Abstract
Direct UV matrix-assisted laser desorption/ionization (MALDI) mass spectrometric analysis of uncomplexed, underivatized, highly sulfated oligosaccharides has been carried out using ionic liquids as matrices. Under conventionally used MALDI time-of-flight experimental conditions, uncomplexed polysulfated oligosaccharides do not produce any signal. We report that 1-methylimidazolium α-cyano-4-hydroxycinnamate and butylammonium 2,5-dihydroxybenzoate ionic liquid matrices allow the detection of picomole amounts of the sodium salts of a disaccharide, sucrose octasulfate, and an octasulfated pentasaccharide, Arixtra. The experimental results indicate that both analytes undergo some degree of thermal fragmentation with a mass loss corresponding to cleavage of O–SO3Na bonds in the matrix upon laser irradiation, reflecting lability of sulfo groups.
An effective matrix-assisted laser desorption/ionization mass spectrometric (MALDI-MS) analysis depends on several steps, among which the sample preparation procedure, especially the selection of suitable matrix, largely determines whether subsequent desorption/ionization and detection successfully follow.1 Significant effort has been put forth toward understanding the mechanism of MALDI2,3 and the role of matrix in the ionization process;4-6 yet matrix selection and formulation remain essentially empirical procedures.
Over the past few years, a growing interest in ionic liquids has resulted in the publication of several reports describing applications of these compounds in the MALDI-MS analysis of a variety of compounds including biomolecules.7-11 Ionic liquids are organic salts generally having melting points below 100 °C.12 This class of compounds have a number of characteristics that make them attractive for use as MALDI matrices, particularly, their high thermal stability, negligible vapor pressure, and virtually universal solvent properties.13,14 In a pioneering study, Armstrong and co-workers evaluated 38 ionic liquids for use as matrices for ultraviolet (UV) MALDI-MS;13 20 of these were shown effective for more than one analyte.7 In comparison to the conventional matrices, the effective ionic liquid matrices (ILMs) afforded greater vacuum stability, lower detection limits, higher homogeneity of the matrix/analyte mixture, and thus better shot-to-shot reproducibility.13 Later, ionic liquids were successfully applied in the MALDI-MS analysis of synthetic oligonucleotides, monosialylated glycans and glycosides, neutral oligosaccharides, poly-(ethylene glycol), glycoconjugates, peptides, proteins, and phospholipids.8-11
The work presented here describes a method for UV-MALDI time-of-flight (TOF) mass spectrometric analysis of highly sulfated oligosaccharides using ILMs. Oligosaccharides are small biopolymers (dimers to decamers) of monosaccharide units joined through glycosidic linkages between the anomeric hydroxyl group of one monosaccharide and a hydroxyl group of another monosaccharide. A great variety of monosaccharide building blocks and the possibility of different positional and stereochemical (α- or β-glycosidic) linkages between these saccharide building blocks result in the tremendous structural complexity of oligosaccharides critical for their diverse biological functions.15,16 All cells are surrounded by a carbohydrate-rich anionic coating known as a glycocalix. Alterations in a cell’s glycocalix, particularly the extended hydrated mesh of negatively charged proteoglycans capable of specific interactions with a large number of proteins and enzymes, have been implicated in many pathological conditions. The central role of the oligosaccharide sequences of the glycosaminoglycan chains of these proteoglycans in cell recognition and signaling renders them a potentially important class of pharmaceutical agents. Anionic polysaccharides have also been successfully tested in vivo as vaccines against malaria, leishmaniasis, HIV, cancer, and tuberculosis.17 Two analytes used in this study are synthetic, highly sulfated oligosaccharides that are established pharmaceutical agents. An octasulfated disaccharide, sodium sucrose octasulfate (SOS), is the active ingredient of the antiulcer drug Sucralfate, and an octasulfated pentasaccharide (Arixtra) is an anticoagulant drug used in prophylaxis of deep vein thrombosis.
While the biological significance of highly sulfated oligosaccharides is widely recognized, relatively little progress has been made to date in developing high-throughput MALDI mass spectrometry-based procedures for characterization of these compounds.18,19 Due to their polydispersity, structural complexity, and labile chemical nature, polysulfated oligosaccharide natural products are challenging analytes. The only currently available method for detection and characterization of highly sulfated oligosaccharides by MALDI-TOF-MS not involving chemical derivatization was developed 11 years ago by Juhasz and Biemann.20 They showed that heparin-derived polysulfated oligosaccharides produced high-quality MALDI mass spectra in a form of noncovalent complexes with basic peptides, and the analyte fragmentation through sulfate loss was almost completely suppressed. The method permitted the detection of a polysulfated oligosaccharide molecular ion in complex with the peptide. While uncomplexed disulfated and trisulfated oligosaccharides required 100 pmol/μL concentration to obtain a signal in the negative-ion mode, a noncovalent complex of octasulfated hexasaccharide and an arginine-containing synthetic peptide produced a strong positive-ion signal at 3 pmol/μL concentration of oligosaccharide. The most efficient formation of a peptide-oligosaccharide complex was observed when the number of arginine residues, n in a peptide with a sequence (Arg-Gly)n exceeded the number of sulfate groups in oligosaccharide by at least one.21 While this method has been pioneered by Sasisekharan and co-workers22 for the analysis of sulfated oligosaccharides, its relatively high sensitivity is compromised by the necessity of having a library of peptides suitable for analysis of polydisperse mixtures, which renders the method time-consuming and material-consuming. The current study describes the first successful analysis of uncomplexed, underivatized, highly sulfated oligosaccharides. UV-MALDI-TOF mass spectrometry was used to analyze the sodium salts of two octasulfated oligosaccharides, SOS and Arixtra. Five ILMs were tested for their effectiveness in facilitating the detection of the two analytes. Each analyte produced signal with multiple ionic liquids, but only two ILMs were determined to be equally efficient for the analysis of both SOS and Arixtra.
EXPERIMENTAL SECTION
Materials
Arixtra (methyl-O-2-deoxy-6-O-sulfo-2-(sulfoamino)-α-d-glucopyranosyl-(1→4)-O-β-d-glucopyranuronosyl-(1→4)-O-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-α-d-glucopyranosyl-(1→4)-O-2-O-sulfo-α-l-idopyranuronosyl-(1→4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-α-d-glucopyranoside, sodium salt) was purchased from Organon Sanofi-Synthelabo (West Orange, NJ). Sucrose octasulfate, sodium salt of pharmaceutical purity, was a gift from Bukh Meditec (Farum, Denmark). Butylamine, 1-methylimidazole, pyridine, 2,5-dihydroxybenzoic acid, α-cyano-4-hydroxycinnamic acid, diammonium hydrogen citrate, ammonium tartrate, and ammonium acetate were purchased from Sigma Aldrich (St. Louis, MO) and used without purification. HPLC-grade water, methanol, and acetonitrile were obtained from Fisher (Fair Lawn, NJ).
Ionic Liquids
Ionic liquids were prepared as described by Armstrong and co-workers.7 1-Methylimidazolium and butylammonium 2,5-dihydroxybenzoates (ImDHB and DHBB), and 1-methylimidazolium, pyridinium, and butylammonium α-cyano-4-hydroxycinnamates (ImCHCA, PyCHCA, and CHCAB) were solids at room temperature and were used without further purification.
Model Analyte Solutions
Octasulfated pentasaccharide, sodium salt (Arixtra, an anticoagulant drug), was purchased as a solution for intravenous injection. Physiological saline was removed by dialysis against water using a membrane with 1000 molecular weight cutoff. Purity and structure of both analytes were confirmed by NMR and ESI-MS.23 Both Arixtra and SOS sodium salts were freeze-dried and subsequently dissolved in water at 0.1–1.0 mM concentrations for use in MALDI-MS experiments.
Matrix Solutions
Ionic liquid (70 mg) was dissolved in 1 mL of 50 vol % aqueous acetonitrile/water for use as a matrix. In the case of PyCHCA and CHCAB, saturated solutions were prepared in 50 vol % aqueous acetonitrile, since the solubility of these compounds in this solvent system was <70 mg/mL. Solutions of ImCHCA and DHBB in 100% methanol were prepared at 70 mg/mL concentration.
MALDI-TOF-MS Analyses
Samples for MALDI-MS analysis were prepared by mixing 1 μL each of 0.1 mM oligosaccharide solution, 10 mM aqueous ammonium acetate, and 70 mg/mL ionic liquid in acetonitrile/water. Samples containing 10 mM ammonium tartrate or diammonium hydrogen citrate were prepared as described above. The resulting mixture (0.1–0.25 μL) was deposited on a stainless steel MALDI target and allowed to dry at room temperature and atmospheric pressure. All samples were also prepared without an ammonium salt additive; in these cases, 1 μL of water was added to the analyte/matrix mixture in place of 1 μL of an ammonium salt.
For the sensitivity determination, a series of dilutions of SOS and Arixtra were prepared, with concentrations 100, 50, 25, and 10 μM (50 μM solution of SOS, sodium salt corresponds to 58 μg/mL, and 50 μM Arixtra corresponds to 86 μg/mL). These samples for MALDI analysis were prepared by mixing two parts of an aqueous analyte solution with one part of the 70 mg/mL matrix solution in 50 vol % aqueous acetonitrile and depositing 0.25 μL of this mixture on a stainless steel MALDI plate.
Mass spectra were acquired on a TofSpec2E MALDI-TOF mass spectrometer (Micromass, Manchester, U.K.) in reflectron mode using default instrument parameters: source 20 kV, reflectron 26 kV, extraction lens 19.95 kV, focus lens 16 kV. Ionization was achieved using pulsed N2 laser at a wavelength of 337 nm. Mass spectra were recorded in both positive-ion and negative-ion modes and processed using MassLynx 3.5 software (Micromass).
RESULTS AND DISCUSSION
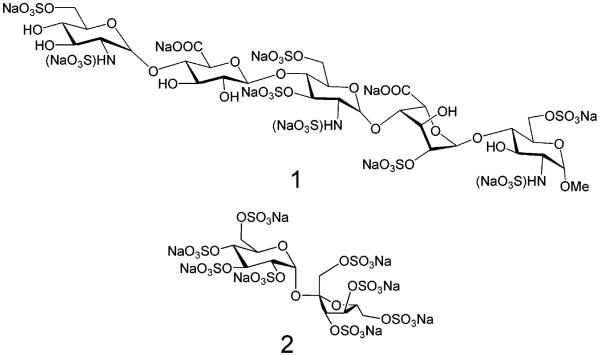
Factors such as the amount of laser energy transmitted onto the sample surface, matrix selection, and sample preparation procedures greatly affect the quality of MALDI mass spectra.1-6 In the process of selecting suitable conditions for the MALDI-TOF-MS detection of the two sulfated oligosaccharide analytes, SOS, sodium salt and Arixtra (Figure 1), several experimental parameters were evaluated.
Figure 1.
Structures of an octasulfated pentasaccharide, Arixtra, sodium salt (1), and an octasulfated disaccharide, SOS, sodium salt (2)
Matrix Selection and Sample Preparation
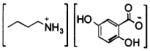
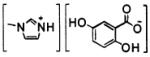
We prepared five ILMs: butylammonium and 1-methylimidazolium 2,5-dihydroxy-benzoates and butylammonium, pyridinium, and 1-methylimidazolium α-cyano-4-hydroxycinnamates (Table 1) and compared their efficiency as matrices in MALDI mass spectra of these two highly sulfated oligosaccharide analytes (Table 2). With the exception of ImDHB, which was prepared to complement ImCHCA, all ILMs were previously reported effective as matrices for the MALDI-MS analysis of peptides, proteins, DNA oligomers, and neutral oligosaccharides.7-11 To ensure validity of the comparison, all samples were spotted on one target plate and analyzed on the same day. This analysis was repeated twice; thus, each signal intensity value in Table 2 was obtained by independent experiments. There was no apparent correlation between the identity of a cation of the ILM and the efficiency of the ILM in these analyses. For each of the four effective ILMs (Table 2), the formation of a uniform glassy film on MALDI target appeared to improve the signal for both analytes. At a given analyte concentration, a thinner sample film (i.e., smaller volumes or more diluted matrix solutions deposited on the target) afforded higher intensity analyte signals than thicker sample films, although below a critical ILM concentration, the analyte signal was lost. We found for SOS and Arixtra, a 70 mg/mL ILM concentration to be optimal.
Table 1.
Compounds Tested as Matrices for MALDI-TOF-MS Analysis of SOS and Arixtra
| structure | abbreviated name |
mp, °C | pKa of cation |
appearance of a sample spot |
|---|---|---|---|---|
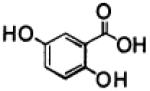
|
DHB | 203 | 2.97 | crystalline solid |
|
DHBB9 | 112–114 | 10.8 | glass film |
|
|
CHCAB9 | 158–161 | 10.8 | crystalline solid |
|
ImDHB | 120–122 | 6.95 | glass film |
|
|
ImCHCA7 | 142–143 | 6.95 | glass film |

|
PyCHCA7 | >227 | 5.25 | crystalline solid |
Table 2.
Efficiency of the Tested ILMs for the UV-MALDI-TOF Detection of Two Model Analytes, SOS and Arixtra
| analyte, 40 pmol |
reflectron mode |
laser energya |
analyte signal intensity in arbitrary unitsb |
||||
|---|---|---|---|---|---|---|---|
| DHBB | ImDHB | ImCHCA | PyCHCA | CHCAB | |||
| SOS | positive | low | – | – | 200, 940 | – | 100, 120 |
| high | 240, 600 | 100, 130 | 340, 1450 | – | 140, 160 | ||
| SOS | negative | low | – | – | 360, 1600 | – | 400, 760 |
| high | 230, 1480 | 300, 380 | 1690, 2160 | – | 200, 540 | ||
| Arixtra | positive | low | – | – | 100, 170 | – | – |
| high | 100, 950 | – | – | – | – | ||
| Arixtra | negative | low | – | – | 470, 560 | – | – |
| high | 1200, 2050 | 100, 160 | 810, 1180 | – | 100, 140 | ||
Low-energy setting corresponded to the 10% transmission, and high-energy setting corresponded to the 25% transmission of the laser energy.
Signal intensity values are reported for [M + Na]+ and [M − Na]− SOS ions, and [M + Na − 2NaSO3 + 2H]+ and [M − Na − 2NaSO3 + 2H]− ions of Arixtra; absence of an analyte signal is denoted (−).
The samples for MALDI-MS analysis of both SOS and Arixtra were prepared using each of the five ILMs with and without the addition of 10 mM ammonium acetate, ammonium tartrate, or diammonium hydrogen citrate. Ammonium salts are believed to form ammonium adducts with negatively charged analytes, preventing the formation of other cation adducts. Subsequent dissociation of the ammonium ion in the TOF analyzer should result in the formation of [M – H]− analyte species in negative-ion mode.24,25 In the current experiments, however, the addition of ammonium salts did not lead to the Na/H exchange in the two octasulfated oligosaccharides. Instead, both SOS and Arixtra appeared as fully sodiated species in the MALDI mass spectra. Furthermore, analytical sensitivity was not enhanced through the addition of ammonium salts (Figure 2); thus ammonium salts were omitted in preparation of samples for further studies in sensitivity of this analysis.
Figure 2.
MALDI-TOF mass spectra of SOS (40 pmol) acquired using 70 mg/mL ImCHCA matrix in positive-ion (panel A) and negative-ion (panel B) reflectron modes. Upper mass spectra in both panels A and B were obtained with samples in the absence of ammonium salt, and the lower mass spectra in both panels were recorded from samples containing ammonium acetate.
Two ILMs were found most suitable for the MALDI-MS analysis of SOS and Arixtra, DHBB and ImCHCA. Of the five ILMs tested, ImCHCA worked equally well at high and low laser energy settings and was the only matrix that allowed detection of Arixtra at the low laser energy settings in negative-ion and positive-ion modes. In addition, ImCHCA offered best shot-to-shot reproducibility and signal stability in the analysis of sample from a single area irradiated by laser. Analyte signal did not deteriorate even during 100-shot acquisition scans. Another important advantage of the ImCHCA matrix was that the analyte signal was observed immediately upon laser irradiation. In contrast, the DHBB matrix required some time to “warm” the sample before analyte signal could be observed at high laser energy settings. An important advantage of DHBB was the suppression of fragmentation through sulfate loss observed for both analytes, as discussed below. By using ImCHCA and DHBB as ILMs, we were able to detect picomole amounts of both SOS and Arixtra.
Laser Energy
Next, the effect of the amount of laser energy transmitted to a sample spot was examined as another parameter potentially impacting the quality of the MALDI-TOF mass spectra. The instrument used in this study, TofSpec 2E (Micromass), is equipped with an N2 337-nm laser, which generates 4-ns-wide pulses having energy of 180 μJ/pulse. The energy produced by the laser is fixed, as well as the area of the sample being irradiated. The amount of energy arriving at the sample surface can be controlled by the simultaneous use of two devices: a stepped filter and a continuously variable iris. The four-stage “coarse” filter can transmit 100, 50, 20, or 5% of the laser energy to the target, while the “fine” iris can further reduce the transmitted energy in a continuous way, allowing any figure of transmission to be achieved. The lowest energy setting sufficient for ionization of an analyte is typically applied because it ensures the strongest analyte signal and maximum resolution. Since one matrix may require significantly higher laser energy than another before the analyte signal can be observed, we used two laser energy settings denoted in Table 2 as “high” and “low”. The low-energy setting corresponded to the coarse filter set to 20% and the fine iris set to 50% (18 μJ/pulse), while the high-energy setting corresponded to a 50% setting on both devices (45 μJ/pulse). The two DHB-derived ILMs required higher energy for analyte ionization than did the two effective CHCA-derived ILMs (Table 2). This result is consistent with the trend observed with crystalline DHB and CHCA.5,26
MALDI-TOF mass spectra of both analytes displayed a series of peaks corresponding to ions of the general formula [M + Na − nSO3Na + nH]+ in positive-ion mode or [M − Na − nSO3Na + nH]− in negative-ion mode. The amount of laser energy transmitted to the sample spot did not affect the number of sulfates lost from SOS and Arixtra. At higher laser energies, all peaks including molecular species had higher intensities than at lower laser energies, while the number of observed fragments remained the same. Both analytes afforded MALDI mass spectra having a series of up to five peaks, corresponding to the molecular species or fragment peaks corresponding to O–SO3Na bond cleavages with an H substituting an SO3Na group (a net loss of 102 mass units). Since both analytes exhibited some degree of fragmentation, a species with the highest m/z value that was detected in all effective matrices was chosen for the comparison of signal strength within studied set of matrices. Thus, the analyte signal intensity values reported in Table 2 correspond to the intensities of [M + Na]+ and [M − Na]− peaks for SOS, sodium salt, and [M + Na − 2NaSO3 + 2H]+ and [M − Na − 2NaSO3 + 2H]− peaks for Arixtra. Each value represents the result of combining of two 100-shot scans. The differences between the MALDI signals obtained in two independent acquisitions for every entry in Table 2 reflect the dependence of MALDI analysis on quality of a sample spot.
Both analytes exhibited stronger signals in negative-ion than in positive-ion reflectron mode when desorbed using the four effective ILMs. This observation can be explained by the highly negatively charged nature of the two polysulfated oligosaccharides. Surprisingly, sucrose octasulfate showed strong [M + Na]+ signal using the ImCHCA and DHBB matrices, whereas Arixtra was more difficult to detect in the positive-ion mode. Molecular species of Arixtra were detected only at relatively high concentration of 40 pmol/spot. Certainly, fragmentation of analyte presents a problem that needs to be addressed if the current method is to be applied for the analysis of unknowns. In earlier work from our laboratory, we demonstrated that it is possible to find optimal conditions for detection of unfragmented highly sulfated oligosaccharides using ESI-MS.23 Even though MALDI mechanisms are quite different from ESI mechanisms, it might also be possible to optimize the MALDI conditions to completely suppress fragmentation.
Sensitivity
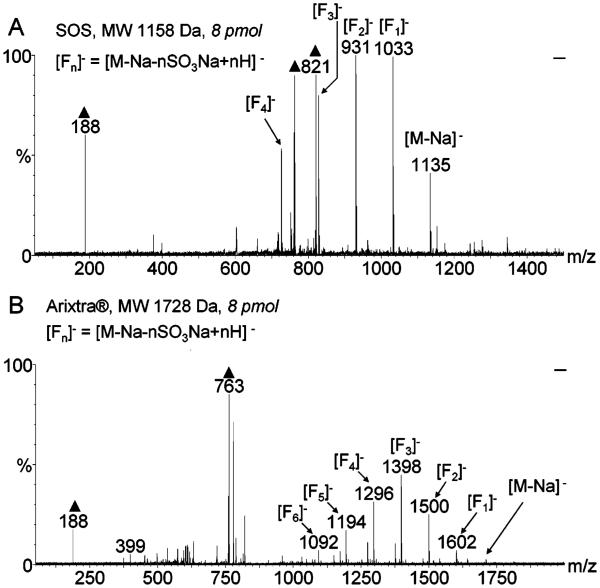
A series of dilutions of SOS and Arixtra were prepared in ImCHCA ILM and analyzed to assess sensitivity of MALDI-TOF-MS. Strong and reproducible analyte signals were acquired with 8 pmol of sample material per target spot (Figure 3). The 8 pmol/spot samples were prepared using 50 μM aqueous analyte solutions and depositing 0.25 μL of the analyte/matrix mixture on a MALDI target. At 25 μM concentration, the analyte signal was not always observed, and at 10 μM concentration, no analyte signal was detected. The use of ImCHCA as ILM for the analysis of SOS and Arixtra afforded sensitivity within a biologically relevant range of analyte concentrations; thus, the current method has potential applications in analysis of polysulfated oligosaccharides derived from biological sources, such as plasma samples from patients receiving these drug products. DHBB was not used in the sensitivity experiments since it required a higher analyte concentration for observing the signal.
Figure 3.
MALDI-TOF mass spectra of the 8 pM samples of SOS (panel A) and Arixtra (panel B) in 70 mg/mL ImCHCA matrix acquired in the negative-ion mode using 45 μJ/pulse laser energy setting. Both analytes were detected without searching for a good spot and produced stable signals during the 100-shot acquisition scans. Peaks due to the matrix are marked with triangles.
Suppression of Fragmentation Resulting from Cleavage of O–SO3 Bonds
In the process of optimizing experimental variables, the two selected matrices ImCHCA and DHBB were dissolved in 100% methanol at 70 mg/mL concentration to determine whether the matrix solvent affects the quality of MALDI mass spectra of SOS and Arixtra. The DHBB/methanol matrix required a greater amount of analyte in the sample and a higher laser energy setting to observe the analyte signal than did the ImCHCA/methanol matrix. However, fragmentation of both SOS and Arixtra was suppressed when these compounds were desorbed using the DHBB/methanol matrix.
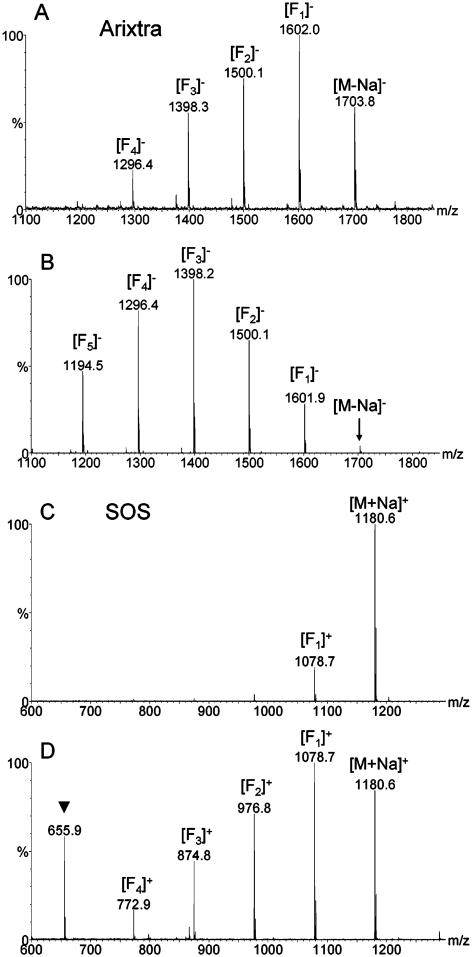
Both negative-ion and positive-ion MALDI mass spectra of Arixtra obtained using the DHBB solution in methanol as the matrix exhibited a lower number of fragment peaks and a more abundant molecular ion peak as compared to the spectra obtained with the ImCHCA solution in methanol (Figure 4A and B). Molecular species of SOS, sodium salt, dominated positive-ion MALDI mass spectra obtained with the methanol solution of DHBB as the matrix, whereas a greater number of fragments were detected with the methanol solution of ImCHCA as the matrix (Figure 4C and D). Negative-ion MALDI mass spectra of SOS, sodium salt, obtained with DHBB/methanol matrix showed a greater number of fragment peaks as compared to the positive-ion spectra, and the sensitivity of the analysis was lower with DHBB matrix than with ImCHCA matrix regardless of the matrix solvent. The fragmentation resulting from cleavage of the O–SO3 bond appears to be analyte-dependent and matrix-dependent but can be suppressed under certain conditions as evident from the MALDI mass spectrum of SOS, sodium salt.
Figure 4.
Negative-ion MALDI mass spectra of Arixtra, sodium salt, obtained with DHBB/methanol matrix (panel A) and ImCHCA/methanol matrix (panel B). Fragments due to the sulfate loss are denoted [Fn]−, where [Fn]− = [M − Na − nSO3Na + nH]−. Positive-ion MALDI mass spectra of SOS, sodium salt obtained with DHBB/methanol matrix (panel C) and ImCHCA/methanol matrix (panel D). Fragments due to the sulfate loss are denoted [Fn]+, where [Fn]+ = [M + Na − nSO3Na + nH]+. Peak due to the matrix is marked with triangle.
CONCLUSIONS
A method for the MALDI-TOF-MS analysis of polysulfated oligosaccharides using two ILMs, ImCHCA and DHBB, is described here having a number of distinctive advantages. First, it allows the detection of highly sulfated oligosaccharides in their underivatized and uncomplexed form. Second, sample preparation is simple and time-efficient, involving mixing an aqueous analyte solution with a solution of ILM. Third, the method requires very small amounts of sample (μg/mL concentrations) for analysis, and similarly, small volumes (0.1–0.25 μL) of analyte/matrix mixture are required for obtaining homogeneous spots on the MALDI target. The homogeneity of such sample spots also makes the current method amenable to automated data acquisition.
The use of DHBB as the matrix allows detecting molecular ions of two highly negatively charged oligosaccharides, SOS and Arixtra, in their uncomplexed underivatized form. However, the issues of sensitivity and fragmentation must be further addressed before the method can be applied in the MALDI-MS analysis of unknown highly sulfated oligosaccharide mixtures. Clearly, fragmentation of analyte due to the losses of sulfo groups negatively impacts the sensitivity of analysis. Therefore, the first priority of future studies will be to establish experimental conditions that facilitate the detection of polysulfated oligosaccharides as intact molecular species.
ACKNOWLEDGMENT
This work was supported by NIH grant GM 038060 and NSF grant CHE-0078056 used for the acquisition of the MALDI-TOF mass spectrometer.
References
- (1).Dreiswerd K. Chem. Rev. 2003;103:395–425. doi: 10.1021/cr010375i. [DOI] [PubMed] [Google Scholar]
- (2).Zenobi R, Knochenmuss R. Mass Spectrom. Rev. 1998;17:337–366. [Google Scholar]
- (3).Gabelica V, Schultz E, Karas M. J. Mass Spectrom. 2004;39:579–593. doi: 10.1002/jms.651. [DOI] [PubMed] [Google Scholar]
- (4).Knochenmuss R, Stortelder A, Breuker K, Zenobi R. J. Mass Spectrom. 2000;35:1237–1245. doi: 10.1002/1096-9888(200011)35:11<1237::AID-JMS74>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- (5).Stevenson E, Breuker K, Zenobi R. J. Mass Spectrom. 2000;35:1035–1041. doi: 10.1002/1096-9888(200008)35:8<1035::AID-JMS34>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- (6).Knochenmuss R, Zenobi R. Chem. Rev. 2003;103:441–452. doi: 10.1021/cr0103773. [DOI] [PubMed] [Google Scholar]
- (7).Armstrong D, Zhang L-K, He L, Gross ML. Anal. Chem. 2001;73:3679–3686. doi: 10.1021/ac010259f. [DOI] [PubMed] [Google Scholar]
- (8).Carda-Broch S, Berthod A, Armstrong DW. Rapid Commun. Mass Spectrom. 2003;17:553–560. doi: 10.1002/rcm.931. [DOI] [PubMed] [Google Scholar]
- (9).Mank M, Stahl B, Boehm G. Anal. Chem. 2004;76:2938–2950. doi: 10.1021/ac030354j. [DOI] [PubMed] [Google Scholar]
- (10).Li YL, Gross ML. J. Am. Soc. Mass Spectrom. 2004;15:1833–1837. doi: 10.1016/j.jasms.2004.08.011. [DOI] [PubMed] [Google Scholar]
- (11).Li YL, Gross ML. J. Am. Soc. Mass Spectrom. 2005;16:679–682. doi: 10.1016/j.jasms.2005.01.017. [DOI] [PubMed] [Google Scholar]
- (12).Hagiwara R, Ito Y. J. Fluorine Chem. 2000;105:221–227. [Google Scholar]
- (13).Armstrong DW, He L, Liu Y-S. Anal. Chem. 1999;71:3873–3876. doi: 10.1021/ac990443p. [DOI] [PubMed] [Google Scholar]
- (14).Anderson JL, Ding J, Welton T, Armstrong DW. J. Am. Chem. Soc. 2002;124:14247–14254. doi: 10.1021/ja028156h. [DOI] [PubMed] [Google Scholar]
- (15).Mulloy B, Linhardt RJ. Curr. Opin. Struct. Biol. 2001;11:623–628. doi: 10.1016/s0959-440x(00)00257-8. [DOI] [PubMed] [Google Scholar]
- (16).Capila I, Linhardt RJ. Angew. Chem., Int. Ed. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- (17).Wertz DB, Seeberger PH. Chem. Eur. J. 2005;11:3194–3206. doi: 10.1002/chem.200500025. [DOI] [PubMed] [Google Scholar]
- (18).Harvey DJ. Mass Spectrom. Rev. 1999;18:349–451. doi: 10.1002/(SICI)1098-2787(1999)18:6<349::AID-MAS1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- (19).Zaia J. Mass Spectrom. Rev. 2004;23:161–227. doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- (20).Juhasz P, Biemann K. Proc. Natl. Acad. Sci. U.S.A. 1994;91:4333–4337. doi: 10.1073/pnas.91.10.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Juhasz P, Biemann K. Carbohydr. Res. 1995;270:131–147. doi: 10.1016/0008-6215(94)00012-5. [DOI] [PubMed] [Google Scholar]
- (22).Venkataraman G, Shriver Z, Raman R, Sasisekharan R. Science. 1999;286:537–542. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- (23).Gunay NS, Tadano-Aritomi K, Toida T, Ishizuka I, Linhardt RJ. Anal. Chem. 2003;75:3226–3231. doi: 10.1021/ac034053l. [DOI] [PubMed] [Google Scholar]
- (24).Lavanant H, Lange C. Rapid Commun. Mass Spectrom. 2002;16:1928–1933. doi: 10.1002/rcm.816. [DOI] [PubMed] [Google Scholar]
- (25).Papac DI, Wong A, Jones AJ. Anal. Chem. 1996;68:3215–3223. doi: 10.1021/ac960324z. [DOI] [PubMed] [Google Scholar]
- (26).Karas M, Bahr U, Strupat K, Hillenkamp F, Tsarbopoulos A, Pramanik BM. Anal. Chem. 1995;67:675–679. doi: 10.1021/ac00085a022. [DOI] [PubMed] [Google Scholar]