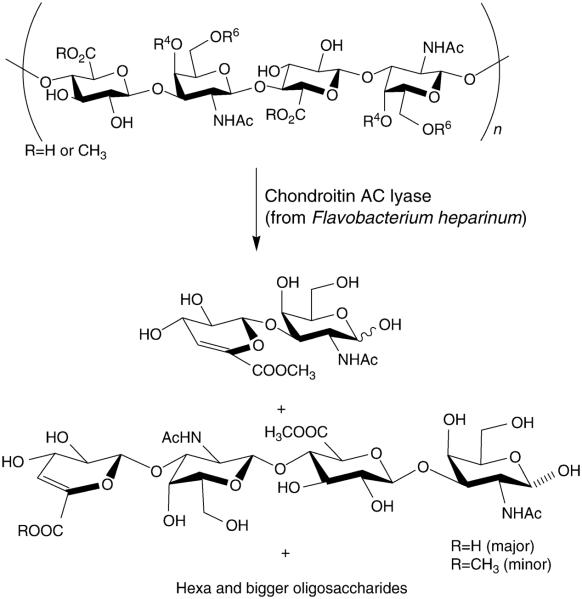
I. Chondroitin Sulfate Glycosaminoglycans
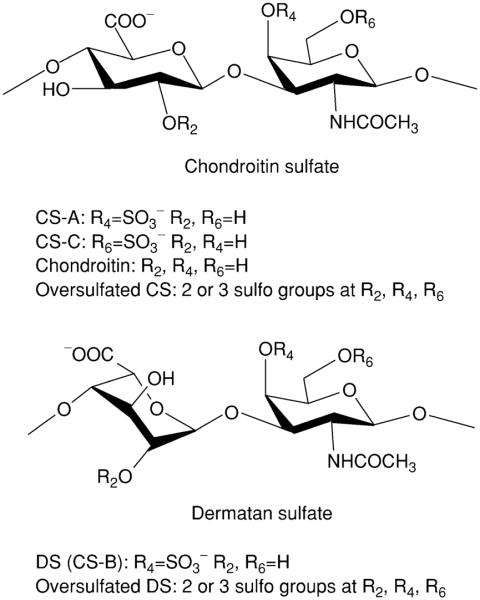
Glycosaminoglycans (GAGs) are a family of highly sulfated, complex mixture of linear polysaccharides that display a wide array of biological activities (Boneu, 1996; Jackson et al., 1991). GAGs can be classified into four basic types—hyaluronan, chondroitin/dermatan sulfates (CS/DS), heparin/heparan sulfate, and keratan sulfate (Capila and Linhardt, 2002; Esko and Selleck, 2002; Iozzo, 1998; Linhardt and Toida, 2004). Chondroitin/dermatan sulfates are the focus of this chapter. Chondroitin/dermatan sulfates are linear, polydisperse GAGs with a repeating core of disaccharide structure composed of a D-glucopyranosyl uronic (GlcAp) acid or L-idopyranosyl uronic (IdoAp) acid glycosidically linked to 2-deoxy, 2-acetamido-D-galactopyranose (GalpNAc) residue (Fig. 1). The major classes of the chondroitin family of GAGs are: chondroitin; chondroitin-4-sulfate (CS-A); dermatan sulfate (CS-B or DS), and chondroitin-6-sulfate (CS-C).
FIGURE 1.
CS, oversulfated CS and chondroitin: the molecular weight ranges from 5000–50,000 (average 25 kDa). DS and oversulfated DS: the molecular weight ranges from 5000–50,000 (average 25 kDa).
GAGs are the sulfated polysaccharide side chains of proteoglycans (PGs). These PGs are ubiquitous in animals and found localized on the external cell membrane and extracellular matrix (ECM) in all tissues (Iozzo, 1998). Despite intensive studies on this class of biopolymers, their precise chemical structures and biological functions are still not well understood. The major families of GAGs differ based on their disaccharide repeating unit, their linkage chemistry, and their sulfation pattern. Our current understanding of GAG structure and the biosynthetic pathway of GAG synthesis suggest the presence of defined sequences that specifically interact with an array of GAG-binding proteins (Esko and Selleck, 2002). Among these proteins are several families of growth factors, chemokines, enzymes, and adhesion proteins (Capila and Linhardt, 2002). The GAG chains of PGs act as receptors in signal transduction, controlling cell growth, differentiation, migration, adhesion, and other important physiological and pathophysiological events (Linhardt and Toida, 2004). CS is the predominant GAG present in aggrecan, the major PG of cartilage (Kresse and Schonherr, 2001). Due to the sulfo and carboxyl moieties of their GAG components, PGs concentrate large amounts of negative charge in the ECM. This has direct osmotic effects on these tissues in which the GAGs are under hydrated due to constraints imposed by the collagen fiber network, giving cartilage its shock-absorbing function (Kempson, 1980). GalNpAc O-sulfonation of chondroitin can occur at the 4- and/or 6-positions (CS-A and CS-C, respectively), and it is not known how the sulfated units are distributed throughout the PG molecules or whether particular regions have different biological functions. CS-A, CS-C, and DS are found within the ECM or on cell membranes attached to a variety of proteins, including decorin, biglycan, and aggrecan (Kresse and Schonherr, 2001).
II. Enzymes Mediating GAG Synthesis
GAG presence is associated with all animals, ranging from C. elegans to man (Esko and Selleck, 2002). These polysaccharide chains are synthesized by a set of specialized enzymes that assemble an initiation tetrasaccharide on specific serine residues of the core protein, followed by successive addition of repeating disaccharide units to the nonreducing end by synthases (Spicer and McDonald, 1998; Yada et al., 2003). GAGs, however, are not unique to eukaryotes. Several specialized microorganisms also produce simpler forms of these polymers. The enzymes mediating GAG synthesis have been characterized, and the ability to express them in large quantities would greatly facilitate the production of defined GAG components (Spicer and McDonald, 1998; Yada et al., 2003).
Glycosaminoglycans are synthesized by the serial addition of UDP-sugars through the action of dedicated membrane anchored glycosyl transferases followed by several sulfotransferases, deacetylases, and epimerases (Esko and Selleck, 2002; Hannesson et al., 1996; Silbert and Sugumaran, 2002; Sugahara and Kitagawa, 2002). Sugar transfer occurs in the Golgi, and the type of GAG added is believed to be dependent on as yet unclear signals present on the core proteins (Iozzo, 1998; Rosenberg et al., 1997). In general, GAGs are added to a specific region of the core protein. In most cases a common tetrasaccharide linkage region is assembled by xylosylation of specific serine residues, followed by addition of two galactose units and glucuronic acid. The next step, addition of N-acetyl hexosamine determines whether the resulting chain will be CS/DS or heparan sulfate (HS). The three-dimensional structures of two of these biosynthetic enzymes have been determined (Negishi et al., 2003). The final glycosyl transferases that add repeating disaccharides to extend the growing GAG chain are of greatest interest. These are bifunctional enzymes that alternatively add GlcUAp and GalNpAc or GlcNpAc. Families of these enzymes have been identified for human CS (Kitagawa et al., 2001, 2003; Uyama et al., 2003; Yada et al., 2003) and HS (Esko and Selleck, 2002) synthesis. A gene coding for chondroitin synthase was identified in the K4 strain of E. coli (Ninomiya et al., 2002). While the mammalian enzymes can only be expressed in tiny amounts, bacterial chondroitin synthase has been produced as a soluble recombinant protein (Yada et al., 2003).
Since no protein core is associated with hyaluronic acid, a different biosynthetic mechanism is in place for this macromolecule. Two classes of hyaluronan synthases (HAS) have been characterized (DeAngelis, 1999). The class I enzymes are integral membrane proteins whereas the class II members are membrane associated through a C-terminal membrane spanning segment. Mammalian HAS belong to class I and appear to be bifunctional enzymes that add alternating UDP sugars to the growing hyaluronan chain, which is extruded onto the cell surface and into the ECM. Three mammalian genes coding for HAS have been identified, and their gene structures are evolutionarily well conserved (Monslow et al., 2003). These isoforms show high-amino acid sequence identity between themselves and to some bacterial enzymes, for example, Streptococcus pyogenes HA synthase. They contain seven putative membrane-spanning regions with a long cytoplasmic loop containing the putative UDP binding and glycosyltransferase catalytic sites (Itano and Kimata, 2002). A soluble and active fragment of human HAS2 was expressed in E. coli (Hoshi et al., 2004), opening the door to functional and structural studies of this enzyme.
Assembly of heparan and chondroitin chains involves two additional modifications—sulfation of specific positions of the N-acetyl hexosamine and the uronic acid units and epimerization of glucuronic acid residues. C5-epimerases responsible for this step in heparan biosynthesis have been characterized (Li et al., 1997), but those responsible for chondroitin to dermatan conversion remain unclear (Seidler et al., 2002). Finally, a series of sulfotransferases acts to modify the CS/DS and HS/heparin families of PGs. The three-dimensional structures of two of these biosynthetic enzymes have been determined (Moon et al., 2004; Thorp et al., 2004).
III. CS Degrading Enzymes
Two chemically distinct enzymatic mechanisms have evolved for the degradation of GAGs, and the enzymes are accordingly classified as either hydrolases or lyases. Henrissat has divided these enzymes into families based on sequence similarity (http://afmb.cnrs-mrs.fr/CAZY/). Cleavage of the hexuronic acid!hexosamine bond always involves a standard glycosidase mechanism of either inverting or retaining type in which the glycosidic bond is hydrolyzed by addition of a water molecule (Fig. 2) (Zechel and Withers, 2000). In contrast, cleavage of the hexosamine!hexuronic acid bond can occur through either a hydrolytic, catalyzed by hydrolases, or an eliminative mechanism, catalyzed by lyases (Fig. 2) (Ernst et al., 1995; Linhardt et al., 1986; Michaud et al., 2003). While polysaccharide hydrolases are found in virtually all organisms, polysaccharide lyases are not found in vertebrates.
FIGURE 2.
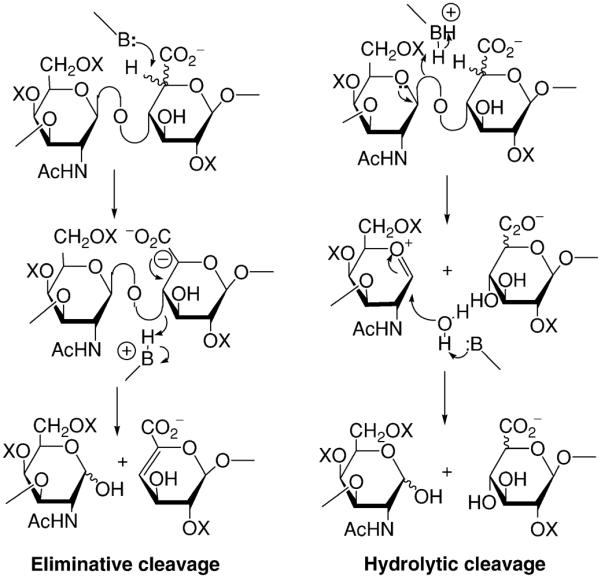
Mechanism for the enzymatic breakdown of GAGs. Lyases catalyze eliminative cleavage and hydrolases catalyze hydrolytic cleavage leading to different oligosaccharide products.
In many tissues GAGs undergo rapid turnover. Hyaluronan degradation plays a major part in the release of PGs from cartilage that occurs in normal development and in arthritis (Sztrolovics et al., 2002). It has been estimated that in the dermis, which contains more than half of the HA in the body, 50–75% of this GAG is turned over every 24 h (Frost et al., 1996; McCourt, 1999). Similarly, cell surface HSPGs have a half-life of 3–8 h (Stringer and Gallagher, 1997), indicating the efficiency with which these molecules can be broken down. Following endocytosis of the substrates into the cell, complete GAG disassembly proceeds by an ordered desulfation/exolytic cleavage to yield monosaccharide products. The consequences of defects in enzymes mediating intracellular GAG degradation are evidenced by the different pathologies of the mucopolysaccharidoses (Leroy and Wiesmann, 1993). Extracellular GAG degradation, although of equal importance, is an incompletely understood process. Extracellular endolytic GAG-degrading enzymes, the hyaluronidases (Frost et al., 1996), and heparanase (Vlodavsky et al., 1999) have been characterized, and they play major roles in normal and pathological turnover of the ECM, and specific inhibitors of these enzymes would have important therapeutic benefits.
Testicular hyaluronidase has been known for many years (Kreil, 1995). This membrane-bound hydrolase cleaves hyaluronate but can also degrade CS and plays an important role in fertilization. It has been shown that this GAG hydrolase is the prototype of a six-member human gene family. Five functional hyaluronidases and an expressed pseudogene have been characterized. Hyal-1, -2, and -3 occur as a cluster on chromosome 3 at position 3p21, while the well-characterized testicular hyaluronidase (also termed PH20) as well as hyal-4 and the pseudogene (hyalP1) are found on chromosome 7q31 (Csóka et al., 1999). While “hyaluronidase” has been generally considered to be a lysosomal enzyme, there is strong evidence for extracellular hyaluronidase activity. Various reports have associated elevated hyaluronidase levels with increased tumorogenicity (Madan et al., 1999a,b; Novak et al., 1999), and it has been demonstrated that hyal-2 is a cell surface receptor for a retrovirus in sheep (Miller, 2003).
A. Microbial CS-Degrading Enzymes (Polysaccharide Lyases)
In contrast to the vertebrates, microorganisms utilize an eliminative mechanism to breakdown GAGs, which involves abstraction of the proton at C-5 of the hexuronic acid by a general base and β-elimination of the 4-O-glycosidic bond with concomitant formation of an unsaturated C4–C5 bond within the hexuronic acid located at the nonreducing end (Fig. 2). The leaving group must be protonated, either by a side chain acting as a general acid or by proton abstraction from a water molecule. Proton abstraction and β-elimination are expected to proceed in a stepwise as opposed to concerted manner (Godavarti and Sasisekharan, 1998; Guthrie and Kluger, 1993). There is an extensive variation in specificity among lyases for different GAG types. Thus, chondroitinase B is specific for cleavage of DS, accepting only an iduronic acid, whereas chondroitinase ABC will accept either glucuronic acid or iduronic acid. Extensive biochemical and mutagenesis studies have been carried out on enzymes obtained from Flavobacterium heparinum (Pedobacter heparinus) that produces two chondroitinases (FlavoAC and FlavoB) (Gu et al., 1995) and on two general specificity chondroitinases from Proteus vulgaris (PvulABCI and PvulABCII) (Hamai et al., 1997). In addition, several hyaluronate lyases contributing to virulence have been characterized from different bacteria and bacteriophage (Hynes and Walton, 2000; Li et al., 2000; Rigden and Jedrzejas, 2003). Of particular interest is the genomic sequence of the commensal bacterium, Bacteroides thetaiotaomicron (Xu et al., 2003). B. thetaiotaomicron and B. stercoris (Ahn et al., 1998; Kim et al., 2000) are dominant members of the intestinal microbiota of humans and other mammals. Most notably the genome of B. thetaiotaomicron shows a markedly expanded repertoire of genes involved in polysaccharide uptake and degradation, specifically for utilizing a large variety of complex polysaccharides as a source of carbon and energy (Xu et al., 2003). Among these are several chondroitinases (BthetABC and BsterABC) and heparinases, which contribute to the nutrition of the host (Ahn et al., 1998; Kim et al., 2000; Xu et al., 2003).
While glycoside hydrolases display an extraordinary variety of folds (Bourne and Henrissat, 2001), only three folds have been identified for GAG lyases, the (α/α)5-toroid, the right-handed β-helix, and a β-sandwich. The intriguing observation of both eliminative and hydrolytic enzymes within the first two-fold families and similarity of the β-sandwich to the N-terminal domain of PvulABCI suggest that binding of linear uronic acid-containing polysaccharide substrates demands special structural features. Of the 15 classified lyase families and over 25 unclassified sequences, the fold has been established for 9 families encompassing several pectate/pectin lyase families, alginate, chondroitin, and rhamnogalacturonan lyases.
B. Purification and Characterization of Chondroitinases
Chondroitin AC and B lyases from Flavobacterium heparinum (FlavoAC and FlavoB) were first purified to homogeneity, and their physical and kinetic constants were determined in the Linhardt laboratory in 1995 (Gu et al., 1995). From the N-terminal sequences that we determined, these enzymes were subsequently cloned and expressed in E. coli (Pojasek et al., 2001). Chondroitinase ABC (BsterABC) was first isolated from B. stercoris by the Kim laboratory in 1998 (Ahn et al., 1998), and the B. thetaiotaomicron chondroitinase ABC (BthetABC) has been cloned and expressed in the Kim laboratory (in preparation). The BsterABC and BthetABC enzymes appear to be structurally and catalytically similar to one another. The P. vulgaris chondroitinase ABC (a mixture of PvulABCI and -II) is the only commercially available chondroitinase ABC preparation (Seikagaku, Tokyo, Japan). The endolytic chondroitinase ABC (PvulABCI) was cloned and expressed in E. coli (Prabhakar et al., 2005). Some of the well-characterized polysaccharide lyases acting on chondroitins are listed in Table I.
TABLE I.
Properties of Polysaccharide Lyases Acting on Chondroitins
| Namea | Substrates | Linkage specificityb | Action pattern | Mr (Da) | KMc,d | Vmaxc,e |
|---|---|---|---|---|---|---|
| Chondroitinase AC (Fh) (Gu et al., 1995) |
CS-A (4S) CS-C (6S) HA | →3)GalNAc(or GlcNAc) 4X,6X(1→4) GlcA(1→ |
Endo | 74,000 | 9.3 | 121 |
| Chondroitinase AC (Aa) (Linhardt, 1994; Lunin et al., 2004) |
CS-A (4S) CS-C (6S) HA | →3)GalNAc(or GlcNAc) 4X,6X(1→4) GlcA(1→ |
Exo | 79,840 | 0.01 | |
| Chondroitinase B (Fh) (Gu et al., 1995) |
DS | →3)GalNAc 4X,6X(1→4)IdoA2X(l→ | Endo | 55,200 | 7.4 | 209 |
| Chondroitinase ABC (Bs) (Hong et al., 2002) |
CS-A (4S) CS-C (6S) DS | →3)GalNAc 4X,6X(1→4)UA2X(1→ | Endo | 116,000 | 45.7 | |
| Chondroitinase ABC I (Pv) (Hamai et al., 1997) |
CS-A (4S) CS-C (6S) DS | →3)GalNAc 4X,6X(1→4)UA2X(1→ | Endo | 100,000 | 66 | 310 |
| Chondroitinase ABC II (Pv) (Hamai et al., 1997) |
CS-A (4S) CS-C (6S) DS | →3)GalNAc 4X,6X(1→4)UA2X(1→ | Exo | 105,000 | 80 | 34 |
| Hyaluronate lyase (Pa) (Ingham et al., 1979) |
HA CS-A (4S) CS-C (6S) | →3)GalNAc(or GlcNAc) 4X,6X(1→4) GlcA(1→ |
85,110 | 7.75 |
Fh: Flavobacterium heparinum; Aa: Arthrobacter aurescens; Pv: Proteus vulgaris; Bs: Bacteroides stercoris.
The primary sites of action are shown. X = SO3− or H, UA = glucuronic or iduronic acid.
Kinetic parameters are given for the primary substrates.
Apparent Km in μM.
Vmax in μmol min−1 mg−1.
C. Chondroitin Lyase Structures and Mechanism
The Cygler laboratory has determined the three-dimensional structures of representatives of chondroitinases B, AC, and ABC (Fig. 3).
FIGURE 3.
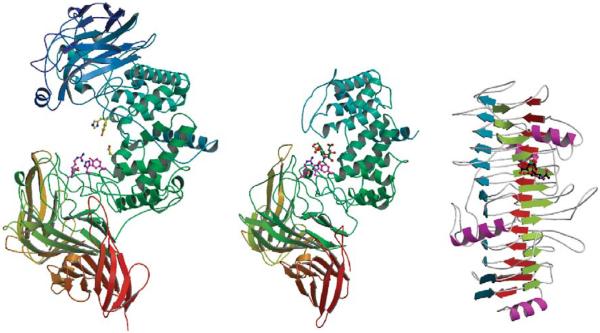
Comparison of crystal structures of chondroitinases PvulABCI (left), FlavoAC (center), and FlavoB (right).
1. Chondroitin AC Lyase
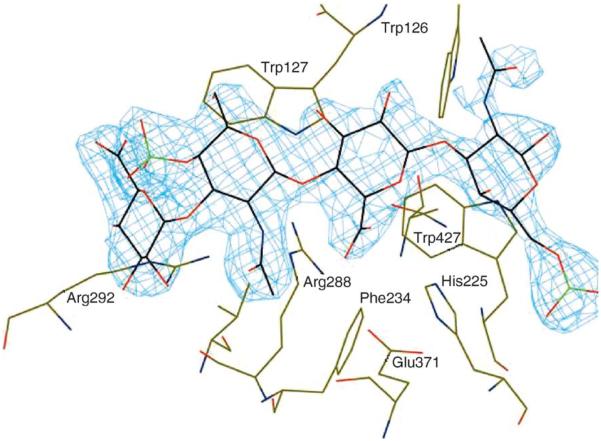
The three-dimensional structures and the enzymatic mechanisms of two chondroitin AC lyases from F. heparinum (FlavoAC) and Arthrobacter aurescens (ArthroAC) were investigated. Both FlavoAC and ArthroAC act on CS-A and CS-C as well as on hyaluronan (Linhardt, 1994). Neither enzyme acts on pure DS, containing only repeating units of →3)GalNpAc4S (1→4)IdoAp(1→, and both AC lyases are inhibited by this GAG (Gu et al., 1993). Studies in the Linhardt laboratory confirmed that both FlavoAC and ArthroAC act on the →3)GalNpAc(4S or 6S)(1→4)GlcAp(1→4) sequences found within CS and in many DS (Gu et al., 1993). FlavoAC is an endolytic, and ArthroAC is an exolytic chondroitin lyase (Jandik et al., 1994). The substrate specificities of both AC lyases have been extensively investigated on natural (Hernáiz and Linhardt, 2001; Linhardt, 1994; Yang et al., 2000) as well as unnatural (Avci et al., 2003) substrates. The structure of FlavoAC was determined at 1.9 Å resolution and revealed a two-domain molecule with the N-terminal α-helical domain and the C-terminal β-sheet domain (Fig. 3) (Féthière et al., 1999). The N-terminal domain is folded into an incomplete double-layered (α/α)5 toroid. This domain contains the catalytic machinery and provides a major part of the substrate-binding site. The C-terminal domain is composed of four antiparallel β-sheets. Since chondroitin AC lyase is inhibited by DS, we investigated complexes of FlavoAC with DS oligosaccharides. The tetrasaccharide binding site [subsites −2, −1, +1, +2 with the cleavage site between −1 and +1 using nomenclature according to Davies and coworkers (Davies et al., 1997)] and four putative catalytic residues—His225, Tyr234, Arg288, and Glu371—have also been identified (Huang et al., 2001). Expression of His225Ala, Tyr234Phe, and Arg288Ala mutants in F. heparinum, by integration of the DNA containing the mutated gene into the genomic DNA of the bacterium, rendered the enzyme inactive (Blain et al., 2002). Candidates for the general base, abstracting the glucuronic acid C-5 proton, were Tyr234 (transiently deprotonated during catalysis) or His225. The Tyr234 was deemed to be the best candidate to protonate the leaving group. Arg288 likely contributes to charge neutralization and stabilization of the enolate anion intermediate during catalysis.
Subsequently, the crystal structure of Tyr234Phe mutant with a CS-A tetrasaccharide was determined, confirming the general features of substrate binding, but this structure was inconclusive in the assignment of the role of general acid to either His225 or Tyr234 due to an enzymatically noncompetent conformation of the substrate (Fig. 4) (Huang et al., 2001).
FIGURE 4.
Chondroitin-4,6-sulfate tetrasaccharide in the active site of FlavoAC Tyr234Phe mutant.
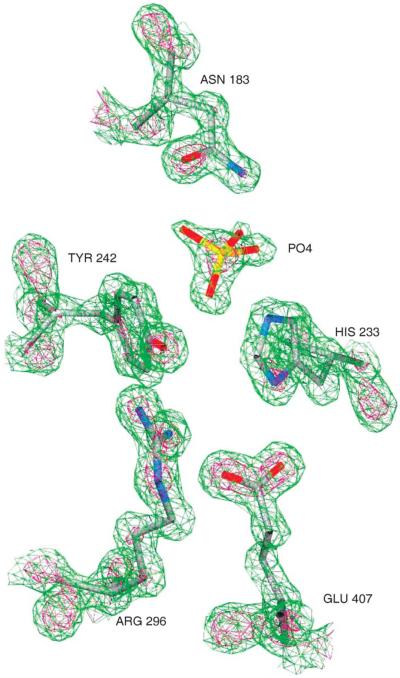
A breakthrough that allowed the assignment of the catalytic general base came from the investigation of chondroitin AC lyase from A. aurescens (ArthroAC). Although the amino acid sequence of this protein was not known at the onset of our investigations, it was likely that it shared homology with FlavoAC. We were fortunate in obtaining crystals diffracting to near atomic (1.25 Å) resolution (Lunin et al., 2004). The resulting electron density maps allowed us to determine the amino acid sequence (Fig. 5), which was confirmed subsequently by mass spectrometry (MS) of tryptic peptides. This sequence showed 24% identity to FlavoAC. Using a series of short soaks of the crystals with CS-A tetrasaccharide, their immediate freezing in liquid nitrogen, and data collection at the synchrotron, we showed that the enzyme acted slowly in the crystal, allowing us to capture the enzymatically active conformation. This data resolved that Tyr242 acts as the general base that abstracts the proton and His233 helps in deprotonation of Tyr242 and in the proper orientation of the glucuronate acidic group. The glucuronate assumes a distorted boat conformation, much like that observed in lysozyme (Fig. 6).
FIGURE 5.
Experimental electron density map of the active site region of ArthroAC. Green contours are drawn at 3σ level, red contours at 5σ level. In the native structure, there is a phosphate ion in the active site. Nitrogen atoms are blue, oxygens are red, carbons are gray, and phosphorus is yellow.
FIGURE 6.
Conformation of the chondroitin-4-sulfate tetrasaccharide substrate bound in the active site of ArthroAC. Omit electron density map is drawn at the 3σ level.
Chondroitin O-methyl ester (C-OMe) is also depolymerized by chondroitin AC lyase from F. heparinum (Avci et al., 2003). The major product isolated from the depolymerization reaction was found to be chondroitin Δdi-O-methyl ester (Fig. 7). Although, in chemical terms, abstraction of an acidic proton at the αa-position of methyl ester group is expected, the esterification of the carboxylate group might alter the interaction of anion-stabilizing elements in an enzymatic reaction, adversely impacting catalysis (Fig. 2). Kinetic studies show that the KM on C-OMe (12.0 mM) is comparable to CS-A (7.0 mM) and lower than that observed on chondroitin (63.0 mM). In contrast, the Vmax on C-OMe (0.3 mmol min−1 mg−1) is significantly lower than on CS-A (2.0 mmol min−1 mg−1) or chondroitin (3.3 mmol min−1 mg−1) suggesting that the binding step is less adversely impacted than catalytic step by methylation of the carboxyl group. The low KM observed for C-OMe (comparable to CS-A) might be ascribed to the contribution of hydrophobic interactions between the methyl ester and the enzyme, replacing ionic interactions lost through the desulfonation and methyl esterification of the substrate. Both chondroitinase AC I from F. heparinum and chondroitinase AC II from A. aurescens were demonstrated by the Toida laboratory to be capable of depolymerizing hyaluronan O-methyl ester (Hirano et al., 2005).
FIGURE 7.
Enzymatic depolymerization of chondroitin O-methyl ester.
2. Chondroitin B Lyase
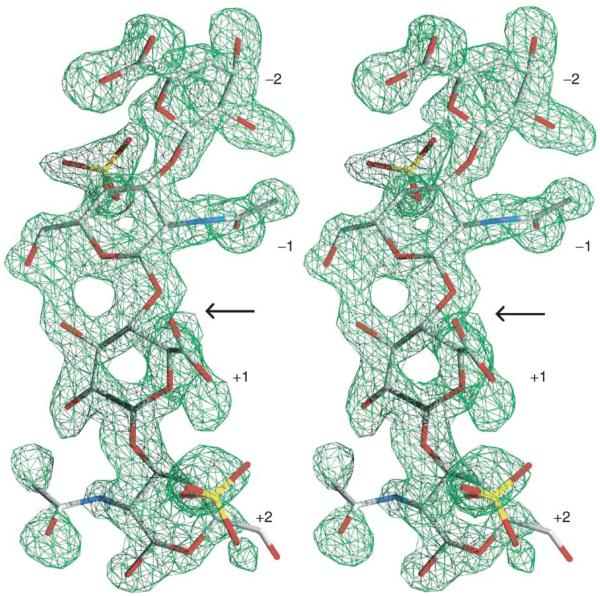
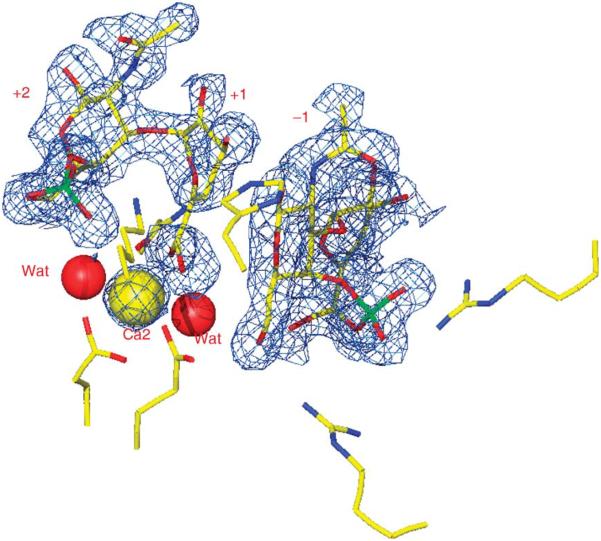
The Cygler laboratory has determined the structure of native F. heparinum chondroitin B lyase (FlavoB) and its complex with the reaction disaccharide product (Huang et al., 1999). The fold of this lyase is completely different to that of FlavoAC and belongs to the β-helix family with 13 coils. The soaking of chondroitin B lyase crystals with a DS tetrasaccharide resulted in a complex with two DS disaccharide reaction products occupying (−2, −1) and (+1, +2) subsites in the substrate-binding site. Unexpectedly, this structure showed the presence of a Ca2+ ion coordinated by conserved acidic residues and by the carboxyl group of the L-iduronic acid at the +1 subsite (Fig. 8).
FIGURE 8.
Three disaccharide products bound in the active site of chondroitinase B. Bound calcium atom is shown as a yellow sphere, two water molecules as red spheres.
Chondroitin B lyase was subsequently shown to absolutely require calcium for its activity, indicating that the protein-Ca2+-oligosaccharide complex is functionally relevant. We proposed that the Ca2+ ion neutralizes the carboxyl moiety of the L-iduronic acid at the cleavage site, while the conserved Lys250 and Arg271 act as general base and acid, respectively. Model building showed that a DS substrate would bind in a bent conformation and that this sugar ring adopts a distorted conformation. The requirement of Ca2+ for catalysis was further investigated in collaboration with Dr Sasisekharan by measuring Kcat and KM as a function of calcium concentration (Michel et al., 2004).
3. Chondroitin ABC I (endo) Lyase
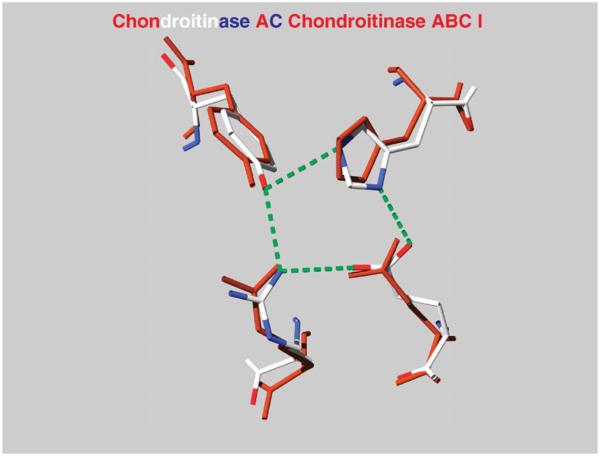
The Cygler laboratory crystallized (Huang et al., 2000) and determined the structure (Huang et al., 2003) of chondroitin ABC I (endo) lyase from P. vulgaris (PvulABCI). This 110 kD protein consists of three domains. The amino acid sequence comparison indicated only that the C-terminal domain is homologous to the C-terminal, noncatalytic domain of FlavoAC, and this was confirmed by the structure. The N-terminal domain has a similar fold to carbohydrate-binding domains of xylanases and some lectins while the middle domain showed, unexpectedly, structural similarity to the catalytic domain of FlavoAC and to hyaluronan lyases. The superposition of these two domains showed the conservation of residues forming the active site tetrad (Fig. 9).
FIGURE 9.
The superposition of the active-site tetrad of FlavoAC and PvulABCI. The Asn175 of FlavoAC and Arg500 of PvulABCI are also shown.
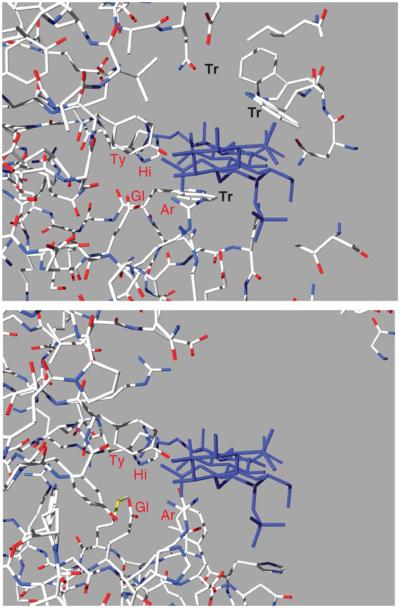
The Asn175 of FlavoAC, which plays an essential role in binding the acidic group of glucuronic acid, is not conserved in PvulABCI. Instead, the side chain of Arg500 may perform this function. We speculated that the charged guanidinium group at the end of a long arm provides the flexibility essential for adapting the enzyme’s catalytic machinery to two possible configurations of the acidic group at the C-5 position of the uronic acid ring. The substrate binding area in this structure is wide open, and we have not yet been able to obtain complexes with oligosaccharides (Fig. 10). It has not yet been possible to unequivocally deduce from this structure residues that contribute to substrate binding and the key protein–substrate interactions. Further efforts toward obtaining PvulABCI complexes with substrate or inhibitors are essential for the understanding of the mechanistic properties of these enzymes and their ability to break down CS and DS.
FIGURE 10.
The disposition of the substrate in FlavoAC (left) was transferred to PvulABCI (right) based on the superposition of the active site tetrad. In this open form of PvulABCI are very few contacts between the enzyme and its substrate.
While the PvulABCI is an endolytic lyase, this bacterium also produces a closely homologous enzyme chondroitin ABC II (exo) lyase (PvulABCII) with similar spectrum of substrate specificities while being an exolytic lyase. We have obtained crystals of this protein and would like to determine its structure to understand the mechanism underlying exolytic vs. endolytic selectivity and to detail the catalytic mechanism.
IV. Analytical Applications
Determination of CS/DS oligosaccharide structure is a formidable analytical problem that has limited structure–activity relationship studies, and the development of improved methods is necessary for further progress. Current approaches involve the preparation of CS/DS oligosaccharides using chondroitin lyases followed by separation techniques including gel permeation chromatography (GPC) (Yang et al., 2000), strong anion exhange-high performance liquid chromatography (SAX-HPLC) (Linhardt et al., 1994; Yang et al., 2000), polyacrylamide gel electrophoresis (PAGE) (Linhardt et al., 1991, 1994), and capillary electrophoresis (CE) (Al-Hakim and Linhardt, 1991; Pervin et al., 1993, 1994). These provide important data on composition and domain structure but generally yield indirect and incomplete sequence information. MS has also been applied to the analysis of CS/DS oligosaccharides (Kitagawa et al., 1997; Lamb et al., 1992; Yang et al., 2000). Fast-atom bombardment (FAB-MS), electrospray ionization (ESI-MS), and matrix-assisted laser desorption ionization (MALDI-MS) are capable of determining the molecular weight of oligosaccharides. Nuclear magnetic resonance (NMR) spectroscopy provides for the accurate determination of the chemical fine structure of small CS/DS oligosaccharides (containing 2–14 saccharide units) (Linhardt et al., 1992; Yang et al., 2000).
A. Oligosaccharide Structure Analysis
GAG lyases are used for the structural analysis of GAGs. In a general oligosaccharide structure analysis procedure first, a small quantity of the polysaccharide is exhaustively depolymerized in the reaction using the proper lyase enzyme (Table II) (Linhardt, 1994). Next, a controlled, partial depolymerization is performed to obtain the UV absorbance value at 232 nm (the nonreducing end uronic acid residue absorbs at 232 nm) corresponding to the maximum number of oligosaccharides that can be detected. The partial depolymerization reaction is next scaled up, and the resulting oligosaccharide mixture is next size fractionated by GPC. This separation affords size-uniform oligosaccharide mixtures corresponding to oligosaccharides ranging in size from disaccharide to octadecasaccharide. PAGE analysis of these size-fractionated oligosaccharides demonstrates the degree of size separation. The size fractions are next purified by semipreparative SAX-HPLC, desalted and lyophilized. Analytical SAX-HPLC is used to assess whether a second semipreparative SAX-HPLC separation step is necessary. Finally, mass spectral analysis and 1D/2D NMR analysis are acquired for their structural elucidation.
TABLE II.
Reaction Conditions for Polysaccharide Lyases Acting on Chondroitins with Optimum Buffers and Reaction Conditions
| Lyase | Buffer | Optimum temperature (°C) |
|---|---|---|
| Chondroitinase ABC | Tris.Cl/sodium acetate, pH 8 | 37 |
| Chondroitinase AC | Tris.Cl/sodium acetate, pH 8 | 37 |
| Chondroitinase B | Ethylenediamine/acetic acid/NaCl, pH 8 | 25 |
| Hyaluronate lyase | Sodium acetate/NaCl, pH 5.2 | 30 |
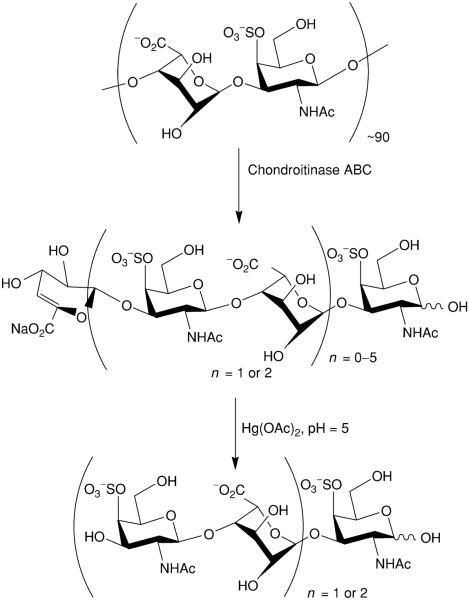
Using chondroitin ABC lyase, we prepared eight oligosaccharides from DS and elucidated their structures (Fig. 11) (Yang et al., 2000). Treatment of tetrasaccharide and hexasaccharide fragments with mercuric acetate afforded trisaccharide and pentasaccharide products, respectively. The purity of the oligosaccharides obtained was confirmed by analytical SAX-HPLC and CE. The molecular mass and degree of sulfation of the eight purified oligosaccharides were elucidated using ESI-MS, and their structures were established with NMR. These DS oligosaccharides are being used to study interaction of the DS with biologically important proteins.
FIGURE 11.
Controlled enzymatic depolymerization of DS by chondroitinase ABC and mercuric acetate treatment to remove the unsaturated nonreducing end residue.
B. Oligosaccharide Mapping
Quantitative saccharide compositional analysis of GAGs can be conveniently performed using high resolution, high sensitivity CE. CS and DS can be depolymerized using chondroitin lyases (or hyaluronate lyases) into eight to nine different disaccharides that can be easily resolved by CE (Al-Hakim and Linhardt, 1991). Sensitive (picomole range) UV detection is possible due to the presence of an unsaturated residue formed in each disaccharide through the eliminase action of the polysaccharide lyases. Oligosaccharide mapping of GAGs is comparable in principle to the peptide mapping of protein and has been widely used for the comparison of GAGs from different tissues or species (Linhardt et al., 1991, 1992; Loganathan et al., 1990). A GAG is first treated, either partially or completely, with various depolymerizing chemicals or enzymes, and PAGE maps are prepared using PAGE and SAX-HPLC (Linhardt et al., 1988). Using this approach we have studied chondroitinase ABC depolymerized mixtures of DS sample obtained from different species and tissue origins. High molecular weight and low-molecular weight DS as well as charge-fractionated DS, having substantially different heparin cofactor-II (HC-II) mediated antithrombin activities were examined. Gradient PAGE was used to study the DS oligosaccharide mixtures prepared by chondroitinase ABC treatment. Commercially available disaccharide standards have been examined by SAX-HPLC, and their retention times compared with the oligosaccharide mixture obtained from chondroitinase ABC treated DS. On the basis of these studies, certain structural features associated with DS have been established (Linhardt et al., 1988).
C. Disaccharide Analysis
CS is widely used as a neutraceutical and pharmaceutical raw materials. As the number of products containing CS increases, stricter and more accurate evaluation should be required for the manufacture of high-quality products. Disaccharide analysis using HPLC should be useful for evaluation of the quality of CS as a pharmaceutical and nutraceutical ingredient. The pretreatment method followed by enzymatic digestion makes it possible to quantify CS content in soft capsules and liquid preparations and should be applicable for the quality control of CS. The present methods can be applied to confirm the purity and label claim of CS in raw materials, pharmaceuticals, and neutraceuticals.
The Kim laboratory performed the quantitative analysis of CS obtained from raw materials and various pharmaceutical preparations (Sim et al., 2005). To quantify CS content in raw materials and in an ophthalmic solution, each test sample and the authentic CS were first digested by chondroitinase ABC. The CS disaccharides produced were analyzed by HPLC, and CS content was quantified by calculating the total peak areas of the disaccharides derived from a CS calibration curve. In the case of soft capsules, CS was first extracted with hexane followed by phenol-chloroform to remove oil and protein ingredients. The extracted CS samples were depolymerized by chondroitinase ABC, and CS content was determined. Quantitative analysis of the disaccharides derived from raw materials and an ophthalmic solution showed the CS contents (%) were 39.5–105.6 and 103.3, respectively. In case of CS analysis in soft capsules and liquid preparations, the overall recovery (%) of the spiked CS was 96.79–103.54 and 97.10–103.17, respectively (Table III). In conclusion, the quantitative analysis of the disaccharides produced by enzymatic digestion can be used in the direct quantitation of CS containing pharmaceutical formulations.
TABLE III.
Quantitation of CS from Pharmaceuticalsa
| Sample | Labeled amount (mg) |
Determined amount (mg) |
Label claim (%) |
|---|---|---|---|
| Ophthalmic solution | 2 | 2.07 | 103.33 |
| Liquid preparation | 30 | 29.25 | 97.48 |
| Soft capsule | 120 | 114.06 | 95.05 |
The CS content was quantified by calculating the total peak areas of the disaccharides derived from a CS calibration curve.
V. Synthetic Applications
A. Oligosaccharide Preparation
Polysaccharide lyases have been used to produce Δ4-uronate disaccharides and higher oligosaccharides from heparin, HS, CS, DS, hyaluronan, and chemically modified GAGs (Linhardt and Al-Hakim, 1991; Pervin et al., 1995; Weiler et al., 1992). Because both the polysaccharide substrates and enzymes are relatively inexpensive, these oligosaccharides can be prepared in large quantities at a low cost. The discovery of new GAGs, such as acharan sulfate (Kim et al., 1996) as well as the mild acid hydrolyzate E. coli polysaccharide K5 (Razi et al., 1995), can afford novel structures in large quantities and at low cost.
B. Chemoenzymatic Synthesis
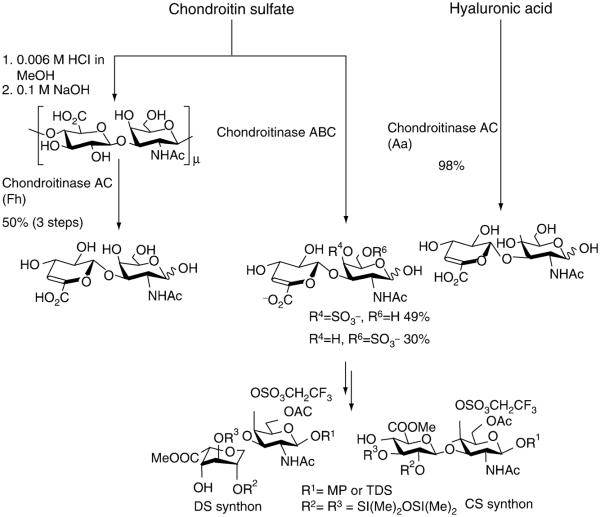
We proposed that the lyase-derived oligosaccharides could be chemically linked together to form larger oligosaccharides with the requisite structure for a wide variety of biological activities. The first objective would be to differentially protect enzymatically prepared desulfated disaccharides and to use these neutral disaccharides to prepare larger target oligosaccharides (Fig. 12) (Islam et al., 2003). The advantages of this approach are (1) disaccharides can be assembled into oligosaccharides with a reduced number of glycosylation reactions and (2) a high level of structural complexity (i.e., stereochemistry, sulfation pattern) is already built into these disaccharides. In addition, we investigated the use of 2,2,2-trifluorodiazoethane as a reagent for sulfo group protection in enzymatically prepared CS disaccharides (Fig. 12) (Avci et al., 2004). This approach was first used for sulfate ester protection in carbohydrates by Flitsch and coworkers (Proud et al., 1997). Once the sulfo groups have been protected, the free hydroxyl and carboxyl groups could be protected in organic solvents used in standard carbohydrate synthesis. This chemistry has been successfully used to selectively protect primary and secondary O- and N- sulfo groups in unprotected sulfated mono- and disaccharides in high yields (Karst et al., 2003, 2004).
FIGURE 12.
Preparation of desulfated and sulfoprotected disaccharide starting materials for the synthesis of CS/DS/HA oligosaccharides.
VI. Therapeutic Applications
Bacterial GAG-degrading enzymes also have direct medical applications. Heparinase is an important reagent that can be used to remove anticoagulant heparin from blood in the prevention of excessive bleeding following coronary artery bypass surgery (Langer et al., 1982). In vitro experiments have revealed that chondroitinases inhibit melanoma invasion, proliferation, and angiogenesis (Denholm et al., 2001). Subretinal injection of chondroitinase ABC (PvulABCI and PvulABCII) promotes retinal reattachment in rabbits (Yao et al., 1992). Chondroitinase ABC (PvulABCI and -II) has also been applied therapeutically to treat invertebral disc protrusion (Kato et al., 1990).
It has been shown that chondroitinase ABC (PvulABCI and -II) promotes the regeneration of central nervous system axons (Bradbury et al., 2002; Pizzorusso et al., 2002). Permanent paralysis can result in adult mammals following spinal cord injuries due to the inability of axons to regenerate (Fawcett and Asher, 1999). A glial scar develops at the site of the central nervous system injury (Fitch and Silver, 1999). This scar is composed of ECM molecules and is particularly rich in CSPGs (Fawcett and Asher, 1999; Fitch and Silver, 1999). In vitro CSPGs inhibit axonal growth (McKeon et al., 1991; Niederost et al., 1999; Smith-Thomas et al., 1994), and in vivo regions rich in CSPGs stop regenerating axons (Davies et al., 1999). Chondroitinase ABC catalyzed removal of CS chains in vitro can reverse this inhibitory activity (Fidler et al., 1999; McKeon et al., 1995; Moon et al., 2001; Zuo et al., 1998). In a recent in vivo study in an adult rat spinal cord injury model, the delivery of chondroitinase ABC directly to the injury site promoted functional recovery (Bradbury et al., 2002). In this study, chondroitinase ABC was delivered intrathecally to lessoned dorsal columns of adult rats. This treatment upregulated regeneration-associated protein in injured neurons, promoting regeneration of both ascended sensory projections and descending corticospinal tract axons. This treatment restored postsynaptic activity below the lesion after electrical stimulation of corticospinal neurons. Chondroitinase ABC also promoted functional recovery of locomotor and proprioceptive behaviors.
VII. Conclusions
Modern analytical methods, including NMR and MS, are widely used for the determination of CS structure. While modern spectroscopic techniques provide limited information on the structure of the intact CS polysaccharide, it is often useful to utilize chondroitin lyases to prepare oligosaccharides for more detailed structural determination. The structure, activity, and specificity of these enzymes were the focus of this chapter. These lyases can be combined with separation methods, such as chromatography and electrophoresis, for the preparation of CS oligosaccharides for biological evaluations as well as for disaccharide analysis, oligosaccharide mapping, and polysaccharide sequencing. These enzymes have also been shown to have direct therapeutic value. This chapter examined the various applications for this important class of enzymes.
References
- Ahn M, Shin K, Kim D-H, Jang E-A, Toida T, Linhardt R, Kim Y. Characterization of a bacteroides species from human intestine that degrades glycosaminoglycans. Can. J. Microbiol. 1998;44:423–429. doi: 10.1139/cjm-44-5-423. [DOI] [PubMed] [Google Scholar]
- Al-Hakim A, Linhardt RJ. Capilary electrophoresis for the analysis of chondroitin sulfate and dermatan sufate derived disaccharides. Anal. Biochem. 1991;195:68–73. doi: 10.1016/0003-2697(91)90296-6. [DOI] [PubMed] [Google Scholar]
- Avci FY, Toida T, Linhardt RJ. Chondroitin O-methyl ester: An unusual substrate for chondroitin AC lyase. Carbohydr. Res. 2003;338:2101–2104. doi: 10.1016/s0008-6215(03)00348-3. [DOI] [PubMed] [Google Scholar]
- Avci FY, Karst N, Islam T, Linhardt RJ. Trifluoroethylsulfonate protected monosaccharides in glycosylation reactions; 228th ACS National Meeting; Philadelphia. 2004. [Google Scholar]
- Blain F, Tkalec AL, Shao Z, Poulin C, Pedneault M, Gu K, Eggimann B, Zimmermann J, Su H. Expression system for high levels of GAG lyase gene expression and study of the hepA upstream region in Flavobacterium heparinum. J. Bacteriol. 2002;184:3242–3252. doi: 10.1128/JB.184.12.3242-3252.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneu B. Glycosaminoglycans: Clinical use. Sem. Thromb. Hemost. 1996;22:209–212. doi: 10.1055/s-2007-999010. [DOI] [PubMed] [Google Scholar]
- Bourne Y, Henrissat B. Glycoside hydrolases and glycosyltransferases: Families and functional modules. Curr. Opin. Struct. Biol. 2001;11:593–600. doi: 10.1016/s0959-440x(00)00253-0. [DOI] [PubMed] [Google Scholar]
- Bradbury EJ, Moon LD, Popat RJ, King VR, Bennet GS, Patel PN, Fawcett JW, McMahon SB. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature. 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- Capila I, Linhardt RJ. Heparin protein interactions. Angew. Chemie Int. Ed. 2002;41:390–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Csóka AB, Scherer SW, Stern R. Expression analysis of six paralogous human hyaluronidase genes clustered on chromosomes 3p21 and 7q31. Genomics. 1999;60:356–361. doi: 10.1006/geno.1999.5876. [DOI] [PubMed] [Google Scholar]
- Davies GJ, Wilson KS, Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SJ, Gaucher DR, Doller C, Silver J. Robust regeneration of adult sensory axons in degenerating white matter of the adult rat spinal cord. J. Neurosis. 1999;19:5810–5822. doi: 10.1523/JNEUROSCI.19-14-05810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis PL. Hyaluronan synthases: Fascinating glycosyltransferases from vertabrates, bactrerial pathogens and algal viruses. Cell Mol. Life Sci. 1999;56:670–682. doi: 10.1007/s000180050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denholm EM, Lin YQ, Silver PJ. Anti-tumor activities of chondroitinase AC and chondroitinase B: Inhibition of angiogenesis, proliferation and invasion. Eur. J. Pharmacol. 2001;416:213–221. doi: 10.1016/s0014-2999(01)00884-6. [DOI] [PubMed] [Google Scholar]
- Ernst S, Langer R, Cooney CL, Sasisekharan R. Enzymatic degradation of glycosaminoglycans. Crit. Rev. Biochem. Mol. Biol. 1995;30:44–45. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Fawcett JW, Asher RA. The glial scar and central nervous system repair. Brain Res. Bull. 1999;49:377–391. doi: 10.1016/s0361-9230(99)00072-6. [DOI] [PubMed] [Google Scholar]
- Féthière J, Eggimann B, Cygler M. Crystal structure of chondroitin AC lyase, a representative of a family of glycosaminoglycan degrading enzymes. J. Mol. Biol. 1999;288:635–647. doi: 10.1006/jmbi.1999.2698. [DOI] [PubMed] [Google Scholar]
- Fidler PS, Schuette K, Asher RA, Dobbertin A, Thornton SR, Calle-Patino Y, Muir E, Levin JM, Geller HM, Rogers J, Faissner A, Fawcett JW. Comparing astrocytic cell lines that are inhibitory or permissive for axon growth: The major axon-inhibitory proteoglycans is NG2. J. Neurosis. 1999;19:8778–8788. doi: 10.1523/JNEUROSCI.19-20-08778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch MT, Silver J. CNS regeneration. In: Tusynski MH, Kordower JH, editors. Basic Science and Clinical Advances. Academic Press; San Diego: 1999. pp. 55–58. [Google Scholar]
- Frost GI, Csoka T, Stern R. The hyaluronidases: A chemical, biological and clinical overview. Trends Glycosci. Glycotechnol. 1996;44:419–434. [Google Scholar]
- Godavarti R, Sasisekharan R. Heparinase I from Flavobacterium heparinum: Role of positive charge in enzymatic activity. J. Biol. Chem. 1998;273:248–255. doi: 10.1074/jbc.273.1.248. [DOI] [PubMed] [Google Scholar]
- Gu K, Liu J, Pervin A, Linhardt RJ. Comparison of the activity of two chondroitin AC lyases on dermatan sulfate. Carbohydr. Res. 1993;244:369–377. doi: 10.1016/0008-6215(83)85014-9. [DOI] [PubMed] [Google Scholar]
- Gu K, Linhardt RJ, Laliberte M, Gu K, Zimmermann J. Purification, characterization and specificity of chondroitin lyases and glycuronidase from Flavobacterium heparinum. Biochem. J. 1995;312:569–577. doi: 10.1042/bj3120569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie JP, Kluger R. Electrostatic stabilization can explain the unexpected acidity of carbon acids in enzyme-catalyzed reactions. J. Am. Chem. Soc. 1993;115:11569–11572. [Google Scholar]
- Hamai A, Hashimoto N, Mochizuki H, Kato F, Makiguchi Y, Horie K, Suzuki S. Two distinct chondroitin sulfate ABC lyases: An endoeliminase yielding tetrasaccharides and an exoeliminase preferentially acting on oligosaccharides. J. Biol. Chem. 1997;272:9123–9130. doi: 10.1074/jbc.272.14.9123. [DOI] [PubMed] [Google Scholar]
- Hannesson HH, Hagner-McWhirter A, Tiedemann K, Lindahl U, Malmstrom A. Biosynthesis of dermatan sulfate: Defructosylated Escherichia coli K4 capsular polysaccharide as a substrate for the D-glucuronyl C-5 epimerase, and an indication of a two-base reaction mechanism. Biochem. J. 1996;313:589–596. doi: 10.1042/bj3130589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernáiz MJ, Linhardt RJ. Degradation of chondroitin and dermatan sulfate with chondroitin lyases. In: Iozzo RV, editor. Methods in Molecular Biology. Humane Press; New York: 2001. [DOI] [PubMed] [Google Scholar]
- Hirano K, Sakai S, Ishikawa T, Avci FY, Linhardt RJ, Toida T. Preparation of hyaluronan Omethylester and its enzymatic degradation. Carbohydr. Res. 2005;338:2101–2104. doi: 10.1016/j.carres.2005.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-W, Kim B-T, Shin H-Y, Kim W-S, Lee K-S, Kim Y-S, Kim D-H. Purification and characterization of novel chondroitin ABC and AC lyases from Bacteroides stercoris HJ-15, a human intestinal anaerobic bacterium. Eur. J. Biochem. 2002;269:2934–2940. doi: 10.1046/j.1432-1033.2002.02967.x. [DOI] [PubMed] [Google Scholar]
- Hoshi H, Nakagawa H, Nishiguchi S, Iwata K, Niikura K, Monde K, Nishimura S. An engineered hyaluronan synthase 2 expressed in Escherichia coli. J. Biol. Chem. 2004;279:2341–2349. doi: 10.1074/jbc.M305723200. [DOI] [PubMed] [Google Scholar]
- Huang W, Matte A, Li Y, Kim YS, Linhardt RJ, Su H, Cygler M. Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccharide product at 1.7 Å resolution. J. Mol. Biol. 1999;294:1257–1269. doi: 10.1006/jmbi.1999.3292. [DOI] [PubMed] [Google Scholar]
- Huang W, Matte A, Suzuki S, Sugiura N, Miyazono H, Cygler M. Crystallization and preliminary x-ray analysis of chondroitin sulfate ABC lyases I and II from Proteus vulgaris. Acta Crystallogr. 2000;D56:904–906. doi: 10.1107/s0907444900005102. [DOI] [PubMed] [Google Scholar]
- Huang W, Boju L, Tkalec L, Su H, Yang HO, Gunay NS, Linhardt RJ, Kim YS, Matte A, Cygler M. Active site of chondroitin AC lyase revealed by the structure of enzyme-oligosaccharide complexes and mutagenesis. Biochemistry. 2001;40:2359–2372. doi: 10.1021/bi0024254. [DOI] [PubMed] [Google Scholar]
- Huang W, Lunin VV, Li Y, Suzuki S, Sugiura N, Miyazono H, Cygler M. Crystal structure of Proteus vulgaris chondroitin sulfate ABC lyase I at 1.9 Å resolution. J. Mol. Biol. 2003;328:623–634. doi: 10.1016/s0022-2836(03)00345-0. [DOI] [PubMed] [Google Scholar]
- Hynes WL, Walton SL. Hyaluronidases of Gram-positive bacteria. FEMS Microbial. Lett. 2000;183:201–207. doi: 10.1111/j.1574-6968.2000.tb08958.x. [DOI] [PubMed] [Google Scholar]
- Ingham E, Holland KT, Gowland G, Cunliffe WJ. Purification and partial characterization of hyaluronate lyase (EC 4.2.2.1) from Propionibacterium acnes. J. Gen. Microbiol. 1979;115:411–418. doi: 10.1099/00221287-115-2-411. [DOI] [PubMed] [Google Scholar]
- Iozzo RV. Matrix proteoglycans: From molecular design to cellular function. Annu. Rev. Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- Islam T, Avci FY, Karst N, Zhang J, Linhardt RJ. Disaccharide approach for the synthesis of glycosaminoglycan oligosaccharides; 226th ACS National Meeting; New York. 2003. [Google Scholar]
- Itano N, Kimata K. Mammalian hyaluronan synthases. IUMB Life. 2002;54:195–199. doi: 10.1080/15216540214929. [DOI] [PubMed] [Google Scholar]
- Jackson RL, Busch SJ, Cardin AD. Glycosaminoglycans: Molecular properties, protein interactions, and role in physiological processes. Physiol. Rev. 1991;71:481–539. doi: 10.1152/physrev.1991.71.2.481. [DOI] [PubMed] [Google Scholar]
- Jandik KA, Gu K, Linhardt RJ. Action pattern of polysaccharide lyases on glycosaminoglycans. Glycobiology. 1994;4:289–296. doi: 10.1093/glycob/4.3.289. [DOI] [PubMed] [Google Scholar]
- Karst NA, Islam TF, Linhardt RJ. Sulfo-protected hexosamine monosaccharides: Potentially versatile building blocks for glycosaminoglycan synthesis. Org. Lett. 2003;5:4839–4842. doi: 10.1021/ol035882w. [DOI] [PubMed] [Google Scholar]
- Karst NA, Islam TF, Avci FY, Linhardt RJ. Trifluoroethylsulfonate protected monosaccharides in glycosylation reactions. Tetrahedron Lett. 2004;45:6433–6437. [Google Scholar]
- Kato F, Iwata H, Hiatus K, Miura T. Experimental chemonucleolysis with chondroitinase ABC. Clin. Ortho. 1990;253:301–308. [PubMed] [Google Scholar]
- Kempson GE. The mechanical properties of articular cartilage. In: Sokoloff L, editor. The Joints and Synovial Fluid. Vol. 2. Academic Press; New York: 1980. pp. 177–238. [Google Scholar]
- Kim B-T, Kim W-S, Kim YS, Linhardt RJ, Kim D-H. Purification and characterization of a novel heparinase from Bacteroides stercoris HJ-15. J. Biochem. Tokyo. 2000;128:323–328. doi: 10.1093/oxfordjournals.jbchem.a022756. [DOI] [PubMed] [Google Scholar]
- Kim YS, Jo YT, Chang IM, Toida T, Park Y, Linhardt RJ. A new glycosaminoglycan from the giant african snail, Achatina fulica. J. Biol. Chem. 1996;271:11750–11755. doi: 10.1074/jbc.271.20.11750. [DOI] [PubMed] [Google Scholar]
- Kim B-T, Kim W-S, Kim YS, Linhardt RJ, Kim D-H. Purification and characterization of a novel heparinase from Bacteroides stercoris HJ-15. J. Biochem. Tokyo. 2000;128:323–328. doi: 10.1093/oxfordjournals.jbchem.a022756. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Tanaka Y, Yamada S, Seno N, Haslam SM, Morris HR, Dell A, Sugahara K. A novel pentasaccharide sequence GlcA(3-sulfate)(beta1-3)GalNAc (4-sulfate)(beta1-4)(Fuc alpha1-3)GlcA(beta1-3)GalNAc(4-sulfate) in the oligosaccharides isolated from king crab cartilage chondroitin sulfate K and its differential susceptibility to chondroitinases and hyaluronidase. Biochemistry. 1997;36:3998–4008. doi: 10.1021/bi962740j. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Uyama T, Sugahara K. Molecular cloning and expression of a human chondroitin synthase. J. Biol. Chem. 2001;276:38721–38726. doi: 10.1074/jbc.M106871200. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Izumikawa T, Uyama T, Sugahara K. Molecular cloning of a chondroitin polymerizing factor that cooperates with chondroitin synthase for chondroitin polymerization. J. Biol. Chem. 2003;278:23666–23671. doi: 10.1074/jbc.M302493200. [DOI] [PubMed] [Google Scholar]
- Kreil G. Hyaluronidases: A group of neglected enzymes. Protein Sci. 1995;4:1666–1669. doi: 10.1002/pro.5560040902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresse H, Schonherr E. Proteoglycans of the extracellular matrix and growth control. J. Cell Physiol. 2001;189:266–274. doi: 10.1002/jcp.10030. [DOI] [PubMed] [Google Scholar]
- Lamb DJ, Wang HM, Mallis LM, Linhardt RJ. Negative-ion fast atom bombardment tandem mass spectrometry to determine sulfate and linkage position in glycosaminoglycan-derived disaccharides. J. Am. Soc. Mass Spectrom. 1992;3:797–803. doi: 10.1016/1044-0305(92)80002-3. [DOI] [PubMed] [Google Scholar]
- Langer R, Linhardt RJ, Hoffberg S, Larsen AK, Cooney CL, Topper D, Klein M. An enzymatic system for removing heparin in extracorporeal therapy. Science. 1982;217:261–263. doi: 10.1126/science.7089564. [DOI] [PubMed] [Google Scholar]
- Leroy JG, Wiesmann U. Disorders of lysosomal enzymes. In: Royce P, Steinmann B, editors. Connective Tissue and its Heritable Disorders: Molecular, Genetic, and Medical Aspects. Wiley; New York: 1993. pp. 613–639. [Google Scholar]
- Li JP, Hagner-McWhirter A, Kjellen L, Palgi J, Jalkanen M, Lindahl U. Biosynthesis of heparin/heparan sulfate: cDNA cloning and expression of D-glucuronyl C5-epimerase from bovine lung. J. Biol. Chem. 1997;272:28158–28163. doi: 10.1074/jbc.272.44.28158. [DOI] [PubMed] [Google Scholar]
- Li S, Kelly SJ, Lamani E, Ferraroni M, Jedrzejas MJ. Structural basis of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. EMBO J. 2000;19:1228–1240. doi: 10.1093/emboj/19.6.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhardt RJ, Galliher P, Cooney C. Plysaccharide lyases. Appl. Biochem. Biotechnol. 1986;12:135–176. doi: 10.1007/BF02798420. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Rice KG, Kim YS, Lohse DL, Wang HM, Loganathan D. Mapping and quantification of the major oligosaccharide components of heparin. Biochem. J. 1988;254:781–787. doi: 10.1042/bj2540781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhardt RJ, Al-Hakim A, Liu J, Hoppensteadt D, Mascelanni P, Fareed J. Structural features of dermatan sulfates and their relationship to anticoagulant and antithrombotic activities. Biochem. Pharm. 1991;42:1609–1619. doi: 10.1016/0006-2952(91)90431-4. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Al-Hakim A. Biocatalysis for the synthesis and modification of biopolymers. In: Dordick JS, editor. Biocatalysis for Industry: Topics in Applied Chemistry. Vol. 5. Plenum Publishing Company; New York: 1991. pp. 83–112. [Google Scholar]
- Linhardt RJ, Ampofo S, Fareed J, Hoppensteadt D, Mulliken J, Folkman J. Isolation and characterization of a human heparin. Biochemistry. 1992;31:12441–12445. doi: 10.1021/bi00164a020. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ. Analysis of glycosaminoglycans with polysaccharide lyases. In: Varki A, editor. Current Protocols in Molecular Biology, Analysis of Glycoconjugates. Vol. 2. Wiley Interscience; Boston: 1994. pp. 17.13.17–17.13.32. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Desai U, Liu J, Pervin A, Hoppensteadt D, Fareed J. Low molecular weight dermatan sulfate as an antithrombotic agent. Biochem. Pharmacol. 1994;47:1241–1252. doi: 10.1016/0006-2952(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Toida T. Role of glycosaminoglycans in cellular communication. Acc. Chem. Res. 2004;37:431–438. doi: 10.1021/ar030138x. [DOI] [PubMed] [Google Scholar]
- Loganathan D, Wang HM, Mallis LM, Linhardt RJ. Structural variation of the antithrombin III binding site and its occurrence in heparin from different sources. Biochemistry. 1990;29:4362–4368. doi: 10.1021/bi00470a015. [DOI] [PubMed] [Google Scholar]
- Lunin VV, Li Y, Linhardt RJ, Miyazano H, Kyogashima M, Kaneko T, Bell AW, Cygler M. High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: An enzyme-substrate complex defines the catalytic mechanism. J. Mol. Biol. 2004;337:367–386. doi: 10.1016/j.jmb.2003.12.071. [DOI] [PubMed] [Google Scholar]
- Madan AK, Yu K, Dhurandhar N, Cullinane C, Pang Y, Beech DJ. Association of hyaluronidase and breast adenocarcinoma invasiveness. Oncol. Rep. 1999a;6:607–609. doi: 10.3892/or.6.3.607. [DOI] [PubMed] [Google Scholar]
- Madan AK, Pang Y, Wilkiemeyer MB, Yu D, Beech DJ. Increased hyaluronidase expression in more aggressive prostate adenocarcinoma. Oncol. Rep. 1999b;6:1431–1433. doi: 10.3892/or.6.6.1431. [DOI] [PubMed] [Google Scholar]
- McCourt PAG. How does the hyaluronan scrap-yard operate. Matrix Biol. 1999;18:427–432. doi: 10.1016/s0945-053x(99)00045-1. [DOI] [PubMed] [Google Scholar]
- McKeon RJ, Schreiber RC, Rudge JS, Silver J. Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes. J. Neurosis. 1991;11:3398–3411. doi: 10.1523/JNEUROSCI.11-11-03398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon RJ, Hoke A, Silver J. Injury-induced proteoglycans inhibit the potential for laminmediated axon growth on astrocytin scars. Exp. Neural. 1995;136:32–43. doi: 10.1006/exnr.1995.1081. [DOI] [PubMed] [Google Scholar]
- Michaud P, Da Costa A, Courtois B, Courtois J. Polysaccharide lyases: Recent developments as biotechnological tools. Crit. Rev. Biotechnol. 2003;23:233–266. doi: 10.1080/07388550390447043. [DOI] [PubMed] [Google Scholar]
- Michel G, Pojasek K, Li Y, Sulfa T, Linhardt RJ, Raman R, Probhakar V, Sasisekharan R, Cygler M. The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a calcium-dependent catalytic machinery. J. Biol. Chem. 2004;279:32882–32896. doi: 10.1074/jbc.M403421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AD. Identification of Hyal2 as the cell-surface receptor for jaagsiekte sheep retrovirus and ovine nasal adenocarcinoma virus. Curr. Top Microbiol. Immunol. 2003;275:179–199. doi: 10.1007/978-3-642-55638-8_7. [DOI] [PubMed] [Google Scholar]
- Monslow J, Williams JD, Nurton N, Guy CA, Price IK, Coleman SL, Williams NM, Buckland PR, Spicer AP, Topley N. The human hyaluronan synthase genes: Genomic structures, proximal promoters and polymorphic microsatellite markers. Int. J. Biochem. Cell Biol. 2003;35:1272–1283. doi: 10.1016/s1357-2725(03)00048-7. [DOI] [PubMed] [Google Scholar]
- Moon AF, Edavettal SC, Krahn JM, Munoz EM, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Structural analysis of the sulfotransferase (3-OST-3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1. J. Biol. Chem. 2004;279:45185–45193. doi: 10.1074/jbc.M405013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon LD, Asher RA, Rhodes KE, Fawcett JW. Regeneration of CNS axons back to their target following treatment of adult rat brain with chondroitinases ABC. Nat. Neurosci. 2001;4:465–466. doi: 10.1038/87415. [DOI] [PubMed] [Google Scholar]
- Negishi M, Dong J, Darden TA, Pedersen LG, Pedersen LC. Glycosaminoglycan biosynthesis: What we can learn from the X-ray crystal structures of glycosyl transferases GlcAT1 and EXTL2. Biochem. Biohys. Res. Commun. 2003;303:393–398. doi: 10.1016/s0006-291x(03)00356-5. [DOI] [PubMed] [Google Scholar]
- Niederost BP, Zimmermann DR, Schwab ME, Bandtlow CE. Bovine CNS myelin contains neurite growth-inhibitory activity associated with chondroitin sulfate proteoglycans. J. Neurosis. 1999;19:8979–8989. doi: 10.1523/JNEUROSCI.19-20-08979.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya T, Sugiura N, Tawada A, Sugimoto K, Watanabe H, Kimata K. Molecular cloning and characterization of chondroitin polymerase from Escherichia coli strain K4. J. Biol. Chem. 2002;277:21567–21575. doi: 10.1074/jbc.M201719200. [DOI] [PubMed] [Google Scholar]
- Novak U, Stylli SS, Kaye AH, Lepperdinger G. Hyaluronidase-2 overexpression accelerates intracerebral but not subcutaneous tumor formation of murine astrocytoma cells. Cancer Res. 1999;59:6246–6250. [PubMed] [Google Scholar]
- Pervin A, Gu K, Linhardt RJ. Capilary electrophoresis to measure sulfoesterase activity on chondroitin sulfate and heparin derived disaccharides. Appl. Theor. Electroph. 1993;3:297–303. [PubMed] [Google Scholar]
- Pervin A, Al-Hakim A, Linhardt RJ. Separation of glycosamionoglycan derived oligosaccharides by capilary electrophoresis using reverse polarity. Anal. Biochem. 1994;221:182–188. doi: 10.1006/abio.1994.1395. [DOI] [PubMed] [Google Scholar]
- Pervin A, Gallo C, Jandik KA, Han XJ, Linhardt RJ. Preparation and structural characterization of large heparin-derived oligosaccharides. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- Pizzorusso T, Medini P, Berardi N, Chierzi S, Fawcett JW, Maffei L. Reactivation of ocular dominance plasticity in the adult visual cortex. Science. 2002;298:1248–1251. doi: 10.1126/science.1072699. [DOI] [PubMed] [Google Scholar]
- Pojasek K, Shriver Z, Kiley P, Venkataraman G, Sasisekharan R. Recombinant expression, purification, and kinetic characterization of chondroitinase AC and chondroitinase B from Flavobacterium heparinum. Biochem. Biophys. Res. Commun. 2001;286:343–351. doi: 10.1006/bbrc.2001.5380. [DOI] [PubMed] [Google Scholar]
- Prabhakar V, Capila I, Bosques CJ, Pojasek K, Sasisekharan R. Chondroitinase ABC I from Proteus vulgaris: Cloning, recombinant expression and active site identification. Biochem. J. 2005;386:103–112. doi: 10.1042/BJ20041222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud AD, Prodger JC, Flitsch SL. Development of a protecting group for sulfate esters. Tetrahedron Lett. 1997;38:7243–7246. [Google Scholar]
- Razi N, Feyzi E, Bjork I, Naggi A, Casu B, Lindahl U. Structural and functional properties of heparin analogs obtained by chemical sulphation of Escherichia coli K5 capsular polysaccharide. Biochem. J. 1995;309:465–472. doi: 10.1042/bj3090465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden DJ, Jedrzejas MJ. Structures of Streptococcus pnemoniae hyaluronate lyase in complex with chondroitin and chondroitin sulfate disaccharides. J. Biol. Chem. 2003;278:50596–50606. doi: 10.1074/jbc.M307596200. [DOI] [PubMed] [Google Scholar]
- Rosenberg RD, Shworak NW, Liu J, Schwartz JJ, Zhang L. Heparan sulfate proteoglycans of the cardiovascular system. Specific structures emerge but how is synthesis regulated? J. Clin. Invest. 1997;99:2062–2070. doi: 10.1172/JCI119377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler DG, Breuer E, Grande-Allen KJ, Hascall VC, Kresse H. Core protein dependence of epimerization of glucuronosyl residues in galactosaminoglycans. J. Biol. Chem. 2002;279:25789–25797. doi: 10.1074/jbc.M208442200. [DOI] [PubMed] [Google Scholar]
- Silbert JE, Sugumaran G. Biosynthesis of chondroitin/dermatan sulfate. IUMB Life. 2002;54:177–186. doi: 10.1080/15216540214923. [DOI] [PubMed] [Google Scholar]
- Sim JS, Jun G, Toida T, Cho SY, Choi DW, Chang SY, Linhardt RJ, Kim YS. Quantitative analysis of chondroitin sulfate in raw materials, ophthalmic solutions, soft capsules and liquid preparations. J. Chromatogr. B. 2005;818:133–139. doi: 10.1016/j.jchromb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Smith-Thomas LC, Fok-seang J, Stevene J, Du J, Muir E, Faissner A, Geller HM, Rogers JH, Fawcett JW. An inhibitor of neurite outgrowth produced by astrocytes. J. Cell Sci. 1994;107:1687–1695. doi: 10.1242/jcs.107.6.1687. [DOI] [PubMed] [Google Scholar]
- Spicer AP, McDonald JA. Characterization and molecular evolution of a vertabrate hyaluronan synthase gene family. J. Biol. Chem. 1998;273:1923–1932. doi: 10.1074/jbc.273.4.1923. [DOI] [PubMed] [Google Scholar]
- Stringer SE, Callagher JT. Heparan sulphate. Int. J. Biochem. Cell Biol. 1997;29:709–714. doi: 10.1016/s1357-2725(96)00170-7. [DOI] [PubMed] [Google Scholar]
- Sugahara K, Kitagawa H. Heparin and heparan sulfate biosynthesis. IUMB Life. 2002;54:163–175. doi: 10.1080/15216540214928. [DOI] [PubMed] [Google Scholar]
- Sztrolovics R, Recklies AD, Roughley PJ, Mort J. Hyaluronate degradation as an alternate mechanism for proteoglycan release from cartilage during interleukin-1β-stimulated catabolism. Biochem. J. 2002;362:473–479. doi: 10.1042/0264-6021:3620473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorp S, Lee KA, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J. Biol. Chem. 2004;279:25789–25797. doi: 10.1074/jbc.M401089200. [DOI] [PubMed] [Google Scholar]
- Uyama T, Kitagawa H, Tanaka J, Tamura J, Ogawa T, Sugahara K. Molecular cloning and expression of a second chondroitin N-acetylgalactosaminyltransferase involved in the initiation and elongation of chondroitin/dermatan sulfate. J. Biol. Chem. 2003;278:3072–3078. doi: 10.1074/jbc.M209446200. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I, Spector L, Pecker I. Mammalian heparanse: Gene cloning, expression and function in tumor progression and metastasis. Nat. Med. 1999;5:793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- Weiler JM, Edens RE, Linhardt RJ, Kapelanski DP. Heparin and modified heparin inhibit complement activation in vivo. J. Immunol. 1992;148:3210–3215. [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Yada T, Sato T, Kaseyama H, Gotoh M, Iwasaki H, Kikuchi N, Kwon YD, Togayachi A, Kudo T, Watanabe H, Narimatsu H, Kimata K. Chondroitin sulfate synthase-3: Molecular cloning and characterization. J. Biol. Chem. 2003;278:39711–39725. doi: 10.1074/jbc.M304421200. [DOI] [PubMed] [Google Scholar]
- Yang HO, Gunay NS, Toida T, Kuberan B, Yu G, Kim YS, Linhardt RJ. Preparation and structural determination of dermatan sulfate-derived oligosaccharides. Glycobiology. 2000;10:1033–1040. doi: 10.1093/glycob/10.10.1033. [DOI] [PubMed] [Google Scholar]
- Yao XY, Hageman GS, Marmor MF. Recovery of retinal adhesion after enzymatic perturbation of the interphotoreceptor matrix. Invest. Ophthalmol. Vis. Sci. 1992;33:498–503. [PubMed] [Google Scholar]
- Zechel DL, Withers SG. Glycosidase mechanism: Anatomy of a finely tuned catalyst. Acc. Chem. Res. 2000;33:11–18. doi: 10.1021/ar970172+. [DOI] [PubMed] [Google Scholar]
- Zuo J, Neubauer D, Dyes K, Ferguson TA, Muir D. Degradation of chondroitin sulfate proteoglycans enhances the neurite-promoting potential of spinal cord tissue. Exp. Neurol. 1998;154:654–662. doi: 10.1006/exnr.1998.6951. [DOI] [PubMed] [Google Scholar]