Abstract
Alterations in the bidirectional interactions between the gut and the nervous system play an important role in IBS pathophysiology and symptom generation. A body of largely preclinical evidence suggests that the gut microbiota can modulate these interactions. Characterizations of alterations of gut microbiota in unselected IBS patients, and assessment of changes in subjective symptoms associated with manipulations of the gut microbiota with prebiotics, probiotics and antibiotics support a small, but poorly defined role of dybiosis in overall IBS symptoms. It remains to be determined if the observed abnormalities are a consequence of altered top down signaling from the brain to the gut and microbiota, if they are secondary to a primary perturbation of the microbiota, and if they play a role in the development of altered brain gut interactions early in life. Different mechanisms may play role in subsets of patients. Characterization of gut microbiome alterations in large cohorts of well phenotyped patients as well as evidence correlating gut metabolites with specific abnormalities in the gut brain axis are required to answer these questions.
INTRODUCTION
Alterations in bidirectional brain gut interactions have been considered a likely pathophysiological construct underlying IBS and related functional GI disorders for some time.1,2 However, considerable controversy remains regarding the involved molecular mechanisms and the precise targets within the brain gut axis that are responsible for such alterations. Similarly, it remains unclear which of the reported changes are primary and which are secondary in the development of symptoms. The gut microbiota and their metabolic products have recently been proposed as one such plausible mechanism, given their demonstrated ability primarily in preclinical studies to influence intestinal permeability 3 and immune function,4 activity in the enteric nervous system (reviewed in5), the HPA axis,6 pain modulation systems7 and the brain (reviewed in 8, 9). Alterations of the normal gut microbiota (“dysbiosis”) have been implicated in putative IBS pathophysiology in terms of enhanced gut permeability,10–12 mucosal immune activation,10–14 visceral hypersensitivity14, 15 and altered intestinal motility.16 However, there is conflicting evidence regarding alterations in the organization and metabolic products of the gut microbiome in patients with chronic abdominal pain and in adult and pediatric IBS,12 and on the beneficial effects of gut microbial manipulations with prebiotics, probiotics, and antibiotics in some IBS patients. Furthermore, it remains unclear if the observed IBS related alterations in the gut microbiome are related to altered intestinal function/physiology and/or changes in brain signaling. For example, there are multiple mechanisms by which the brain (via the hypothalamic pituitary [HPA] axis, the autonomic nervous system (ANS), and ANS modulation of the enteric nervous system) can influence the context and the intestinal environment in which the microbiota live, by influencing regional gut motility patterns, epithelial permeability, luminal secretions, mucosal immune function and possibly intraluminal release of neurotransmitters from enteroendocrine and other cells in the gut (reviewed in 1, 2). There are also a limited number of intriguing original preclinical study reports to support an influence of the gut microbiota on brain development and behaviors and on the adult brain,6, 17–19 including characterization of neuroactive metabolites which may underlie this influence.8 A recent study has demonstrated for the first time in healthy human subjects that perturbation of the normal gut microbiota with a probiotic can influence brain function. 20 The knowledge of the healthy human microbiome is rapidly advancing and these reference data sets will enable scientists to distinguish physiological from pathological changes associated with the intestinal microbiome.21
In this article, we will review the published literature which supports a role for altered gut microbiota in symptoms and pathophysiology of IBS, the most prevalent and best studied functional GI disorder. We will first focus on findings in human patients by critically reviewing reported evidence in support of alterations in gut microbiota (dysbiosis) and related metabolites in IBS patients. We will review the evidence supporting a possible causative role of the reported dysbiosis in IBS symptoms, based on symptomatic responses to modulation of the gut microbiome by diet, by pre- and probiotics, and by antibiotics. We will then review the possible consequences of dysbiosis on the gut brain axis and resulting IBS symptoms, and the possible causes of the dysbiosis. It needs to be emphasized that given the unique nature of bidirectional brain gut interactions, it remains impossible at this point to determine from published cross sectional studies the causality between observed alterations in gut microbiota, intestinal function, the brain and IBS symptoms. However, with the rapidly evolving technologies to characterize the gut microbiome and its metabolic products, as well as brain signatures which may be related to the dysbiosis, important breakthroughs in the characterization of the role of the gut microbiota in modulating gut brain interactions and in the pathophysiology of functional gastrointestinal disorders (FGIDs) can be expected.
Clinical evidence for alterations in the gut microbiome in IBS patients
There has been a rapid evolution of analytical techniques to characterize different aspects of the gut microbiome.22–24 While cultured species represents merely 20–30% of identified gut phylotypes (dominated by Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria in the human colon), these techniques remain crucial for investigating microbial diversity and for the selection of key functional groups.12 Culture independent approaches include 16S rRNA gene based analyses (identifying which microbes are present in the GI tract), metagenomic approaches (addressing which microbial genes are present), and metatranscriptomics, metaproteonomics, and metabolomic techniques (addressing the functional consequences of the microbiome). Currently published studies in IBS subjects have used a variety of these techniques12 and Table 1).
Table 1.
Bacterial classification shifts in human IBS
| Classification Method | Microbiome Shift | Study Subjects |
|---|---|---|
| Culture |
 Bifidobacteria (genus which is part of the phylum Actinobacteria); Lactobaccili (which are part of the phylum Firmicutes); Anaerobes Bifidobacteria (genus which is part of the phylum Actinobacteria); Lactobaccili (which are part of the phylum Firmicutes); Anaerobes |
Balsari et al (1982)109 (IBS = 20; Con = 20) Si et al (2004)110 (IBS = 25; Con = 25) Mättö et al (2005)111 (IBS = 26; Con = 25) Carroll et al (2010)112 (IBS-D = 10; Con = 10) |
 Enterobacteria; Aerobes Enterobacteria; Aerobes | ||
| PCR-DGGE/qPCR |
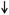 Anaerobes; Lactobaccilli in IBS-D Anaerobes; Lactobaccilli in IBS-D |
Mättö et al (2005)111 (IBS = 26; Con = 25) Malinen et al (2005)113 (IBS = 27; Con = 22) |
 Aerobes Aerobes | ||
| FISH |
 Bifidobacteria Bifidobacteria |
Kerckhoffs et al (2009)114 (IBS = 41; Con = 26) |
 Firmicutes (phylum which includes Lactobacilli) Firmicutes (phylum which includes Lactobacilli) | ||
| Microarray |
 Bacteroidetes (phylum which includes the genus Bacteroides) ; Bifidobacteria Bacteroidetes (phylum which includes the genus Bacteroides) ; Bifidobacteria |
Rajilic-Stojanoovic (2011) 115 (IBS = 62; Con = 42) Saulnier et al (2011)116 (Pediatric IBS = 22; Con = 22) |
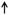 Firmicutes Firmicutes | ||
| 16S-yrosequencing |
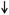 Bacteroidetes; Bifidobacteria; Actinobacteria Bacteroidetes; Bifidobacteria; Actinobacteria |
Krogius-Kurikka et al (2009)117 (IBS-D = 10; Con = 23) Rajilic-Stojanoovic (2011)115 (IBS = 62; Con = 42) Saulnier et al (2011)116 (Pediatric IBS = 22; Con = 22) Jeffery (2012)38 (IBS = 37; Con = 20) |
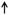 Firmicutes; Proteobacteria Firmicutes; Proteobacteria |
The table summarizes general bacterial genus and phyla shifts in stool specimens across a range of different classification methods in adult and pediatric IBS cohorts. Data from 13 (out of a total 22) published reports where there is a general consensus on microbiota composition changes in IBS are included. qPCR, quantitative polymerase chain reaction; DGGE, denaturing gradient gel electrophoresis; FISH, fluorescence in situ hybridization.
Composition and organization of gut microbiota in IBS
Seven studies have evaluated shifts in microbial small bowel community composition in the upper bowel (summarized in 12, 25) of a total of 314 subjects meeting IBS diagnostic criteria based on culture results, with two additional studies reporting results from molecular studies (Table 1).26, 27 A larger number of studies (n=22 in a total of 827 subjects) have reported significant microbial shifts in fecal microbial community composition between healthy controls and IBS patients, based on disease subtypes (IBS-D; IBS-C; IBS-M), age (pediatric versus adult), and compartment (mucosa versus stool).12 Despite a lack of consensus on the wide range of gut microbial differences between IBS subjects and healthy controls and the specific microbial changes that may be correlated to disease outcome, some general trends are starting to emerge as depicted in Table 1. In contrast to culture based studies of small bowel microbiota where no consistent differences are evident between IBS and controls, recent molecular based methods of mucosal brushings or luminal aspirates suggest decreased diversity in small bowel microbiota with increased abundance of gram negative organisms in IBS.26, 27 Based on analysis of fecal samples, regardless of analytical methodology used, a number of studies reported decreased relative abundance of the genera Bifidobacterium and Lactobacillus, and increased Firmicutes:Bacteroidetes ratios at the phylum level. If these IBS related microbial patterns can be confirmed in future studies, one may speculate about some of the many possible causes for such changes - stress and diet. As discussed below, a temporary reduction in lactobacilli has been reported in animal models of early life stress, which may be related to stress induced changes in intestinal transit.28–30 On the other hand, Firmicutes is the dominant phylum in adult microbiota consuming a western diet (high in animal fat and protein), while Bacteroidetes was the dominant phylyum in a pediatric population consuming a plant fiber based, agrarian diet.31 The dominant genera within Bacteroidetes may differ since Prevotella was more abundant in an African population consuming a plant-based diet, whereas Bacteroides was more abundant in a North American population consuming a different plan-based diet.32 Long-term food preferences appear to shape enterotypes, which are primarily defined by bacterial genera as the key principal components. Diet induced shifts have also been reported at the genus level: Within the phylum Bacteroidetes, individuals on a Western diet have a Bacteroides-enriched enterotype as opposed to those living on an agrarian diet which have a Prevotella-enriched enterotype. When viewed together with the reported shifts in gut microbial composition in pediatric and adult IBS patients at the phylum level, one could speculate that the findings of an enhanced Firmicutes: Bacteroidetes ratio may in part be due to factors related to the typical Western diet.33 However, even though the reported prevalence rates are higher in some countries consuming Western diets, in particular the US, UK and Italy, few reliable data are available to support the concept that IBS prevalence differs significantly between individuals living in countries or cultural settings (urban vs rural) consuming a typical Western diet and those living on an agrarian type diet.34 The question if dietary habits are associated with IBS patterns of dysbiosis, should be addressed in future studies.
Unfortunately little work to date has examined the mucosa-associated microbiota in health or in IBS. In healthy individuals and patients with IBS, the mucosa-associated microbiota determined from duodenal brushings or rectal biopsies differs from that found in feces.26, 35 These mucosal-fecal microbiota differences have been described as being greater than the fecal microbial differences between patients with IBS and controls.36, 37 Differences between microbiota associated with mucosa and those found in feces are likely critical to understanding the relationship between dysbiosis and IBS because of the closer communication between signaling systems between mucosa associated microbiota and the epithelium.2
Widely conflicting reports of microbial dysbiosis exist in IBS and a number of significant variables likely contribute to theses discrepancies that limit our understanding of the collective literature. Many clinical studies have used markedly different experimental approaches to define microbial communities. In many cases there are poorly defined clinical cohorts, presence of psychological comorbidities, inadequate numbers, and/or lack of repeat studies in the same patient study population. Generally, there is a lack of temporal stool sampling that coincides with clinical symptoms, and most studies have not considered dietary variations or drug use (e.g. antibiotics, proton pump inhibitors) that may directly influence microbial community composition, or indirectly through alteration of gut motility, immune activity, or other functions such as mucosal permeability. Furthermore, there is currently no way to decide if the observed alterations in microbial communities in IBS are a primary abnormality responsible for IBS related symptoms or if the observed changes are secondary to IBS related alterations in various gut functions (regional motility, secretion), or if both of these mechanisms contribute to the persistence of altered bidirectional brain gut interactions.
Larger and more homogeneous sample sizes, control for differences in physiologic parameters (e.g., diet, transit, gut permeability, immune status), and stratification of patients based on greater phenotypic discrimination (e.g., postinfectious, symptom duration, age, sex) will be necessary to observe reproducible differences between microbial communities from IBS subjects and healthy control subjects. This is supported by a recent study demonstrating that two IBS microbial clusters, not related to IBS-D or IBS-C cohorts, show significant compositional differences to healthy controls, with a third IBS cluster sharing an identical enterotype with controls.38
Gut microbiota related metabolites in IBS
Recent studies suggest that significant changes in microbiota related metabolites can occur without detectable changes in the organizational structure of the gut microbiota39, and that host genotype (FUT2 gene status) can affect the metabolic response of the gut microbiota to the diet.40 Despite the central role of the gut microbiota in producing the short chain fatty acids (SCFAs) butyrate, propionate, and acetate, there is disagreement regarding whether their fecal concentrations differ between IBS and controls and whether changes in concentration are related to IBS symptoms.41–45 In addition to reported IBS related alterations in SCFAs, alterations in other metabolites including choline, taurine and branched chain fatty acids,42 lysophosphatidylcholine,46 2(3H)-furanone, and volatile organic metabolites (VOM)47 Esters of SCFAs and cyclohexanecarboxylic acid and its derivatives were significantly associated with IBS-D, and key VOMs were able to predict a diagnosis of IBS.46, 47 Inconsistencies among reported studies on microbial metabolites in IBS likely relate, in part, to such variables as methodology, patient selection and control for diet and concurrent medications.
Evidence to support a causative role of dysbiosis in IBS symptoms
Even before the recent explosion in knowledge about the gut microbiome and its implication as a factor in IBS pathophysiology, various dietary modifications have been suggested to treat IBS symptoms including the traditional (inconsistent) teaching to avoid fiber rich diets while other recommendations have include the intake of various fiber supplements. Such decade old treatment recommendations, poorly substantiated by controlled trials, can now be reevaluated in the context of how such interventions may affect alterations in gut microbiota in IBS. Of the many dietary recommendations given to IBS patients over the years, the one that has received most recent attention is the low fermentable substrate diet (low FODMAPs concept). This diet recommends reduced consumption of foods containing oligosaccharides, disaccharides, monosaccharides and polyols that are poorly absorbed in the small intestine and are fermented by gut bacteria in the large intestine. A recently published randomized controlled trialin IBS patients comparing a FODMAP diet to a regular diet reported a reduction of IBS symptoms in the FODMAP diet group.48 Whether a similar mechanism also explains the reported response in IBS to a gluten free diet remains to be determined.49
Prebiotics
There are only four randomized trials (one single blind) of prebiotics in IBS and no systematic reviews or metaanalyses.50 Given the adverse effects of a high carbohydrate diet, including one containing fermentable oligosaccharides, disaccharides, monosaccharides, and polyols (FODMAPs) in some patients with IBS, it is not surprising that high intake of prebiotics worsened or did not improve symptoms in three studies.51, 52
Probiotics
Based on several meta-analyses probiotics appear to provide some benefit in IBS (Table 2).53 However, these metaanalyses highlight the problems interpreting results from probiotic studies in IBS. These include inadequate sample size, poor study design (e.g., crossover with inadequate washout between study periods), inclusion of different IBS subtypes, and the use of multiple strains and doses across studies. Ortiz-Lucas et al.54 evaluated 10 studies adequate for meta-analysis with a focus on the specific organisms potentially effective. A significant benefit on pain relief was found for B. breve, B. longum, and L. acidophilus (Table 2).54 No effects on stool frequency or consistency were found.54 Horvath et al. reviewed three studies of L. rhamnosus in children with IBS and reported a significant benefit for pain improvement (Table 2).55
Table 2.
Meta-analyses of Probiotics in IBS
| Author | Outcome | n | Outcome | NNT |
|---|---|---|---|---|
| Ortiz-Lucas 201354 | Abdominal pain | 862 | SMD = −0.24; 95% CI −0.16 to 0.51* | |
| Enck 201057 | Global symptoms | Dichotomous - 1838 | OR = 2.24; 95% CI 1.51 to 2.75 | 8 |
| Moayyedi 2010118 | Global symptoms Abdominal pain |
Dichotomous - 918 Continuous - 1351 Continuous - 834 |
RR = 0.71; 95% CI0.57 to 0.88 SMD = −0.34; 95% CI −0.06 to −.07 SMD = −0.51; 95% − 0.91 to −0.09 |
4 |
| Hoveyda 2009119 | Global symptoms Abdominal pain |
Dichotomous - 895 Continuous - 657 Dichotomous – 398 |
OR = 1.6; 95% CI 1.2 to 2.2 SMD = 0.23; 95% CI 0.07 to 0.38 OR = 2.88; 95% CI 1.84 to 4.5 |
|
| McFarland 2008120 | Global symptoms Abdominal pain |
Dichotomous – 1254 Dichotomous - 1039 |
RR = 0.77; 95% CI 0.62 to 0.94 RR = 0.78; 95% CI 0.69 to 0.88 |
7.3 8.9 |
| Horvath 201155 | Abdominal pain in children treated with L. rhamnosus | No pain or improved pain - 167 | RR = 1.7; 95% CI 1.27 to 2.27 | 4 |
Average of responses to specific organisms
NNT = number needed to treat, SMD = standardized mean difference, RR = relative risk, OR = odds ratio
Antibiotics
As demonstrated in both preclinical and clinical studies, the gut microbial composition can be altered by treatment with antibiotics. The best data to evaluate the effect of an antibiotic on IBS symptoms comes from studies with the non-absorbable antibiotic rifaximin. In a recent systematic review and metaanalysis on the use of rifaximin in IBS,56 with the primary outcome improvement in global IBS symptoms, five studies (n=1803) reported an overall small improvement after treatment (OR = 1.57; 95% CI 1.22 to 2.01) with a NNT of 10.2.56 Using that NNT, rifaximin would be less effective than peppermint oil, psychotherapy, tricyclic antidepressants, spasmolytics, selective serotonin reuptake inhibitors, and probiotics based on a review of metaanalyses of treatments for IBS in adults.57 A double blind, placebo controlled trial of rifaximin in children with pediatric Rome-defined abdominal pain functional GI disorders (n=75) found no benefit.58 Evidence has been put forth suggesting that rifaximin works by treating small intestinal bacterial overgrowth (SIBO) in IBS.59 Although rifaximin is bacteriocidal against a broad array of enteric pathogens (gram negative/positive, aerobic/anaerobic), two weeks of treatment did not affect fecal coliform median log counts,60 and other anti-inflammatory mechanisms of actions have been demonstrated.61
In summary, given the limited data available from high quality randomized clinical trials to assess the effectiveness of prebiotics, probiotics and antibiotics in IBS patients, published data and metaanalyses to date suggest that alterations in gut microbiota may play a relatively small role in generating GI symptoms. However, the interventional data viewed together with the likely heterogeneity of mechanisms contributing to IBS symptoms, suggest that subsets of patients with specific microbiota-related alterations (e.g. deficiencies or excesses of certain microorganisms or metabolites which play a role in causing distinct brain gut abnormalities) may significantly benefit from specific interventions aimed at normalizing a particular dysbiotic state. The identification of such patient subsets with distinct patterns of dysbiosis will require large scale studies in well phenotyped patients with functional GI disorders.,
Possible causes of dysbiosis in IBS
Enteric infections
Persistence of IBS-like symptoms (so called postinfectious IBS) which has been reported in a small (8–15%) percentage of patients following an initial episode of documented bacterial or viral enteric infections supports a possible role of perturbations of the gut microbiome by pathogens in the development of altered brain gut interactions associated with IBS like symptoms. Common bacterial causes of traveler’s diarrhea (E. coli, Salmonella and Campylobacter) are particularly strongly associated with postinfectious IBS, and both biological and psychological risk factors have been identified.62 Biological factors include duration and severity of diarrhea, microbial toxin production, gut pathology and inflammation. Psychological factors include a high somatization score in affected patients (e.g. a high number of somatic symptoms the subject has experienced prior to the infection), trait anxiety, and the presence of a major psychosocial stressor around the time of the infection. When these factors are viewed together, it is intriguing to speculate that increased levels of stress related catecholamines in the stool (as observed in animal models of stress) result in increased virulence of the respective pathogen (reviewed in Rhee, et. al.,2 thereby increasing the duration and severity of the infection. Such a prolonged infection with or without additional antibiotic treatment may shift the gut microbiota to an “IBS-prone enterotypes.” Other possible mechanisms include persistently increased mast cell numbers and alterations in neuropeptide homeostasis as a result of infection induced enteroendocrine cell hyperplasia. Increased serotonin containing cells are a hallmark of IBS-D, and increased serotonin release from such cells is likely to alter signaling within the enteric nervous system and gut to brain signaling (reviewed in Hughes et al.14).
Antibiotics
Both clinical and published evidence suggest that in some individuals, previous antibiotic therapy for non-GI related indications may increase the new onset or symptom flare of existing IBS symptoms,63, 64 and that antibiotic treatment may increase the development of long term post infectious IBS symptoms.65 However, in contrast to other health problems that have seen a dramatic recent increase in developed countries and which have been associated with increased antibiotic use in childhood,66, 67 no such epidemiologic changes in IBS prevalence have been reported, arguing against a major causative role of antibiotic use in IBS pathophysiology.
Top down modulation of the gut microbiota by the CNS
CNS modulation of the gastrointestinal tract via the autonomic nervous system (ANS) and the hypothalamus–pituitary–adrenal (HPA) axis can influence enteric microbiota both indirectly, via changes in their environment, and directly, via host–enteric microbiota signaling.(reviewed in 2) Both branches of the ANS have a prominent role in the modulation of gut functions, such as regional motility, secretion of acid, bicarbonates and mucus, epithelial fluid handling, gut permeability and mucosal immune response (Fig. 1AB)(reviewed elsewhere68). The majority of these functions, except for sympathetically and cortisol mediated immune modulation are affected via sympathetic and parasympathetic influences on circuits of the enteric nervous system. Regional and global changes in gastrointestinal transit can have profound effects on the delivery of important nutrients to the enteric microbiota (such as prebiotics, including resistant starches and certain dietary fibers) pH, and on the luminal environment in healthy and diseased states. For example, impaired intestinal transit caused by compromised, migrating motor complexes (a motor pattern characteristic of the fasting state of the gastrointestinal tract that is under parasympathetic control), is associated with bacterial overgrowth in the small intestine.69 A reduced number of giant, migrating contractions in the colon has been reported in slow transit constipation,70 and may play a role in the subset of IBS-C patients with slow transit, while accelerated intestinal transit, with an increased number of giant, migrating contractions, is seen in many diarrheal states, including diarrhea-predominant IBS.71 The ANS mediated modulation of mucus secretion is likely to have important effects on the size and quality of the intestinal mucus layer, an important habitat for the biofilm, where the majority of the enteric microbiota reside.72 The ANS also affects epithelial mechanisms involved in immune activation of the gut, either directly, through modulation of the response of the gut immune cells (for example, macrophages and mast cells) to luminal bacteria, through secretion of antimicrobial peptides73 or indirectly, through alteration of the access of luminal bacteria to gut immunocytes. For example, several preclinical studies have demonstrated that stressful stimuli can enhance the permeability of the intestinal epithelium, facilitating translocation of luminal organisms and triggering an immune response in the intestinal mucosa.74–79
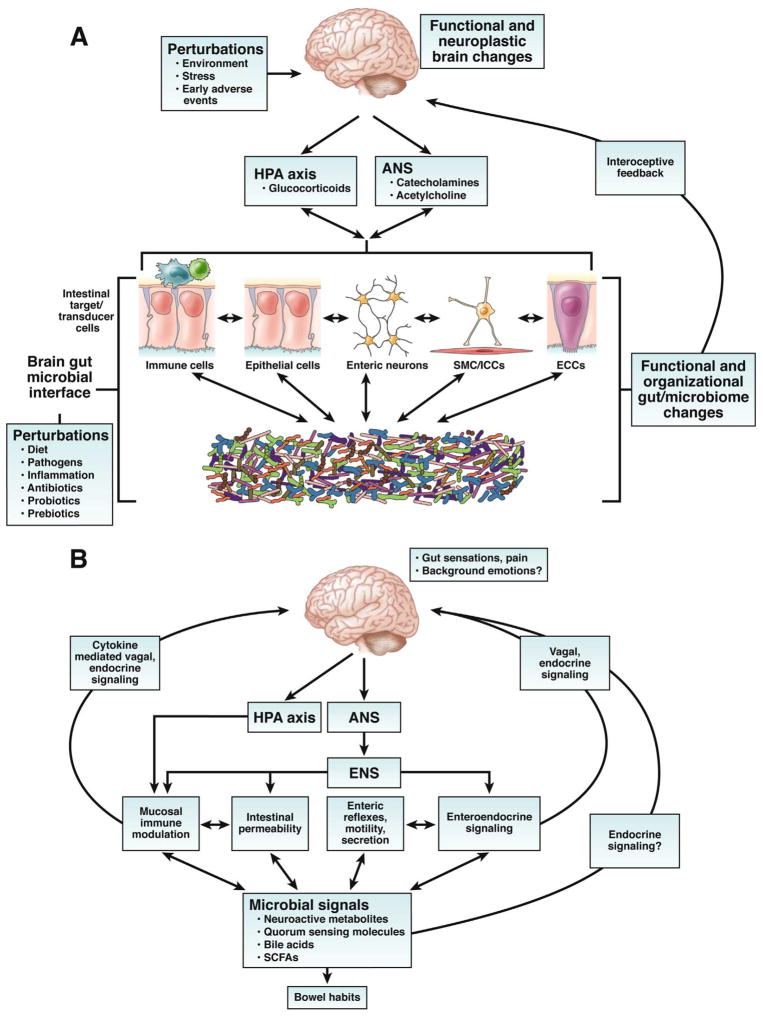
Figure 1. Bidirectional brain gut microbial interactions.
A. Key components of brain gut microbial axis. A network of specialized target/transducer cells in the gut wall functions as an interface between the organism and the gut lumen. In response to external and bodily demands, the brain modulates individual cells (ECC – enterochromaffin cells; SMC – smooth muscle cells; ICC – interstitial cells of Cajal) within this network via the branches of the autonomic nervous system (ANS) (sympathetic and parasympathetic/vagal efferents) and the hypothalamic pituitary adrenal (HPA) axis. Such modulation can be transient (e.g. in response to transient perturbations) or longlasting (in response to chronically altered brain output). The microbiota are in constant bidirectional communication with this interface via multiple signaling pathways, and this communication is modulated in response to perturbations of the microbiota, or the brain. The integrated output of the brain gut microbial interface is transmitted to the brain via multiple afferent signaling pathways, including endocrine and neurocrine (vagal, spinal afferents) pathways. While acute alterations in this interoceptive feedback result in transient functional brain changes, chronic alterations are associated with neuroplastic brain changes.
B. Functional and symptom-related consequences of brain gut microbial interactions. Several intestinal processes with possible relevance for IBS symptoms can be modulated both the brain (via the ANS, including its enteric nervous system [ENS] branch) and by signals from the microbiota. Microbiota generated molecules can signal to the brain indicrectly via activation of vagal (and possibly spinal) afferent nerve pathways, by microbiota stimulated cytokine and neurotransmitter release from immune or enteroendocrine cells, or such signals may reach the brain via an endocrine route. Microbiota gut brain signaling may contribute to the generation of abdominal pain and discomfort, while microbiota mediated modulation of enteric reflexes is likely to play a role in the pathophysiology of altered bowel habits.
Considerable evidence supports a role of stress and its mediators in modulating the gut microbiome.28, 80, 81 Both pre and postnatal stressors in animal models have been been shown to modulate the composition and total biomass of the enteric microbiota.28, 30 In newborn animals, the reported shedding of lactobacilli may have been related to the stress induced acceleration of intestinal transit, since normal bacterial levels were restored 1 week after separation.28 In adult mice, a psychosocial stressor decreased the relative abundance of the genus Bacteroidetes, while increasing the relative abundance of the genus Clostridia.82 It remains to be determined if the reported reductions in the abundance of Lactobacilli/Bifidobacteria and Bacteroidetes reported in several preclinical studies and in several IBS studies (see Table 1) may both be a consequence of stress induced acceleration of intestinal transit, or other stress system mediated effects on the gut microbiota.
Intraluminal release of neurotransmitters
In addition to CNS-induced changes in the gut environment in which the microbiota reside, there is a another, neuroendocrine communication system which allows for bidirectional signaling with the gut microbes, which has been referred to as microbial endocrinology.83 Several signaling molecules used by the host for neuronal and neuroendocrine signaling (including but not limited to catecholamines, serotonin, dynorphin, and cytokines) may also be released into the gut lumen by neurons, immune cells, enterochromaffin cells and possibly, gut microbes themselves, with the CNS likely having an important role in the release of these molecules.84–86 Of particular interest for mechanisms with potential relevance for the pathophysiology of a stress sensitive disorder like IBS is the observation that different types of stressors can result in increased luminal levels of catecholamines, including norepinephrine.87, 88 Recent evidence suggests that the gut microbiota derived beta-glucuronidase may also play an important role in the generation of free, e.g. unconjugated form of norepinephrine.89 It has long been known that some pathogens can change their proliferative activity in response to exogenous catecholamines in vitro.90 For example, norepinephrine can stimulate the growth of several strains of enteric pathogens (reviewed elsewhere88) and magnifies the virulent properties of Campylobacter jejuni.91
Bottom up modulation of the gut brain axis by IBS related dysbiosis
There are multiple ways, levels, and signaling mechanisms by which gut microbiota can influence the activity and responsiveness of the gut brain axis, including the brain. Such influences may occur early in life and affect the development of the nervous system, the brain gut axis and the HPA axis, or they may occur in the adult organism and modulate fully developed circuits (reviewed in 5, 8, 9). However, the fact that more review articles, and reports in the lay press on this topic have appeared in the last 5 years than original articles confirming many of the initial observations, suggests caution when extrapolating from existing data to unsubstantiated speculations.
Role of gut microbiota in brain development
A limited number of preclinical studies have demonstrated that the development of brain mechanisms related to hyperalgesia,7 HPA axis,6 affective behavior17, 18 and associated brain biochemistry 17 depends on an intact gut microbiome (Table 3). However, as these observations were obtained in non-physiological conditions (germ-free status) which may affect specific maternal rodent behaviors which have been shown to be associated with epigenetic changes in stress related genes92 required for the normal development of the central nervous system,93 premature conclusions about similar effects occurring in humans should be avoided.
Table 3.
Reported effects of gut microbiota on brain and behavior in newborn and adult rodents.
Role of gut microbiota derived metabolites and signaling molecules in modulating the gut brain axis in the adult
A number of candidate signaling molecules have been identified by which the gut microbiota may communicate with the host, including communications of the microbiota with the enteric nervous system and the brain. Quorum sensing molecules used by microbes to communicate with each other (including metabolites and neurotransmitter homologues) have also been shown to be recognized by the host and may influence enteroendocrine, immune cells, and nerve endings in the gut (reviewed in 2). Metabolites produced by gut microbes (including short chain fatty acids (SCFA), many neuroactive substances including GABA, tryptophan, serotonin and catecholamines (reviewed in 94), and metabolites of bile acids and neurotransmitters), and cytokines released in response microbe host interactions 82 can signal via specific receptors on local cells within the gut, or signal by neurocrine (afferent vagal pathways) and endocrine mechanisms to long distance targets beyond the GI tract, including vagal afferents in the portal vein, and the brain. Thus, regardless of the sequence of events leading to a dysbiotic state in some IBS patients (see preceding section), the altered microbial community is likely to exert a modulating effect on bidirectional communication within the gut brain axis. In the following we will highlight a few metabolites which may have a direct relevance to IBS pathophysiology.
Short chain fatty acids (SCFA)
Fermentable carbohydrates entering the colon are converted to the three primary SCFAs, acetate, propionate, and butyrate which have been demonstrated to exert a number of physiologic effects including reducing food intake, improving glucose tolerance, enhancing lymphocyte and neutrophil function, and activating pathways important in epithelial cell signaling.16, 95–99 Recent data, primarily from animal experiments, suggest that SCFA mediate many of their effects through G-protein-coupled receptors, specifically GPR43, GPR109A, and GPR41 (for reviews see Bindels et al. and Ganapathy et al.100, 101 Evidence suggests that signaling through these receptors as well as transport of SCFA by SLC5A8, and the resultant physiological effects is affected by dietary intake of fermentable fiber 100 Gut microbiota appear capable of regulating gene expression of SCLC5A8 and GPR109A.102 GPR43 is found in mast cells and may be one mechanism whereby SCFA can alter serotonin (5-HT) release.103 Most recently, work suggests that GPR41 and GPR43 serve as sensors for SCFA in enteroendocrine cells, but only GPR41 serves this role in neuronal cells of the submucosal and myenteric ganglia.104 However, despite the potential relevance of altered SCFAs in IBS pathophysiology, few studies are available to support such a role based on therapeutic interventions aimed at this signaling mechanism.105, 106
Bile acids
Primary bile acids are biosynthesized in the liver by the oxidation of cholesterol, conjugated to either glycine or taurine and secreted into the gut via the bile duct. In the gut, glycine and taurine residues are removed, and some of the bile acids are converted into secondary bile acids by the gut microbiota. It is conceivable that microbial dysbiosis observed in some IBS patients may be associated with a shift in microbes that metabolize and conjugate primary bile acids, as preliminary studies have suggested.107 Bile acids have been proposed to constitute a primary cause of disease symptoms in approximately 30% of adult IBS-D patients42, 107 and recent evidence are consistent with a role of bile acids in pediatric IBS-C patients as well. 108.
Implications/future directions
Despite the exciting new and rapidly growing insights into the interactions of the gut microbiota with the gut, the enteric nervous system and the brain, the role of alterations in these interactions in the elusive pathophysiology of IBS remains to be determined. Current evidence is most supportive of a top down modulation of the gut microbiota by the brain through the ANS, and possibly the HPA axis. However, these brain induced microbial alterations may alter the complex signaling of the microbiota to enteric neurons modulating gut functions (e.g. motility and secretion), and to the brain, modulating back ground emotions and visceral perception. Carefully designed translational studies in both human subjects and preclinical models are required to establish the causality of these events. Combining multimodal brain imaging techniques with detailed characterization of gut microbioal signaling systems in healthy adult and pediatric subjects, and well phenotyped patient populations has promise to answer the question what role the gut microbiome plays in determining brain structure and function. Regardless of the precise causality underlying a dysbiotic state in IBS, the gut microbiome has become a promising target for therapeutic interventions.
Acknowledgments
This work was in part supported by National Institutes of Health grants DK 64531 (EAM) and DK 48351 (EAM). The authors thank Dr. J. Versalovic for valuable scientific advice and C. Liu for invaluable editorial assistance.
References
- 1.Mayer EA. Gut feelings: the emerging biology of gut-brain communication. Nat Rev Neurosci. 2011;12:453–66. doi: 10.1038/nrn3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rhee SH, Pothoulakis C, Mayer EA. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol. 2009;6:306–14. doi: 10.1038/nrgastro.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frazier TH, DiBaise JK, McClain CJ. Gut microbiota, intestinal permeability, obesity-induced inflammation, and liver injury. JPEN J Parenter Enteral Nutr. 2011;35:14S–20S. doi: 10.1177/0148607111413772. [DOI] [PubMed] [Google Scholar]
- 4.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsythe P, Kunze WA. Voices from within: gut microbes and the CNS. Cell Mol Life Sci. 2013;70:55–69. doi: 10.1007/s00018-012-1028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558:263–75. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaral FA, Sachs D, Costa VV, Fagundes CT, Cisalpino D, Cunha TM, Ferreira SH, Cunha FQ, Silva TA, Nicoli JR, Vieira LQ, Souza DG, Teixeira MM. Commensal microbiota is fundamental for the development of inflammatory pain. Proc Natl Acad Sci U S A. 2008;105:2193–7. doi: 10.1073/pnas.0711891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–12. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 9.Bercik P, Collins SM, Verdu EF. Microbes and the gut-brain axis. Neurogastroenterol Motil. 2012;24:405–13. doi: 10.1111/j.1365-2982.2012.01906.x. [DOI] [PubMed] [Google Scholar]
- 10.Camilleri M, Lasch K, Zhou W. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. The confluence of increased permeability, inflammation, and pain in irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2012;303:G775–85. doi: 10.1152/ajpgi.00155.2012. [DOI] [PubMed] [Google Scholar]
- 11.Matricon J, Meleine M, Gelot A, Piche T, Dapoigny M, Muller E, Ardid D. Review article: Associations between immune activation, intestinal permeability and the irritable bowel syndrome. Aliment Pharmacol Ther. 2012;36:1009–31. doi: 10.1111/apt.12080. [DOI] [PubMed] [Google Scholar]
- 12.Simren M, Barbara G, Flint HJ, Spiegel BM, Spiller RC, Vanner S, Verdu EF, Whorwell PJ, Zoetendal EG, Rome Foundation C. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159–76. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ringel Y, Maharshak N. Intestinal microbiota and immune function in the pathogenesis of irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2013;305:G529–41. doi: 10.1152/ajpgi.00207.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes PA, Zola H, Penttila IA, Blackshaw LA, Andrews JM, Krumbiegel D. Immune activation in irritable bowel syndrome: can neuroimmune interactions explain symptoms? Am J Gastroenterol. 2013;108:1066–74. doi: 10.1038/ajg.2013.120. [DOI] [PubMed] [Google Scholar]
- 15.Valdez-Morales EE, Overington J, Guerrero-Alba R, Ochoa-Cortes F, Ibeakanma CO, Spreadbury I, Bunnett NW, Beyak M, Vanner SJ. Sensitization of Peripheral Sensory Nerves by Mediators From Colonic Biopsies of Diarrhea-Predominant Irritable Bowel Syndrome Patients: A Role for PAR2. Am J Gastroenterol. 2013;108:1634–43. doi: 10.1038/ajg.2013.241. [DOI] [PubMed] [Google Scholar]
- 16.Cani PD, Everard A, Duparc T. Gut microbiota, enteroendocrine functions and metabolism. Curr Opin Pharmacol. 2013 doi: 10.1016/j.coph.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108:3047–52. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil. 2011;23:255–64. e119. doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 19.Gareau MG, Wine E, Rodrigues DM, Cho JH, Whary MT, Philpott DJ, Macqueen G, Sherman PM. Bacterial infection causes stress-induced memory dysfunction in mice. Gut. 2011;60:307–17. doi: 10.1136/gut.2009.202515. [DOI] [PubMed] [Google Scholar]
- 20.Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, Guyonnet D, Legrain-Raspaud S, Trotin B, Naliboff B, Mayer EA. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144:1394–401. 1401e1–4. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Backhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, Versalovic J, Young V, Finlay BB. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12:611–22. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jorgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clement K, Dore J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, Bork P, Wang J, Ehrlich SD, Pedersen O, Guedon E, Delorme C, Layec S, Khaci G, van de Guchte M, Vandemeulebrouck G, Jamet A, Dervyn R, Sanchez N, Maguin E, Haimet F, Winogradski Y, Cultrone A, Leclerc M, Juste C, Blottiere H, Pelletier E, LePaslier D, Artiguenave F, Bruls T, Weissenbach J, Turner K, Parkhill J, Antolin M, Manichanh C, Casellas F, Boruel N, Varela E, Torrejon A, Guarner F, Denariaz G, Derrien M, van Hylckama Vlieg JE, Veiga P, Oozeer R, Knol J, Rescigno M, Brechot C, M’Rini C, Merieux A, Yamada T. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–6. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 23.Methe BA, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C, Gevers D, Petrosino J, Abubucker S, Badger JH, Chinwalla AT, Ear AM, FitzGerald MG, Fulton RS, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, et al. A framework for human microbiome research. Nature. 2012;486:215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li K, Bihan M, Yooseph S, Methe BA. Analyses of the microbial diversity across the human microbiome. PLoS One. 2012;7:e32118. doi: 10.1371/journal.pone.0032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang ZK, Yang YS. Upper gastrointestinal microbiota and digestive diseases. World J Gastroenterol. 2013;19:1541–50. doi: 10.3748/wjg.v19.i10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerckhoffs AP, Ben-Amor K, Samsom M, van der Rest ME, de Vogel J, Knol J, Akkermans LM. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol. 2011;60:236–45. doi: 10.1099/jmm.0.022848-0. [DOI] [PubMed] [Google Scholar]
- 27.Pimentel MFV, Giamarellos-Bourboulis EJ, Pyleris E, Pistiki K, Tang J, Lee C, Harkins T, Kim G, Weitsman S, Barlow GM, Chang C. The First Large Scale Deep Sequencing of the Duodenal Microbiome in Irritable Bowel Syndrome Reveals Striking Differences Compared to Healthy Controls. Gastroenterology. 2013;144:S59. [Google Scholar]
- 28.Bailey MT, Coe CL. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev Psychobiol. 1999;35:146–55. [PubMed] [Google Scholar]
- 29.Bailey MT, Karaszewski JW, Lubach GR, Coe CL, Lyte M. In vivo adaptation of attenuated Salmonella typhimurium results in increased growth upon exposure to norepinephrine. Physiol Behav. 1999;67:359–64. doi: 10.1016/s0031-9384(99)00087-6. [DOI] [PubMed] [Google Scholar]
- 30.Bailey MT, Lubach GR, Coe CL. Prenatal stress alters bacterial colonization of the gut in infant monkeys. J Pediatr Gastroenterol Nutr. 2004;38:414–21. doi: 10.1097/00005176-200404000-00009. [DOI] [PubMed] [Google Scholar]
- 31.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeffery IB, O’Toole PW. Diet-microbiota interactions and their implications for healthy living. Nutrients. 2013;5:234–52. doi: 10.3390/nu5010234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gwee KA. Irritable bowel syndrome in developing countries--a disorder of civilization or colonization? Neurogastroenterol Motil. 2005;17:317–24. doi: 10.1111/j.1365-2982.2005.00627.x. [DOI] [PubMed] [Google Scholar]
- 35.Durban A, Abellan JJ, Jimenez-Hernandez N, Ponce M, Ponce J, Sala T, D’Auria G, Latorre A, Moya A. Assessing gut microbial diversity from feces and rectal mucosa. Microb Ecol. 2011;61:123–33. doi: 10.1007/s00248-010-9738-y. [DOI] [PubMed] [Google Scholar]
- 36.Carroll IM, Ringel-Kulka T, Keku TO, Chang YH, Packey CD, Sartor RB, Ringel Y. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301:G799–807. doi: 10.1152/ajpgi.00154.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durban A, Abellan JJ, Jimenez-Hernandez N, Salgado P, Ponce M, Ponce J, Garrigues V, Latorre A, Moya A. Structural alterations of faecal and mucosa-associated bacterial communities in irritable bowel syndrome. Environ Microbiol Rep. 2012;4:242–247. doi: 10.1111/j.1758-2229.2012.00327.x. [DOI] [PubMed] [Google Scholar]
- 38.Jeffery IB, O’Toole PW, Ohman L, Claesson MJ, Deane J, Quigley EM, Simren M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 39.McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kashyap PC, Marcobal A, Ursell LK, Smits SA, Sonnenburg ED, Costello EK, Higginbottom SK, Domino SE, Holmes SP, Relman DA, Knight R, Gordon JI, Sonnenburg JL. Genetically dictated change in host mucus carbohydrate landscape exerts a diet-dependent effect on the gut microbiota. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1306070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kopecny JSJ. Cellulolytic bacteria in the human gut and irritable bowel syndrome. Acta Vet Brno. 2002;71:421–427. [Google Scholar]
- 42.Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011;10:4208–18. doi: 10.1021/pr2003598. [DOI] [PubMed] [Google Scholar]
- 43.Mortensen PB, Andersen JR, Arffmann S, Krag E. Short-chain fatty acids and the irritable bowel syndrome: the effect of wheat bran. Scand J Gastroenterol. 1987;22:185–92. doi: 10.3109/00365528708991878. [DOI] [PubMed] [Google Scholar]
- 44.Tana C, Umesaki Y, Imaoka A, Handa T, Kanazawa M, Fukudo S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–515. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 45.Treem WR, Ahsan N, Kastoff G, Hyams JS. Fecal short-chain fatty acids in patients with diarrhea-predominant irritable bowel syndrome: in vitro studies of carbohydrate fermentation. J Pediatr Gastroenterol Nutr. 1996;23:280–6. doi: 10.1097/00005176-199610000-00013. [DOI] [PubMed] [Google Scholar]
- 46.Kajander K, Myllyluoma E, Kyronpalo S, Rasmussen M, Sipponen P, Mattila I, Seppanen-Laakso T, Vapaatalo H, Oresic M, Korpela R. Elevated pro-inflammatory and lipotoxic mucosal lipids characterise irritable bowel syndrome. World J Gastroenterol. 2009;15:6068–74. doi: 10.3748/wjg.15.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahmed I, Greenwood R, de Costello BL, Ratcliffe NM, Probert CS. An investigation of fecal volatile organic metabolites in irritable bowel syndrome. PLoS One. 2013;8:e58204. doi: 10.1371/journal.pone.0058204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halmos EP, Power VA, Shepherd SJ, Gibson PR, Muir JG. A Diet Low in FODMAPs Reduces Symptoms of Irritable Bowel Syndrome. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 49.Biesiekierski JR, Peters SL, Newnham ED, Rosella O, Muir JG, Gibson PR. No effects of gluten in patients with self-reported non-celiac gluten sensitivity after dietary reduction of fermentable, poorly absorbed, short-chain carbohydrates. Gastroenterology. 2013;145:320–8. e1–3. doi: 10.1053/j.gastro.2013.04.051. [DOI] [PubMed] [Google Scholar]
- 50.Whelan K. Probiotics and prebiotics in the management of irritable bowel syndrome: a review of recent clinical trials and systematic reviews. Curr Opin Clin Nutr Metab Care. 2011;14:581–7. doi: 10.1097/MCO.0b013e32834b8082. [DOI] [PubMed] [Google Scholar]
- 51.Olesen M, Gudmand-Hoyer E. Efficacy, safety, and tolerability of fructooligosaccharides in the treatment of irritable bowel syndrome. Am J Clin Nutr. 2000;72:1570–5. doi: 10.1093/ajcn/72.6.1570. [DOI] [PubMed] [Google Scholar]
- 52.Silk DB, Davis A, Vulevic J, Tzortzis G, Gibson GR. Clinical trial: the effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment Pharmacol Ther. 2009;29:508–18. doi: 10.1111/j.1365-2036.2008.03911.x. [DOI] [PubMed] [Google Scholar]
- 53.Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, Sherman PM, Mayer EA. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62:787–96. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ortiz-Lucas M, Tobias A, Saz P, Sebastian JJ. Effect of probiotic species on irritable bowel syndrome symptoms: A bring up to date meta-analysis. Rev Esp Enferm Dig. 2013;105:19–36. doi: 10.4321/s1130-01082013000100005. [DOI] [PubMed] [Google Scholar]
- 55.Horvath A, Dziechciarz P, Szajewska H. Meta-analysis: Lactobacillus rhamnosus GG for abdominal pain-related functional gastrointestinal disorders in childhood. Aliment Pharmacol Ther. 2011;33:1302–10. doi: 10.1111/j.1365-2036.2011.04665.x. [DOI] [PubMed] [Google Scholar]
- 56.Menees SB, Maneerattannaporn M, Kim HM, Chey WD. The efficacy and safety of rifaximin for the irritable bowel syndrome: a systematic review and meta-analysis. Am J Gastroenterol. 2012;107:28–35. doi: 10.1038/ajg.2011.355. quiz 36. [DOI] [PubMed] [Google Scholar]
- 57.Enck P, Junne F, Klosterhalfen S, Zipfel S, Martens U. Therapy options in irritable bowel syndrome. Eur J Gastroenterol Hepatol. 2010;22:1402–11. doi: 10.1097/MEG.0b013e3283405a17. [DOI] [PubMed] [Google Scholar]
- 58.Collins BS, Lin HC. Double-blind, placebo-controlled antibiotic treatment study of small intestinal bacterial overgrowth in children with chronic abdominal pain. J Pediatr Gastroenterol Nutr. 2011;52:382–6. doi: 10.1097/MPG.0b013e3181effa3b. [DOI] [PubMed] [Google Scholar]
- 59.Sachdev AH, Pimentel M. Antibiotics for irritable bowel syndrome: rationale and current evidence. Curr Gastroenterol Rep. 2012;14:439–45. doi: 10.1007/s11894-012-0284-2. [DOI] [PubMed] [Google Scholar]
- 60.DuPont HL, Jiang ZD, Okhuysen PC, Ericsson CD, de la Cabada FJ, Ke S, DuPont MW, Martinez-Sandoval F. A randomized, double-blind, placebo-controlled trial of rifaximin to prevent travelers’ diarrhea. Ann Intern Med. 2005;142:805–12. doi: 10.7326/0003-4819-142-10-200505170-00005. [DOI] [PubMed] [Google Scholar]
- 61.Cheng J, Shah YM, Gonzalez FJ. Pregnane X receptor as a target for treatment of inflammatory bowel disorders. Trends Pharmacol Sci. 2012;33:323–30. doi: 10.1016/j.tips.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spiller RC. Postinfectious irritable bowel syndrome. Gastroenterology. 2003;124:1662–71. doi: 10.1016/s0016-5085(03)00324-x. [DOI] [PubMed] [Google Scholar]
- 63.Mendall MA, Kumar D. Antibiotic use, childhood affluence and irritable bowel syndrome (IBS) Eur J Gastroenterol Hepatol. 1998;10:59–62. doi: 10.1097/00042737-199801000-00011. [DOI] [PubMed] [Google Scholar]
- 64.Maxwell PR, Rink E, Kumar D, Mendall MA. Antibiotics increase functional abdominal symptoms. Am J Gastroenterol. 2002;97:104–8. doi: 10.1111/j.1572-0241.2002.05428.x. [DOI] [PubMed] [Google Scholar]
- 65.Barbara G, Stanghellini V, Berti-Ceroni C, De Giorgio R, Salvioli B, Corradi F, Cremon C, Corinaldesi R. Role of antibiotic therapy on long-term germ excretion in faeces and digestive symptoms after Salmonella infection. Aliment Pharmacol Ther. 2000;14:1127–31. doi: 10.1046/j.1365-2036.2000.00818.x. [DOI] [PubMed] [Google Scholar]
- 66.Marild K, Ye W, Lebwohl B, Green PH, Blaser MJ, Card T, Ludvigsson JF. Antibiotic exposure and the development of coeliac disease: a nationwide case-control study. BMC Gastroenterol. 2013;13:109. doi: 10.1186/1471-230X-13-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trasande L, Blustein J, Liu M, Corwin E, Cox LM, Blaser MJ. Infant antibiotic exposures and early-life body mass. Int J Obes (Lond) 2013;37:16–23. doi: 10.1038/ijo.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayer EA. The neurobiology of stress and gastrointestinal disease. Gut. 2000;47:861–9. doi: 10.1136/gut.47.6.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Felius ID, Akkermans LM, Bosscha K, Verheem A, Harmsen W, Visser MR, Gooszen HG. Interdigestive small bowel motility and duodenal bacterial overgrowth in experimental acute pancreatitis. Neurogastroenterol Motil. 2003;15:267–76. doi: 10.1046/j.1365-2982.2003.00410.x. [DOI] [PubMed] [Google Scholar]
- 70.Lembo A, Camilleri M. Chronic constipation. New England Journal of Medicine. 2003;349:1360–1368. doi: 10.1056/NEJMra020995. [DOI] [PubMed] [Google Scholar]
- 71.Chey WY, Jin HO, Lee MH, Sun SW, Lee KY. Colonic motility abnormality in patients with irritable bowel syndrome exhibiting abdominal pain and diarrhea. American Journal of Gastroenterology. 2001;96:1499–1506. doi: 10.1111/j.1572-0241.2001.03804.x. [DOI] [PubMed] [Google Scholar]
- 72.Macfarlane S, Dillon JF. Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol. 2007;102:1187–96. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- 73.Alonso C, Guilarte M, Vicario M, Ramos L, Ramadan Z, Antolin M, Martinez C, Rezzi S, Saperas E, Kochhar S, Santos J, Malagelada JR. Maladaptive intestinal epithelial responses to life stress may predispose healthy women to gut mucosal inflammation. Gastroenterology. 2008;135:163–172. e1. doi: 10.1053/j.gastro.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 74.Groot J, Bijlsma P, Van Kalkeren A, Kiliaan A, Saunders P, Perdue M. Stress-induced decrease of the intestinal barrier function. The role of muscarinic receptor activation. Ann N Y Acad Sci. 2000;915:237–46. doi: 10.1111/j.1749-6632.2000.tb05247.x. [DOI] [PubMed] [Google Scholar]
- 75.Jacob C, Yang PC, Darmoul D, Amadesi S, Saito T, Cottrell GS, Coelho AM, Singh P, Grady EF, Perdue M, Bunnett NW. Mast cell tryptase controls paracellular permeability of the intestine. Role of protease-activated receptor 2 and beta-arrestins. J Biol Chem. 2005;280:31936–48. doi: 10.1074/jbc.M506338200. [DOI] [PubMed] [Google Scholar]
- 76.Kiliaan AJ, Saunders PR, Bijlsma PB, Berin MC, Taminiau JA, Groot JA, Perdue MH. Stress stimulates transepithelial macromolecular uptake in rat jejunum. American Journal of Physiology. 1998;275:G1037–G1044. doi: 10.1152/ajpgi.1998.275.5.G1037. [DOI] [PubMed] [Google Scholar]
- 77.Soderholm JD, Yates DA, Gareau MG, Yang PC, MacQueen G, Perdue MH. Neonatal maternal separation predisposes adult rats to colonic barrier dysfunction in response to mild stress. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2002;283:G1257–G1263. doi: 10.1152/ajpgi.00314.2002. [DOI] [PubMed] [Google Scholar]
- 78.Yates DA, Santos J, Söderholm JD, Perdue MH. Adaptation of stress-induced mucosal pathophysiology in rat colon involves opioid pathways. American Journal of Physiology - Gastrointestinal and Liver Physiology. 2001;281:G124–G128. doi: 10.1152/ajpgi.2001.281.1.G124. [DOI] [PubMed] [Google Scholar]
- 79.Keita AV, Soderholm JD. The intestinal barrier and its regulation by neuroimmune factors. Neurogastroenterol Motil. 2010;22:718–33. doi: 10.1111/j.1365-2982.2010.01498.x. [DOI] [PubMed] [Google Scholar]
- 80.Bailey MT, Dowd SE, Parry NM, Galley JD, Schauer DB, Lyte M. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infect Immun. 2010;78:1509–19. doi: 10.1128/IAI.00862-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tannock GW, Savage DC. Influences of dietary and environmental stress on microbial populations in the murine gastrointestinal tract. Infect Immun. 1974;9:591–8. doi: 10.1128/iai.9.3.591-598.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bailey MT, Dowd SE, Galley JD, Hufnagle AR, Allen RG, Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav Immun. 2011;25:397–407. doi: 10.1016/j.bbi.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lyte M, Freestone PPEE. Microbial Endocrinology: interkingdom signaling in health and disease. Springer Publishers; 2010. [Google Scholar]
- 84.Santos J, Saperas E, Nogueiras C, Mourelle M, Antolín M, Cadahia A, Malagelada JR. Release of mast cell mediators into the jejunum by cold pain stress in humans. Gastroenterology. 1998;114:640–648. doi: 10.1016/s0016-5085(98)70577-3. [DOI] [PubMed] [Google Scholar]
- 85.Stephens RL, Tache Y. Intracisternal injection of a TRH analogue stimulates gastric luminal serotonin release in rats. Am J Physiol. 1989;256:G377–83. doi: 10.1152/ajpgi.1989.256.2.G377. [DOI] [PubMed] [Google Scholar]
- 86.Yang H, Stephens RL, Tache Y. TRH analogue microinjected into specific medullary nuclei stimulates gastric serotonin secretion in rats. Am J Physiol. 1992;262:G216–22. doi: 10.1152/ajpgi.1992.262.2.G216. [DOI] [PubMed] [Google Scholar]
- 87.Alverdy J, Holbrook C, Rocha F, Seiden L, Wu RL, Musch M, Chang E, Ohman D, Suh S. Gut-derived sepsis occurs when the right pathogen with the right virulence genes meets the right host: evidence for in vivo virulence expression in Pseudomonas aeruginosa. Ann Surg. 2000;232:480–9. doi: 10.1097/00000658-200010000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hughes DT, Sperandio V. Inter-kingdom signalling: communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–20. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Asano Y, Hiramoto T, Nishino R, Aiba Y, Kimura T, Yoshihara K, Koga Y, Sudo N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1288–95. doi: 10.1152/ajpgi.00341.2012. [DOI] [PubMed] [Google Scholar]
- 90.Lyte M. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol. 2004;12:14–20. doi: 10.1016/j.tim.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 91.Cogan TA, Thomas AO, Rees LE, Taylor AH, Jepson MA, Williams PH, Ketley J, Humphrey TJ. Norepinephrine increases the pathogenic potential of Campylobacter jejuni. Gut. 2007;56:1060–5. doi: 10.1136/gut.2006.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fish EW, Shahrokh D, Bagot R, Caldji C, Bredy T, Szyf M, Meaney MJ. Epigenetic programming of stress responses through variations in maternal care. Ann N Y Acad Sci. 2004;1036:167–80. doi: 10.1196/annals.1330.011. [DOI] [PubMed] [Google Scholar]
- 93.Meaney MJ. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annual Review of Neuroscience. 2001;24:1161–1192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- 94.Dinan TG, Stanton C, Cryan JF. Psychobiotics: a novel class of psychotropic. Biol Psychiatry. 2013;74:720–6. doi: 10.1016/j.biopsych.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 95.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–7. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vinolo MA, Ferguson GJ, Kulkarni S, Damoulakis G, Anderson K, Bohlooly YM, Stephens L, Hawkins PT, Curi R. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS One. 2011;6:e21205. doi: 10.1371/journal.pone.0021205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Layden BT, Angueira AR, Brodsky M, Durai V, Lowe WL., Jr Short chain fatty acids and their receptors: new metabolic targets. Transl Res. 2013;161:131–40. doi: 10.1016/j.trsl.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 99.Nepelska M, Cultrone A, Beguet-Crespel F, Le Roux K, Dore J, Arulampalam V, Blottiere HM. Butyrate produced by commensal bacteria potentiates phorbol esters induced AP-1 response in human intestinal epithelial cells. PLoS One. 2012;7:e52869. doi: 10.1371/journal.pone.0052869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ganapathy V, Thangaraju M, Prasad PD, Martin PM, Singh N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr Opin Pharmacol. 2013 doi: 10.1016/j.coph.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 101.Bindels LB, Dewulf EM, Delzenne NM. GPR43/FFA2: physiopathological relevance and therapeutic prospects. Trends Pharmacol Sci. 2013;34:226–32. doi: 10.1016/j.tips.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 102.Cresci GA, Thangaraju M, Mellinger JD, Liu K, Ganapathy V. Colonic gene expression in conventional and germ-free mice with a focus on the butyrate receptor GPR109A and the butyrate transporter SLC5A8. J Gastrointest Surg. 2010;14:449–61. doi: 10.1007/s11605-009-1045-x. [DOI] [PubMed] [Google Scholar]
- 103.Karaki S, Mitsui R, Hayashi H, Kato I, Sugiya H, Iwanaga T, Furness JB, Kuwahara A. Short-chain fatty acid receptor, GPR43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res. 2006;324:353–60. doi: 10.1007/s00441-005-0140-x. [DOI] [PubMed] [Google Scholar]
- 104.Nohr MK, Pedersen MH, Gille A, Egerod KL, Engelstoft MS, Husted AS, Sichlau RM, Grunddal KV, Seier Poulsen S, Han S, Jones RM, Offermanns S, Schwartz TW. GPR41/FFAR3 and GPR43/FFAR2 as Cosensors for Short-Chain Fatty Acids in Enteroendocrine Cells vs FFAR3 in Enteric Neurons and FFAR2 in Enteric Leukocytes. Endocrinology. 2013;154:3552–64. doi: 10.1210/en.2013-1142. [DOI] [PubMed] [Google Scholar]
- 105.Vanhoutvin SA, Troost FJ, Kilkens TO, Lindsey PJ, Hamer HM, Jonkers DM, Venema K, Brummer RJ. The effects of butyrate enemas on visceral perception in healthy volunteers. Neurogastroenterol Motil. 2009;21:952–e76. doi: 10.1111/j.1365-2982.2009.01324.x. [DOI] [PubMed] [Google Scholar]
- 106.Banasiewicz T, Krokowicz L, Stojcev Z, Kaczmarek BF, Kaczmarek E, Maik J, Marciniak R, Krokowicz P, Walkowiak J, Drews M. Microencapsulated sodium butyrate reduces the frequency of abdominal pain in patients with irritable bowel syndrome. Colorectal Dis. 2013;15:204–9. doi: 10.1111/j.1463-1318.2012.03152.x. [DOI] [PubMed] [Google Scholar]
- 107.Camilleri M. Peripheral mechanisms in irritable bowel syndrome. N Engl J Med. 2012;367:1626–35. doi: 10.1056/NEJMra1207068. [DOI] [PubMed] [Google Scholar]
- 108.Abrahamsson H, Ostlund-Lindqvist AM, Nilsson R, Simren M, Gillberg PG. Altered bile acid metabolism in patients with constipation-predominant irritable bowel syndrome and functional constipation. Scand J Gastroenterol. 2008;43:1483–8. doi: 10.1080/00365520802321212. [DOI] [PubMed] [Google Scholar]
- 109.Balsari A, Ceccarelli A, Dubini F, Fesce E, Poli G. The fecal microbial population in the irritable bowel syndrome. Microbiologica. 1982;5:185–194. [PubMed] [Google Scholar]
- 110.Si JM, Yu YC, Fan YJ, Chen SJ. Intestinal microecology and quality of life in irritable bowel syndrome patients. World J Gastroenterol. 2004;10:1802–5. doi: 10.3748/wjg.v10.i12.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Matto J, Maunuksela L, Kajander K, Palva A, Korpela R, Kassinen A, Saarela M. Composition and temporal stability of gastrointestinal microbiota in irritable bowel syndrome--a longitudinal study in IBS and control subjects. FEMS Immunol Med Microbiol. 2005;43:213–22. doi: 10.1016/j.femsim.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 112.Carroll IM, Chang YH, Park J, Sartor RB, Ringel Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog. 2010;2:19. doi: 10.1186/1757-4749-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Malinen E, Rinttila T, Kajander K, Matto J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–82. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 114.Kerckhoffs AP, Samsom M, van der Rest ME, de Vogel J, Knol J, Ben-Amor K, Akkermans LM. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol. 2009;15:2887–92. doi: 10.3748/wjg.15.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rajilic-Stojanovic M, Biagi E, Heilig HG, Kajander K, Kekkonen RA, Tims S, de Vos WM. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 116.Saulnier DM, Riehle K, Mistretta TA, Diaz MA, Mandal D, Raza S, Weidler EM, Qin X, Coarfa C, Milosavljevic A, Petrosino JF, Highlander S, Gibbs R, Lynch SV, Shulman RJ, Versalovic J. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology. 2011;141:1782–91. doi: 10.1053/j.gastro.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Krogius-Kurikka L, Lyra A, Malinen E, Aarnikunnas J, Tuimala J, Paulin L, Makivuokko H, Kajander K, Palva A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moayyedi P, Ford AC, Talley NJ, Cremonini F, Foxx-Orenstein AE, Brandt LJ, Quigley EM. The efficacy of probiotics in the treatment of irritable bowel syndrome: a systematic review. Gut. 2010;59:325–32. doi: 10.1136/gut.2008.167270. [DOI] [PubMed] [Google Scholar]
- 119.Hoveyda N, Heneghan C, Mahtani KR, Perera R, Roberts N, Glasziou P. A systematic review and meta-analysis: probiotics in the treatment of irritable bowel syndrome. BMC Gastroenterol. 2009;9:15. doi: 10.1186/1471-230X-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McFarland LV, Dublin S. Meta-analysis of probiotics for the treatment of irritable bowel syndrome. World J Gastroenterol. 2008;14:2650–61. doi: 10.3748/wjg.14.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kamiya T, Wang L, Forsythe P, Goettsche G, Mao Y, Wang Y, Tougas G, Bienenstock J. Inhibitory effects of Lactobacillus reuteri on visceral pain induced by colorectal distension in Sprague-Dawley rats. Gut. 2006;55:191–6. doi: 10.1136/gut.2005.070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rousseaux C, Thuru X, Gelot A, Barnich N, Neut C, Dubuquoy L, Dubuquoy C, Merour E, Geboes K, Chamaillard M, Ouwehand A, Leyer G, Carcano D, Colombel JF, Ardid D, Desreumaux P. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat Med. 2007;13:35–7. doi: 10.1038/nm1521. [DOI] [PubMed] [Google Scholar]
- 123.Verdu EF, Bercik P, Verma-Gandhu M, Huang XX, Blennerhassett P, Jackson W, Mao Y, Wang L, Rochat F, Collins SM. Specific probiotic therapy attenuates antibiotic induced visceral hypersensitivity in mice. Gut. 2006;55:182–90. doi: 10.1136/gut.2005.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bercik P, Verdu EF, Foster JA, Macri J, Potter M, Huang X, Malinowski P, Jackson W, Blennerhassett P, Neufeld KA, Lu J, Khan WI, Corthesy-Theulaz I, Cherbut C, Bergonzelli GE, Collins SM. Chronic gastrointestinal inflammation induces anxiety-like behavior and alters central nervous system biochemistry in mice. Gastroenterology. 2010;139:2102–2112. e1. doi: 10.1053/j.gastro.2010.06.063. [DOI] [PubMed] [Google Scholar]
- 125.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108:16050–5. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]