive NO in the brain has been associated with several acute and chronic human neurodegenerative diseases, tipping the nitrosylation/denitrosylation homeostatic balance towards increased nitrosylation (Hara and Snyder, 2007; Nakamura et al., 2013). Accordingly, we observed increased levels of SNO-MEF2 in human stroke and AD brains, indicating that this redox switch is active in human brain. Thus, we find that dysregulation of a single transcription factor family via S-nitrosylation of an evolutionally conserved Cys can adversely impact both neuronal cell survival and adult neurogenesis, providing a common mechanism for prior empirical observations associated with increased NO in these neurodegenerative conditions. While additional aberrant nitrosylation reactions may also contribute to these effects (Nakamura et al., 2013), the fact that a single Cys residue in a single family of transcription factors plays a role in both neurodegeneration and neurogenesis points to potential evolutionary as well as therapeutic consequences.
Consideration of the temporal expression of NO, and hence of SNO-MEF2 formation, may also have important implications for neuronal cell death and neurogenesis in various disease states. For example, during an acute stroke, NO levels are initially high, associated with the onset of neuronal cell death, but then quickly fall, prior to the onset of increased neurogenesis, which occurs several days post stroke (Huang et al., 1994; Ohab et al., 2006). In AD, there is progressive generation of Aβ oligomers, which induce NO, coinciding with increasing neuronal cell death. Additionally, most reports suggest that neurogenesis decreases as AD progresses (Winner et al., 2011), again coinciding with increased Aβ production and consequent generation of NO. Hence, the time course of SNO-MEF2 generation may help explain these phenomena, at least in part.
Moreover, we also relate our results, showing the relation of SNO-MEF2 to neuronal cell death and dysregulated neurogenesis, to findings in the human brain. Specifically, to determine whether the levels of SNO-MEF2 in human diseased brains are potentially important, we used a published technique to calculate the ratio of SNO-MEF2 (determined by biotin switch assay) to total MEF2 (from immunoblots) (Uehara et al., 2006; Cho et al., 2009). This ratio in human neurodegenerative brain was comparable to that encountered in our cell-based and animal models of disease (Figure S6F), consistent with the notion that pathophysiologically relevant amounts of SNO-MEF2 are present in human AD brains. Intriguingly, a large genome-wide association study (GWAS) showed that the MEF2C locus is associated with AD (Lambert et al., 2013). This finding raises the possibility that formation of SNO-MEF2, by inhibiting physiological function, may lower the threshold for or even mimic the effect of genetic alteration (Nakamura et al., 2013). Additionally, our results suggest that SNO-MEF2 may serve as a biomarker for AD as well as other neurodegenerative conditions. Moreover, in mouse models of disease in vivo, the non-nitrosylatable MEF2(C39A) mutant construct rescued neurons from apoptotic cell death and improved neurogenesis. In this light, the pocket surrounding Cys39 on MEF2 may be a potential target for small molecules that could protect this residue from aberrant S-nitrosylation; such protection from redox-related reactions on this transcription factor may prove to be a useful therapeutic intervention for a variety of neurodegenerative disorders.
EXPERIMENTAL PROCEDURES
Plasmids and Lentiviral Vectors
Bcl-xL-promoter luciferase reporter was provided by Gabriel Nunez (University of Michigan) (Grillot et al., 1997). SEP3 cDNA was provided by Martin F. Yanofsky (UCSD). High-titer lentiviral constructs (~1010 transducing units/ml) for in vivo injection were generated at the Viral Core of the Salk Institute for Biological Studies. For details on plasmids and constructs, see Supplemental Experimental Procedures.
Cell Culture and Cell Analysis
Human embryonic kidney (HEK) 293T, HEK293 cells stably expressing neuronal NOS, cerebrocortical neurons, and adult NSCs were cultured as previously described (Gage et al., 1995; Palmer et al., 1997; Okamoto et al., 2002; Uehara et al., 2006). For details on assays for apoptosis and cell differentiation, see Supplemental Experimental Procedures.
In Vivo Mouse Models and Analysis
The intraluminal filament model of middle cerebral artery (MCA) occlusion (stroke) was used as described previously (Gu et al., 2002; Satoh et al., 2006). We analyzed SNO-MEF2 formation 1 h after occlusion, and cell death 1 d after a 90 min occlusion. As an AD model, Tg2576 mice were utilized. To assess the effect of MEF2A on adult neurogenesis, 4 to 6-mo old male MEF2A KO or WT littermate mice were used. For details of lentiviral injection, and analysis of apoptosis, neuronal differentiation, and human brain tissues, see Supplemental Experimental Procedures.
Analysis of S-Nitrosylation of MEF2
The biotin switch and DAN assays were performed as previously described (Gu et al., 2002; Jaffrey and Snyder, 2001; Nakamura et al., 2010; Wink et al., 1999; Yao et al., 2004). For top-down mass spectrometry analysis of MEF2C, an LTQ-Orbitrap-XL mass spectrometer with ETD was used. For details of each analysis, see Supplemental Experimental Procedures.
Geometry optimization of the S-nitrosylated MEF2-DNA complex
For quantum mechanical calculations of the NO-modified cysteine residue, a molecular model was constructed using the GAMESS program for molecular dynamics (Schmidt et al., 1993), the RESP method (Bayly et al., 1993), R.E.D.III program (Grivel et al.), AMBER, a classical molecular mechanical program (Case et al., 2005), and its force field (Cornell et al., 1995). For details, see Supplemental Experimental Procedures.
Statistical Analysis
Population distributions of the data were examined by the D’Agostino-Pearson omnibus normality test. If the data did not show a normal distribution, non-parametric tests (Mann-Whitney and Kruskal-Wallis) were used for single comparisons and for multiple comparisons, respectively. Otherwise, a two-tailed Student’s t-test was applied for single comparisons and an ANOVA with a Scheffé's or Dunnett’s post hoc test was used for multiple comparisons. All experiments were performed in a blinded fashion. Sample sizes were based on Power Analyses performed using our prior data. Statistical significance was a priori set at a p value < 0.05.
Other standard methods, including EMSA, ChIP, and microarray profiling, are described in the Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
-
➢
S-Nitrosylation of MEF2 at conserved cysteine residues occurs in plants and animals
-
➢
S-Nitrosylation of MEF2 inhibits its DNA-binding and transcriptional activity
-
➢
S-Nitrosylation of MEF2 inhibits both neurogenesis and neuronal survival
ACKNOWLEDGMENTS
We thank T. Fang and T. F. Newmeyer for cortical cultures, M. F. Yanofsky (UCSD) for SEP3 cDNA, G. Nuñez (University of Michigan) for Bcl-xL promoter constructs, and E. Masliah (UCSD) for human brain samples. This study was supported in part by a Postdoctoral Fellowship for Research Abroad from the Japan Society for Promotion of Science to T.N., the California HIV/AIDS Research Program, a Shiley-Marcos Alzheimer’s Disease Research Center (UCSD) Pilot Award, NIH grant R21 MH102672 to S.-i.O., and NIH grants P01 HD29587, P01 ES016738, and P30 NS076411 to S.A.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
mRNA profiling data have been deposited in the NCBI GEO under accession number GSE57184.
SUPPLEMENTAL INFORMATION
Supplemental Information includes 6 figures, Supplemental Experimental Procedures, and one dataset and can be found with this article online at doi:xxxxxxxxxx.
AUTHOR CONTRIBUTIONS
This study was designed, directed, and coordinated by S.-i.O., T.N., and S.A.L. The experiments were performed by S.-i.O., T.N., S.F.C., E.K., and C.B. with the assistance of R.Z., R.N., A.C., and A.H. P.C. and A.G. modeled the S-nitrosylated MEF2/DNA complex. X.H. analyzed microarray data. L.L, X.H., and J.R.Y. conducted mass spectrometric analysis. D.H., F.H., and S.S.-B. contributed to in vitro experiments, and A.N., S.S., H.L., J.D.Z., J.C., Z.G., and S.M contributed to in vivo experiments. S.-i.O. and S.A.L. wrote the manuscript.
REFERENCES
- Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- Bonfoco E, Krainc D, Ankarcrona M, Nicotera P, Lipton SA. Apoptosis and necrosis: two distinct events induced, respectively, by mild and intense insults with N-methyl-D-aspartate or nitric oxide/superoxide in cortical cell cultures. Proc. Natl. Acad. Sci. USA. 1995;92:7162–7166. doi: 10.1073/pnas.92.16.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, Lipton SA. S-Nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell WD, Cieplak P, Bayly CI, Gould IKM, Merz J, Ferguson D, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- Dawson VL, Dawson TM, London ED, Bredt DS, Snyder SH. Nitric oxide mediates glutamate neurotoxicity in primary cortical cultures. Proc. Natl. Acad. Sci. USA. 1991;88:6368–6371. doi: 10.1073/pnas.88.14.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Vizarra P, Fernandez AP, Castro-Blanco S, Encinas JM, Serrano J, Bentura ML, Munoz P, Martinez-Murillo R, Rodrigo J. Expression of nitric oxide system in clinically evaluated cases of Alzheimer's disease. Neurobiol. Dis. 2004;15:287–305. doi: 10.1016/j.nbd.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Gage FH, Coates PW, Palmer TD, Kuhn HG, Fisher LJ, Suhonen JO, Peterson DA, Suhr ST, Ray J. Survival and differentiation of adult neuronal progenitor cells transplanted to the adult brain. Proc. Natl. Acad. Sci. USA. 1995;92:11879–11883. doi: 10.1073/pnas.92.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco TM, Hodara R, Parastatidis I, Heijnen HF, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc. Natl. Acad. Sci. USA. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillot DA, Gonzalez-Garcia M, Ekhterae D, Duan L, Inohara N, Ohta S, Seldin MF, Nunez G. Genomic organization, promoter region analysis, and chromosome localization of the mouse bcl-x gene. J. Immunol. 1997;158:4750–4757. [PubMed] [Google Scholar]
- Grivel N, Cieplak P, Dupradeau F-Y. REDIII, (RESP ESP charge Derive program) http://wwwq4md-forcefieldtoolsorg/RED/
- Gu Z, Kaul M, Yan B, Kridel SJ, Cui J, Strongin A, Smith JW, Liddington RC, Lipton SA. S-Nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- Han A, He J, Wu Y, Liu JO, Chen L. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J. Mol. Biol. 2005;345:91–102. doi: 10.1016/j.jmb.2004.10.033. [DOI] [PubMed] [Google Scholar]
- Han A, Pan F, Stroud JC, Youn HD, Liu JO, Chen L. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature. 2003;422:730–734. doi: 10.1038/nature01555. [DOI] [PubMed] [Google Scholar]
- Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc. Natl. Acad. Sci. USA. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara MR, Snyder SH. Cell signaling and neuronal death. Annu. Rev. Pharmacol. Toxicol. 2007;47:117–141. doi: 10.1146/annurev.pharmtox.47.120505.105311. [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al. Nitric oxide represses the Arabidopsis floral transition. Science. 2004;305:1968–1971. doi: 10.1126/science.1098837. [DOI] [PubMed] [Google Scholar]
- Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell. Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Huang K, Louis JM, Donaldson L, Lim FL, Sharrocks AD, Clore GM. Solution structure of the MEF2A-DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. Embo J. 2000;19:2615–2628. doi: 10.1093/emboj/19.11.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Huang PL, Panahian N, Dalkara T, Fishman MC, Moskowitz MA. Effects of cerebral ischemia in mice deficient in neuronal nitric oxide synthase. Science. 1994;265:1883–1885. doi: 10.1126/science.7522345. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell. Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001:pl1–pl9. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- Kader A, Frazzini VI, Solomon RA, Trifiletti RR. Nitric oxide production during focal cerebral ischemia in rats. Stroke. 1993;24:1709–1716. doi: 10.1161/01.str.24.11.1709. [DOI] [PubMed] [Google Scholar]
- Krainc D, Bai G, Okamoto S, Carles M, Kusiak JW, Brent RN, Lipton SA. Synergistic activation of the N-methyl-D-aspartate receptor subunit 1 promoter by myocyte enhancer factor 2C and Sp1. J. Biol. Chem. 1998;273:26218–26224. doi: 10.1074/jbc.273.40.26218. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, Destefano AL, Bis JC, Beecham GW, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat. Genet. 2013 doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer D, Krainc D, Yu YT, McDermott J, Breitbart RE, Heng J, Neve RL, Kosofsky B, Nadal-Ginard B, Lipton SA. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc. Natl. Acad. Sci. USA. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, Nakanishi N, et al. Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc. Natl. Acad. Sci. USA. 2008;105:9397–9402. doi: 10.1073/pnas.0802876105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Kim WR, Ming GL, Song H. Activity-dependent extrinsic regulation of adult olfactory bulb and hippocampal neurogenesis. Ann. N.Y. Acad. Sci. 2009;1170:664–673. doi: 10.1111/j.1749-6632.2009.04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Lopez B, Romero-Grimaldi C, Noval JA, Murillo-Carretero M, Matarredona ER, Estrada C. Nitric oxide is a physiological inhibitor of neurogenesis in the adult mouse subventricular zone and olfactory bulb. J. Neurosci. 2004;24:85–95. doi: 10.1523/JNEUROSCI.1574-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Tu S, Akhtar MW, Sunico CR, Okamoto S, Lipton SA. Aberrant protein S-nitrosylation in neurodegenerative diseases. Neuron. 2013;78:596–614. doi: 10.1016/j.neuron.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Wang L, Wong CC, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, et al. Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol. Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001;2:186–195. doi: 10.1038/35056041. [DOI] [PubMed] [Google Scholar]
- Ohab JJ, Fleming S, Blesch A, Carmichael ST. A neurovascular niche for neurogenesis after stroke. J. Neurosci. 2006;26:13007–13016. doi: 10.1523/JNEUROSCI.4323-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S-i, Krainc D, Sherman K, Lipton SA. Antiapoptotic role of the p38 mitogen-activated protein kinase-myocyte enhancer factor 2 transcription factor pathway during neuronal differentiation. Proc. Natl. Acad. Sci. USA. 2000;97:7561–7566. doi: 10.1073/pnas.130502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S-i, Li Z, Ju C, Schölzke MN, Mathews E, Cui J, Salvesen GS, Bossy-Wetzel E, Lipton SA. Dominant-interfering forms of MEF2 generated by caspase cleavage contribute to NMDA-induced neuronal apoptosis. Proc. Natl. Acad. Sci. USA. 2002;99:3974–3979. doi: 10.1073/pnas.022036399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer MA, Stasiv Y, Benraiss A, Chmielnicki E, Grinberg A, Westphal H, Goldman SA, Enikolopov G. Nitric oxide negatively regulates mammalian adult neurogenesis. Proc. Natl. Acad. Sci. USA. 2003;100:9566–9571. doi: 10.1073/pnas.1633579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer TD, Takahashi J, Gage FH. The adult rat hippocampus contains primordial neural stem cells. Mol. Cell. Neurosci. 1997;8:389–404. doi: 10.1006/mcne.1996.0595. [DOI] [PubMed] [Google Scholar]
- Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, Lee B, Lopez K, Nutter A, Shan B, Molokanova E, Zhang Y, Han X, Nakamura T, Masliah E, Yates JR, 3rd, Nakanishi N, Andreyev AY, Okamoto S-i, Jaenisch R, Ambasudhan R, Lipton SA. Isogenic human iPSC Parkinson's model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 2013;155:1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santelli E, Richmond TJ. Crystal structure of MEF2A core bound to DNA at 1.5 A resolution. J. Mol. Biol. 2000;297:437–449. doi: 10.1006/jmbi.2000.3568. [DOI] [PubMed] [Google Scholar]
- Satoh T, Okamoto SI, Cui J, Watanabe Y, Furuta K, Suzuki M, Tohyama K, Lipton SA. Activation of the Keap1/Nrf2 pathway for neuroprotection by electrophilic [correction of electrophillic] phase II inducers. Proc. Natl. Acad. Sci. USA. 2006;103:768–773. doi: 10.1073/pnas.0505723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, et al. General atomic and molecular electronic structure system. J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Dudekula DB, Ko MS. CisView: a browser and database of cis-regulatory modules predicted in the mouse genome. DNA Res. 2006;13:123–134. doi: 10.1093/dnares/dsl005. [DOI] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur. J. Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: translocation, regulation, and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Uehara T, Nakamura T, Yao D, Shi ZQ, Gu Z, Ma Y, Masliah E, Nomura Y, Lipton SA. S-Nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- Vodovotz Y, Lucia MS, Flanders KC, Chesler L, Xie QW, Smith TW, Weidner J, Mumford R, Webber R, Nathan C, et al. Inducible nitric oxide synthase in tangle-bearing neurons of patients with Alzheimer's disease. J. Exp. Med. 1996;184:1425–1433. doi: 10.1084/jem.184.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink DA, Vodovotz Y, Grisham MB, DeGraff W, Cook JC, Pacelli R, Krishna M, Mitchell JB. Antioxidant effects of nitric oxide. Methods Enzymol. 1999;301:413–424. doi: 10.1016/s0076-6879(99)01105-2. [DOI] [PubMed] [Google Scholar]
- Winner B, Kohl Z, Gage FH. Neurodegenerative disease and adult neurogenesis. Eur. J. Neurosci. 2011;33:1139–1151. doi: 10.1111/j.1460-9568.2011.07613.x. [DOI] [PubMed] [Google Scholar]
- Yao D, Gu Z, Nakamura T, Shi ZQ, Ma Y, Gaston B, Palmer LA, Rockenstein EM, Zhang Z, Masliah E, et al. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA. 2004;101:10810–10814. doi: 10.1073/pnas.0404161101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, Zou Y, He W, Gage FH, Evans RM. A role for adult TLX-positive neural stem cells in learning and behaviour. Nature. 2008;451:1004–1007. doi: 10.1038/nature06562. [DOI] [PubMed] [Google Scholar]
- Zhang ZG, Chopp M, Bailey F, Malinski T. Nitric oxide changes in the rat brain after transient middle cerebral artery occlusion. J. Neurol. Sci. 1995;128:22–27. doi: 10.1016/0022-510x(94)00216-b. [DOI] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.