Abstract
Neural Crest (NC) cells are a multipotent migratory stem cell population unique to vertebrates, which contributes extensively to the formation of a wide array of neural and non-neural structures in the embryo. NC cells originate in the ectoderm at the border of the neural tube, undergo an epithelial-mesenchymal transition and acquire outstanding individual and collective migratory properties that allow them to disseminate and differentiate to different parts of the body. This exquisite capacity to switch from an epithelium to motile cells represents both a puzzling biological issue and an attractive model to address the basic mechanisms of cell migration and their alteration during cancer progression. Here we review how signaling pathways controlled by Rho GTPases, key players in cell adhesion, contraction, migration and polarity, contribute to control the different phases of NC development.
Keywords: Neural Crest, Rho GTPases, collective migration, EMT, signaling pathways
Introducing the Neural Crest
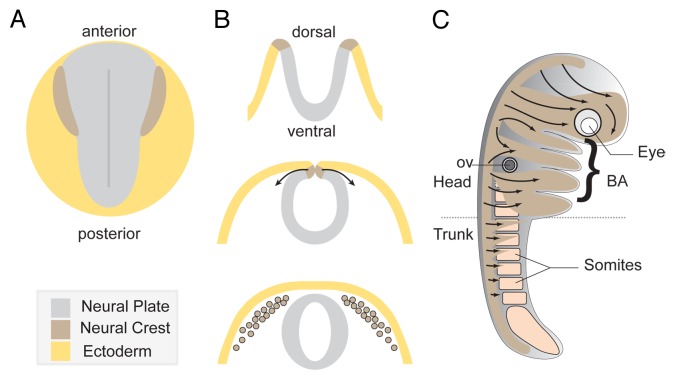
The Neural Crest (NC) is an ectodermal cell population that is induced within the neural plate border region during neurulation1 (Fig. 1A). NC cells are multipotent stem cells that generate an astonishing array of derivatives. In particular, they make an outstanding contribution to head and neck structures.2-4 NC cells produce most of the cephalic bones and cartilages and contribute to the meninges that sheathe the brain. NC cells also give rise to connective tissues of the cephalic blood vessels, tendons and smooth muscles and help to organize the outflow track of the heart.5 They generate pigment cells, all the glia and neurons of the gut and trunk peripheral nervous systems (PNS) as well as most of the cephalic PNS together with placodes.2,3 NC cells also contribute to peri-ocular structures and numerous cephalic organs including the middle and inner ear, the teeth, and the tongue among others.6,7
Figure 1. Overview of Neural Crest development. (A) Diagram depicting a dorsal view of a vertebrate embryo at early neurula stage. (B) Neural tube formation and neural crest delamination. Neural crest cells exit the dorsal region of the closing neural tube. (C) Summary of the main neural crest streams traveling through the embryo. Cephalic NC cells invade the face and the branchial arches while trunk neural crest cells migrate under the skin and through the somites.
NC cells are only found in vertebrates and their emergence had a huge impact on the evolution of the chordate phylum. Comparisons of NC development in higher vertebrates (mouse, chicken, Xenopus and zebrafish) and lower vertebrates (lampreys) as well as studies of common ancestors (amphioxus and ascidians) have given great insight into NC evolution. NC cells most likely emerged from a primitive pigment cell population that acquired motile properties and progressively co-opted genes and functions during evolution.2,8,9 Their massive contribution to the organization of the head of the vertebrate has led to the “new head” idea which proposes that the new organization of sensory structures, rendered possible by the emergence of NC cells and placodes, allowed the acquisition of a predatory lifestyle, unavailable to passive filter-feeders like urochordates or cephalochordates.10,11
NC induction is a complex process involving interactions between the neural plate, the non-neural ectoderm and the underlying mesoderm.1,12-14 Ligands from the Bone Morphogenetic Protein (BMP) family coming from the non-neural ectoderm are counter-balanced by anti-BMP signals produced at the midline (i.e Noggin, Chordin, Nodal). This antagonism establishes a medio-lateral gradient of BMP activity such that low levels of BMP signaling are found within the neural plate and high levels in the adjacent ectoderm. Then, cells that experience intermediate levels of BMP are induced to become NC cells by Wnt ligands coming from the non-neural ectoderm and the mesoderm. NC induction also involves other pathways such as Fibroblast Growth Factor (FGF) and Notch/Delta. FGF signaling acts upstream of Wnt,15 whereas the Notch pathway modulates BMP signaling.16-19
This combination of inductive signals triggers the expression of several genes that can be subdivided into 2 main categories. First, a series of neural plate border markers are upregulated at the interface between the non-neural and neural ectoderm. Among these genes, we find transcription factors from the Msx, Pax and Zic families as well as AP2, Dlx5, Gbx2, and Hairy2.1,20 Then, Neural Crest specific markers such Snail1/2, Twist, Foxd3, and SoxE genes are induced.14
After induction, NC cells migrate extensively, before which they detach from the neurectoderm in a process known as delamination (Fig. 1B). This step involves an epithelial-mesenchymal transition (EMT) that includes a loss of apico-basal polarity, a modification of cell-cell and cell-matrix properties, a local degradation of extracellular matrix (ECM) or membrane receptors and acquisition of motility.21-25 EMT is controlled by an array of transcription factors downstream of Wnt and BMP signaling. The main regulators of NC EMT include Snail1/2, Twist, Foxd3, and Sox9/1026-32 and are found in most animal models although their role in EMT has not necessarily been addressed. In cephalic regions, studies on the chick embryos have highlighted additional factors have been identified such as LSox5,33 Ets1,34-36 and p53.37 Together, these factors control the various aspects of EMT required for delamination to proceed. In particular, they control changes in cell adhesion properties.38,39 NC cells have to separate from neurectoderm and switching to a different repertoire of adhesion molecules is an efficient way of preventing NC and neuroepithelial cells from mixing with one another. NC cells downregulate E-Cadherin and subsequently downregulate N-Cadherin4 but the timing of these successive losses of cadherin expression may vary along the antero-posterior axis and across species. In addition, they initiate expression of multiple type-II cadherins (i.e., Cadherin 6/6B/7/11) which mediate weaker adhesion forces.21,25,38,40 This cadherin switch liberates NC cells from other cell populations and contributes to the onset of NC migration, aided and abetted by local matrix rearrangements and integrin activation.
NC cells go on to generate the astonishing list of derivatives mentioned above and to colonize almost every part of the embryo. The pathways involved in NC differentiation are beyond the scope of this review and relevant information can be found elsewhere.4,41,42 The cell and molecular mechanisms of NC cell migration will be briefly described in the next section.
NC cells are an excellent model with which to study classical questions of developmental biology, from induction and patterning to stem cell properties and differentiation to cell migration. Interestingly, the main steps of NC development are also reminiscent of the series of events that occur during cancer metastasis. Cells within a given organ acquire new capabilities; they separate from their original tissue; migrate and finally settle elsewhere. Many genes essential for NC development (i.e., Snail, Twist, Sox, and Ets factors) are key players in cancer progression too.43-48 Thus, NC cells are a good model to study the normal function of these genes. Several cancers actually arise from NC-derived cell types: Skin49 and uveal50 melanoma (melanocytes), neuroblastoma51 (adrenal gland or nerve of the sympathetic nervous system), pheochromocytomas52 (chromaffin cells), schwannoma53 and neurofibrosarcoma54 (Schwann cells) . Furthermore, several cell lines have been made from tumors of NC origin. The most common include PC12 (rat pheochromocytoma), N1E-115 (mouse neuroblastoma) as well as countless melanoma cell lines from various model organisms.
Overview of Neural Crest cell migration
Neural Crest guidance and migration pathways
Migratory NC cells follow well-defined routes throughout the embryo4,55,56 (Fig. 1C). These paths are lined with permissive extracellular matrix molecules,57 mostly fibronectin, laminins and collagens to which NC cells bind via integrins and syndecans.58 Access to a given area is controlled by negative and positive cues. The main negative regulators act in 2 ways: (1) they prevent entry into a given territory or (2) they prevent some NC cells to join a specific NC sub-population. These 2 levels of control generate of pattern of discrete streams from the continuous wave of NC cells that delaminate from the neuroepithelium.
In the head, NC cells are subdivided into 3 main streams found in all species: (1) An anterior stream that goes around the eye and invades the anterior-most region of the face and first branchial arch (BA1); (2) A pre-otic stream that mostly migrates to BA2 and (3) a post-otic stream that invades BA3 and 4 (Fig. 1C) but also contains cells that will migrate further ventrally toward the heart and along the gut.4 In the trunk, the main pattern is observed when cells cross the somites (Fig. 1C). In each animal model studied, migratory NC cells avoid a part of the somite. They pass through the anterior half in mice and chick embryos whereas they migrate along the medial or caudal part in fish and frogs, respectively.48 Numerous negative cues have been identified based on functional assays and/or loss-of-functions performed in chick and mouse and the main players are members of the semaphorin,59-65 Eph-ephrin,66-72 Slit-Robo,73-75 and endothelin76 families of signaling molecules. Some of these molecules have dual roles, acting as negative regulators for some NC cells and as positive cues for others.
The generation of the 3 cephalic NC streams is mostly due to the presence of type3-Semaphorins coming from the rhombencephalon and the otic vesicle which block migration in the mesenchyme adjacent to rhombomeres 3 and 5.64,65 In parallel, Eph-ephrin signaling controls cell sorting among NC cells but also between NC cells and their local environment to ensure that only cells with a specific Eph-ephrin repertoire are able to go through a given territory.68,71,72 The downstream effectors of the inhibitory cues remain poorly understood but in vitro studies indicate that they may act by blocking adhesion to the matrix. Similar signaling pathways and mechanisms pattern trunk NC migration.56
NC cells are further guided toward specific areas by positive regulators of cell migration that act as chemokinetic or chemotactic factors. Vascular Endothelium Growth Factor A (VEGFA),77 FGF2/8,78-80 Stromal cell-derived factor-1 (Sdf1/Cxcl12),81-89 Platelet-derived Growth Factors (PDGF),90,91 Glial cell-Derived Neurotrophic Factor (GDNF),92-94 Endothelin-3 (ET-3)95,96and semaphorin 3C60 have been found to act as positive cues for NC migration. Sdf1 controls the overall dorso-ventral migration of cephalic NC cells,81,84,86 the targeted migration of dorsal root89 or sympathetic85 ganglia precursors, melanocyte migration toward the hair follicle87 and early cardiac NC migration.82 VEGFA and FGF signaling are required for migration into the branchial arches.77,78 GDNF and ET-3 control enteric NC migration along the gut41,97 whereas FGF2/8 controls NC migration toward the face.78,80
Mechanisms of solitary and collective migration in NC cells
An interesting aspect of NC migration is that the various NC subpopulations exhibit a range of different modes of migration with varying degrees of cooperation. Some, like melanocytes, migrate as single cells whereas cephalic NC cells migrate in a collective manner, either as loosely connected chains as in chick NC cells or as a pseudoepithelium, as seen in early migratory cephalic Xenopus NC cells. Other types, such as enteric NC cells, have intermediate phenotypes with a mix of directional spreading, individual movement and collective behavior.
Importantly, all NC subpopulations studied so far respond to physical cell–cell contact regardless of their modes of migration. Time-lapse movies performed on mouse,97 chick,98,99 Xenopus84,100 and zebrafish NC100 cells have shown that when two NC cells collide they retract their cell protrusions and momentarily stop migrating. This behavior is reminiscent of contact inhibition of locomotion (CIL), which was first described in fibroblasts.101,102 Indeed, further experiments in fish and frogs confirmed that NC cells exhibit CIL.81,84 Since CIL repolarizes cells away from the contact with one another, it tends to promote cell dispersion. As such it is one of the main driving forces of NC migration.
However, NC cells do not simply disperse and most NC subpopulations travel in large cohorts. Some, like cephalic NC cells, clearly undergo collective cell migration and remain close to one another throughout migration. Intriguingly, CIL is counter-balanced by another mechanism, coined co-attraction, which acts at a distance to maintain cell cohesiveness.103 In Xenopus, each NC cell expresses both complement factor C3a and its receptor C3aR. C3a diffuses and establishes a short-range gradient around each NC cell. Thus, NC cells are able to sense each other from a distance and are drawn toward one another.103 The co-existence of CIL and co-attraction means that cephalic NC cells do not disperse excessively in spite of their inability to maintain stable cell-cell junctions. Observations made in chick embryos98,99 suggest that NC cells may also follow one another in this model but the molecular effectors remain to be found.
Rho Signaling in Neural Crest Cell Development
Introducing the Rho family of GTPases
Rho GTPases are key regulators of basic cell dynamics (cell-cell and cell-ECM adhesion, polarity, migration, contraction) and have been implicated in many biological processes, especially cell migration and differentiation.104 Given the outstanding dynamics of NC cells, it was thus likely that these regulators were at play in NC development.
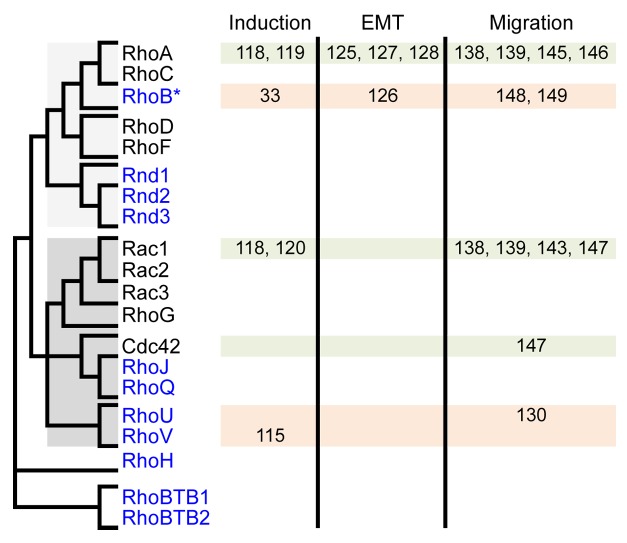
Rho GTPases delineate a Ras-like family that emerged early in eukaryotes.105 In vertebrates, the Rho family contains about 20 members that cluster in 8 sub-families (Fig. 2). RhoA-C, Rac and Cdc42, the most ancient members, are present in all fungi and metazoans and have been extensively studied in many animal and cellular models. They constitute the “big three” GTPases that control basic F-actin-dependent structures to modify cell shape, adhesion and migration in response to external cues: RhoA-C promotes cell-ECM adhesion and contraction through the formation of focal adhesions and actomyosin bundles, whereas Rac and Cdc42 act in the opposite way by favoring actin polymerization and branching over cell contraction and thus allowing extension of membrane protrusions, i.e., lamellipodia for Rac and filopodia for Cdc42. At the biochemical level, Rho GTPases behave as molecular switches by oscillating between inactive GDP-bound and active GTP-bound conformations. Only when bound to GTP do they gain the ability to bind to and activate a set of downstream effector targets, which mediate their cellular effects.104
Figure 2. GTPases of the Rho family and their implication in early phases of NC development. Rho members are shown on the left and their relatedness is summarized by a simplified dendrogram derived from Boureux, 2007.105 Rho members regulated at the transcriptional level are in blue. They are constitutively active, either because they are deficient for GTPase activity (Rnd1–3, RhoH, RhoBTB1–2) or because they exchange their nucleotide spontaneously (RhoJ, RhoQ, RhoU, RhoV). RhoB represents a particular case, being induced at the transcriptional level yet controlled biochemically by regulators also acting on other members. The implication of Rho members in early phases of NC development (induction, EMT and migration) is indicated with the corresponding reference numbers.
For about half of the Rho members (RhoA, C; Rac1, 2, 3; Cdc42; RhoQ; RhoD, F), activity is controlled either positively by RhoGEFs, which catalyze guanine nucleotide exchange reaction thus favoring GTP loading, or negatively by RhoGAPs, which boost the intrinsic GTPase activity and promote the return to the inactive GDP-bound form. In contrast, the other members (Rnd1, 2, 3,106 the splicing Rac1b variant,107 RhoJ,108 RhoBTB1/2, RhoH109 or RhoU/V110) either exchange guanine nucleotides spontaneously or are devoid of GTPase activity. These members, also known as atypical Rho GTPases,111 are therefore constitutively bound to GTP. Contrary to canonical Rho members, the biochemical activity of atypical members is thus tightly linked to their levels of expression. This particularity constitutes a plus for developmental studies, since the spatiotemporal activity of atypical Rho GTPases can be readily detected by mRNA in situ hybridization. Furthermore, since activity correlates with expression, the use of morpholino antisense oligonucleotides is likely to efficiently inhibit their activity as compared with canonical GTPases.
The following sections will discuss the involvement of Rho signaling in NC induction, delamination and migration.
Rho signaling in NC induction
As mentioned above, NC induction is a complex process triggered by BMP, Wnt, FGF or Notch/Delta emanating from mesoderm and neural- and non-neural ectoderm. Combination of these external cues induces a cascade of transcription factors specifying and maintaining the NC precursors (see Steventon 2005112 and Klymkowsky 2010113 for reviews). Among NC specifiers are Hairy, Msx1, Dlx, or AP-2, initially expressed in the non-neural ectoderm and later restricted to the neural folds, and Pax3, Zic1 or c-Myc, expressed in the neural folds. Maintenance of NC precursors is controlled by Snail1–2, FoxD3, Id2/3, Sox5, 9 and 10, and Twist1. Rho factors are also involved in the cascade leading to NC precursors. Here we will expand on the involvement of the RhoGEF Lfc and the Rho members RhoV, RhoA, and Rac1.
Lfc
Ectopic expression of the RhoGEF Lfc, normally controlled by BMP-4 in the neural plate, elicited expansion of the neural plate territory at the expense of NC cells.114 However, it is not known whether Lfc affects NC specification directly or indirectly through its effect on the neural plate.
RhoV
In Xenopus, RhoV (an atypical Rho GTPase closely related to Rac and Cdc42105) is induced very early in NC, detected at stage 12 in prospective NC domains lateral to the neural plate in a canonical (β-catenin-dependent) Wnt manner.115 RhoV is also induced early in NC in the chick, detected in the neural folds from HH stage 7.116 RhoV is no longer detected in early migratory NC cells (stage 22) that are driven by non-canonical (β-catenin-independent) Wnt signaling.117 RhoV depletion inhibited expression of several NC specifiers (Snail2, Sox9–10, Twist) in a cell autonomous fashion and restricted the prospective NC territory at the benefit of the neural plate. Conversely, RhoV overexpression increased NC specifiers' expression and expanded the prospective NC territory at the expense of the neural plate. Overall these data paint a picture whereby RhoV favors the acquisition of NC identity in response to canonical Wnt signaling. It must be stressed that in contrast to Rac1 and RhoA, whose activation is controlled by RhoGEFs, RhoV is spontaneously active and its activity therefore correlates with its level of expression.
RhoA and Rac1
RhoA and Rac1 are ubiquitously expressed Rho members whose activities are controlled biochemically and cannot thus be inferred from expression levels measured by in situ hybridization or conventional immuno-histochemistry. However, their activities can be manipulated by ectopic expression of constitutively active (CA) or dominant negative (DN) mutants, analogous to the Ras mutants G12V/Q61L and T17N, respectively.104 Activation of RhoA- and Rac1-controlled pathways showed opposite effects in NC induction118; NC prospective territory and expression of NC specifiers varied proportionally to Rac1 activity and inversely proportionally to RhoA activity. Snail2 showed the most pronounced sensitivity to either GTPase as it was almost completely repressed upon Rac1 inhibition or RhoA activation. The effect of RhoA inhibition was similar to that of its downstream effector ROCK, which increased Snail2 gene transcription in Xenopus118 and increased the proportion of NC-like progenitors in human ES cells.119 By contrast, the inhibition of the p21 activated kinase 1 (PAK1), a major effector of RhoU–V, Rac1–3, and Cdc42 family members, had no effect on Snail2, Sox9, or AP2 expression120 indicating that this kinase does not mediate the effects of Rac1 or RhoV during NC induction.
All these observations raise the question of the mechanisms through which Rho signaling might affect gene expression during NC induction. This may be associated with the common outcomes of Rho activities on cell physiology, such as cell adhesion, polarity, lateral inhibition or cell cycle dynamics, all known to be critical for NC induction.113,121,122 It may also pertain to less known outcomes, like the role of Rac1 on c-myc phosphorylation and degradation during keratinocyte differentiation.123 This remains a wide open field.
Rho signaling in NC delamination
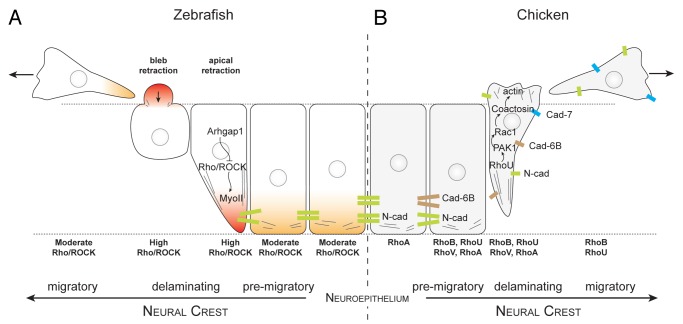
In vivo imaging at trunk levels in chick and fish embryos showed that NC cells undergo morphological changes characteristic of EMT.124,125 Premigratory NC cells lose their adherens junctions, round up at the basal edge of the neuroepithelium, produce membrane blebs and detach from the basal surface. The free apical cell tail retracts, the NC cell body translocates out of the epithelium and apico-basal polarity is lost (Fig. 3). Interestingly, these events need not occur in a particular order. For instance, many NC cells exit the neural tube without downregulating cell-cell adhesion first.124,125 This reflects the fact that EMT is controlled by an array of transcription factors acting in parallel rather than in a linear fashion. Rho-controlled pathways are prime candidates for regulating these cellular events in NC cell EMT since cell rounding and blebbing are known to be controlled by RhoA, ROCK and myosin-II in other cell systems. Indeed, inhibition of RhoA, B and C activity by C3 exotoxin prevented delamination of trunk NC cells in the chick126 and the pharmacological inhibition of ROCK or myosin-II led to a substantial decrease in NC blebbing and EMT in zebrafish cranial NC.124,125 The requirement of Rho/ROCK activity is therefore consistent with the notion that cell rounding and blebbing are required at the onset of delamination to allow NC cell detachment from the neural tube.
Figure 3. Rho signaling during neural crest delamination in zebrafish and chick embryos. (A) Current view of zebrafish neural crest delamination. Arhgap1 restricts Rho activity to the apical side. NC cells exit the neural tube by retracting their apical side and forming blebs on their basal side. (B) Current view of chick neural crest delamination. Changes in cell-cell adhesion repertoire occur concomitantly with changes in cell contractility. NC cells may exit by first disrupting their cell-cell adhesion and then acquire motility. Alternatively, they may force their way out by forming protrusions and mechanically disrupting their junction with other neuroepithelial cells by pulling the cell body. Delaminating chick NC cells express RhoA, B, U and V. The respective roles of the Rho proteins are open to conjecture although it is likely that RhoU controls protrusion formation via PAK1/Rac1 whereas RhoB appears to be involved in cell-cell adhesion disassembly.
There is some controversy however as opposite observations were reported in quail embryos, in which Rho or ROCK inhibition facilitated EMT of trunk NC cells.127 The conflict may lie either in the use of distinct Rho C3 exoenzymes and ROCK inhibitors, which may have differential potencies, or in the different developmental stages at which explants were assayed in the 2 studies (stage 10 vs. stage 12–13). One likely explanation is that cell-cell adhesion and contractility require different levels of Rho activity, which cannot be precisely controlled in loss-of-function experiments. On the other hand, overexpression of RhoB in chick triggered NC cell rounding, which led to the idea that RhoB is important for the disassembly of the cell-cell adhesion complex.28 Whether this is due to an increased contractility or to a competition with RhoA at cell-cell junctions remains unknown. However, RhoB overexpression did not prime NC delamination or motility but triggered NC cell apoptosis, indicating that the downregulation of cell-cell adhesion and the acquisition of motility are largely independent.
A recent study revisited this issue in zebrafish and showed that Rho/ROCK activation is restricted to the apical subcellular region in an Arhgap1-dependent manner and is essential for detachment from the neuroepithelium128 (Fig. 3A). The actual regulators of actomyosin contractility in chick NC remain elusive. However, data collected on RhoU, PAK1, and coactosin116,129,130 suggest that these factors could be involved in lamellipodia formation during delamination and migration (Fig. 3B).
Overall, data in fish and chick embryos support the notion that, at trunk level, the Rho/ROCK pathway promotes EMT through apical actomyosin contraction during a very narrow spatiotemporal window whereas Rac activity is likely to be mostly required for newly acquired motile properties.
At cephalic levels, NC departure is a lot more dramatic with multi-layered groups of cells migrating out of the neural tube at once. Several cranial specific factors such as Ets134,131 and LSox533 have been identified but their downstream effectors are still elusive. The expression of Snail2 is maintained throughout the whole delamination phase whereas it is quickly lost at trunk levels and this may also contribute to enhance EMT at cephalic levels. However, we do not have information about Rho GTPases activity or subcellular localization during cephalic crest emigration. In chick, RhoB is clearly expressed in emigrating cephalic NC cells132 but its function has not been studied at early stages. With dozens of NC cells undergoing EMT at the same time, the requirements for cell contractility and retraction of the apical tail may be very different from those in the trunk. In chick, the integrity of the dorsal neuroepithelium is not preserved during delamination since dorsal neural tube closure and NC departure are happening at the same time.133 In Xenopus and mouse, cephalic NC departure takes place while the neural plate is still open and thus NC cells may be leaving a more intact epithelial sheet. However, cranial NC cells in mouse have a very mesenchymal phenotype from the onset of migration134 whereas in Xenopus they start as a pseudoepithelial sheet.135,136 All these observations suggest that contractility, cell-cell adhesion or polarity may be controlled differently in mouse, chick, and Xenopus but in absence of experimental data one can only speculate about potential differences in terms of Rho GTPases activities in cephalic NC cells.
Rho signaling and NC migration
Once delaminated, NC cells must gain and maintain persistent migration and navigate along specific paths to reach their final destinations. This requires the coordination of multiple processes, e.g., sensing of complex chemo-attractive and repulsive cues and their integration into appropriate cellular adhesive and migratory properties. As a consequence of EMT, NC cells extend protrusions, i.e., lamellipodia and filopodia, and acquire a fibroblast-like motile behavior. This type of motility has been extensively studied in many biological systems and relies on the coordinated antagonistic activities of RhoA and Rac1.
RhoA and Rac1 in NC cell migration
Although ubiquitously expressed in all cells of Xenopus embryos, Rac1 has a more pronounced expression in regions, such as migrating NC, that undergo intense cell movements.137 Expression of DN-Rac1 mutants in quail embryos inhibited NC migration but had no effect on specification or delamination. This is consistent with the fact that lamellipodia, major cell outcomes of Rac1 activity, are only formed after EMT completion.138 Interestingly, expression of CA-Rac1 mutants also inhibited NC cell migration. This likely results from the isotropic distribution of CA-Rac1 in transfected cells, whereas endogenous active Rac1 is normally distributed along a gradient from leading to trailing edge, as demonstrated in vivo and ex vivo by FRET analysis in Xenopus embryos.139
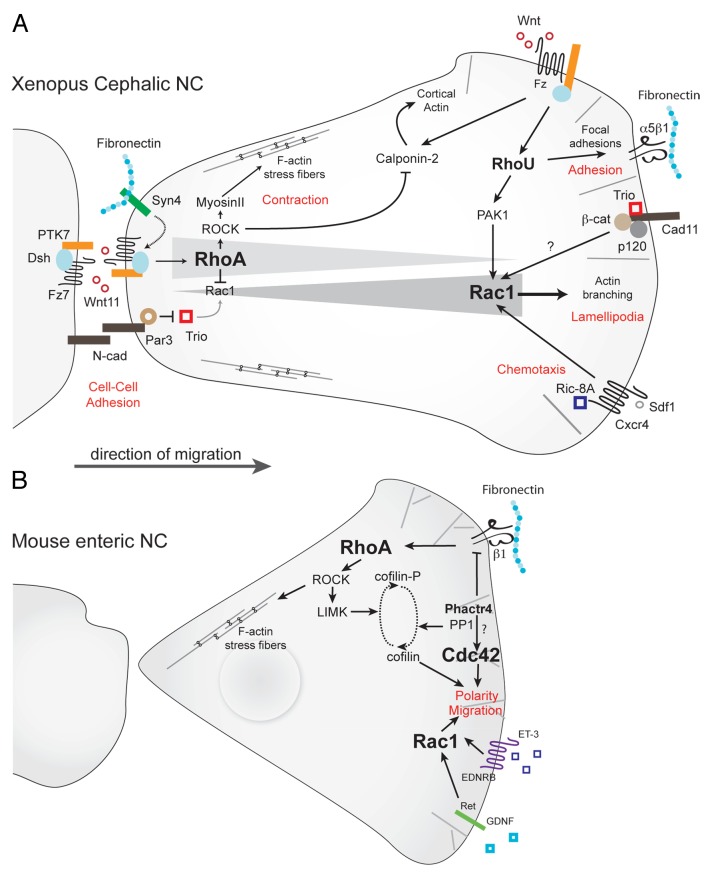
The restricted cellular distribution of active Rac1 is controlled by multiple antagonistic signals: at the trailing edge, Rac1 activity is downregulated by cell-cell contacts, such as those mediated by N-cadherin.47,84 Cell-matrix interactions via Syndecan-4 signaling also contribute to lower Rac1 activity.139 A molecular mechanism evidenced for the reduction of Rac1 signaling is the inhibition of the RhoGEF Trio through its interaction with Par3.140 Rac1 activity is also indirectly reduced through RhoA activation by the non-canonical Wnt/PCP pathway via Wnt11, Frizzled7, PTK-7 and Dishevelled81,100,117,139,141,142 (Fig. 4A).
Figure 4. Rho signaling in neural crest migration. (A) Rho signaling in migratory Xenopus cephalic neural crest cells. RhoA/ROCK signaling predominates at the rear of the cells. Wnt/PCP effectors (Wnt11/Fz7/PTK7/Dsh) activate RhoA whereas Par3 lowers Rac1 activity by inhibiting Trio. At the cell leading edge, Rac1 activity is triggered by Cadherin-11/Trio, RhoU via PAK1 and Sdf1/Cxcr4 via Ric-8A. The non-canonical Wnt pathway induces RhoU, which is required for cell adhesion, and Calponin-2, which antagonizes Rho-induced stress fibers. (B) In mouse enteric NC cells, integrin signaling activates Rho/ROCK pathways whereas GDNF and ET-3 activate Rac1/Cdc42 activities. Phactr4 controls lamellipodia formation at the leading edge cell through inhibition of integrin signaling toward RhoA/LIMK and cofilin dephosphorylation. Potential pathways are indicated by question marks.
At the leading edge, Rac1 is activated by Trio, itself activated by the pro-migratory Cadherin-11143 and can be further activated by external guidance cues; (1) Exposure to Sdf1 elicits a pronounced activation of Rac1 at the leading edge of Xenopus NC cells through Cxcr4 stimulation.84 Sdf1-mediated Rac1 activation requires a pre-established gradient of Rac1 activity within the cell, mostly controlled by cell-cell interactions,84 as discussed above; (2) Rac1 is also activated by C3a/C3aR signaling as part of the co-attraction mechanism that maintains migratory NC cells at relatively constant density103 (see section 2). Whether Rac1 activation downstream of C3a/C3aR or Sdf1/Cxcr4 requires Cadherin-11 and/or Trio remains unknown; (3) In enteric NC cells, proposed chemoattractants such as Glial cell line-Derived Neurotrophic Factor (GDNF) and Endothelin-3 (ET-3) also activate Rac1144 whereas integrin signaling seems to mostly activate the RhoA/ROCK signaling.145
RhoA signaling also controls NC collective migration. In the chick embryo, post-otic NC cells were less directional when RhoA was over activated, while they migrated more slowly upon RhoA inhibition.146 A similar observation was reported in the mouse enteric NC (Fig. 4B), in which the level of RhoA/ROCK activity at the leading edge is negatively controlled by Phactr4 (PHosphatase and ACTin Regulator 4).145In both cases collective migration was inhibited.145,146 This effect is likely due to the fact that affecting RhoA levels strongly modifies cell shape and prevents NC cells from efficiently interacting with one another.
Altogether Rho and Rac signals integrate to control cell polarity (Fig. 4). The role of Rac1 however remains to be examined in mammals in more details, since its inactivation in mice was reported to have no detectable effect on early steps of NC development.147 Whether the impact of RhoA on cell polarity also influences chemotactic abilities in chick and mouse NC cells remains to be investigated.
RhoB and RhoU in NC cell migration
In Xenopus and mice, RhoB mRNA expression was detected in NC cells all along their migration paths,148,149 whereas in the chick, RhoB expression was restricted to pre-and early migrating cells.126 The exact role of RhoB in NC migration requires confirmation by loss-of-function experiments that specifically inhibit RhoB as the C3 exotoxin used in Liu et al.126 inactivates the three RhoA, B, and C. On the other hand, RhoB overexpression into the neural tube is not sufficient to endow cells with motile properties28; therefore it must be acting in concert with other factors.
In Xenopus and chick, the atypical RhoU mRNA is induced in NC at the onset of migration.130 RhoU most likely promotes actin polymerization through PAK1109 (Fig. 4A). In Xenopus embryos, RhoU depletion or high expression both inhibited NC migration and led to a strong reduction in size of the NC-derived structures. RhoU could rescue RhoV depletion in NC specification115 whereas RhoV did not rescue RhoU depletion in NC migration.130 The major difference between these 2 closely related GTPases are (1) RhoU displays an N-terminal SH3-binding domain and (ii) they are induced by distinct pathways (canonical Wnt for RhoV and non-canonical Wnt for RhoU). This supports a scenario according to which RhoU/RhoV duplication in vertebrates was rapidly followed by functional and temporal specialization in NC. Whereas RhoU depletion inhibited cell adhesion and protrusions in Xenopus NC explants, a moderate overexpression did not affect cell adhesion nor migration speed. However, it strongly increased the number of cells leaving the explants in a PAK1- and Rac1-dependent manner but inhibited the persistence of their migration. The hypothesized role of RhoU in NC migration is consistent with its cellular effects on cell adhesion in fibroblastic and epithelial cells.150-152
Concluding Remarks
NC development represents an outstanding dynamic process in which key issues of healthy and cancer cell biology are at play: interaction with the microenvironment to form a presumptive territory, control of EMT and maintenance of an epithelial structure, connection between cell proliferation, cell sorting, cell fate and survival, control of multipotency and cell differentiation, individual and collective cell migration. The currently identified functions of Rho GTPases in NC development mainly pertain to the control of contraction and adhesion and the establishment and maintenance of cell polarity, thus influencing downstream processes such as cell-cell interaction, long-distance migration and cell guidance. This leaves major unanswered issues on how Rho signaling controls the early steps of NC induction and opens new questions on the spatiotemporal activation of Rho signaling: as we have discussed above, subtle variations in Rho activity levels during EMT are linked to different cell outcomes. This likely relies on dynamic changes in activity of RhoGEFs or RhoGAPs, whose identification should help understand how the level of active Rho influences cell-cell adhesion and motility. Getting deeper knowledge on the function Rho GTPases in such dynamic processes will also require new, sensitive and tunable tools.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique (CNRS) and grants from the Ligue Nationale contre le Cancer (P.F.) and the Fondation pour la Recherche Medicale (FRM/AJE201224) (E.T.).
References
- 1.Milet C, Monsoro-Burq AH. Neural crest induction at the neural plate border in vertebrates. Dev Biol. 2012;366:22–33. doi: 10.1016/j.ydbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Hall B. The neural crest and neural crest cells in vertebrate development and evolution. New York: Springer, 2009. [Google Scholar]
- 3.Le Douarin N, Kalcheim C. The neural crest. Cambridge, UK; New York, NY, USA: Cambridge University Press, 1999. [Google Scholar]
- 4.Mayor R, Theveneau E. The neural crest. Development. 2013;140:2247–51. doi: 10.1242/dev.091751. [DOI] [PubMed] [Google Scholar]
- 5.Kirby ML, Hutson MR. Factors controlling cardiac neural crest cell migration. Cell Adh Migr. 2010;4:609–21. doi: 10.4161/cam.4.4.13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupin E, Creuzet S, Le Douarin NM. The contribution of the neural crest to the vertebrate body. Adv Exp Med Biol. 2006;589:96–119. doi: 10.1007/978-0-387-46954-6_6. [DOI] [PubMed] [Google Scholar]
- 7.Liu HX, Komatsu Y, Mishina Y, Mistretta CM. Neural crest contribution to lingual mesenchyme, epithelium and developing taste papillae and taste buds. Dev Biol. 2012;368:294–303. doi: 10.1016/j.ydbio.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikitina N, Sauka-Spengler T, Bronner-Fraser M. Chapter 1. Gene regulatory networks in neural crest development and evolution. Curr Top Dev Biol. 2009;86:1–14. doi: 10.1016/S0070-2153(09)01001-1. [DOI] [PubMed] [Google Scholar]
- 9.Sauka-Spengler T, Bronner-Fraser M. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis. 2008;46:673–82. doi: 10.1002/dvg.20436. [DOI] [PubMed] [Google Scholar]
- 10.Northcutt RG, Gans C. The genesis of neural crest and epidermal placodes: a reinterpretation of vertebrate origins. Q Rev Biol. 1983;58:1–28. doi: 10.1086/413055. [DOI] [PubMed] [Google Scholar]
- 11.Gans C, Northcutt RG. Neural crest and the origin of vertebrates: a new head. Science. 1983;220:268–73. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- 12.Steventon B, Mayor R. Early neural crest induction requires an initial inhibition of Wnt signals. Dev Biol. 2012;365:196–207. doi: 10.1016/j.ydbio.2012.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–68. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- 15.Hong CS, Park BY, Saint-Jeannet JP. Fgf8a induces neural crest indirectly through the activation of Wnt8 in the paraxial mesoderm. Development. 2008;135:3903–10. doi: 10.1242/dev.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glavic A, Silva F, Aybar MJ, Bastidas F, Mayor R. Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development. 2004;131:347–59. doi: 10.1242/dev.00945. [DOI] [PubMed] [Google Scholar]
- 17.Endo Y, Osumi N, Wakamatsu Y. Bimodal functions of Notch-mediated signaling are involved in neural crest formation during avian ectoderm development. Development. 2002;129:863–73. doi: 10.1242/dev.129.4.863. [DOI] [PubMed] [Google Scholar]
- 18.Cornell RA, Eisen JS. Delta signaling mediates segregation of neural crest and spinal sensory neurons from zebrafish lateral neural plate. Development. 2000;127:2873–82. doi: 10.1242/dev.127.13.2873. [DOI] [PubMed] [Google Scholar]
- 19.Stuhlmiller TJ, García-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 2012;69:3715–37. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plouhinec JL, Roche DD, Pegoraro C, Figueiredo AL, Maczkowiak F, Brunet LJ, Milet C, Vert JP, Pollet N, Harland RM, et al. Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers. Dev Biol. 2014;386:461–72. doi: 10.1016/j.ydbio.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Theveneau E, Mayor R. Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Dev Biol. 2012;366:34–54. doi: 10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 22.Strobl-Mazzulla PH, Bronner ME. Epithelial to mesenchymal transition: new and old insights from the classical neural crest model. Semin Cancer Biol. 2012;22:411–6. doi: 10.1016/j.semcancer.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471–86. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- 24.Kerosuo L, Bronner-Fraser M. What is bad in cancer is good in the embryo: importance of EMT in neural crest development. Semin Cell Dev Biol. 2012;23:320–32. doi: 10.1016/j.semcdb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duband JL. Diversity in the molecular and cellular strategies of epithelium-to-mesenchyme transitions: Insights from the neural crest. Cell Adh Migr. 2010;4:458–82. doi: 10.4161/cam.4.3.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barriga EH, Maxwell PH, Reyes AE, Mayor R. The hypoxia factor Hif-1α controls neural crest chemotaxis and epithelial to mesenchymal transition. J Cell Biol. 2013;201:759–76. doi: 10.1083/jcb.201212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Development. 2003;130:5681–93. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- 28.Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell. 2005;8:179–92. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 29.Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–38. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- 30.Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–79. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- 31.Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134:1481–90. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nieto MA, Sargent MG, Wilkinson DG, Cooke J. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 1994;264:835–9. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Alcala S, Nieto MA, Barbas JA. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–65. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- 34.Théveneau E, Duband JL, Altabef M. Ets-1 confers cranial features on neural crest delamination. PLoS One. 2007;2:e1142. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simões-Costa MS, McKeown SJ, Tan-Cabugao J, Sauka-Spengler T, Bronner ME. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS Genet. 2012;8:e1003142. doi: 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Betancur P, Sauka-Spengler T, Bronner M. A Sox10 enhancer element common to the otic placode and neural crest is activated by tissue-specific paralogs. Development. 2011;138:3689–98. doi: 10.1242/dev.057836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinon A, Molchadsky A, Nathan E, Yovel G, Rotter V, Sarig R, Tzahor E. p53 coordinates cranial neural crest cell growth and epithelial-mesenchymal transition/delamination processes. Development. 2011;138:1827–38. doi: 10.1242/dev.053645. [DOI] [PubMed] [Google Scholar]
- 38.McKeown SJ, Wallace AS, Anderson RB. Expression and function of cell adhesion molecules during neural crest migration. Dev Biol. 2013;373:244–57. doi: 10.1016/j.ydbio.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 39.Dady A, Blavet C, Duband JL. Timing and kinetics of E- to N-cadherin switch during neurulation in the avian embryo. Dev Dyn. 2012;241:1333–49. doi: 10.1002/dvdy.23813. [DOI] [PubMed] [Google Scholar]
- 40.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–35. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 41.Sasselli V, Pachnis V, Burns AJ. The enteric nervous system. Dev Biol. 2012;366:64–73. doi: 10.1016/j.ydbio.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 42.Pavan WJ, Raible DW. Specification of neural crest into sensory neuron and melanocyte lineages. Dev Biol. 2012;366:55–63. doi: 10.1016/j.ydbio.2012.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 44.Gray RS, Cheung KJ, Ewald AJ. Cellular mechanisms regulating epithelial morphogenesis and cancer invasion. Curr Opin Cell Biol. 2010;22:640–50. doi: 10.1016/j.ceb.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuji T, Ibaragi S, Hu GF. Epithelial-mesenchymal transition and cell cooperativity in metastasis. Cancer Res. 2009;69:7135–9. doi: 10.1158/0008-5472.CAN-09-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Theveneau E, Mayor R. Collective cell migration of epithelial and mesenchymal cells. Cell Mol Life Sci. 2013;70:3481–92. doi: 10.1007/s00018-012-1251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theveneau E, Mayor R. Cadherins in collective cell migration of mesenchymal cells. Curr Opin Cell Biol. 2012;24:677–84. doi: 10.1016/j.ceb.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nat Rev Cancer. 2003;3:559–70. doi: 10.1038/nrc1145. [DOI] [PubMed] [Google Scholar]
- 50.Couturier J, Saule S. Genetic determinants of uveal melanoma. Dev Ophthalmol. 2012;49:150–65. doi: 10.1159/000328270. [DOI] [PubMed] [Google Scholar]
- 51.Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolačkov K, Tupikowski K, Bednarek-Tupikowska G. Genetic aspects of pheochromocytoma. Adv Clin Exp Med. 2012;21:821–9. [PubMed] [Google Scholar]
- 53.Carroll SL. Molecular mechanisms promoting the pathogenesis of Schwann cell neoplasms. Acta Neuropathol. 2012;123:321–48. doi: 10.1007/s00401-011-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta G, Maniker A. Malignant peripheral nerve sheath tumors. Neurosurg Focus. 2007;22:E12. doi: 10.3171/foc.2007.22.6.13. [DOI] [PubMed] [Google Scholar]
- 55.Kulesa PM, Bailey CM, Kasemeier-Kulesa JC, McLennan R. Cranial neural crest migration: new rules for an old road. Dev Biol. 2010;344:543–54. doi: 10.1016/j.ydbio.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gammill LS, Roffers-Agarwal J. Division of labor during trunk neural crest development. Dev Biol. 2010;344:555–65. doi: 10.1016/j.ydbio.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perris R, Perissinotto D. Role of the extracellular matrix during neural crest cell migration. Mech Dev. 2000;95:3–21. doi: 10.1016/S0925-4773(00)00365-8. [DOI] [PubMed] [Google Scholar]
- 58.Duband JL. Neural crest delamination and migration: integrating regulations of cell interactions, locomotion, survival and fate. Adv Exp Med Biol. 2006;589:45–77. doi: 10.1007/978-0-387-46954-6_4. [DOI] [PubMed] [Google Scholar]
- 59.Lwigale PY, Bronner-Fraser M. Semaphorin3A/neuropilin-1 signaling acts as a molecular switch regulating neural crest migration during cornea development. Dev Biol. 2009;336:257–65. doi: 10.1016/j.ydbio.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toyofuku T, Yoshida J, Sugimoto T, Yamamoto M, Makino N, Takamatsu H, Takegahara N, Suto F, Hori M, Fujisawa H, et al. Repulsive and attractive semaphorins cooperate to direct the navigation of cardiac neural crest cells. Dev Biol. 2008;321:251–62. doi: 10.1016/j.ydbio.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 61.Schwarz Q, Vieira JM, Howard B, Eickholt BJ, Ruhrberg C. Neuropilin 1 and 2 control cranial gangliogenesis and axon guidance through neural crest cells. Development. 2008;135:1605–13. doi: 10.1242/dev.015412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gammill LS, Gonzalez C, Bronner-Fraser M. Neuropilin 2/semaphorin 3F signaling is essential for cranial neural crest migration and trigeminal ganglion condensation. Dev Neurobiol. 2007;67:47–56. doi: 10.1002/dneu.20326. [DOI] [PubMed] [Google Scholar]
- 63.Gammill LS, Gonzalez C, Gu C, Bronner-Fraser M. Guidance of trunk neural crest migration requires neuropilin 2/semaphorin 3F signaling. Development. 2006;133:99–106. doi: 10.1242/dev.02187. [DOI] [PubMed] [Google Scholar]
- 64.Yu HH, Moens CB. Semaphorin signaling guides cranial neural crest cell migration in zebrafish. Dev Biol. 2005;280:373–85. doi: 10.1016/j.ydbio.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 65.Eickholt BJ, Mackenzie SL, Graham A, Walsh FS, Doherty P. Evidence for collapsin-1 functioning in the control of neural crest migration in both trunk and hindbrain regions. Development. 1999;126:2181–9. doi: 10.1242/dev.126.10.2181. [DOI] [PubMed] [Google Scholar]
- 66.Mellott DO, Burke RD. Divergent roles for Eph and ephrin in avian cranial neural crest. BMC Dev Biol. 2008;8:56. doi: 10.1186/1471-213X-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kasemeier-Kulesa JC, Bradley R, Pasquale EB, Lefcort F, Kulesa PM. Eph/ephrins and N-cadherin coordinate to control the pattern of sympathetic ganglia. Development. 2006;133:4839–47. doi: 10.1242/dev.02662. [DOI] [PubMed] [Google Scholar]
- 68.Davy A, Aubin J, Soriano P. Ephrin-B1 forward and reverse signaling are required during mouse development. Genes Dev. 2004;18:572–83. doi: 10.1101/gad.1171704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baker RK, Antin PB. Ephs and ephrins during early stages of chick embryogenesis. Dev Dyn. 2003;228:128–42. doi: 10.1002/dvdy.10354. [DOI] [PubMed] [Google Scholar]
- 70.Santiago A, Erickson CA. Ephrin-B ligands play a dual role in the control of neural crest cell migration. Development. 2002;129:3621–32. doi: 10.1242/dev.129.15.3621. [DOI] [PubMed] [Google Scholar]
- 71.Smith A, Robinson V, Patel K, Wilkinson DG. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr Biol. 1997;7:561–70. doi: 10.1016/S0960-9822(06)00255-7. [DOI] [PubMed] [Google Scholar]
- 72.Krull CE, Lansford R, Gale NW, Collazo A, Marcelle C, Yancopoulos GD, Fraser SE, Bronner-Fraser M. Interactions of Eph-related receptors and ligands confer rostrocaudal pattern to trunk neural crest migration. Curr Biol. 1997;7:571–80. doi: 10.1016/S0960-9822(06)00256-9. [DOI] [PubMed] [Google Scholar]
- 73.Shiau CE, Bronner-Fraser M. N-cadherin acts in concert with Slit1-Robo2 signaling in regulating aggregation of placode-derived cranial sensory neurons. Development. 2009;136:4155–64. doi: 10.1242/dev.034355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jia L, Cheng L, Raper J. Slit/Robo signaling is necessary to confine early neural crest cells to the ventral migratory pathway in the trunk. Dev Biol. 2005;282:411–21. doi: 10.1016/j.ydbio.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 75.De Bellard ME, Rao Y, Bronner-Fraser M. Dual function of Slit2 in repulsion and enhanced migration of trunk, but not vagal, neural crest cells. J Cell Biol. 2003;162:269–79. doi: 10.1083/jcb.200301041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harris ML, Hall R, Erickson CA. Directing pathfinding along the dorsolateral path - the role of EDNRB2 and EphB2 in overcoming inhibition. Development. 2008;135:4113–22. doi: 10.1242/dev.023119. [DOI] [PubMed] [Google Scholar]
- 77.McLennan R, Teddy JM, Kasemeier-Kulesa JC, Romine MH, Kulesa PM. Vascular endothelial growth factor (VEGF) regulates cranial neural crest migration in vivo. Dev Biol. 2010;339:114–25. doi: 10.1016/j.ydbio.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kubota Y, Ito K. Chemotactic migration of mesencephalic neural crest cells in the mouse. Dev Dyn. 2000;217:170–9. doi: 10.1002/(SICI)1097-0177(200002)217:2<170::AID-DVDY4>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 79.Sato A, Scholl AM, Kuhn EN, Stadt HA, Decker JR, Pegram K, Hutson MR, Kirby ML. FGF8 signaling is chemotactic for cardiac neural crest cells. Dev Biol. 2011;354:18–30. doi: 10.1016/j.ydbio.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Creuzet S, Schuler B, Couly G, Le Douarin NM. Reciprocal relationships between Fgf8 and neural crest cells in facial and forebrain development. Proc Natl Acad Sci U S A. 2004;101:4843–7. doi: 10.1073/pnas.0400869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Theveneau E, Steventon B, Scarpa E, Garcia S, Trepat X, Streit A, Mayor R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat Cell Biol. 2013;15:763–72. doi: 10.1038/ncb2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Escot S, Blavet C, Härtle S, Duband JL, Fournier-Thibault C. Misregulation of SDF1-CXCR4 signaling impairs early cardiac neural crest cell migration leading to conotruncal defects. Circ Res. 2013;113:505–16. doi: 10.1161/CIRCRESAHA.113.301333. [DOI] [PubMed] [Google Scholar]
- 83.Saito D, Takase Y, Murai H, Takahashi Y. The dorsal aorta initiates a molecular cascade that instructs sympatho-adrenal specification. Science. 2012;336:1578–81. doi: 10.1126/science.1222369. [DOI] [PubMed] [Google Scholar]
- 84.Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kasemeier-Kulesa JC, McLennan R, Romine MH, Kulesa PM, Lefcort F. CXCR4 controls ventral migration of sympathetic precursor cells. J Neurosci. 2010;30:13078–88. doi: 10.1523/JNEUROSCI.0892-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Olesnicky Killian EC, Birkholz DA, Artinger KB. A role for chemokine signaling in neural crest cell migration and craniofacial development. Dev Biol. 2009;333:161–72. doi: 10.1016/j.ydbio.2009.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Belmadani A, Jung H, Ren D, Miller RJ. The chemokine SDF-1/CXCL12 regulates the migration of melanocyte progenitors in mouse hair follicles. Differentiation. 2009;77:395–411. doi: 10.1016/j.diff.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Svetic V, Hollway GE, Elworthy S, Chipperfield TR, Davison C, Adams RJ, Eisen JS, Ingham PW, Currie PD, Kelsh RN. Sdf1a patterns zebrafish melanophores and links the somite and melanophore pattern defects in choker mutants. Development. 2007;134:1011–22. doi: 10.1242/dev.02789. [DOI] [PubMed] [Google Scholar]
- 89.Belmadani A, Tran PB, Ren D, Assimacopoulos S, Grove EA, Miller RJ. The chemokine stromal cell-derived factor-1 regulates the migration of sensory neuron progenitors. J Neurosci. 2005;25:3995–4003. doi: 10.1523/JNEUROSCI.4631-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eberhart JK, He X, Swartz ME, Yan YL, Song H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, Postlethwait JH. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290–8. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richarte AM, Mead HB, Tallquist MD. Cooperation between the PDGF receptors in cardiac neural crest cell migration. Dev Biol. 2007;306:785–96. doi: 10.1016/j.ydbio.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mwizerwa O, Das P, Nagy N, Akbareian SE, Mably JD, Goldstein AM. Gdnf is mitogenic, neurotrophic, and chemoattractive to enteric neural crest cells in the embryonic colon. Dev Dyn. 2011;240:1402–11. doi: 10.1002/dvdy.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cornejo M, Nambi D, Walheim C, Somerville M, Walker J, Kim L, Ollison L, Diamante G, Vyawahare S, de Bellard ME. Effect of NRG1, GDNF, EGF and NGF in the migration of a Schwann cell precursor line. Neurochem Res. 2010;35:1643–51. doi: 10.1007/s11064-010-0225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Young HM, Hearn CJ, Farlie PG, Canty AJ, Thomas PQ, Newgreen DF. GDNF is a chemoattractant for enteric neural cells. Dev Biol. 2001;229:503–16. doi: 10.1006/dbio.2000.0100. [DOI] [PubMed] [Google Scholar]
- 95.Pla P, Alberti C, Solov’eva O, Pasdar M, Kunisada T, Larue L. Ednrb2 orients cell migration towards the dorsolateral neural crest pathway and promotes melanocyte differentiation. Pigment Cell Res. 2005;18:181–7. doi: 10.1111/j.1600-0749.2005.00230.x. [DOI] [PubMed] [Google Scholar]
- 96.Lee HO, Levorse JM, Shin MK. The endothelin receptor-B is required for the migration of neural crest-derived melanocyte and enteric neuron precursors. Dev Biol. 2003;259:162–75. doi: 10.1016/S0012-1606(03)00160-X. [DOI] [PubMed] [Google Scholar]
- 97.Young HM, Anderson RB, Anderson CR. Guidance cues involved in the development of the peripheral autonomic nervous system. Auton Neurosci. 2004;112:1–14. doi: 10.1016/j.autneu.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 98.Teddy JM, Kulesa PM. In vivo evidence for short- and long-range cell communication in cranial neural crest cells. Development. 2004;131:6141–51. doi: 10.1242/dev.01534. [DOI] [PubMed] [Google Scholar]
- 99.Kulesa PM, Fraser SE. In ovo time-lapse analysis of chick hindbrain neural crest cell migration shows cell interactions during migration to the branchial arches. Development. 2000;127:1161–72. doi: 10.1242/dev.127.6.1161. [DOI] [PubMed] [Google Scholar]
- 100.Carmona-Fontaine C, Matthews HK, Kuriyama S, Moreno M, Dunn GA, Parsons M, Stern CD, Mayor R. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–61. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abercrombie M, Heaysman JE. Observations on the social behaviour of cells in tissue culture. I. Speed of movement of chick heart fibroblasts in relation to their mutual contacts. Exp Cell Res. 1953;5:111–31. doi: 10.1016/0014-4827(53)90098-6. [DOI] [PubMed] [Google Scholar]
- 102.Mayor R, Carmona-Fontaine C. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 2010;20:319–28. doi: 10.1016/j.tcb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carmona-Fontaine C, Theveneau E, Tzekou A, Tada M, Woods M, Page KM, Parsons M, Lambris JD, Mayor R. Complement fragment C3a controls mutual cell attraction during collective cell migration. Dev Cell. 2011;21:1026–37. doi: 10.1016/j.devcel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40:1378–82. doi: 10.1042/BST20120103. [DOI] [PubMed] [Google Scholar]
- 105.Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol. 2007;24:203–16. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J, Settleman J. Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Mol Cell Biol. 1996;16:2689–99. doi: 10.1128/mcb.16.6.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jordan P, Brazåo R, Boavida MG, Gespach C, Chastre E. Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene. 1999;18:6835–9. doi: 10.1038/sj.onc.1203233. [DOI] [PubMed] [Google Scholar]
- 108.Vignal E, De Toledo M, Comunale F, Ladopoulou A, Gauthier-Rouvière C, Blangy A, Fort P. Characterization of TCL, a new GTPase of the rho family related to TC10 andCcdc42. J Biol Chem. 2000;275:36457–64. doi: 10.1074/jbc.M003487200. [DOI] [PubMed] [Google Scholar]
- 109.Aspenström P, Fransson A, Saras J. Rho GTPases have diverse effects on the organization of the actin filament system. Biochem J. 2004;377:327–37. doi: 10.1042/BJ20031041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shutes A, Berzat AC, Cox AD, Der CJ. Atypical mechanism of regulation of the Wrch-1 Rho family small GTPase. Curr Biol. 2004;14:2052–6. doi: 10.1016/j.cub.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 111.Aspenström P, Ruusala A, Pacholsky D. Taking Rho GTPases to the next level: the cellular functions of atypical Rho GTPases. Exp Cell Res. 2007;313:3673–9. doi: 10.1016/j.yexcr.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 112.Steventon B, Carmona-Fontaine C, Mayor R. Genetic network during neural crest induction: from cell specification to cell survival. Semin Cell Dev Biol. 2005;16:647–54. doi: 10.1016/j.semcdb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 113.Klymkowsky MW, Rossi CC, Artinger KB. Mechanisms driving neural crest induction and migration in the zebrafish and Xenopus laevis. Cell Adh Migr. 2010;4:595–608. doi: 10.4161/cam.4.4.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morgan R, Hooiveld MH, Durston AJ. A novel guanine exchange factor increases the competence of early ectoderm to respond to neural induction. Mech Dev. 1999;88:67–72. doi: 10.1016/S0925-4773(99)00172-0. [DOI] [PubMed] [Google Scholar]
- 115.Guémar L, de Santa Barbara P, Vignal E, Maurel B, Fort P, Faure S. The small GTPase RhoV is an essential regulator of neural crest induction in Xenopus. Dev Biol. 2007;310:113–28. doi: 10.1016/j.ydbio.2007.07.031. [DOI] [PubMed] [Google Scholar]
- 116.Notarnicola C, Le Guen L, Fort P, Faure S, de Santa Barbara P. Dynamic expression patterns of RhoV/Chp and RhoU/Wrch during chicken embryonic development. Dev Dyn. 2008;237:1165–71. doi: 10.1002/dvdy.21507. [DOI] [PubMed] [Google Scholar]
- 117.De Calisto J, Araya C, Marchant L, Riaz CF, Mayor R. Essential role of non-canonical Wnt signalling in neural crest migration. Development. 2005;132:2587–97. doi: 10.1242/dev.01857. [DOI] [PubMed] [Google Scholar]
- 118.Broders-Bondon F, Chesneau A, Romero-Oliva F, Mazabraud A, Mayor R, Thiery JP. Regulation of XSnail2 expression by Rho GTPases. Dev Dyn. 2007;236:2555–66. doi: 10.1002/dvdy.21273. [DOI] [PubMed] [Google Scholar]
- 119.Hotta R, Pepdjonovic L, Anderson RB, Zhang D, Bergner AJ, Leung J, Pébay A, Young HM, Newgreen DF, Dottori M. Small-molecule induction of neural crest-like cells derived from human neural progenitors. Stem Cells. 2009;27:2896–905. doi: 10.1002/stem.208. [DOI] [PubMed] [Google Scholar]
- 120.Bisson N, Wedlich D, Moss T. The p21-activated kinase Pak1 regulates induction and migration of the neural crest in Xenopus. Cell Cycle. 2012;11:1316–24. doi: 10.4161/cc.19685. [DOI] [PubMed] [Google Scholar]
- 121.Kerosuo L, Bronner ME. Biphasic influence of Miz1 on neural crest development by regulating cell survival and apical adhesion complex formation in the developing neural tube. Mol Biol Cell. 2014;25:347–55. doi: 10.1091/mbc.E13-06-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gammill LS, Bronner-Fraser M. Genomic analysis of neural crest induction. Development. 2002;129:5731–41. doi: 10.1242/dev.00175. [DOI] [PubMed] [Google Scholar]
- 123.Nikolova E, Mitev V, Minner F, Deroanne CF, Poumay Y. The inhibition of the expression of the small Rho GTPase Rac1 induces differentiation with no effect on cell proliferation in growing human adult keratinocytes. J Cell Biochem. 2008;103:857–64. doi: 10.1002/jcb.21455. [DOI] [PubMed] [Google Scholar]
- 124.Ahlstrom JD, Erickson CA. The neural crest epithelial-mesenchymal transition in 4D: a ‘tail’ of multiple non-obligatory cellular mechanisms. Development. 2009;136:1801–12. doi: 10.1242/dev.034785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Berndt JD, Clay MR, Langenberg T, Halloran MC. Rho-kinase and myosin II affect dynamic neural crest cell behaviors during epithelial to mesenchymal transition in vivo. Dev Biol. 2008;324:236–44. doi: 10.1016/j.ydbio.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu JP, Jessell TM. A role for rhoB in the delamination of neural crest cells from the dorsal neural tube. Development. 1998;125:5055–67. doi: 10.1242/dev.125.24.5055. [DOI] [PubMed] [Google Scholar]
- 127.Groysman M, Shoval I, Kalcheim C. A negative modulatory role for rho and rho-associated kinase signaling in delamination of neural crest cells. Neural Dev. 2008;3:27. doi: 10.1186/1749-8104-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Clay MR, Halloran MC. Rho activation is apically restricted by Arhgap1 in neural crest cells and drives epithelial-to-mesenchymal transition. Development. 2013;140:3198–209. doi: 10.1242/dev.095448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hou X, Katahira T, Ohashi K, Mizuno K, Sugiyama S, Nakamura H. Coactosin accelerates cell dynamism by promoting actin polymerization. Dev Biol. 2013;379:53–63. doi: 10.1016/j.ydbio.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 130.Fort P, Guémar L, Vignal E, Morin N, Notarnicola C, de Santa Barbara P, Faure S. Activity of the RhoU/Wrch1 GTPase is critical for cranial neural crest cell migration. Dev Biol. 2011;350:451–63. doi: 10.1016/j.ydbio.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 131.Tahtakran SA, Selleck MA. Ets-1 expression is associated with cranial neural crest migration and vasculogenesis in the chick embryo. Gene Expr Patterns. 2003;3:455–8. doi: 10.1016/S1567-133X(03)00065-6. [DOI] [PubMed] [Google Scholar]
- 132.Coles EG, Taneyhill LA, Bronner-Fraser M. A critical role for Cadherin6B in regulating avian neural crest emigration. Dev Biol. 2007;312:533–44. doi: 10.1016/j.ydbio.2007.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fernández Caso M, De Paz P, Fernandez Alvarez JG, Chamorro C, Villar JM. Delamination of neuroepithelium and nonneural ectoderm and its relation to the convergence step in chick neurulation. J Anat. 1992;180:143–53. [PMC free article] [PubMed] [Google Scholar]
- 134.Innes PB. The ultrastructure of early cephalic neural crest cell migration in the mouse. Anat Embryol (Berl) 1985;172:33–8. doi: 10.1007/BF00318941. [DOI] [PubMed] [Google Scholar]
- 135.Alfandari D, Cousin H, Marsden M. Mechanism of Xenopus cranial neural crest cell migration. Cell Adh Migr. 2010;4:553–60. doi: 10.4161/cam.4.4.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sadaghiani B, Thiébaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- 137.Lucas JM, Nikolic I, Hens MD. cDNA cloning, sequence comparison, and developmental expression of Xenopus rac1. Mech Dev. 2002;115:113–6. doi: 10.1016/S0925-4773(02)00117-X. [DOI] [PubMed] [Google Scholar]
- 138.Shoval I, Kalcheim C. Antagonistic activities of Rho and Rac GTPases underlie the transition from neural crest delamination to migration. Dev Dyn. 2012;241:1155–68. doi: 10.1002/dvdy.23799. [DOI] [PubMed] [Google Scholar]
- 139.Matthews HK, Marchant L, Carmona-Fontaine C, Kuriyama S, Larraín J, Holt MR, Parsons M, Mayor R. Directional migration of neural crest cells in vivo is regulated by Syndecan-4/Rac1 and non-canonical Wnt signaling/RhoA. Development. 2008;135:1771–80. doi: 10.1242/dev.017350. [DOI] [PubMed] [Google Scholar]
- 140.Moore R, Theveneau E, Pozzi S, Alexandre P, Kashef J, Richardson J, Linker C, Mayor R. Par3 controls neural crest migration by promoting microtubule catastrophe during contact inhibition of locomotion. 2013;140:4763–75. doi: 10.1242/dev.098509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shnitsar I, Borchers A. PTK7 recruits dsh to regulate neural crest migration. Development. 2008;135:4015–24. doi: 10.1242/dev.023556. [DOI] [PubMed] [Google Scholar]
- 142.Wagner G, Peradziryi H, Wehner P, Borchers A. PlexinA1 interacts with PTK7 and is required for neural crest migration. Biochem Biophys Res Commun. 2010;402:402–7. doi: 10.1016/j.bbrc.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 143.Kashef J, Köhler A, Kuriyama S, Alfandari D, Mayor R, Wedlich D. Cadherin-11 regulates protrusive activity in Xenopus cranial neural crest cells upstream of Trio and the small GTPases. Genes Dev. 2009;23:1393–8. doi: 10.1101/gad.519409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Goto A, Sumiyama K, Kamioka Y, Nakasyo E, Ito K, Iwasaki M, Enomoto H, Matsuda M. GDNF and endothelin 3 regulate migration of enteric neural crest-derived cells via protein kinase A and Rac1. J Neurosci. 2013;33:4901–12. doi: 10.1523/JNEUROSCI.4828-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhang Y, Kim TH, Niswander L. Phactr4 regulates directional migration of enteric neural crest through PP1, integrin signaling, and cofilin activity. Genes Dev. 2012;26:69–81. doi: 10.1101/gad.179283.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rupp PA, Kulesa PM. A role for RhoA in the two-phase migratory pattern of post-otic neural crest cells. Dev Biol. 2007;311:159–71. doi: 10.1016/j.ydbio.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 147.Fuchs S, Herzog D, Sumara G, Büchmann-Møller S, Civenni G, Wu X, Chrostek-Grashoff A, Suter U, Ricci R, Relvas JB, et al. Stage-specific control of neural crest stem cell proliferation by the small rho GTPases Cdc42 and Rac1. Cell Stem Cell. 2009;4:236–47. doi: 10.1016/j.stem.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 148.Henderson DJ, Ybot-Gonzalez P, Copp AJ. RhoB is expressed in migrating neural crest and endocardial cushions of the developing mouse embryo. Mech Dev. 2000;95:211–4. doi: 10.1016/S0925-4773(00)00333-6. [DOI] [PubMed] [Google Scholar]
- 149.Vignal E, de Santa Barbara P, Guémar L, Donnay J-M, Fort P, Faure S. Expression of RhoB in the developing Xenopus laevis embryo. Gene Expr Patterns. 2007;7:282–8. doi: 10.1016/j.modgep.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 150.Brady DC, Alan JK, Madigan JP, Fanning AS, Cox AD. The transforming Rho family GTPase Wrch-1 disrupts epithelial cell tight junctions and epithelial morphogenesis. Mol Cell Biol. 2009;29:1035–49. doi: 10.1128/MCB.00336-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chuang YY, Valster A, Coniglio SJ, Backer JM, Symons M. The atypical Rho family GTPase Wrch-1 regulates focal adhesion formation and cell migration. J Cell Sci. 2007;120:1927–34. doi: 10.1242/jcs.03456. [DOI] [PubMed] [Google Scholar]
- 152.Ory S, Brazier H, Blangy A. Identification of a bipartite focal adhesion localization signal in RhoU/Wrch-1, a Rho family GTPase that regulates cell adhesion and migration. Biol Cell. 2007;99:701–16. doi: 10.1042/BC20070058. [DOI] [PubMed] [Google Scholar]