Abstract
Eri1 is an evolutionarily conserved 3′–5′ exoribonuclease that participates in 5.8S rRNA 3′ end processing and turnover of replication-dependent histone mRNAs. Over the course of evolution, Eri1 has also been recruited into a variety of conserved and species-specific regulatory small RNA pathways that include endogenous small interfering RNAs and microRNAs. Recent advances in Eri1 biology illustrate the importance of RNA metabolism in epigenetic gene regulation and illuminate common principles and players in RNA biogenesis and turnover. In this review, we highlight Eri1 as a member of a growing class of ribosome- and histone mRNA-associated proteins that have been recruited into divergent RNA metabolic pathways. We summarize recent advances in the understanding of Eri1 function in these pathways and discuss how Eri1 impacts gene expression and physiology in a variety of eukaryotic species. This emerging view highlights the possibility for crosstalk and coregulation of diverse cellular processes regulated by RNA.
Keywords: microRNA (miRNA), RNA interference (RNAi), histone mRNA, ribosomal RNA (rRNA), epigenetic regulation, gene expression
Eri1 Structure and Substrate Specificity
RNA has long been understood to impact epigenetic gene regulation. Thus RNAses and other RNA metabolizing enzymes provide an additional layer of regulatory control. Eri1 (also known as Thex1) is a 3′–5′ exoribonuclease of the DEDDh family that includes poly(A) ribonuclease, RNase T, and the ε proofreading subunit of Escherichia coli DNA polymerase III (ε186) [1]. Eri1 impacts gene expression in multiple organisms by participating in small regulatory RNA pathways. In Caenorhabditis elegans, ERI-1 forms a complex with Dicer that generates specific classes of endogenous small interfering RNAs (siRNAs), whereas Schizosaccharomyces pombe Eri1 regulates the turnover of chromatin-associated siRNAs. In the mouse, Eri1 negatively regulates global microRNA abundance. In addition to these small RNA targets, Eri1 has conserved roles in 3′ end trimming of the 5.8s rRNA and turnover of replication-dependent histone mRNAs. Though quite diverse, these substrates share a common structural motif: an RNA duplex bearing unpaired 3′ nucleotides that are subject to exonucleolytic activity.
Like other DEDD exonucleases, Eri1 has a conserved catalytic core consisting of four acidic amino acid residues, DEDD, that span across three conserved sequence motifs and coordinate Mg2+ cations (Fig. 1a). The DEDDh subfamily has a unique motif III variant, H-x(4)-D, named exo IIIε after ε186 [2]. This conserved histidine deprotonates Mg2+-coordinated water, thereby converting it into a nucleophile that cleaves terminal oligoribonucleotide phosphodiester bonds [3]. The enzymatic pocket accommodates only one to two nucleotides, which explains why double stranded RNA (dsRNA) is a poor Eri1 substrate, while 3′ single stranded RNA (ssRNA) overhangs are efficiently cleaved [4,5].
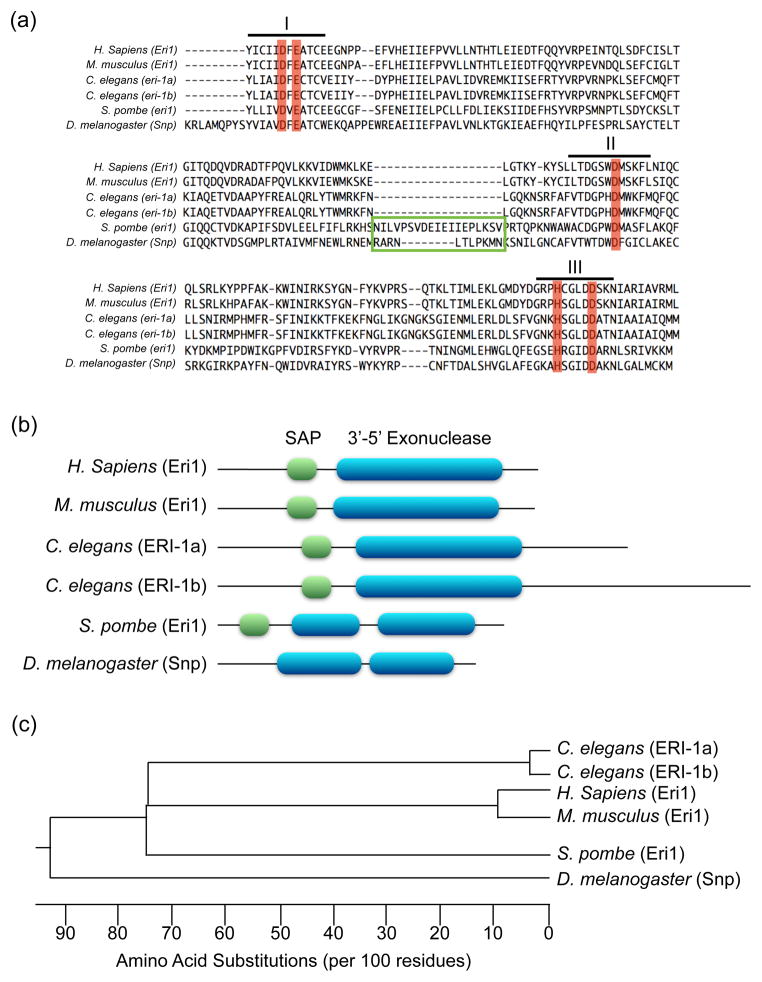
Figure 1.
Eri1 protein conservation among eukaryotic species. (a) ClustalW alignment of Eri1 family exonuclease domains. Three conserved sequence motifs are indicated with roman numerals. DEDDh residues that coordinate Mg2+ and H2O at the enzymatic active site are highlighted in red. The green box indicates non-conserved amino acid inserts in the enzymatic domain. (b) Location of conserved Eri1 SAP and DEDDh exonuclease domains. Note that C. elegans express two ERI-1 isoforms, ERI-1a and ERI-1b, which arise from alternative splicing. The ERI-1b splice variant is poorly conserved and contains a C-terminal extension that is required for interaction with the C. elegans Dicer-containing ERI complex. Drosophila melanogaster Snipper (Snp) is a relative of Eri1 that lacks a SAP domain. (c) Phylogenetic tree produced from ClustalW alignment of eukaryotic Eri1 protein sequences.
One feature of Eri1 that distinguishes it from other DEDDh family members is a conserved SAP (SAF-box, Acinus and PIAS) domain (Fig. 1b–c), which binds double-stranded nucleic acids via conserved amphipathic helices [6]. Eukaryotic SAP domains anchor nucleic acids at protein C or N termini, which are then brought in close proximity to other RNA binding or metabolizing domains [7]. As expected, Eri1 SAP mutants less efficiently process 5.8s rRNA and histone mRNAs [4,8,9]. It is unclear if the SAP domain primarily anchors Eri1 to RNA or if it additionally recruits proteins to RNA-dependent complexes that modify substrate processing. A recent crystal structure shows that Eri1 SAP binding induces a conformational change in the histone mRNA 3′ stem-loop [10]. This may explain why Eri1 and Stem Loop Binding Protein (SLBP) bind cooperatively to opposite sides of the 3′ histone stem-loop [4,8]. Alternatively, the SAP domain may prevent helicase or nuclease unwinding and degradation of RNA.
As a broadly expressed exoribonuclease, Eri1 affects both transcriptional gene regulation, through targeting non-coding RNAs involved in heterochromatin formation, and post-transcriptional gene regulation, by targeting regulatory small RNAs or by degrading mRNA transcripts directly. This review summarizes recent advances in Eri1 substrate identification and explores how Eri1 exemplifies a broader class of RNA modifying enzymes recruited into diverse regulatory small RNA pathways.
Eri1 Regulation of Small RNAs
RNA interference (RNAi) was initially described in C. elegans as sequence-dependent silencing of endogenous mRNAs by exogenous dsRNA [11–13]. This requires generation of small ~22 nt RNA species that silence either translation or transcription based on binding to specific Argonaute complexes [14]. In the cytoplasmic RNA-induced silencing complex (RISC), Argonaute uses one small RNA strand to guide degradation or translational repression of cognate mRNAs. In the nucleus of S. pombe and C. elegans, small RNAs guide silencing complexes to genomic regions. In metazoan somatic cells, the regulatory small RNA repertoire is dominated by short interfering RNAs (siRNAs) and microRNAs (miRNAs), which silence targets by similar mechanisms but differ in their precursors, duplex mismatch frequency, and Argonaute specificity. siRNAs are generated from long dsRNA precursors by Dicer cleavage. miRNAs originate from long hairpin-containing primary transcripts that undergo successive cleavage by the Microprocessor complex (comprised of DGCR8 and the RNase III enzyme Drosha) and Dicer. Arabidopsis and C. elegans have abundant miRNA and siRNA species. In contrast, miRNAs dominate the small RNA repertoire in most mammalian cells with the exception of germ cells, which additionally contain siRNAs and piRNAs [15,16].
Small RNA-induced silencing efficiency is tuned by numerous mechanisms, including small RNA amplification and degradation. C. elegans eri-1 was discovered in a screen for mutants demonstrating an enhanced RNAi (i.e. “ERI”) phenotype in neurons, which are partially refractory to RNAi [5]. In physiologic conditions Eri1 degrades 3′ overhangs from RNA duplexes but has little activity towards RNA with blunted ends.
C. elegans ERI-1 is a potent modulator of exogenous and endogenous RNAi pathways that converge on Dicer [17,18]. Proteomic analysis of Dicer-associated proteins identified a complex of four binding partners previously shown to display “ERI” phenotypes [19]. This complex contained ERI-1, the RNA-dependent RNA polymerase RRF-3, the novel protein ERI-3, and the Tudor domain-containing protein ERI-5. eri-1, dicer1, rrf-3, and eri-3 mutants fail to generate selective classes of endogenous siRNAs (endo-siRNAs), leading to a corresponding increase in mRNA targets. eri-1 and rrf-3 mutant animals specifically fail to accumulate 26G endo-siRNAs that regulate spermatogenesis and zygotic development and give rise to 22G RNAs [20,21]. eri-1 and rrf-3 mutants up-regulate transcripts complementary to siRNAs [18]. Unexpectedly, only ten genes were up-regulated in both eri-1 and rrf-3 mutants, suggesting function in both overlapping and distinct endogenous RNAi pathways. ERI-1 was subsequently found to associate with dsRNA binding factor RDE-4, the microRNA-binding Argonaute proteins ALG-1 and 2, ERI-9, and the RNA phosphatase PIR-1 [17,22,23].
ERI-1 is a selective RNase that promotes the biogenesis of some endo-siRNAs while inhibiting the abundance of others, including exogenous siRNAs. The “ERI” phenotype observed in eri-1 mutants may reflect RNAi pathway competition. In this model, the absence of ERI-1-dependent Dicer recruitment into endogenous RNAi pathways enhances exogenous RNAi. The opposite is also true. Wild-type animals fed exogenous siRNAs showed reduced abundance of at least one endo-siRNA [17]. In an alternative model for the ERI phenotype, exogenous and endogenous siRNAs may compete for limiting RISC proteins, such as Argonautes.
Both human [24] and mouse [25] cells with reduced Eri1 also display an ERI phenotype. Although competition for limiting Dicer protein may explain the phenotype in C. elegans eri-1 mutants, mammals likely have a distinct mechanism. Even though the ERI-1a splice variant is deeply conserved, only ERI-1b, a unique C. elegans variant, binds Dicer. ERI-1b contains a 400 nt C-terminal extension with no conserved domains (Fig. 1b), and its ectopic expression is sufficient to rescue the ERI phenotype in eri-1 mutants [5]. In contrast, ectopic ERI-1a expression cannot rescue ERI phenotypes [26]. Another reason the C. elegans Dicer competition model may not broadly apply is that the endo-siRNAs affected by the absence of Eri1, like 26G endo-siRNAs, lack mammalian homologs. To date no endo-siRNAs have been identified in somatic mammalian cells; however, Dicer competition between small RNA pathways may occur in oocytes, which express endo-siRNAs. Although the role of ERI-1b in C. elegans endo-siRNA biogenesis is poorly understood, it appears to anchor Dicer to mRNAs (Fig. 2a) [17]. These mRNAs may become substrates for RRF-3, which has been proposed to generate long dsRNAs processed by Dicer [17]; however, evidence for this specific model is currently lacking. Importantly, ERI-1 is found in complexes that do not include Dicer but do include the microRNA Argonautes ALG-1/2 [22]. These interactions hint at the existence of Dicer-independent small RNA pathways regulated by ERI-1.
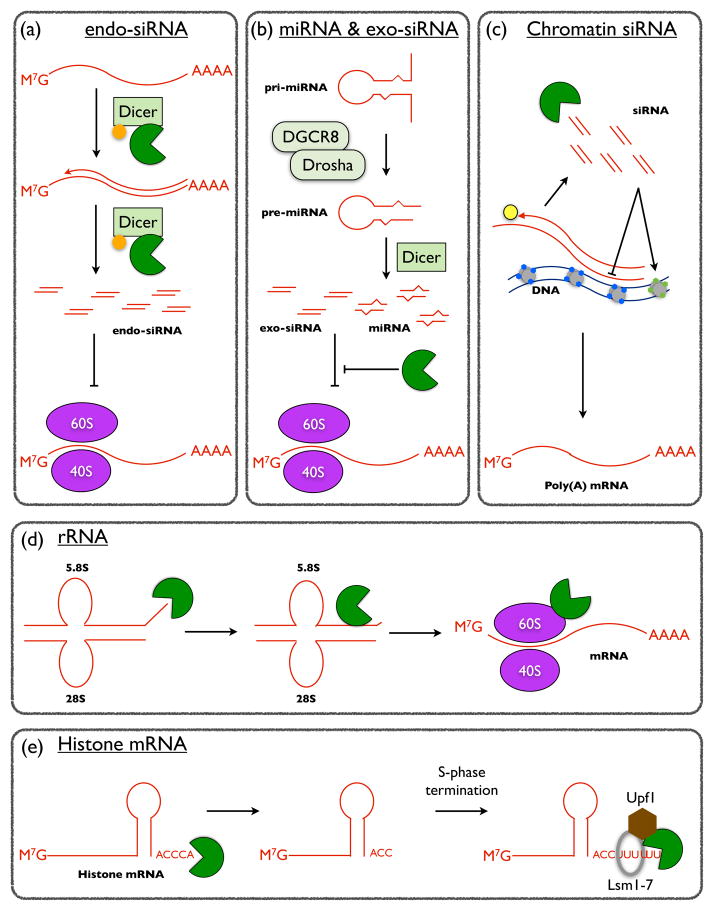
Figure 2.
Summary of the effects of Eri1 on cellular RNAs. (a)C. elegans DCR-1 forms a complex with the ERI-1b isoform (green) and the RNA-dependent RNA polymerase RRF-3 (orange circle). This complex is purported to sequester Dicer activity to promote the biogenesis of some endogenous siRNAs while preventing the generation of others. (b) Metazoan Eri1 inhibits RNAi mediated by exogenous siRNAs. In addition, mammalian Eri1 negatively regulates the abundance of mature miRNAs. Both siRNAs and miRNAs repress gene expression predominantly by destabilizing mRNA transcripts, but also by inhibiting translation. Therefore, the absence of Eri1 leads to increased exogenous siRNA- and miRNA-mediated silencing of cognate mRNA targets. (c) In S. pombe, Eri1 negatively regulates endogenous siRNAs that participate in nascent transcript decay (co-transcriptional gene silencing) and heterochromatin formation (transcriptional gene silencing) by recruiting the RITS complex (not shown) to genomic regions in cis. In the absence of Eri1, siRNAs target alleles in trans, leading to promiscuous heritable gene silencing. Euchromatin (gray circles with blue diamonds) and heterochromatin (gray circles with green diamonds) are represented. An RNA-directed RNA polymerase Complex is shown (yellow circle). (D) Eri1 has a conserved function in 5.8S rRNA maturation by trimming several unpaired nucleotides from the 3′ end. After trimming Eri1 remains associated the 40S and 60S subunits as well as with monosomes and, to a lesser extent, actively translating polysomes. (E) In mammalian cells, Eri1 trims two nucleotides from the 3′ end of replication-dependent histone mRNAs. At the end of S-phase, canonical histone mRNAs become oligouridylated, which leads to recruitment of Eri1, the heptameric Lsm1-7 decapping complex, and the RNA helicase Upf1, all of which contribute to subsequent histone mRNA degradation through the 3′ stem-loop.
A better understanding of the small RNA profile of Eri1-deficient mammalian cells was recently obtained by sequencing small RNA libraries from Eri1−/− and wild-type mouse lymphocytes. Although eri-1 mutant C. elegans show at least a five-fold decrease in specific endo-siRNAs [27], no such Eri1-dependent small RNAs were identified in mouse [28]. However, there was a two-fold increase in all miRNAs (Fig. 2b) [29]. This observation is consistent with the previous finding that eri-1 mutant C. elegans express increased levels of pre-and mature miR-238 [18]. It is unclear if this finding extends to other miRNA species as only miR-238 was assayed. However, given recent findings in mice, it is likely that C. elegans ERI-1 impacts the abundance of multiple or possibly all miRNA species. Compared to C. elegans eri-1 mutants, loss of Eri1 in the mouse does not affect accumulation of any other abundant small RNA populations aside from miRNAs. Thus, Eri1 competition for Dicer activity may explain why eri-1-deficient C. elegans species have decreased levels of some endo-siRNAs and increased levels of at least one miRNA species; however, a Dicer competition model is unlikely to explain the observed increase in miRNAs seen in Eri1-deficient mice. One exception would be if mouse Eri1 sequestered Dicer activity to promote the biogenesis of larger RNA species not detected by small RNA sequencing.
Biochemical studies show that Eri1 cleaves 3′ overhangs from dsRNA, but it is unclear how this activity reduces miRNA abundance. Eri1 may degrade precursor or mature miRNAs, similar to the small RNA exonucleases SDN1 in Arabidopsis and XRN-2 in C. elegans [30,31]. In this scenario, Eri1 would have to function in tandem with a helicase that exposes single strands from dsRNA. Alternatively, Eri1 may cleave 3′ overhangs from pre-miRNAs or miRNAs, thereby preventing their export into the cytosol [32,33] or loading onto RISC, respectively [34–36]. Eri1 trimming might instead prime miRNA duplexes for uridylation by a terminal uridylyl transferase (TUTase) such as Zcchc11, which was recently shown to uridylate miRNAs and their precursors [37–40]. Interestingly, Zcchc11 also uridylates and destabilizes histone mRNA, which is another Eri1 substrate [41].
In contrast to metazoan Eri1, which enhances post-transcriptional gene silencing by inhibiting siRNAs and miRNAs, S. pombe Eri1 negatively regulates small RNAs that promote transcriptional and cotranscriptional gene silencing (Fig. 2c). This observation complements the finding that RNAi in S. pombe acts primarily to silence heterochromatin-bound RNAs and read through antisense transcripts [42]. In S. pombe, heterochromatin is formed predominantly at centromeres and telomeres. These contain numerous repetitive sequences whose transcripts are processed by Dicer into siRNAs that target the RNA-induced initiation of transcriptional gene silencing (RITS) complex to the genomic repeats from which they originate. The RITS complex mediates nascent transcript degradation followed by recruitment of histone methyltransferases and chromo-domain proteins that nucleate and maintain heterochromatin formation [43,44]. Eri1 deficiency leads to accumulation of centromeric siRNAs and a concomitant increase in H3-K9 methylation and heterochromatic silencing [45,46]. Eri1 likely degrades siRNA duplexes directly, as demonstrated by in vitro assays [45]. Note that this activity is distinctly different from metazoan Eri1, which is strongly inhibited by dsRNA [4,5,8,9].
S. pombe Eri1 is crucial for limiting the extent and heritability of transcriptional gene silencing. A fundamental difference between RNAi-mediated post-transcriptional gene silencing in C. elegans versus RNAi-induced chromatin silencing in S. pombe is that the former functions in trans whereas the later acts strictly in cis. Artificial tethering of RITS to nascent ura4 transcripts produces ura4+ siRNAs that fail to silence trans alleles of ura4 [46]. In the absence of Eri1, however, trans alleles readily undergo RNAi-induced silencing. Thus S. pombe Eri1 functions to spatially restrict RNAi-mediated heterochromatin formation in cis.
Eri1 has a broadly conserved role in regulating small RNA abundance despite the divergence of small regulatory RNA populations in different organisms and cell types. The diverse RNA populations altered in the absence of Eri1 differ in their biogenesis and Argonaute association, however, in their mature form, all share a characteristic two nucleotide 3′ overhang. One unifying molecular mechanism for this broad range of activity is that Eri1 simply trims overhanging nucleotides from the 3′ end of small RNA duplexes and in doing so prevents recognition by Argonaute and/or Dicer PAZ domains. As a result, Eri1-targeted small RNAs are not processed and loaded onto the RISC complex efficiently and so are subject to exonucleolytic activity and faster rates of turnover. Natural variations on this common theme may result from diverse species-specific and cell type-specific RNA decay pathways and the observation that Eri1 exists in multiple different complexes. For example, the C. elegans-specific ERI-1b isoform forms a complex with Dicer to generate specific endo-siRNA populations [17] while both C. elegans isoforms inhibit exogenous RNAi, likely by a conserved mechanism. In contrast, mammalian Eri1 exists in a larger protein complex that mediates oligouridylation-dependent histone mRNA decay (see Eri1 regulation of histone mRNA section below). This observation raises the possibility that Eri1 additionally regulates the degradation of specific small RNAs and their precursors bearing a oligouridine tag.
It is unclear if Eri1 constitutively represses regulatory small RNAs in most cell types or if there are physiologic conditions where it is up- or down-regulated to tune sensitivity to RNAi-mediated silencing. Interestingly, Eri1 is up-regulated in response to high doses of exogenous siRNAs [25,47], suggesting that it may buffer different RNAi pathways.
Eri1 Regulation of Ribosomal RNA
The most conserved RNA target of Eri1 is the 5.8S ribosomal RNA (rRNA) (Fig. 2d). Although most ribosome processing happens in the nucleolus, Eri1-mediated 5.8S rRNA trimming is likely cytoplasmic [5,8,9,26,45]. The 5.8S rRNA 3′ end forms a duplex reminiscent of pre-miRNAs by base pairing with the 5′ end of the 28S rRNA, which leaves a 3′ ssRNA overhang processed by Eri1 into a duplex with 1–2 nt 3′ overhang. dsRNA, which inhibits Eri1 enzymatic activity, prevents overprocessing of the duplex. The ability of Eri1 to process rRNA requires exonuclease activity and is deeply conserved from fission yeast to mammals [9,26]. In C. elegans eri-1 mutants, ectopic expression of ERI-1a or ERI-1b is sufficient to rescue 5.8s rRNA length defects. Thus both isoforms mediate rRNA processing while having divergent roles in RNAi [26]. Although the function of 5.8S rRNA 3′ trimming is poorly understood, it is likely important because it is conserved even in S. cerevisiae, where eri-1 has been lost but an unrelated endonuclease, Ngl2, performs the same function [48].
One hypothesis is that Eri1 may bind intact, cytoplasmic ribosomes as a way of localizing its other functions, such as inhibition of RNAi, to the basal translation machinery. Interestingly, Eri1 is just one of several rRNA processing proteins recruited into regulatory small RNA pathways (Box 1, Table 1). Alternatively, Eri1’s function in ribosome maturation and miRNA turnover may be purely coincidental as 3′ end trimming is a relatively common post-transcriptional modification required for the biogenesis of many unrelated noncoding RNAs. To date, a connection between Eri1-dependent rRNA processing and altered epigenetic gene regulation remains purely speculative. The loss of Eri1, as a ribosomal protein, may compromise global translation rates by altering ribosome folding and/or recruitment of ribosome-associated proteins. Alternatively, Eri1 deficiency may lead to transcript-specific defects in translation, such as those observed in Rpl38-deficient mice [49]. Interestingly, Eri1 remains attached to actively translating polysomes even after 5.8S trimming [9]. Some evidence suggests that miRNAs may function by redistributing targeted mRNAs from polysomes to more slowly-sedimenting ribosomal particles not actively involved in translation [50]. Although the mechanism of Eri1 miRNA regulation is not fully understood, it is intriguing to speculate that it may inhibit miRNA-mRNA target interactions on polysomes. Ribosome profiling, which uses high-throughput sequencing to identify ribosome positions on mRNAs, may provide mechanistic insights into the effects of Eri1 on translation.
Box 1. Evolutionary recruitment of rRNA processing proteins into small regulatory RNA pathways.
Eri1 makes up a class of core rRNA processing enzymes recruited into regulatory small RNA pathways. The large Microprocessor complex containing p68 and p72 (two ATP-dependent DEAD-box RNA helicases) processes pre-miRNAs from pri-miRNAs and participates in pre-rRNA processing [70,71]. p68 or p72 mutations lead to reductions in 5.8S rRNA and selective miRNAs. DGCR8, another microprocessor subunit, may indirectly influence rRNA maturation by mediating cleavage of snoRNAs [72], which are small RNAs that guide chemical modification of rRNA, snRNA, and tRNA. In the absence of DGCR8 there is marked accumulation of mature snoRNAs, though the functional consequence on ribosome maturation is unknown. Depletion of Dicer or Ago2 leads to the accumulation of 5.8S precursor rRNAs, although this mechanism is poorly understood [73]. Interestingly, a direct role for Dicer outside of pre-miRNA processing has been observed in Candida albicans, where CaDCR1 mutants displayed defects in pre-rRNA spacer cleavage and U4 snRNA 3′ end processing [74].
Some rRNA-associated enzymes participate in the biogenesis of non-canonical small RNAs. For example, Rrp6, an exosome subunit and 3′–5′ exonuclease, functions in 3′ end maturation of both the 5.8s rRNA (at a stage preceding Eri1 function [75]) and tailed mirtrons, which are a subset of miRNAs spliced from introns. Mirtrons bypass Drosha cleavage by relying on the spliceosome to generate precursor ends. After splicing and debranching, Rrp6 is necessary and sufficient for 3′ tail removal from pre-miRNA-like hairpins [76]. Further work is required to determine if this Rrp6 activity exists independently of exosome function. There is also precedent for viral co-optation of host RNA processing enzymes for the biogenesis of novel small RNAs, such as miRNAs encoded by the murine gamma-herpesvirus 68. These miRNAs are transcribed by PolIII near viral tRNA-like sequences so that resultant pri-miRNAs bear a 5′ tRNA moiety and are not processed by Drosha but rather the host tRNA endonuclease tRNaseZ [77].
Like Eri1, many of these proteins have been recruited into additional RNA processing pathways independent of their roles in rRNA or small RNA biogenesis. For example, in addition to binding pri-miRNAs and snoRNAs, DGCR8 localizes with long non-coding RNAs and several hundred mRNAs [72,78–80]. DGCR8 cooperates with Drosha to destabilize many of these transcripts, including the DGCR8 mRNA itself. In contrast, DGCR8 affects snoRNAs independently of Drosha. These findings are consistent with the notion that deeply conserved RNA processing proteins exist in different macromolecular complexes that spatially and temporally compartmentalize diverse RNA processing functions. In some cases, however, the dual recruitment of enzymes into rRNA and small regulatory RNA processing pathways may spatially couple RNAi activity with target transcripts loaded onto basal translation machinery. The functional significance of this coupling remains unclear for many of these enzymes and may be purely coincidental.
Table 1.
Proteins recruited into rRNA, histone mRNA, poly(A) mRNA and/or small regulatory RNA processing or biogenesis pathways
| Protein | Protein Activity | rRNA | Histone mRNA | Poly(A) RNAs | Small Regulatory RNA | Refs |
|---|---|---|---|---|---|---|
| Eri1 | 3′–5′ exoribonuclease | + | + | miRNA, endo-siRNA | [4,5,9,17,18,26,28,58] | |
| p68 | RNA helicase | + | Pri-miRNA | [70,71] | ||
| p72 | RNA helicase | + | Pri-miRNA | [70,71] | ||
| caDcr1 | Endonuclease | + | siRNA | [74] | ||
| DGCR8 | dsRNA binding | + | Pri-miRNA | [72,78–80] | ||
| Drosha | Endonuclease | + | Pri-miRNA | [83–85] | ||
| Rrp6 | 3′–5′ exoribonuclease | + | + | + | Tailed mirtron | [75,76,86,87] |
| Ars2 | RNA binding | + | + | miRNA | [81–84,97] | |
| Zcchc11 | TUTase | + | Pre-miRNA, miRNA | [37–41,94] | ||
| Dis3l2 | 3′–5′ exoribonuclease | + | Pre-let-7 | [91–93] | ||
| PARN | Poly(A) exoribonuclease | + | Pre-miR-451 | [95] | ||
| Triman | 3′–5′ exoribonuclease | priRNA, siRNAs | [96] |
Eri1 Regulation of Histone mRNA
Mammalian Eri1 was first described as 3′ histone mRNA exonuclease (3′hExo) because of its ability to degrade replication-dependent histone mRNAs. The abundance of these mRNAs is tightly linked to DNA replication so that large numbers of histones are produced in S-phase to accommodate DNA packaging into nucleosomes. Histone mRNA abundance then falls rapidly in G2. Failure to coordinate histone gene expression with cell cycle progression results in genomic instability and cell cycle arrest [51]. This coordination relies on a unique cis-regulatory stem-loop in the histone 3′UTR, which is the only non-adenylated eukaryotic 3′UTR [52]. Histone mRNA 3′ ends are first formed by U7 snRNP-mediated endonucleolytic cleavage of a precursor mRNA [53]. Cleavage generates a 3′ conserved stem-loop flanked by a 4–5 nt ssRNA overhang. This stem-loop marks the mRNA for rapid degradation at the end of S-phase. SLBP is a key protein recruited to the histone 3′ UTR, where it stabilizes and promotes the translation of histone mRNA. At the end of S-phase, SLBP is degraded by the proteasome, permitting rapid degradation of histone mRNAs [54].
The complex responsible for rapid degradation was hypothesized to contain a 3′–5′ ribosome-associated exoribonuclease with Mg2+-dependent activity [55]. Interestingly, early studies showed that the location of the stem-loop relative to the stop codon is important for rapid mRNA turnover because efficient degradation is lost if the stop codon is more than 45–80 nt away [56,57]. This observation suggested that stem-loop decay may require a terminating ribosome in close proximity, which would explain why translation is required for rapid histone mRNA degradation [56]. Eri1 was identified as a likely candidate for the 3′–5′ exonuclease based on in vivo binding to histone stem-loops and in vitro substrate specificity [4].
Subsequent studies revealed that Eri1 and SLBP cooperatively bind the 3′ and 5′ stem-loop arms, respectively, and that Eri1 binding requires a 3′ unpaired ACCCA consensus sequence [4,8]. A recent crystal structure of the full ternary complex shows no direct contacts between SLBP and Eri1 [10]. Except for two conserved uridine nucleotides in the loop that mediate binding specificity, SLBP and Eri1 primarily recognize the stem-loop shape. Binding of one protein likely leads to a structural change in the loop that promotes cooperative binding of the other. The crystal structure reveals that Eri1 can trim ssRNA only long enough to reach the enzymatic active site [10]. These structural constraints are consistent with prior findings that Eri1 removes only three of the five single-stranded nucleotides from the stem-loop 3′ end [4]. After three terminal nucleotides are trimmed, the 3′ overhang is too short to reach the enzymatic active site. Thus the ternary complex limits the extent of Eri1 trimming.
Recent work in Eri1-deficient mouse cells revealed that Eri1 was critically important for the rapid degradation of oligouridylated histone mRNAs at the end of S-phase [58]. In the absence of Eri1, there was marked accumulation of oligouridylated histone mRNAs in hydroxyurea-treated cells. Small concentrations of Eri1 are sufficient for this function as 80–90% knockdown of Eri1 had no discernable effect on histone mRNA stability [59]. In Eri1-deficient cells, histone mRNA abundance still declined over two-fold at the end of S-phase, suggesting a partially compensatory, Eri1-independent turnover mechanism. Eri1 trims two nucleotides from the histone mRNA 3′ end prior to degradation at the end of S-phase (Fig. 2e), similar to its role in 5.8S maturation [58]. Although the function of this trimming is not fully understood, these findings are consistent with prior observations that mature histone mRNA 3′ ends are two nucleotides shorter than histone pre-mRNAs [59].
Mammalian Eri1 may exist in a larger complex that triggers histone mRNA degradation. Mouse Eri1 shows RNA-independent co-immunoprecipitation with Lsm1 and Lsm4 [58], which comprise the heptameric Lsm1-7 complex that preferentially binds oligo-U tails [60]. Eri1 additionally associates with the RNA helicase Upf1 in an RNA-dependent manner. Like Eri1, both Lsm1 and Upf1 are critical for the efficient decay of oligouridylated histone mRNAs at the end of S-phase [41,59]. In contrast, human 3′hExo co-immunoprecipitates with Lsm6 and Lsm4, which additionally binds SLBP [61].
These data offer the following model for Eri1 participation in histone mRNA decay (Fig. 2e): first, Eri1 is deposited on the 3′ stem-loop, where it trims two unpaired nucleotides. At the end of S-phase, SLBP is degraded by the proteasome, and a terminal uridylyl transferase (TUTase), most likely Zcchc11 [41], oligouridylates the 3′ end. A combination of the uridine tag and direct interactions between Eri1 and SLBP with Lsm subunits leads to recruitment of the Lsm1-7 complex, which initiates mRNA decapping and destabilization. Decapping notably requires Eri1 function or occurs on a minority of mRNA transcripts [58,59]. The Lsm1-7 complex and Eri1 bind to the oligouridylated tail, where Eri1 degrades the histone mRNA through the stem-loop in a step-wise manner that likely requires the helicase Upf1 to expose ssRNA. 3′–5′ exosome-mediated decay may contribute to clearance of histone mRNAs lacking intact stem-loops [59]. Interestingly, Eri1 is just one member of a growing family of enzymes that regulates both histone mRNAs (Box 2, Table 1) or poly(A) transcripts (Box 3, Table 1) and plays a role in the turnover of small regulatory RNAs.
Box 2. Evolutionary recruitment of histone mRNA processing proteins into small regulatory RNA pathways.
Eri1 joins a larger group of histone mRNA modifying proteins that influence small regulatory RNA pathways. Ars2 is a component of the nuclear cap-binding complex that promotes pri-miRNA processing and histone pre-mRNA 3′ end formation [81–85]. Ars2 mediates normal cell proliferation and efficient production of let-7, miR-21, and miR-155, which participate in cellular transformation [85]. Ars2 deficient cells show reductions in these miRNAs and an accumulation of histone mRNAs with inappropriately polyadenylated 3′ ends [83]. During cell proliferation, the co-transcriptional recruitment of Ars2 to pre-RNAs may bridge transcription and pre-mRNA processing. As an additional example, Rrp6, an exonuclease that processes tailed mirtrons and rRNA (see rRNA section above) is part of the exosome complex that mediates efficient 3′–5′ decay of histone mRNAs [86,87]. It is unknown if Rrp6 has constitutive activity towards these diverse substrates or if its activity is coordinated in a cell-cycle-dependent manner.
Pre-miRNAs and the 3′ ends of histone mRNAs share in common a stem-loop structure and a 3′ overhang that are important for maturation and turnover. Exciting new work shows that the decay of several different classes of RNAs is associated with untemplated 3′ addition of one or more uridine nucleotides. These RNAs include replication-dependent histone mRNAs [59], fission yeast mRNA [88], U6 snRNA [89], miRNAs, and pre-miRNAs [38,39,90]. Three new studies support a widespread role for this decay mechanism in eukaryotic RNA turnover and identify Dis3L2 as the major 3′–5′ exonuclease that mediates oligouridylated RNA degradation. One study identified Dis3L2 as the exonuclease that degrades uridylated pre-let-7 and in doing so maintains pluripotency of stem cells by mediating lin28-induced inhibition of let-7 [91]. The remaining two studies identified a role for Dis3L2-mediated decay of widespread uridylated S. pombe [92] and human [93] mRNA transcripts, respectively. Interestingly, Zcchc11, a TUTase that targets histone 3′ ends, has also been shown to uridylate pre-let-7, miR-122, and miR-26a [37–40,94].
Although Eri1 can trim 3′ oligouridylated histone mRNAs [58], it is unclear to what extent it directly targets other classes of oligouridylated RNAs. High-throughput sequencing techniques like HITS-CLIP will offer powerful methods for empirically identifying the breadth of Eri1 targets. Although the function of uridylation-mediated RNA decay remains unclear, it is likely a dual-purpose signal that prevents targeting by some nucleases and stimulates the activity of others. Interestingly, both Dis3L2 and Eri1 have been shown to co-localize with polysomes [9,93], which raises the intriguing possibility that both may efficiently target transcripts for co-translational decay.
Box 3. Evolutionary recruitment of poly(A) transcript processing proteins into small regulatory RNA pathways.
Enzymes involved in turnover of polyadenylated transcripts have been repeatedly recruited into species-specific small RNA pathways. Poly(A)-specific ribonuclease (PARN), which is broadly conserved in eukaryotes and implicated in poly(A) tail trimming and mRNA decay, was recently shown to mediate 3′ end processing of pre-miR-451 [95]. mir-451 is an erythropoietic miRNA conserved in vertebrates and, unlike canonical miRNAs, is generated by a Dicer-independent mechanism that relies on Argonaute2 slicing activity. PARN completes the last 3′ maturation step whereby seven nucleotides are trimmed to give mature ~23 nt miR-451. Though miR-451 is subject to 3′ oligouridylation prior to trimming, these uridines have no effect on the kinetics of 3′ end trimming and, surprisingly, trimming is not required for mRNA target silencing. In fission yeast, the 3′ trimming of Dicer-independent primal small RNAs (priRNAs) is completed by a previously uncharacterized 3–5′ exonuclease called Triman, which belongs to the CAF1 family that is conserved in most eukaryotes [96]. Both of Triman’s known small RNA targets, priRNAs and siRNAs that drive heterochromatin formation, are S. pombe-specific and not conserved in metazoans. Of note, Eri1 negatively regulates this class of siRNAs in S. pombe (see Small regulatory RNA section), suggesting opposing roles for 3′–5′ exonucleases in siRNA biogenesis and decay [45]. Similar to miR-451 in vertebrates, priRNAs are generated by a trimming and protection mechanism that requires the joint activity of Triman and Argonaute. Although Triman is unable to generate RNAs of specific lengths, the presence of Argonaute protects small RNAs from overtrimming, likely by sequestering 3′ RNA ends in the PAZ domain away from the Triman catalytic site. Together these examples demonstrate how canonical enzymes in the miRNA biogenesis pathway, such as Argonaute, recruit conserved exonucleases that direct the processing of non-canonical small RNAs.
Eri1 conservation and evolutionary loss
Species lacking Eri1 provide as much insight into its evolutionary history as those that have evolved novel Eri1 functions. The study of Saccharomyces RNAi provides insight into the co-evolution of Eri1-mediated rRNA processing and small RNA regulation. In eukaryotes, RNAi serves as a kind of genetic immune system that defends against viral nucleic acids and rogue mobile segments including transposons and retroelements [62]. Yet this potent defense mechanism comes at an evolutionary cost. In fungi, RNAi is selectively lost in S. cerevisiae and other sensu stricto clade members, while it is retained in S. castellii and S. pombe [63]. This loss confers an evolutionary advantage in being able to maintain killer, an endemic dsRNA virus system [64]. Killer encodes a protein toxin fatal to neighboring cells while conferring protection to the infected host. Fungi harboring functional RNAi machinery degrade killer dsRNA, rendering themselves resistant to infection yet susceptible to lethal toxins produced by neighboring cells. Although RNAi may confer a selective disadvantage in the short term, RNAi-harboring species likely show more robust long term survival as they are resistant to transposon-induced genomic damage [64].
The negative selective pressure on RNAi explains why it is lost from some species while in others it is restricted by Eri1-like factors. Interestingly, eri1 has been lost in S. cerevisiae, which lacks RNAi machinery, but it is maintained in the RNAi competent S. pombe [5]. Although killer virus has not been detected in S. pombe, it is tempting to speculate that Eri1 dampens RNAi and renders fission yeast transiently susceptible to infection with killer or a similar virus, thus conferring protection against viral toxins. A similar protective mechanism may exist in C. elegans, where the presence of wild-type ERI-1 inhibits antiviral defense, thus rendering animals more susceptible to potentially beneficial viral infections [65]. Notably, since Eri1 has been lost in S. cerevisiae, alternative nucleases have subsumed some of its conserved functions, namely 5.8S 3′ trimming and rapid histone mRNA decay [48,66].
Drosophila melanogaster is one of the only known eukaryotic species with functional RNAi machinery yet no clear Eri1 homolog. The closest Eri1 family member is Snipper (Snp), so named for its potent, promiscuous exonuclease activity [67]. Unlike other Eri1 homologs, Snp mutants fail to accumulate replication-dependent histone mRNAs and show no alteration in exogenous RNAi. Altered Snp substrate specificity can be explained by a 13 amino acid exonuclease domain insertion and the absence of a SAP domain (Fig. 1a–b), which calls into question if Snp is a true Eri1 homolog. The exonucleases that turnover canonical histone mRNAs at the end of S-phase and trim 5.8S rRNA remain to be defined. Interestingly, the Drosophila “5.8S” actually consists of a shortened 5.8S rRNA paired with 2S rRNA to form a 5.8S-like structure seen in other eukaryotes [68]. Thus, 5.8S 3′ end processing has diverged in Drosophila, which coincidentally also lack Eri1. Further work must determine the extent of Eri1 conservation and substrate specificity across metazoan species.
Eri1 at the crossroads of multiple RNA processing pathways
Together the diverse RNAs regulated by Eri1 paint a portrait of a deeply conserved exoribonuclease that functions as an important modulator of epigenetic gene expression. Eri1 has been repeatedly recruited into species- and cell-type-specific RNA metabolic pathways over the course of evolution. The recruitment of one enzyme into multiple RNA processing pathways may allow for crosstalk and coregulation of diverse cellular processes. In one scenario, these unique pathways may compete for Eri1, allowing the exonuclease to buffer pathway activities. This model makes sense in a context where Eri1 is rate-limiting and substrate processing is exquisitely sensitive to Eri1 dosage. This is not the case for 5.8S rRNA, which requires only small amounts of Eri1 for 3′ end processing [9]. miRNAs, however, may be more sensitive to Eri1 levels, as evidenced by the fact that Eri1 heterozygotes show alterations in miRNA abundance [28]. There is a precedent for this type of competition in C. elegans, where numerous small regulatory RNA pathways compete for components of the RNAi machinery, perhaps including Eri1 [17,18].
As an alternative to this model, the role of Eri1 in one pathway may promote its function in another. For example, the association of Eri1 with the ribosome may be required to spatially couple translation termination with histone mRNA decay. Eri1 ribosome association may also place it in appropriate proximity to affect small RNA deposition on mRNA targets. Interestingly, a recent report found that reduced expression of many ribosome-associated proteins diminished miRNA-mediated silencing in human cells, suggesting a precedent for the basal translation machinery to affect RNAi [69]. It is not known whether other ribosome-associated proteins can influence overall miRNA abundance, as is observed for Eri1. An interesting future area of research is to determine if there are specific Eri1 mutants that restrict its activity to specific RNA processing pathways. Such mutants would allow researchers to directly probe whether Eri1’s activity in one pathway impacts its activity in another. If so, it would suggest that these RNA pathways share an evolutionary or co-regulatory relationship bound by Eri1. The continued study of Eri1 biology will undoubtedly lead us to a better understanding of the importance RNA metabolism plays in epigenetic gene regulation.
Highlights.
Eri1 processes ribosomal RNA, histone mRNA, and small regulatory RNAs
Eri1 exemplifies a growing class of multifunctional small RNA processing proteins
Eri1 has been evolutionarily recruited into diverse small regulatory RNA pathways
Eri1 may coordinate the activity of diverse cellular processes regulated by RNA
Acknowledgments
Work in the Ansel lab was supported by the NIH (CA179512, HL109102, HL107202), the Burroughs Wellcome Fund, and the Leukemia & Lymphoma Society. We apologize to investigators whose work we could not discuss due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes MH, et al. The 3′–5′ exonuclease site of DNA polymerase III from gram-positive bacteria: definition of a novel motif structure. Gene. 1995;165:45–50. doi: 10.1016/0378-1119(95)00530-j. [DOI] [PubMed] [Google Scholar]
- 3.Cheng Y, Patel DJ. Crystallographic structure of the nuclease domain of 3′hExo, a DEDDh family member, bound to rAMP. J Mol Biol. 2004;343:305–312. doi: 10.1016/j.jmb.2004.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominski Z, et al. A 3′ exonuclease that specifically interacts with the 3′ end of histone mRNA. Mol Cell. 2003;12:295–305. doi: 10.1016/s1097-2765(03)00278-8. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy S, et al. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 6.Kipp M, et al. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol Cell Biol. 2000;20:7480–7489. doi: 10.1128/mcb.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 8.Yang XC, et al. Characterization of 3′hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J Biol Chem. 2006;281:30447–30454. doi: 10.1074/jbc.M602947200. [DOI] [PubMed] [Google Scholar]
- 9.Ansel KM, et al. Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat Struct Mol Biol. 2008;15:523–530. doi: 10.1038/nsmb.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan D, et al. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science. 2013;339:318–321. doi: 10.1126/science.1228705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee RC, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 12.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 13.Wightman B, et al. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 14.Grishok A. Biology and Mechanisms of Short RNAs in Caenorhabditis elegans. Adv Genet. 2013;83:1–69. doi: 10.1016/B978-0-12-407675-4.00001-8. [DOI] [PubMed] [Google Scholar]
- 15.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 17.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 18.Lee RC, et al. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmer F, et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 20.Han T, et al. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18674–18679. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu W, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–244. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thivierge C, et al. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat Struct Mol Biol. 2012;19:90–97. doi: 10.1038/nsmb.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavelec DM, et al. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics. 2009;183:1283–1295. doi: 10.1534/genetics.109.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bühler M, et al. Transcriptional silencing of nonsense codon-containing immunoglobulin minigenes. Molecular Cell. 2005;18:307–317. doi: 10.1016/j.molcel.2005.03.030. [DOI] [PubMed] [Google Scholar]
- 25.Hong J, et al. High doses of siRNAs induce eri-1 and adar-1 gene expression and reduce the efficiency of RNA interference in the mouse. Biochem J. 2005;390:675–679. doi: 10.1042/BJ20050647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabel HW, Ruvkun G. The exonuclease ERI-1 has a conserved dual role in 5.8S rRNA processing and RNAi. Nat Struct Mol Biol. 2008;15:531–533. doi: 10.1038/nsmb.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gent JI, et al. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 2010;37:679–689. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas MF, et al. Eri1 regulates microRNA homeostasis and mouse lymphocyte development and anti-viral function. Blood. 2012 doi: 10.1182/blood-2011-11-394072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee S, Grosshans H. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature. 2009;461:546–549. doi: 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 32.Yi R, et al. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada C, et al. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 34.Lingel A, et al. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 35.Song JJ, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 36.Ma JB, et al. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagan JP, et al. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Jones MR, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehrbach NJ, et al. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt MJ, et al. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA. 2011;17:39–44. doi: 10.1261/rna.2252511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holoch D, Moazed D. RNAi in fission yeast finds new targets and new ways of targeting at the nuclear periphery. Genes Dev. 2012;26:741–745. doi: 10.1101/gad.191155.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyes-Turcu FE, Grewal SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22:156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iida T, et al. Conserved ribonuclease, Eri1, negatively regulates heterochromatin assembly in fission yeast. Curr Biol. 2006;16:1459–1464. doi: 10.1016/j.cub.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 46.Bühler M, et al. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 47.Bian Y, et al. High-dose siRNAs upregulate mouse Eri-1 at both transcription and posttranscription levels. PLoS One. 2011;6:e26466. doi: 10.1371/journal.pone.0026466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faber AW, et al. Ngl2p is a Ccr4p-like RNA nuclease essential for the final step in 3′-end processing of 5.8S rRNA in Saccharomyces cerevisiae. RNA. 2002;8:1095–1101. doi: 10.1017/s1355838202021027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kondrashov N, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 51.Han M, et al. Histone H2B repression causes cell-cycle-specific arrest in yeast: effects on chromosomal segregation, replication, and transcription. Cell. 1987;48:589–597. doi: 10.1016/0092-8674(87)90237-6. [DOI] [PubMed] [Google Scholar]
- 52.Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 53.Mowry KL, Steitz JA. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA’s. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- 54.Whitfield ML, et al. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ross J, et al. Properties of the exonuclease activity that degrades H4 histone mRNA. J Biol Chem. 1987;262:9374–9381. [PubMed] [Google Scholar]
- 56.Graves RA, et al. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- 57.Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol. 2005;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoefig KP, et al. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol. 2013;20:73–81. doi: 10.1038/nsmb.2450. [DOI] [PubMed] [Google Scholar]
- 59.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sobti M, et al. Engineered rings of mixed yeast Lsm proteins show differential interactions with translation factors and U-rich RNA. Biochemistry. 2010;49:2335–2345. doi: 10.1021/bi901767w. [DOI] [PubMed] [Google Scholar]
- 61.Sabath I, et al. 3′-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA. 2013;19:1726–1744. doi: 10.1261/rna.040360.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drinnenberg I, et al. RNAi in Budding Yeast. Science. 2009 doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drinnenberg IA, et al. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilkins C, et al. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 66.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 67.Kupsco JM, et al. Genetic and biochemical characterization of Drosophila Snipper: A promiscuous member of the metazoan 3′hExo/ERI-1 family of 3′ to 5′ exonucleases. RNA. 2006;12:2103–2117. doi: 10.1261/rna.186706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pavlakis GN, et al. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979;7:2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Janas MM, et al. Reduced Expression of Ribosomal Proteins Relieves MicroRNA-Mediated Repression. Mol Cell. 2012;46:171–186. doi: 10.1016/j.molcel.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 70.Wu H, et al. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J Biol Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 71.Fukuda T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 72.Macias S, et al. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012;19:760–766. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liang XH, Crooke ST. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 2011;39:4875–4889. doi: 10.1093/nar/gkr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bernstein DA, et al. Candida albicans Dicer (CaDcr1) is required for efficient ribosomal and spliceosomal RNA maturation. Proc Natl Acad Sci U S A. 2011;109:523–528. doi: 10.1073/pnas.1118859109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Briggs MW, et al. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 76.Flynt AS, et al. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bogerd HP, et al. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han J, et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kadener S, et al. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Triboulet R, et al. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dong Z, et al. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci U S A. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Machida S, et al. Molecular insights into miRNA processing by Arabidopsis thaliana SERRATE. Nucleic Acids Res. 2011;39:7828–7836. doi: 10.1093/nar/gkr428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gruber JJ, et al. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol Cell. 2012;45:87–98. doi: 10.1016/j.molcel.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sabin LR, et al. Ars2 regulates both miRNA- and siRNA- dependent silencing and suppresses RNA virus infection in Drosophila. Cell. 2009;138:340–351. doi: 10.1016/j.cell.2009.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gruber JJ, et al. Ars2 links the nuclear cap-binding complex to RNA interference and cell proliferation. Cell. 2009;138:328–339. doi: 10.1016/j.cell.2009.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reis CC, Campbell JL. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics. 2007;175:993–1010. doi: 10.1534/genetics.106.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Canavan R, Bond U. Deletion of the nuclear exosome component RRP6 leads to continued accumulation of the histone mRNA HTB1 in S-phase of the cell cycle in Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:6268–6279. doi: 10.1093/nar/gkm691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hirai H, et al. Uridylation of U6 RNA in a nuclear extract in Ehrlich ascites tumor cells. J Biochem. 1988;104:991–994. doi: 10.1093/oxfordjournals.jbchem.a122597. [DOI] [PubMed] [Google Scholar]
- 90.Choi YS, et al. Widespread RNA 3′-end oligouridylation in mammals. RNA. 2012;18:394–401. doi: 10.1261/rna.029306.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chang HM, et al. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Malecki M, et al. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lubas M, et al. Exonuclease hDIS3L2 specifies an exosome-independent 3′–5′ degradation pathway of human cytoplasmic mRNA. EMBO J. 2013;32:1855–1868. doi: 10.1038/emboj.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 95.Yoda M, et al. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marasovic M, et al. Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol Cell. 2013;52:173–183. doi: 10.1016/j.molcel.2013.08.046. [DOI] [PubMed] [Google Scholar]
- 97.Nielsen AF, et al. Ars2 and the Cap-Binding Complex Team up for Silencing. Cell. 2009;138:224–226. doi: 10.1016/j.cell.2009.07.009. [DOI] [PubMed] [Google Scholar]