Abstract
MicroRNAs are important post-transcriptional regulators of skeletal biology, and miRNA-based therapeutics have the potential to aid bone repair. However, efficient tools to deliver miRNA mimics or inhibitors to specific target tissues are limited. Polymeric nanofibers closely mimic natural extracellular matrix morphology, and are attractive candidates to support delivery of cells and bone-anabolic reagents. We hypothesized that gelatin nanofibers could be used for the localized transient delivery of miRNA-based therapeutics, using miR-29a inhibitor as a prototype to increase extracellular matrix (ECM) deposition. miR-29 family members are negative regulators of ECM synthesis, targeting the mRNAs of selected collagens and osteonectin/SPARC. Inhibiting miR-29 activity may therefore, increase extracellular matrix production by cells.
miR-29a inhibitor loaded gelatin nanofibers, prepared by electrospinning, demonstrated continuous release of miRNA inhibitor over 72 hours. Pre-osteoblastic murine MC3T3-E1 cell line seeded on miR-29a inhibitor loaded nanofibers synthesized more osteonectin, indicating efficient inhibitor delivery. These cells also displayed increased Igf1 and Tgfb1 mRNA. Moreover, primary bone marrow stromal cells from transgenic pOBCol3.6cyan reporter mice, grown on miR-29a inhibitor scaffolds, displayed increased col3.6 cyan expression as well as collagen production. This study demonstrated that ECM mimicking nanostructured scaffolds, in conjunction with bioactive miRNA-based therapeutics, may serve as a novel platform for developing biologically active localized cell delivery systems.
1. Introduction
MicroRNAs (miRNAs, miR) are endogenously expressed small non-coding RNAs (18–25 nucleotides) that function as post-transcriptional regulators of gene expression. For the most part, miRNAs interact with complementary regions on target mRNAs, frequently in the 3′ untranslated region (3′ UTR), and cause mRNA destabilization and/or translational repression [1]. Since miRNAs act in the cytoplasm as post-transcriptional regulators, miRNA-based therapeutics have the capacity to regulate gene expression without entering the nucleus [1]. miRNA-based therapeutics are emerging as novel strategies for treating cancer [2, 3], inflammation [4], fibrosis [5], hepatitis C [6], cardiovascular, and metabolic diseases [7].
miRNAs are also key components of the gene expression networks that regulate bone formation and remodeling [1, 8, 9]. Among these, the miR-29 family (miR-29a, miR-29b, miR-29c) is one of the most widely investigated within the field of skeletal biology, and they are vital positive regulators of osteoblast differentiation. The miR-29 family members share a high level of sequence identity, particularly in the seed-binding region (miRNA bases 2–8) important for nucleating interaction of the miRNA with mRNA targets. This sequence conservation suggests that miR-29 family members share target mRNAs and bioactivity.
Transfection of cells with synthetic RNAs, designed to mimic the activity of miR-29 family members or to inhibit their activity, demonstrated that miR-29 family members are potent negative regulators of extracellular matrix synthesis in multiple tissue types [5, 8, 10]. Extracellular matrix synthesis is essential for osteogenic differentiation. Matrix production is one of the early steps of this process, followed by matrix maturation and mineralization [11]. During early stages of osteogenesis, matrix proteins such as osteonectin/SPARC (secreted protein acidic and rich in cysteine) and type I collagen are highly expressed. Osteonectin promotes collagen fiber assembly and is one of the most abundant noncollagenous extracellular matrix proteins in bone [12]. Osteonectin and collagen 1A1 mRNAs are direct targets of miR-29a, and transfection of cells with miR-29a inhibitor results in increased synthesis of osteonectin and type I collagen [5, 8].
In vitro, expression of miR-29 family members is low during early osteoblastic differentiation, when there is abundant extracellular matrix synthesis. Later, as the osteoblasts mature and the matrix is mineralizing, the expression of miR-29 family members increases [8]. In this later phase of differentiation, miR-29 family members potentiate osteoblastogenesis by down regulating several inhibitors of this process, including negative regulators of Wnt signaling [13][8]. We hypothesized that localized transient delivery of miR-29a inhibitor from nanofibers would increase the synthesis of extracellular matrix proteins by the cells to enhance early stages of osteogenesis.
Currently, miRNA-based therapeutics are administrated systemically in vivo [14–16]. However, systemic administration requires large doses of small RNAs, such as siRNA and miRNAs, to stimulate bone formation [15]. Moreover, this systemic administration of large doses of miRNA-based therapeutics carries a high risk for off target, undesired effects, because miRNAs can target multiple mRNAs in an array of tissue types. Therefore, it is likely difficult to restrict the cell types and/or tissues exposed to a systemically administered therapeutic miRNA. Therefore, we reasoned that localized miRNA delivery systems would hold significant advantages for localized tissue regeneration.
In this regard, electrospun nanofiber scaffolds are attractive as synthetic extracellular matrix analogues and as vehicles for localized delivery of therapeutics [17, 18]. Nanofabrication techniques such as electrospinning, phase separation and self-assembly have been developed to form unique nanofibrous structures from both natural and synthetic polymers [3]. Among these, electrospinning represents a versatile and economical technique to produce nanostructured scaffolds with fiber diameters ranging from approximately 1–1000 nm [3]. The high surface area to volume ratio of the nanofibers, combined with their microporous structure, favors cell adhesion, proliferation, migration, and differentiation, all of which are highly desired properties for tissue engineering applications. [3]. Moreover, the electrospinning process allows for encapsulation of biologically active molecules, such as drugs [19] or growth factors [20], within the fibers to modulate cellular function.
The purpose of this study was to evaluate the feasibility of developing miR-29a inhibitor loaded nanofiber matrix and to determine the efficacy of the fibers to enhance extracellular matrix synthesis in cells via localized miR-29a inhibitor delivery. The effect of miR-29a inhibitor incorporation in gelatin nanofiber morphology and diameter was examined. The biological activity of the miR-29a inhibitor loaded gelatin nanofibers was evaluated by quantifying the changes in expression of a miR-29 target gene, osteonectin, in pre-osteoblastic cells and by evaluating the cell fate of primary bone marrow stromal cells.
Materials and Methods
2.0 Materials
The miRNA inhibitors used were small chemically modified single stranded hairpin oligonucleotides designed to bind and sequester endogenous miRNA activity. The RNA inhibitors for miR-29a, a miRNA inhibitor negative control (scramble, SCR), (Meridian® miRNA hairpin inhibitors), miRNAs labeled with Dy547, and TransIT-TKO® Reagent were purchased from Thermo Scientific (Waltham, MA). Trifluoroethanol (TFE), ascorbic acid and gelatin were purchased from Sigma-Aldrich Co. (St. Louis, MI). MC3T3-E1 cells were obtained from the American Type Culture Collection (ATCC, Arlington, VA). Alpha Minimal Essential Medium (αMEM), fetal bovine serum (FBS), phosphate buffer saline (PBS), PicoGreen Assay, penicillin-streptomycin and trypsin-EDTA were purchased from Invitrogen Corp. (Carlsbad, CA). CellTiter 96® Non-Radioactive Cell Proliferation Assay was purchased from Promega (Madison, WI.). The pOBCol3.6 GFPcyan blue reporter mice [21] were a gift from Dr. David Rowe, Center for Regenerative Medicine and Skeletal Development at the University of Connecticut Health Center.
2.1 Electrospinning of Gelatin and miRNA Loaded Gelatin Nanofibers
Gelatin was dissolved in TFE to obtain a 7.5 % (w/v) solution. miR-29a inhibitor or scramble miRNA (negative control) were mixed with transfection reagent TKO at a ratio of 1:1. The miR-29a inhibitor:TKO or scramble miRNA:TKO (negative control) complexes were then added to the gelatin solution to obtain a final miRNA concentrations of 500 nM. The mixtures were vortexed for 1 min to ensure homogeneous distribution of miRNA complex in the solution. Gelatin solutions, without the addition of miRNA/TKO complex, were used as a non-loaded control. Electrospinning was then performed in a custom made chamber where a high voltage of approximately 10.5 kV was applied using ES40 high voltage source GAMMA, High Voltage Research (Ormond Beach, FL). The positive voltage was supplied to the solution by a high voltage wire connected to the tip of the syringe needle. The distance between the syringe tip and collector was approximately 10 cm, and the solution flow rate was kept constant at 0.8 mL/h using a KD Scientific syringe pump. Electrically grounded aluminum film was used as the collector.
2.2 Nanofiber Cross linking
The nanofiber scaffolds were cross linked using various concentrations of glutaraldehyde (GA) (2 mL) vapor at room temperature for 15 minutes in sealed 10 cm chambers. The fibers were lyophilized overnight. For cell studies, nanofiber scaffolds (35–50 μm in thickness) were collected on 12.5 mm diameter glass cover slips, cross linked with 2% GA and sterilized by UV light for 30 minutes.
2.3 Morphological Characterization of Nanofibrous Structure
The morphology of the miRNA loaded and unloaded gelatin nanofibers was determined by Field Emission Scanning Electron Microscopy (FESEM 6335), operated at an accelerating voltage of 10kV and 12μA. Prior to imaging, the samples were mounted on aluminum stubs and platinum coated for improved conductivity. Fiber diameters were determined from the SEM images using Image-J (National Institutes of Health (NIH), http://rsb.info.nih.gov/ij/) image processing software. At least 200 fibers were considered to calculate the average diameter from 3 samples.
2.4 In vitro release of miR-29a Inhibitor from Gelatin Nanofibers
Release kinetics of miR-29a inhibitor was determined by incubating (1 × 1 cm) scaffolds (n=4) in 300μL PBS (pH 7.4) at 37°C for up to 72 hours. Released miRNA inhibitor was quantified by NanoDrop spectrophotometry at 260 nm. The result is reported as cumulative release in ng/mL.
2.5 Preparation of Fluorescently labeled miRNA Loaded Gelatin Nanofibers
In order to confirm the encapsulation of miRNAs within the nanofibrous matrix, Dy547 labeled miRNAs were used. The Dy547 labeled scramble miRNA:TKO complex was loaded into gelatin solution as previously described and electrospun using the aforementioned parameters. The fibers were then visualized using a Zeiss Observer-Z1 microscope, Carl Zeiss, Inc. (Thornwood, NY).
2.6 MC3T3-E1 cell culture
MC3T3-E1 osteoblast-like cells (passages 22–23) were cultured in αMEM/10%FBS/1%Pen-Strep (basal media) in 75cm2 dishes, in a 37°C in a humidified CO2 incubator. Cells were subcultured by treatment with trypsin-EDTA.
2.7 Cell Viability and Cytotoxicity
MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay was used to determine cellular viability. Cells were seeded at a density of 3.5 ×104 cells/well on gelatin nanofibers, gelatin loaded with scramble and gelatin loaded with miR-29a inhibitor nanofibers, in 24 well dishes, allowed to adhere for 24 hours, and washed with PBS. The cells were then cultured for 4 hours at 37°C in a humidified CO2 incubator in basal media in the presence of MTS reagent, followed by measuring the optical density at 490 nm.
2.8 Bioactivity Analysis
2.8.1 Western blot analysis of osteonectin expression
To determine the bioactivity and cellular uptake of miR-29a inhibitor released from the nanofibers, expression of the miR-29 target osteonectin was quantified by Western blot analysis. MC3T3-E1 cells were seeded on glass cover slips and/or nanofiber matrices at 3.5 ×104 cells/well in 24 well dishes, in basal medium, for 24 hours. Culture medium was then replaced with serum-free medium, and the medium was harvested after 6 hours. Protein in the media was precipitated by the addition of ½ volume 10% trichloroacetic acid (TCA), resuspended in reducing sample buffer (62.5 mM Tris pH 6.8, 10% glycerol, 2% SDS, 5% beta mercaptoethanol and bromophenol blue), subjected to electrophoresis through a 10.5% SDS–polyacrylamide gel, and transferred to a PVDF membrane (Millipore, Billerica, MA). Membranes were blocked overnight in 3% BSA in Tris-buffered saline (TBST, 0.1% Tween), and were probed with a rabbit anti-bovine osteonectin primary antibody (BON-1; gift of Dr. L. Fisher, NIDCR, NIH) [22], followed by goat anti-rabbit-horseradish peroxidase conjugated secondary antibody (Sigma). Bands were visualized by chemiluminescence (Perkin-Elmer) and fluorography. Triplicate cultures were analyzed. Relative band densities in scanned images were analyzed with Image J software.
2.8.2 Determination of DNA Content
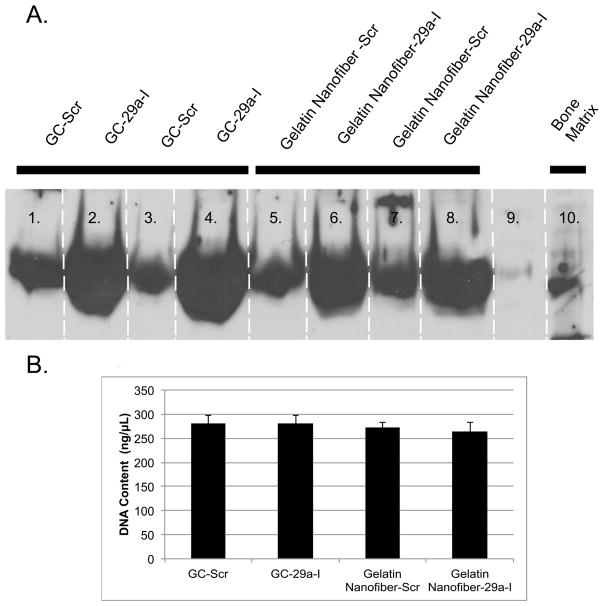
Cell number was estimated by DNA quantification using PicoGreen Assay after 24h of culture. MC3T3-E1 cells were seeded at a density of 3.5 ×104 cells/well (n=4) on glass cover slips, gelatin nanofibers, gelatin loaded with scramble and gelatin loaded with miR-29a inhibitor nanofibers, in 24 well dishes, allowed to adhere for 24 hours. Cells seeded on glass coverslips were allowed to adhere for 6 hours and then treated with 50 nM scramble-TKO complex or miR-29a inhibitor-TKO complex. All groups were allowed to grow for 24 hours and the cells were then washed twice with ice-cold PBS. Cells were lysed in 1% Triton X-100 buffer on ice for 10 minutes and subsequently freeze-thawed three times. The lysate was transferred to a microcentrifuge tube and centrifuged for 10 minutes at 4°C. The supernatant was moved to a fresh tube and DNA content was quantified by Pico Green dsDNA assay using NanoDrop spectrophotometry at 520 nm. The results are reported as ng/mL.
2.8.3 RNA Analysis
RNA was isolated from cultured cells using the RNeasy kit (Qiagen, Valencia, CA). Reverse transcription was carried out using RNA to cDNA EcoDry kit (Clontech, Mountain View, CA) according to manufacturer’s protocol. Igf1 andTgfb1 mRNA levels were determined by real-time PCR using iQ SYBR Green Supermix in an iCycler iQ5 real-time PCR detection system (Bio-Rad, Hercules, CA). GAPDH was used as the housekeeping gene.
2.8.4 Bone Marrow Stromal Cell Cultures
The UCHC Institutional Animal Care and Use Committee approved all aspects of the experimental protocol. Femurs and tibias from 6 to 8 week-old male pOBCol3.6 GFPcyan blue reporter mice were dissected from the surrounding tissues. The epiphyseal growth plates were removed and the marrow was collected by flushing with complete medium from a 25-gauge needle. Cells were plated and allowed to grow for 3 days. On day 3, half of the medium was replaced with fresh medium. Cells were allowed to grow for 5 days, and then re-plated for experiments at a density of 3.5 ×104 cells/well in 24 well dishes in basal media supplemented with 50 μg/ml of ascorbic acid. Bone marrow stromal cells were cultured for 8 days, with a media change every 3 days. Transgenic expression of Col 3.6 cyan blue [21, 23] was followed by fluorescent microscopy using Ziess Observer Z.1 inverted microscope.
2.8.5 Hydroxyproline Assay
Collagen is enriched in the amino acid hydroxyproline, and hydroxyproline levels are frequently used as an indicator of collagen content. BMSCs were cultured on glass coverslips, gelatin-SCR, or gelatin-29a inhibitor nanofibers for 8 days, and then hydroxyproline content was determined. Samples were washed in PBS, lysed in 100 μL of water. The lysate was subsequently transferred into polypropylene tubes and hydrolyzed in 6 M HCl at 120°C for 3 hours. Samples were then oxidized by Chloramine T, incubating at room temperature. After which, DMAB reagent was added to the samples and incubated for 90 minutes at 60°C. The hydroxyproline concentration was measured by spectrophotometry at an absorbance of 545 nm. Background absorbance from glass coverslips, scramble loaded gelatin and miR-29a inhibitor loaded nanofibers were subtracted from the corresponding absorbance readings to obtain the corrected value.
2.9 Statistical analysis
Data were statistically analyzed and expressed as mean± standard deviation (SD). One way ANOVA followed by Tukey’s test or Student’s t-test was performed.
3.0 Results and Discussion
3.1 Morphological Characterization of Nanofibrous Structure
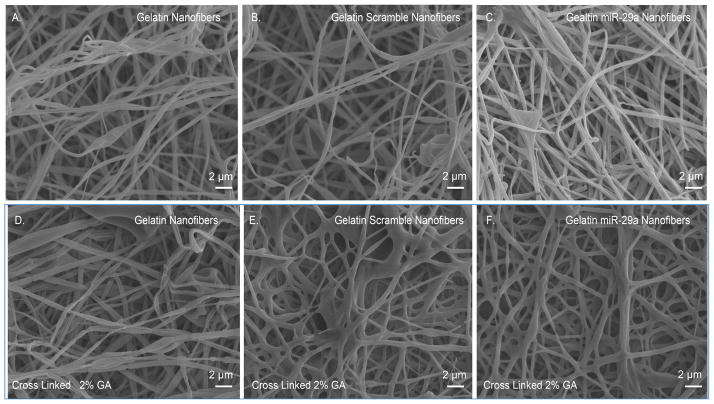
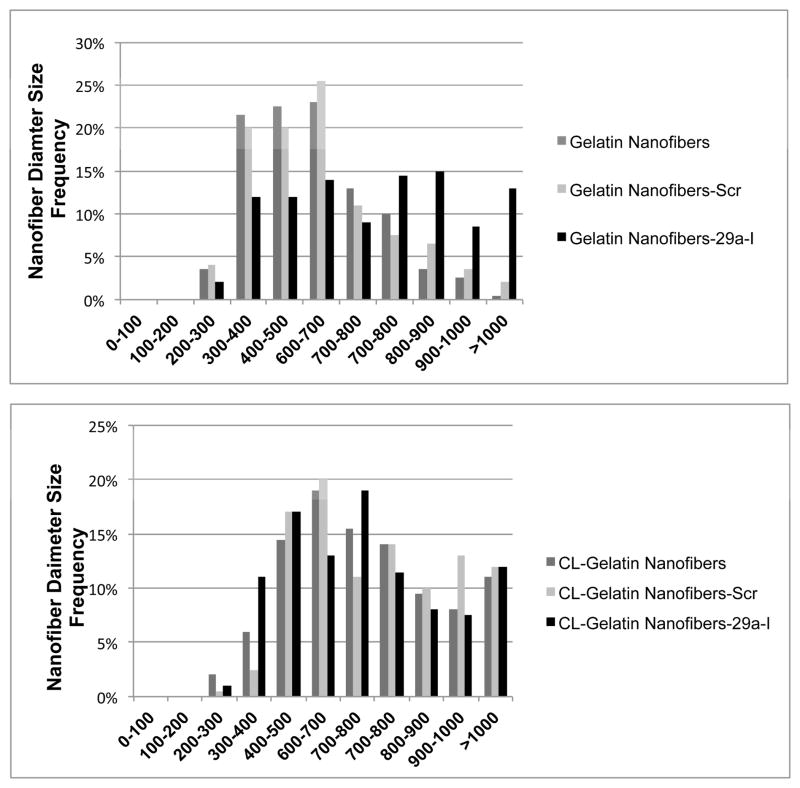
In order to maintain gelatin nanofiber structural integrity in aqueous solution, gelatin nanofibers must be cross linked. Among cross linking methods, glutaraldehyde (GA) vapor cross linking is the most commonly used [24, 25]. However, high concentrations of GA may lead to toxic effects, if residual GA is present during cell culture [26]. Therefore, preliminary studies were performed to identify the minimum amount of GA required for gelatin nanofiber cross linking (Supplemental Figure 1). Gelatin nanofibers were exposed to 2%, 5%, 10%, 15%, 20%, 25% and 50% GA vapors for 15 minutes, and then visualized by SEM. The increase in GA concentrations did not significantly affect the nanofiber morphology or diameter size. Irrespective of cross linking time, the nanofibers were stable in cell culture media for 7 days (data not shown). Hence, 2% GA concentration was used for cross linking the nanofiber scaffolds for all the subsequent studies. Figure 1A shows the SEM micrographs of unloaded gelatin nanofibers indicating a defect free structure. Addition of scramble or miR-29a inhibitors did not cause beading or defects in the nanofibers (Figure 1B, 1C). These results indicate that the miRNAs or TKO reagent do not affect nanofiber spinnability at the concentrations studied. Figures 1D–1F show unloaded and miRNA loaded gelatin nanofibers cross linked with 2% GA vapors for 15 min. As expected, the cross linking method did not adversely affect the morphology of miRNA loaded nanofibers. Figure 2 shows the diameter distribution of unloaded and miRNA loaded gelatin nanofibers before and after cross linking with 2% GA vapor for 15 min. The water content of the GA vapor could increase the diameter of cross linked fibers [26]. In the present study, although a shift in the fiber diameter was observed with cross linked fibers, the diameters of both non cross linked and cross linked nanofibers remained in the 200 –1000 nm range.
Figure 1. Scanning electron micrographs of electrospun gelatin, scramble and miR-29a inhibitor nanofiber mats.
A.) gelatin nanofibers B.) gelatin nanofibres loaded with scramble miRNA C.) gelatin nanofibers loaded with miR-29a inhibitor D.) cross linked gelatin nanofibers E.) cross linked gelatin nanofibers loaded with scramble miRNA and F.) cross linked gelatin nanofibers loaded with miR-29a inhibitor (scale bar 2 μm).
Figure 2. Analysis of nanofiber diameter size.
Histogram showing diameter distribution of nanofibers as measured from SEM images. A.) Non-cross linked nanofibers and B.) cross linked (CL) nanofibers.
3.2 Detection of Encapsulated miRNAs in Gelatin Nanofibers
Figure 3A–F shows the DIC and fluorescence microscopy images of gelatin nanofibers in the presence or absence Dy547-labeled miRNAs. Auto-fluorescence was not detected in the gelatin nanofibers (Figure 3A,3C). In contrast, a uniform red fluorescence was observed from the gelatin nanofibers loaded with Dy547-labeled miRNA, demonstrating uniform loading of the miRNA throughout the fibers (Figure 3D,3F).
Figure 3. Fluorescence micrographs of Dy547 conjugated miRNA incorporated into gelatin nanofibers.
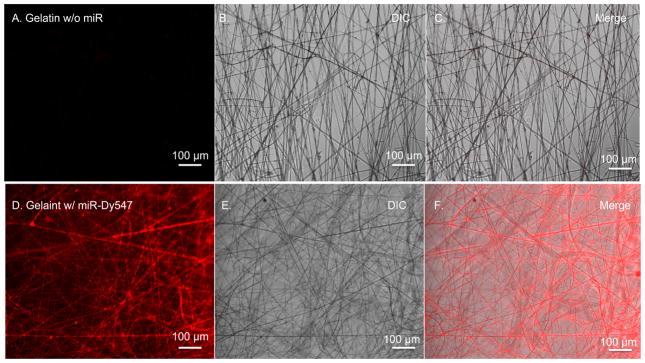
A–C.) Unloaded gelatin nanofibers and D–F.) gelatin nanofibers loaded with fluorescently labeled miRNAs. Differential interference contrast (DIC) image, fluorescent miRNAs (red) (scale 100 μm).
3.3 In vitro Release of miR-29a Inhibitor from Gelatin Nanofibers
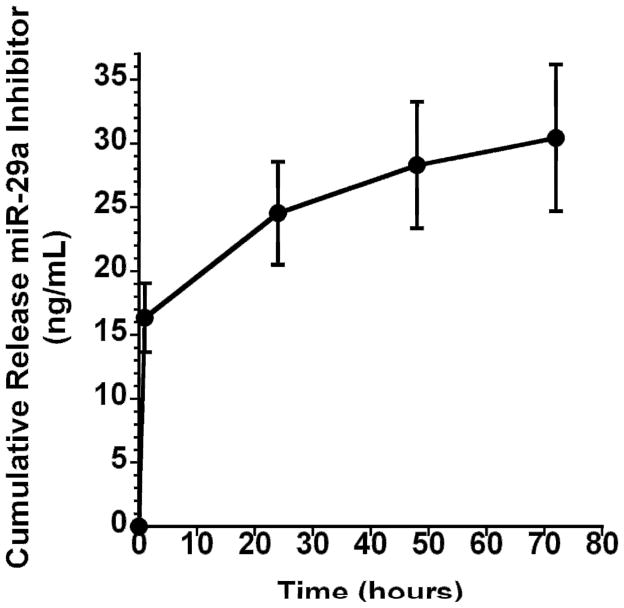
Conventionally, when cells are transiently transfected in tissue culture, they are exposed to one treatment of miRNA-transfection reagent complex for 24–72 hours. To create an optimal transient delivery vehicle, it is important to understand how the miRNAs are released from nanofibers; therefore, a short-term release study was performed. Figure 4 demonstrates the release kinetics of miR-29a inhibitor from gelatin nanofibers. miR-29a inhibitor loaded nanofibers were incubated in PBS at 37° C for up to 72 hours. The cross linked gelatin nanofibers showed an initial burst release of 15 ng/mL miRNA inhibitor within the first 2 hours, followed by the continued release of an additional 10 ng/mL in the next 22 hours. Between 24 and 72 hours, the fibers released an additional 5 ng/mL. Since release of miR-29a inhibitor from the nanofibers revealed an initial burst followed by sustained release for up to 72h, this transfection system may largely resemble transfection in a tissue culture plate.
Figure 4.
Cumulative Release of miR-29a inhibitor from gelatin nanofibers as a function of time.
Composite nanofibers of gelatin with poly caprolactone [27, 28] or poly(l-lactic acid)-co-poly-(ε-caprolactone) [29, 30] have been used to encapsulate large molecules such as fibroblast growth factor 2 (FGF2) [31] with relative ease. With regard to delivery of small RNAs, siRNAs encapsulated in caprolactone and ethyl ethylene phosphate nanofibers demonstrated an initial burst release upon immersion, followed by a sustained delivery [32]. Our data suggest that the electrospun gelatin nanofibers exhibited microRNA release kinetics with characteristic burst release similar to the copolymer delivery systems. In addition, gelatin is a natural biodegradable polymer derived from collagen, it is readily resorbed in the body, and has demonstrated ability to support cellular adhesion [33], proliferation [25], and differentiation [34, 35]. Thus, gelatin is a highly desirable substrate to serve as a local miRNA delivery system to support tissue regeneration.
3.4 Viability of MC3T3-E1 Cells on miR-29a Inhibitor Loaded Gelatin Nanofibers
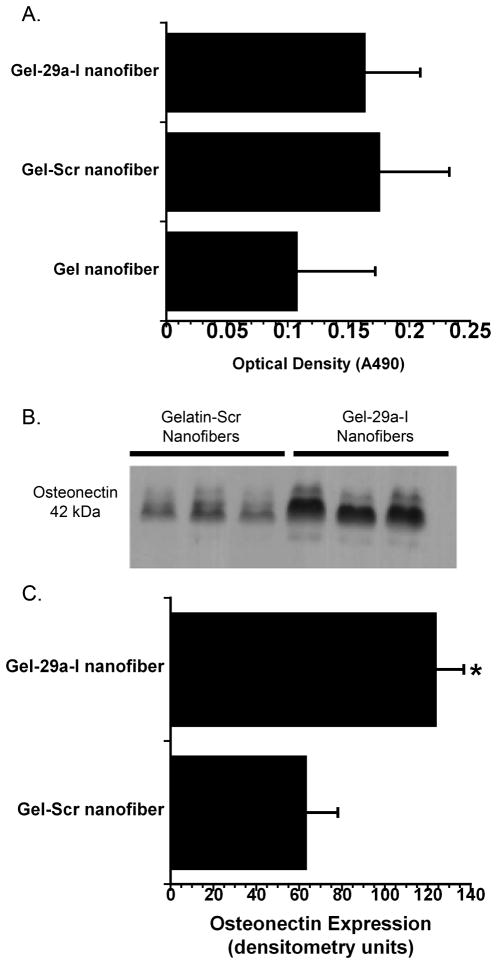
To determine whether the TKO-miRNA inhibitor delivery from gelatin nanofibers had an adverse effect on cell viability, MTS assay was performed using the murine pre-osteoblastic cell line MC3T3 E1. Cells were seeded on gelatin nanofibers, gelatin nanofibers loaded with scramble: TKO, and gelatin nanofibers loaded with miR-29a inhibitor: TKO (Figure 5A). After 24 hours of culture, there were no significant differences in cell viability among any of the nanofibrous groups. Since this demonstrated that TKO or miRs did not affect cell viability, in subsequent experiments, we only compared miR-29a inhibitor nanofiber bioactivity to that containing the non-targeting control, scramble.
Figure 5. Cell number and osteonectin protein production from MC3T3-E1 cells seeded on scramble or miR-29a inhibitor loaded nanofibers.
A.) MTS assay for cell viability comparing all nanofibrous groups B.) Western blot analysis of osteonectin production 24h after cells were seeded on gelatin nanofibers loaded with scramble or gelatin loaded with miR-29a inhibitor. C.) Quantified osteonectin levels. (n=3, * p<0.01).
Currently, there is a large variety of commercially available lipid-based transfection reagents used for increasing the efficacy of siRNA and miRNA delivery. In this study, we chose to use TKO, a proprietary transfection reagent shown to enhance the efficacy of miRNA and siRNA delivery to BMSCs and the multipotent murine mesenchymal cell line C3H10T1/2 [36]. Additionally, TKO was previously shown to enhance siRNA delivery from synthetic nanofiber matrices. Although transfection reagents such as liposomes can be toxic to cells [37], our work demonstrated that TKO reagent, used as described, does not adversely affect the viability of MC3T3-E1 cells (Figure 5A).
3.5 Bioactivity of miR-29a Inhibitor Loaded Gelatin Nanofibers
3.5.1 miR-29a Inhibitor Transfection via Gelatin Nanofibers
To determine whether the miRNA inhibitor released from nanofiber matrices was biologically active for transfecting cells, the expression of the miR-29 target osteonectin was analyzed. For these studies, MC3T3-E1 cells were cultured on nanofibers containing miR-29a inhibitor or scramble for 24 hours. The quantity of osteonectin released into the medium was evaluated by Western blot analysis (Figure 5B,5C). Osteonectin production was significantly enhanced in cells seeded on miR-29a inhibitor loaded nanofibers as compared to scramble loaded gelatin nanofibers. This indicates that the miR-29a inhibitor released from the nanofibers is bioactive, suggesting that the miR-29a inhibitor-loaded scaffolds may have the capacity to induce the expression of other miR-29 family target molecules, such as collagens.
3.5.2 Comparison of 2D Transfection vs. 3D Nanofibrous Transfection
We then investigated the relative efficacy of miRNA inhibitor transfection, mediated by gelatin nanofibers, compared with a conventional, 2D/solution based transfection system. Here, equal numbers of MC3T3-E1 cells were seeded on uncoated cover slips or cover slips coated with nanofibers loaded with the miR-29a-TKO complex. Cells on the uncoated cover slips were exposed to transfection solution containing the same amount of miRNA inhibitor-TKO complex as that contained within the nanofibers. Western blot analysis for osteonectin confirmed that cells cultured on uncoated cover slips and transfected with a scrambled miRNA inhibitor had osteonectin levels similar to that of cells cultured on the scrambled inhibitor loaded nanofibers. In contrast, cells cultured on uncoated cover slips and transfected with miR-29a inhibitor displayed increased osteonectin levels, similar to that of cells grown on miR-29a inhibitor loaded nanofibers (Figure 6A). To ensure that increased osteonectin levels were not due to differences in cell number, DNA was quantified in the cell layers. Significant differences in cell number were not detected when MC3T3-E1 cells were grown for 24 hours on glass coverslips or on the nanofiber groups tested (Figure 6B). In this study, we demonstrated that the transfection mediated by miR-29a inhibitor nanofibers is analogous to 2D transfection in vitro.
Figure 6. Osteonectin protein secreted from transfected MC3T3-E1 cells seeded on 2-D cover slips or miR-29a inhibitor loaded gelatin nanofibers.
A.)Western blot analysis of osteonectin was performed 24h after cells were seeded on scaffolds and B.) DNA content of cells cultured on glass coverslips and miRNA loaded nanofibers for 24h. No statistically significance differences were observed between groups.
3.5.3 mRNA Expression in MC3T3-E1 Cells Seeded on miR-29a Inhibitor Nanofibers
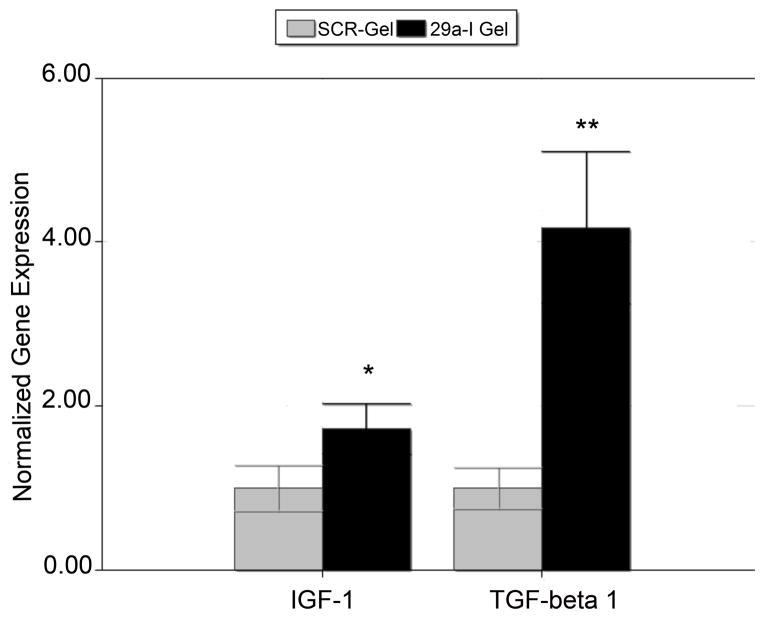
After confirming the biological activity and transfectability of miR-29a inhibitor released from nanofibers, we determined whether miR-29a inhibitor altered the expression of genes important for matrix production. MC3T3-E1 cells were seeded on miR-29a inhibitor nanofibers or scramble loaded nanofibers for 24 hours, and then RNA was isolated and analyzed by qRT-PCR. mRNA levels of both Igf1 and Tgfb1 were significantly up regulated in cells grown on the miR-29a inhibitor loaded scaffolds compared to controls (Figure 7). Insulin-like Growth Factor 1 (IGF1) is an autocrine, paracrine and endocrine growth factor that plays an important anabolic role in bone [38] IGF1 stimulates osteoblast proliferation and survival, and promotes differentiation. Moreover, IGF1 stimulates matrix production by bone cells, and Igf1 mRNA is a direct miR-29 target [39]. miR-29 inhibitor-mediated increase in Igf1 could contribute to the production of matrix stimulated by miR-29a inhibitor scaffolds.
Figure 7. Gene expression in MC3T3-E1 cells seeded on scramble or miR-29a inhibitor loaded nanofibers.
(n=4; Igf1*p<0.05 and Tgfb1 **p<0.01)
Transforming Growth Factor β 1 (TGF-β1) is mitogenic for osteoblast precursors and is a potent inducer of extracellular matrix synthesis [40–42]. This pro-fibrotic growth factor has been shown to decrease the expression of miR-29 family members [10, 43, 44]. In the present study Tgfb1 mRNA was significantly up regulated by miR-29a inhibitor. However, we do not know yet whether Tgfb1 mRNA is a direct miR-29 target or if the up regulation of Tgfb1 mRNA is an indirect effect of a gene expression program triggered by the actions of the miR-29 inhibitor. The up regulation of Tgfb1 and Igf1 mRNA, as well as osteonectin expression in MC3T3-E1 cells, demonstrates the capacity for miR-29a inhibitor loaded nanofibers to enhance extracellular matrix synthesis.
3.5.4 Enhanced Matrix Synthesis by BMSCs
To confirm that the miR-29a inhibitor nanofibrous system could stimulate collagen production and has the capacity to transfect primary cells, we used bone marrow stromal cells (BMSCs) from pOBCol3.6 GFPcyan blue reporter mice (Col 3.6 cyan blue)[21, 23, 45]. These transgenic mice express the GFPcyan reporter gene under the control of a 3.6kb segment of the rat Col1a1 promoter/enhancer (pOBCol3.6). This reporter mouse allows for tracing the biological response of cells within a heterogeneous population of BMSCs by monitoring col 3.6 cyan blue expression over time [23]. Although the cyan blue reporter is expressed in several mesenchymal lineage-derived cell types, its expression is strongest in a population of cells that exhibit commitment to the osteoblastic lineage, and in mature, differentiated osteoblasts. Here we used this marker gene to determine whether miR-29a inhibitor released from nanofibers could affect BMSC fate.
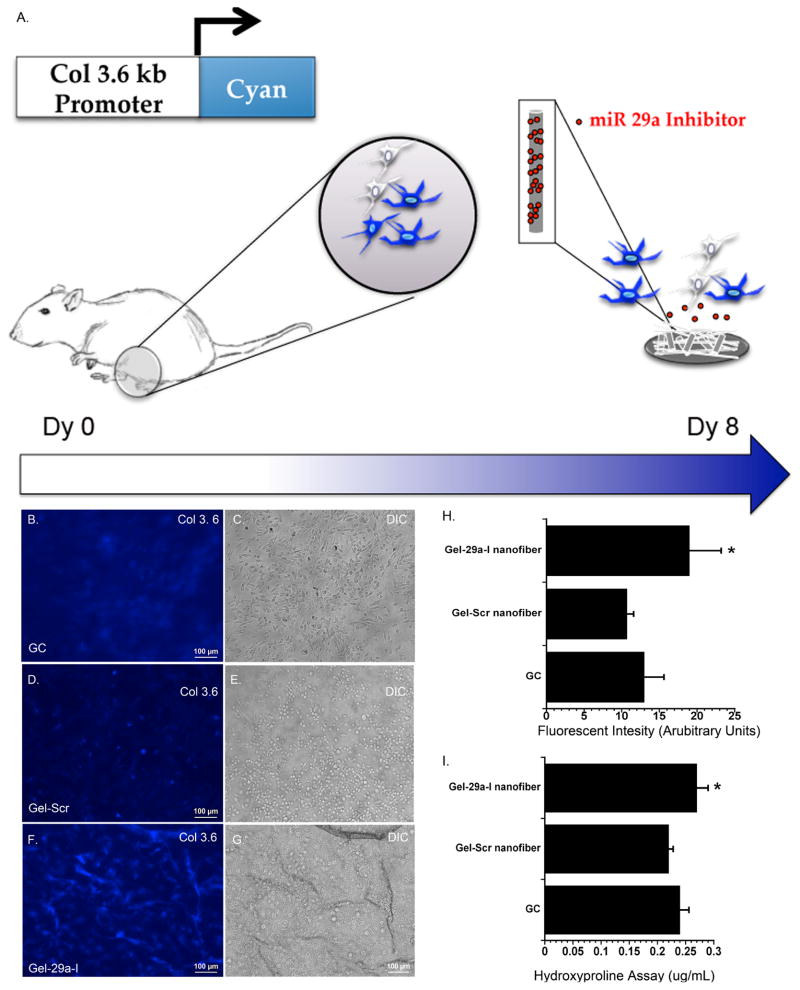
Figure 8B–D, shows fluorescence micrographs of BMSCs from Col 3.6 cyan reporter mice cultured for 8 days on miR-29a inhibitor loaded nanofibers, scramble-loaded nanofibers, or cells cultured on uncoated cover slips. The morphology of cells seeded on glass cover slips (Figure 8E) appeared to be different from those seeded on gelatin nanofibers (Figure 8F,G). The cells seeded on cover slips appeared flat, and Col 3.6 cyan blue fluorescence was diffuse (Figure 8B,E). Cells seeded on gelatin scramble loaded nanofibers also displayed diffuse blue fluorescence, but with select cells in each field displaying a brighter fluorescent signal (Figure 8C). The effect of gelatin nanofibers on cellular morphology requires further investigation. In contrast, cells seeded on miR-29a inhibitor nanofibers appeared to have increased Col 3.6 cyan blue expression, with a distinctly higher percentage of the cells in each field displaying a bright fluorescent signal (Figure 8D). When total fluorescence was quantified, the intensity was significantly higher in cultures grown on miR-29a inhibitor nanofibers, compared with either control (Figure 8H).
Figure 8. Col 3.6 cyan blue expression and collagen content from BMSCs seeded on scaffolds.
A.) Schematic of bone marrow stromal cultures isolated from femurs and tibia of pOBCol3.6 cyan blue mice following 8 days of culture. Fluorescence micrographs of cells cultured on B.) glass cover slips (GC), C.) gelatin nanofibers loaded with scramble D.) gelatin nanofibers loaded with miR-29a inhibitor. E, F and G are DIC images corresponding to the side panel (scale 100 μm). H.) Quantified fluorescence intensity. I.) Hydroxyproline content of cultures after 8 days of culture. (n=3; * p<0.05).
To determine whether miR-29a inhibitor affected collagen deposition in BMSCs, we quantified hydroxyproline levels in the cell layer after 8 days of culture on glass, miR-29a inhibitor nanofibers or scramble control nanofibers. Figure 8I shows BMSCs seeded on miR-29a inhibitor loaded scaffolds had an enhanced collagen deposition compared to BMSC seeded on gelatin loaded scramble nanofibers. It is possible that the increased production of extracellular matrix proteins, mediated by the miR-29a inhibitor, could contribute to the increased expression of the Col 3.6 cyan reporter gene. Overall, these studies show the ability of this miRNA delivery system to transfect primary cells, supporting the potential use of miR-29a inhibitor loaded nanofibers with clinically relevant cells for tissue engineering applications.
In summary, we demonstrated the feasibility of developing a scaffold capable of delivering miRNA-based therapeutics to enhance extracellular matrix production in pre-osteoblast cells and primary BMSCs. SEM micrographs demonstrated the feasibility of obtaining bead/defect-free fibrous structures with diameters in the nanometer range. Fibers exhibited sustained release of miRNA over 72 hours. Further, we demonstrated good cytocompatibility of the miRNA loaded nanofibers. Additionally, miR-29a inhibitor loaded scaffolds increased osteonectin production and levels of Igf1 and Tgfb1 mRNA. Lastly, Col 3.6 cyan blue BMSCs cultured on miR-29a inhibitor loaded nanofibers demonstrated increased collagen and higher expression of the cyan blue reporter gene demonstrating successful transfection in primary bone marrow cells.
4.0 CONCLUSIONS
Collectively, this study demonstrates the feasibility of producing miR-29a inhibitor loaded nanofibers as an extracellular matrix stimulating scaffold for tissue engineering. The unique extracellular matrix mimicking nanofiber scaffolds, combined with their ability to present miRNA-based therapeutics in a sustained and bioactive manner, may serve as a novel platform for tissue engineering.
Supplementary Material
Gelatin nanofibers cross linked using 2%, 5%, and 25% for 15 minutes.
Supplemental Table 1. Sequences of microRNAs used in the study.
Acknowledgments
We thank Dr. Larry Fisher (NIDCR, NIH) for the gift of the BON-1 antibody, and Dr. David Rowe (University of Connecticut Health Center) for the gift of the col3.6cyan mice. Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Numbers R044877 (to AMD) and AR061575 (to LSN).
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kapinas K, Delany AM. MicroRNA biogenesis and regulation of bone remodeling. Arthritis research & therapy. 2011;13:220. doi: 10.1186/ar3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Antonellis P, Liguori L, Falanga A, Carotenuto M, Ferrucci V, Andolfo I, et al. MicroRNA 199b-5p delivery through stable nucleic acid lipid particles (SNALPs) in tumorigenic cell lines. Naunyn-Schmiedeberg’s archives of pharmacology. 2013;386:287–302. doi: 10.1007/s00210-013-0837-4. [DOI] [PubMed] [Google Scholar]
- 3.Nair LS, Laurencin CT. Nanotechnology and Tissue Enginnering: The Scaffold. United States: CRS Press, Taylor and Francis Group; 2008. [Google Scholar]
- 4.Gong Z, Zhang S, Tang K, Li X, Xiang B, Xiang J, et al. MicroRNAs and nonresolving inflammation-related cancer. Zhong nan da xue xue bao Yi xue ban = Journal of Central South University Medical sciences. 2013;38:639–44. doi: 10.3969/j.issn.1672-7347.2013.06.014. [DOI] [PubMed] [Google Scholar]
- 5.He Y, Huang C, Lin X, Li J. MicroRNA-29 family, a crucial therapeutic target for fibrosis diseases. Biochimie. 2013;95:1355–9. doi: 10.1016/j.biochi.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Kumar A. MicroRNA in HCV infection and liver cancer. Biochimica et biophysica acta. 2011;1809:694–9. doi: 10.1016/j.bbagrm.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 7.Hulsmans M, Holvoet P. MicroRNAs as early biomarkers in obesity and related metabolic and cardiovascular diseases. Current pharmaceutical design. 2013;19:5704–17. doi: 10.2174/13816128113199990364. [DOI] [PubMed] [Google Scholar]
- 8.Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. Journal of cellular biochemistry. 2009;108:216–24. doi: 10.1002/jcb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. The Journal of biological chemistry. 2010;285:25221–31. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M, et al. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. Journal of the American Society of Nephrology : JASN. 2012;23:252–65. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentili C, Cancedda R. Cartilage and bone extracellular matrix. Current pharmaceutical design. 2009;15:1334–48. doi: 10.2174/138161209787846739. [DOI] [PubMed] [Google Scholar]
- 12.Alford AI, Hankenson KD. Matricellular proteins: Extracellular modulators of bone development, remodeling, and regeneration. Bone. 2006;38:749–57. doi: 10.1016/j.bone.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, et al. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. The Journal of biological chemistry. 2009;284:15676–84. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godinho BM, Ogier JR, Darcy R, O’Driscoll CM, Cryan JF. Self-assembling modified beta-cyclodextrin nanoparticles as neuronal siRNA delivery vectors: focus on Huntington’s disease. Molecular pharmaceutics. 2013;10:640–9. doi: 10.1021/mp3003946. [DOI] [PubMed] [Google Scholar]
- 15.Zhang G, Guo B, Wu H, Tang T, Zhang BT, Zheng L, et al. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nature medicine. 2012;18:307–14. doi: 10.1038/nm.2617. [DOI] [PubMed] [Google Scholar]
- 16.Kozielski KL, Tzeng SY, Green JJ. Bioengineered nanoparticles for siRNA delivery. Wiley interdisciplinary reviews. Nanomedicine and nanobiotechnology. 2013;5:449–68. doi: 10.1002/wnan.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chew SY, Wen Y, Dzenis Y, Leong KW. The role of electrospinning in the emerging field of nanomedicine. Current pharmaceutical design. 2006;12:4751–70. doi: 10.2174/138161206779026326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nair LS, Laurencin CT. Polymers as biomaterials for tissue engineering and controlled drug delivery. Advances in biochemical engineering/biotechnology. 2006;102:47–90. doi: 10.1007/b137240. [DOI] [PubMed] [Google Scholar]
- 19.Singh H, James E, Kan H-M, Nair LS. Fabrication and Evaluation of Resveratrol Loaded Polymeric Nanofibers. 2012;2:228–235. [Google Scholar]
- 20.James EN, Nair LS. Development and Characterization of Lactoferrin Loaded Poly(ε-Caprolactone) Nanofibers. 2014;10:500–507. doi: 10.1166/jbn.2014.1717. [DOI] [PubMed] [Google Scholar]
- 21.Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, et al. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 22.Ingram RT, Clarke BL, Fisher LW, Fitzpatrick LA. Distribution of noncollagenous proteins in the matrix of adult human bone: evidence of anatomic and functional heterogeneity. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 1993;8:1019–29. doi: 10.1002/jbmr.5650080902. [DOI] [PubMed] [Google Scholar]
- 23.Bilic-Curcic I, Kronenberg M, Jiang X, Bellizzi J, Mina M, Marijanovic I, et al. Visualizing levels of osteoblast differentiation by a two-color promoter-GFP strategy: Type I collagen-GFPcyan and osteocalcin-GFPtpz. Genesis (New York, NY : 2000) 2005;43:87–98. doi: 10.1002/gene.20156. [DOI] [PubMed] [Google Scholar]
- 24.Ratanavaraporn J, Rangkupan R, Jeeratawatchai H, Kanokpanont S, Damrongsakkul S. Influences of physical and chemical crosslinking techniques on electrospun type A and B gelatin fiber mats. International journal of biological macromolecules. 2010;47:431–8. doi: 10.1016/j.ijbiomac.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Zhang YZ, Venugopal J, Huang ZM, Lim CT, Ramakrishna S. Crosslinking of the electrospun gelatin nanofibers. Polymer. 2006;47:2911–7. [Google Scholar]
- 26.Wu SC, Chang WH, Dong GC, Chen KY, Chen YS, Yao CH. Cell adhesion and proliferation enhancement by gelatin naofiber scaffolds. Journal of Bioactive and Compatible Polymers. 2011;26:566–77. [Google Scholar]
- 27.Lee J, Yoo JJ, Atala A, Lee SJ. The effect of controlled release of PDGF-BB from heparin-conjugated electrospun PCL/gelatin scaffolds on cellular bioactivity and infiltration. Biomaterials. 2012;33:6709–20. doi: 10.1016/j.biomaterials.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuppan P, Sethuraman S, Krishnan UM. PCL and PCL-gelatin nanofibers as esophageal tissue scaffolds: optimization, characterization and cell-matrix interactions. Journal of biomedical nanotechnology. 2013;9:1540–55. doi: 10.1166/jbn.2013.1653. [DOI] [PubMed] [Google Scholar]
- 29.Fan H, Tao H, Wu Y, Hu Y, Yan Y, Luo Z. TGF-beta3 immobilized PLGA-gelatin/chondroitin sulfate/hyaluronic acid hybrid scaffold for cartilage regeneration. Journal of biomedical materials research Part A. 2010;95:982–92. doi: 10.1002/jbm.a.32899. [DOI] [PubMed] [Google Scholar]
- 30.Hu J, Wei J, Liu W, Chen Y. Preparation and characterization of electrospun PLGA/gelatin nanofibers as a drug delivery system by emulsion electrospinning. Journal of biomaterials science Polymer edition. 2013;24:972–85. doi: 10.1080/09205063.2012.728193. [DOI] [PubMed] [Google Scholar]
- 31.Kim MS, Shin YM, Lee JH, Kim SI, Nam YS, Shin CS, et al. Release kinetics and in vitro bioactivity of basic fibroblast growth factor: effect of the thickness of fibrous matrices. Macromolecular bioscience. 2011;11:122–30. doi: 10.1002/mabi.201000222. [DOI] [PubMed] [Google Scholar]
- 32.Cao H, Jiang X, Chai C, Chew SY. RNA interference by nanofiber-based siRNA delivery system. Journal of controlled release : official journal of the Controlled Release Society. 2010;144:203–12. doi: 10.1016/j.jconrel.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Ouyang H, Lim CT, Ramakrishna S, Huang ZM. Electrospinning of gelatin fibers and gelatin/PCL composite fibrous scaffolds. Journal of biomedical materials research Part B, Applied biomaterials. 2005;72:156–65. doi: 10.1002/jbm.b.30128. [DOI] [PubMed] [Google Scholar]
- 34.Sisson K, Zhang C, Farach-Carson MC, Chase DB, Rabolt JF. Fiber diameters control osteoblastic cell migration and differentiation in electrospun gelatin. Journal of biomedical materials research Part A. 2010;94:1312–20. doi: 10.1002/jbm.a.32756. [DOI] [PubMed] [Google Scholar]
- 35.Sachar A, Strom TA, Serrano MJ, Benson MD, Opperman LA, Svoboda KK, et al. Osteoblasts responses to three-dimensional nanofibrous gelatin scaffolds. Journal of biomedical materials research Part A. 2012;100:3029–41. doi: 10.1002/jbm.a.34253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang QQ, Otto TC, Lane MD. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9607–11. doi: 10.1073/pnas.0403100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlsen TA, Brinchmann JE. Liposome delivery of microRNA-145 to mesenchymal stem cells leads to immunological off-target effects mediated by RIG-I. Molecular therapy : the journal of the American Society of Gene Therapy. 2013;21:1169–81. doi: 10.1038/mt.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yakar S, Courtland HW, Clemmons D. IGF-1 and bone: New discoveries from mouse models. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2010;25:2543–52. doi: 10.1002/jbmr.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith SS, Kessler CB, Shenoy V, Rosen CJ, Delany AM. IGF-I 3′ untranslated region: strain-specific polymorphisms and motifs regulating IGF-I in osteoblasts. Endocrinology. 2013;154:253–62. doi: 10.1210/en.2012-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang SY, Alliston T. Regulation of postnatal bone homeostasis by TGF[beta] doi: 10.1038/bonekey.2012.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizuno M, Kuboki Y. TGF-beta accelerated the osteogenic differentiation of bone marrow cells induced by collagen matrix. Biochemical and biophysical research communications. 1995;211:1091–8. doi: 10.1006/bbrc.1995.1923. [DOI] [PubMed] [Google Scholar]
- 42.Alliston T. TGF-beta regulation of osteoblast differentiation and bone matrix properties. Journal of musculoskeletal & neuronal interactions. 2006;6:349–50. [PubMed] [Google Scholar]
- 43.Qin W, Chung AC, Huang XR, Meng XM, Hui DS, Yu CM, et al. TGF-beta/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. Journal of the American Society of Nephrology : JASN. 2011;22:1462–74. doi: 10.1681/ASN.2010121308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou L, Wang L, Lu L, Jiang P, Sun H, Wang H. Inhibition of miR-29 by TGF-beta-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PloS one. 2012;7:e33766. doi: 10.1371/journal.pone.0033766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavlin D, Lichtler AC, Bedalov A, Kream BE, Harrison JR, Thomas HF, et al. Differential utilization of regulatory domains within the alpha 1(I) collagen promoter in osseous and fibroblastic cells. The Journal of cell biology. 1992;116:227–36. doi: 10.1083/jcb.116.1.227. 2%, 5%, and 25% for 15 minutes (Top Panel) and 25 minutes (bottom panel) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gelatin nanofibers cross linked using 2%, 5%, and 25% for 15 minutes.
Supplemental Table 1. Sequences of microRNAs used in the study.