Abstract
Oxysterols are intermediates in the synthesis of bile acids and steroid hormones from cholesterol and function as ligands for liver X receptor (LXR). Bile salt export pump (BSEP) is responsible for canalicular secretion of bile acids and is tightly regulated by its substrates bile acids through nuclear receptor farnesoid X receptor (FXR). In a microarray study using human hepatocytes, BSEP was markedly induced not only by chenodeoxycholic acid (CDCA) but also by oxysterol 22(R)-hydroxycholesterol [22(R)-OHC]. We hypothesized that the expression of BSEP was induced by oxysterols through activation of LXR. To test the hypothesis, human primary hepatocytes or hepatoma cells were treated with 22(R)-OHC, and expression of BSEP was determined. The level of BSEP mRNA was increased as much as 5-fold upon oxysterol induction. In contrast to our hypothesis, the oxysterol-induced up-regulation of BSEP is mediated through FXR but not LXR. BSEP promoter activity was markedly induced by 22(R)-OHC in the presence of FXR but not LXRs. Mutation of the FXR element IR1 in the BSEP promoter significantly reduced its ability to respond to oxysterol induction. To determine whether 22(R)-OHC and CDCA bind to similar structural features of FXR, site-directed mutagenesis was performed in the FXR ligand binding domain. Mutation of residues R331 and I352 abolished activation mediated by CDCA and 22(R)-OHC. In contrast, substitution of residues L340 and R351 differentiated CDCA- and 22(R)-OHC-mediated activation. In conclusion, oxysterol 22(R)-OHC functions as an FXR ligand to induce BSEP expression and differs in the binding with FXR from CDCA.
Oxysterols are intermediates in the synthesis of bile acids and steroid hormones from cholesterol (Schroepfer, 2000; Russell, 2003; Auchus, 2004). Oxysterols rather than cholesterol itself serve as biological sensors for cholesterol levels in the body and function as potent endogenous ligands for nuclear receptor liver X receptor (LXR) (Janowski et al., 1996; Peet et al., 1998a). Activation of LXR by oxysterols plays a central role in the regulation of cholesterol metabolism and transport (Peet et al., 1998b; Chiang et al., 2001; Davis et al., 2002; Edwards et al., 2002; Repa et al., 2002).
Molecular and genetic studies have established that bile salt export pump (BSEP) is responsible for the secretion of bile acids from the liver (Strautnieks et al., 1998; Jansen et al., 1999; Noe et al., 2002). Lack of functional BSEP is linked directly to the progressive familial intrahepatic cholestasis type II, a rapidly progressing and lethal hereditary disease (Strautnieks et al., 1998; Jansen et al., 1999). Impairment of BSEP function or expression mediated by endobiotics (Yu et al., 2002; Crocenzi et al., 2003) or xenobiotics leads to cholestatic liver injury (Horikawa et al., 2003; Roman et al., 2003).
Bile salts are physiological detergents and function as solubilizers of cholesterol in bile and emulsifiers of lipids in intestine, playing important physiological roles in transport, adsorption, and elimination of cholesterol and lipids (Fuchs, 2003). On the other hand, excessive accumulation of bile acids is toxic to hepatocytes. Bile acid homeostasis is maintained through the bile enterohepatic circulation (Fuchs, 2003; Kullak-Ublick et al., 2004). The rate-limiting step in bile enterohepatic circulation is the canalicular secretion by BSEP (Meier and Stieger, 2002; Fuchs, 2003; Kullak-Ublick et al., 2004).
The expression of BSEP is profoundly altered by both endobiotics (Ananthanarayanan et al., 2001; Plass et al., 2002; Yu et al., 2002) and xenobiotics (Cui et al., 2003; Owsley and Chiang, 2003). Bile acids, such as chenodeoxycholic acid (CDCA), markedly increase the expression of BSEP (Ananthanarayanan et al., 2001; Plass et al., 2002), whereas xenobiotic guggulsterone antagonizes CDCA-mediated up-regulation of BSEP (Cui et al., 2003; Owsley and Chiang, 2003). Bile acid-mediated up-regulation of BSEP expression is achieved through activation of nuclear receptor farsenoid X receptor (FXR) (Ananthanarayanan et al., 2001; Plass et al., 2002), which plays a vital role in maintaining cholesterol and bile acid homeostasis (Chiang, 2002; Redinger, 2003; Rizzo et al., 2005). A cis-acting element, IR1, in the BSEP promoter has been identified as a FXR-specific binding site (Ananthanarayanan et al., 2001; Plass et al., 2002).
The FXR signaling is functionally related to LXR signaling. Both signaling pathways are involved in regulating a number of target genes important in the synthesis, metabolism, and transport of cholesterol and bile acids. For example, cholesterol-7α-hydroxylase (CYP7A1), a rate-limiting enzyme in the conversion of cholesterol to bile acids in the classic pathway of bile acid synthesis, is directly up-regulated through LXR pathway in rodents (Chiang et al., 2001; Menke et al., 2002) but indirectly down-regulated through FXR pathway in both rodents and human (Chiang et al., 2000; Lu et al., 2000; Davis et al., 2002). The species difference in the regulation of human CYP7A1 by LXR is noted because of the lack of an LXR-responsive element in human CYP7A1 (Chiang et al., 2001). Conversion of cholesterol to bile acids represents a major pathway for cholesterol elimination from the body. On the other hand, secretion or adsorption of cholesterol requires bile acids for cholesterol solubilization.
In a microarray study using human primary hepatocytes to identify transcriptionally regulated target genes by bile acid CDCA and oxysterol 22(R)-OHC, it was unexpectedly observed that BSEP was markedly induced by 22(R)-OHC. The observation is consistent with a recent study that found a high-cholesterol diet results in an increase in bile acid pool size and fecal secretion (Tiemann et al., 2004), indicating an elevated canalicular bile acid secretion by BSEP. We hypothesized that the expression of BSEP was induced by oxysterols through activation of LXR. To test the hypothesis, a reporter harboring the BSEP promoter was constructed and characterized for its responsiveness to 22(R)-OHC in the presence of LXR or FXR. In contrast to our hypothesis, the activation of BSEP promoter by 22(R)-OHC was achieved through FXR but not LXR. Disruption of the IR1 element in the BSEP promoter significantly reduced its ability to respond to oxy-sterol induction. In addition, mutational analysis of the FXR ligand binding domain (LBD) revealed that different structural features of FXR were involved in CDCA- and 22(R)-OHC-mediated activation.
Materials and Methods
Chemicals and Supplies
CDCA, 22(R)-OHC, rifampicin, actinomycin D, dimethyl sulfoxide (DMSO), and Williams’ Medium E were purchased from Sigma (St. Louis, MO). DMEM, LipofectAMINE, and Plus Reagent were from Invitrogen (Carlsbad, CA). Kits for luciferase detection and the null-Renilla luciferase plasmid were from Promega (Madison, WI). Delipidated and normal fetal bovine sera and 100× nonessential amino acids were from HyClone (Logan, UT). Unless otherwise specified, all other reagents were purchased from Fisher Scientific (Pittsburgh, PA). Oligonucleotides for PCR amplification and site-directed mutagenesis were chemically synthesized by Invitrogen. Restriction enzymes were purchased from New England Biolabs (Ipswich, MA).
Plasmid Constructs
BSEP promoter reporter was constructed by cloning a genomic DNA fragment upstream of the transcription start site (+85 to −2525) into the luciferase vector pGL4.10 (Promega). The DNA fragment was PCR-amplified using human genomic DNA as template and a pair of primers with the following sequences: 5′-AATTGCTAGCAGGAGTGGTCCTCAAGCTTCAGCT-3′ (forward) and 5′-AATTCTCGAGGGAAGCCAGAGGAAATAATGGACTCC-3 (reverse). NheI and XhoI restriction sites (underlined) were incorporated into the forward and reverse primers, respectively, to facilitate the cloning of the resulting PCR fragment. The PCR amplification was performed in a total volume of 25 μl containing 10 ng of human genomic DNA templates, 200 nM forward and reverse primer, and 2.5 units of high-fidelity Taq polymerase (Invitrogen). The thermal cycling parameters include an initial denaturing at 94°C for 3 min, followed by 30 cycles at 94°C for 45 s, 65°C for 45 s, and 68°C for 3 min, and a final extension at 68°C for 10 min. The PCR products were subjected to restriction digestion, gel purification, and ligation. The resulting pBSEP-Luc reporter construct was sequence-verified. Expression construct of nuclear receptor pregnane X receptor (PXR) and its target reporter construct pCYP3A4DP-Luc were described previously (Song et al., 2004). Expression plasmids for human nuclear receptors LXRα, LXRβ, and FXR were kindly provided by Dr. D. Mangelsdorf (University of Texas Southwestern Medical Center, Dallas, TX). LXR target gene reporters pTK-Luc-2LXRE-3 and pGL-TK-2610 (LXR) were kindly provided by Dr. P. Edwards (University of California Los Angeles School of Medicine, Los Angeles, CA) and Dr. B. Goodwin (GlaxoSmithkline Inc., Research Triangle Park, NC), respectively.
Site-Directed Mutagenesis
Mutations were introduced by a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The mutagenesis oligonucleotides were listed in Table 1 with the substituted nucleotides in bold and the IR1 element underlined. Multiple nucleotide substitutions in the IR1 element were introduced to completely disrupt the site. Four and three nucleotides were substituted in the first half and second half, respectively. Four amino acid residues in the FXR LBD, including R331, L340, R351, and I352, were selected for mutagenesis. Those amino acid residues are in the binding pocket with ligand 6α-ethyl-chenodeoxycholic acid (6ECDCA) for rat FXR (Mi et al., 2003) or fexaramine for human FXR (Downes et al., 2003) in the crystal structures. The mutagenesis reactions were performed essentially according to the manufacturer’s manual. Briefly, the sense and antisense oligonucleotides were annealed to the pBSEP-Luc construct for IR1 mutation or FXR expression construct for FXR LBD mutation, followed by a total of 18 cycles at 94°C for 45 s, 55°C for 1 min, and 68°C for 8 min. The resultant PCR-amplified constructs were then digested with DpnI to remove the nonmutated parent constructs. The mutated PCR-amplified constructs were used to transform XL1-Blue bacteria. The resulting mutants were subjected to sequence analysis to confirm the desired substitutions being made without introducing errors.
TABLE 1.
Sequences of mutagenesis oligonucleotides
| Oligos | Sequences (5′ to 3′) |
|---|---|
| IR1-sensea,b | GTCTTGGGCTGCCCTTAGACGCGTGAATTCTTAGGCAAATAGATAAT |
| IR1-antisense | ATTATCTATTTGCCTAAGAATTCACGCGTCTAAGGGCAGCCCAAGAC |
| R331L-sense | TGCGGTTGAAGCTATGTTCCTTCTTTCAGCTGAGATTTTCAATAAGAAAC |
| R331L-antisense | GTTTCTTATTGAAAATCTCAGCTGAAAGAAGGAACATAGCTTCAACCGCA |
| L340Y-sense | CAGCTGAGATTTTCAATAAGAAATATCCGTCTGGGCATTCTGACCTATTGG |
| L340Y-antisense | CCAATAGGTCAGAATGCCCAGACGGATATTTCTTATTGAAAATCTCAGCTG |
| R351L-sense | GGGCATTCTGACCTATTGGAAGAACTAATTCGAAATAGTGGTATCTCTGAT |
| R351L-antisense | ATCAGAGATACCACTATTTCGAATTAGTTCTTCCAATAGGTCAGAATGCCC |
| I352Y-sense | CATTCTGACCTATTGGAAGAAAGATATCGAAATAGTGGTATCTCTGATGA |
| I352Y-antisense | TCATCAGAGATACCACTATTTCGATATCTTTCTTCCAATAGGTCAGAATG |
The IR1 element in the BSEP promoter is underlined.
The mutated nucleotides are in bold.
Human Primary Hepatocyte and Hepatoma Cell Culture and Treatments
Plated human primary hepatocytes (in six-well plates) were obtained from Liver Tissues Procurement and Distribution System (University of Minnesota, Minneapolis, MN). Upon arrival, cell supernatants were replaced with rich Williams’ Medium E containing 1% penicillin/streptomycin. After incubation at 37°C with 5% CO2 for 6 h, cells were treated with 10 μM oxysterol 22(R)-OHC, 10 μM CDCA, 10 μM rifampicin, 5 μM actinomycin D, 0.1% solvent ethanol or DMSO, and a mixture of 10 μM 22(R)-OHC and 5 μM actinomycin D. The treatment was continued for 30 h with a change of fresh treatment medium at 24 h.
Hepatoma Huh 7 cells purchased from American Type Culture Collection were maintained in DMEM containing 10% fetal bovine serum, 1% penicillin/streptomycin, and 1× nonessential amino acids. Cells were seeded at a density of 2 × 105 cells/well (12-well plates) in a medium containing 10% delipidated fetal bovine serum. Sixteen hours postseeding, cells were treated with appropriate chemicals at the concentrations specified above. Unless specified, all the treatments were continued for 30 h.
RNA Isolation
Cell monolayers were lysed by adding 400 μl/well (12-well plate) or 800 μl/well (six-well plate) of RNA-Bee solution (Tel-Test, Inc., Friendswood, TX). The cell lysates were transferred to fresh tubes, and 80 or 160 μl of chloroform were added into each tube for the six-well plate or 12-well plate samples, respectively. After vigorously shaking for 15 s, the tubes were centrifuged at 12,000 rpm for 15 min at 4°C. The supernatants were transferred to fresh tubes, and equal volume of isopropanol was added into each tube to precipitate total RNA. After incubating at −70°C for 20 min, RNA precipitants were pelleted by centrifugation at 13,000 rpm for 15 min at 4°C. The RNA pellets were washed once with 75% ethanol, air-dried at room temperature, and dissolved in 25 μl of diethyl pyrocarbonate water. The RNA concentrations were determined by NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE).
Reverse Transcription Coupled-Polymerase Chain Reaction (RT-PCR)
The mRNA levels of BSEP were determined by RT-PCR with a ThermoScript I kit (Invitrogen). Total RNA (1 μg) was subjected to the synthesis of the first strand cDNA in a total volume of 25 μl with random primers and ThermoScript I reverse transcriptase. The reactions were incubated initially at 25°C for 10 min, and then at 50°C for 50 min, followed by inactivation of the reaction at 70°C for 10 min. The cDNAs were then diluted five times with water and subjected to PCR amplification (5 μl of the diluted cDNA). The cycling parameters were 95°C for 30 s, 52°C for 30 s, and 72°C for 30 s for a total of 30 to 38 cycles, depending on the relative abundance of BSEP mRNA in different samples. The primers for BSEP amplification were 5′-AGGAGAATAATGAGAATGGAAATAGGG-3′ (forward) and 5′-GTTGCAAAGGCTTCCAAACAAGGAGAGGCA-3′ (reverse), and the primers for glyceraldehyde-3-phosphate dehydrogenase amplification were 5′-CAGTCCATGCCATCACTGCCA-3′ (forward) and 5′-AGTGTAGCCCAGGATGCCCTT-3′ (reverse). The PCR-amplified products were resolved by agarose gel electrophoresis, detected by a Typhoon 9410 Image Scanner (GE Healthcare, Piscataway, NJ), and quantitated by KODAK 1D Image Analysis software (Eastman Kodak, Rochester, NY).
Transient Transfection and Cell Treatments
Huh 7 cells were plated in 24-well plates in DMEM supplemented with 10% delipidated fetal bovine serum at a density of 5 × 104 cells per well and cultured overnight. Transient transfection was conducted by lipofection with LipofectAMINE and Plus Reagent (Invitrogen) as described previously (Song et al., 2004). For all the transfections, standard amounts of plasmid DNA used per well were 100 ng for BSEP promoter construct, 100 ng for nuclear receptor expression plasmids, and 10 ng for the null-Renilla luciferase plasmid as an internal control. After cells were transfected for 3 h, 0.75 ml of fresh medium was added into each well, and cells were incubated overnight. Then, cell supernatants were replaced with treatment medium containing appropriate chemicals at a concentration specified in the figure legends. The treatment was continued for 30 h unless specified.
Reporter Luciferase Assay
The reporter enzyme activities were assayed with the Dual-Luciferase Reporter assay system as described previously (Song et al., 2004). Treated Huh 7 cells were washed once with phosphate-buffered saline and lysed by adding 100 μl of passive lysis buffer (Promega) with gentle rocking for 30 min. Ten microliters of cell lysates were transferred to a 96-well reader plate, and luciferase activities were measured by an EG&G Berthold microplate luminometer (Perkin Elmer, Boston, MA). The Dual-Luciferase Reporter assay system contained two substrates that were used to determine the activity of two luciferases sequentially. The firefly luciferase activity, which represented the reporter activity, was initiated by mixing aliquot of lysates (10 μl) with Luciferase Assay Reagent II (Promega). Then, the firefly luminescence was quenched, and the Renilla luminescence was simultaneously activated by adding Stop and Glo reagent (Promega) to the sample tube. The firefly luminescence was normalized based on the Renilla luminescence signal, and the ratio of treatment over control served as -fold activation. Data are presented as mean ± standard deviation (S.D.) of at least three separate experiments.
Results
Expression of BSEP was Induced by Oxysterol 22(R)-OH
Oxysterols are intermediates, whereas bile acids are the ultimate metabolites of cholesterol. BSEP is responsible for bile secretion of bile acids, a rate-limiting step in the bile enterohepatic circulation. Oxysterol 22(R)-OHC is a potent agonist of nuclear receptor LXR. In a preliminary microarray experiment using human primary hepatocytes to identify transcriptionally regulated target genes by bile acid CDCA and oxysterol 22(R)-OHC, BSEP expression was induced not only by CDCA, as expected, but also unexpectedly by oxysterol 22(R)-OHC (data not shown). This unexpected finding prompted us to further investigate the underlying mechanism.
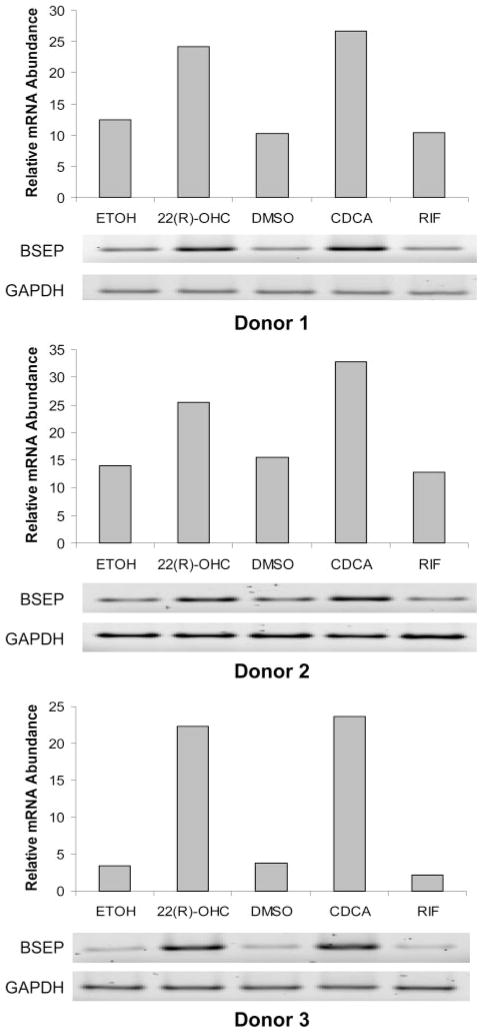
To confirm oxysterol 22(R)-OHC induced BSEP expression, human primary hepatocytes were treated with 10 μM 22(R)-OHC for 30 h, and the level of BSEP mRNA was determined by RT-PCR. As controls, hepatocytes treated with 10 μM CDCA and 10 μM rifampicin were included in the experiments. As shown in Fig. 1, a marked increase in BSEP mRNA was detected in hepatocytes treated with 22(R)-OHC, a level comparable with that detected in the cells treated with CDCA. However, treatment of hepatocytes with antibiotic rifampicin did not significantly alter the level of BSEP mRNA, indicating that BSEP is not a target of nuclear receptor PXR as rifampicin is a potent agonist for PXR. The marked induction was consistently detected in all three donors, although a slight variation was observed with a range from 2- to 5-fold increase in mRNA level.
Fig. 1.
BSEP mRNA expression was induced by oxysterol 22(R)-OHC. Human primary hepatocytes derived from three individual donors were treated with 0.1% ethanol (ETOH), 10 μM 22(R)-OHC, 0.1% DMSO, 10 μM CDCA, and rifampicin (RIF) for 30 h. Total cellular RNAs were isolated and subjected to RT-PCR using BSEP-specific primers. The PCR cycles were 30 for donors 1 and 3 and 35 for donor 2. Detection of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was included as an internal control. The PCR-amplified products were resolved by agarose gel electrophoresis (1.8%), detected by a Typhoon 9410 Image Scanner, and quantitated by KODAK 1D Image Analysis Software.
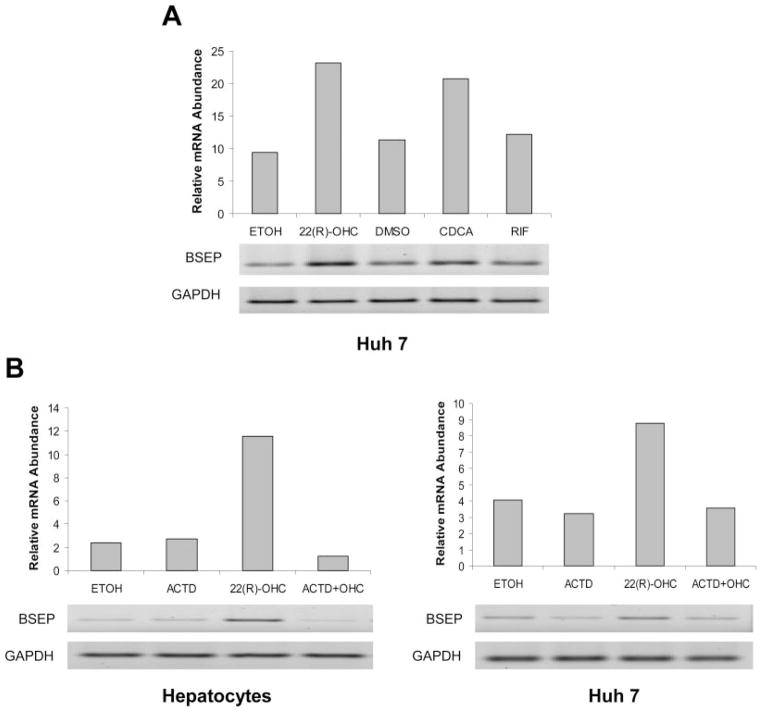
To test whether hepatoma Huh 7 cells respond to 22(R)-OHC similarly as human primary hepatocytes, cells were treated with oxysterol 22(R)-OHC, CDCA, and rifampicin. As shown in Fig. 2A, an approximately 2-fold induction was detected in Huh 7 cells treated with either oxysterol 22(R)-OHC or CDCA. Similar to the data obtained from hepatocytes, no obvious alteration of BSEP expression was detected in cells treated with rifampicin. In summary, the data demonstrated that BSEP mRNA was induced by oxysterol 22(R)-OHC in human primary hepatocytes as well as hepatoma Huh 7 cells.
Fig. 2.
A, induction of BSEP mRNA expression by 22(R)-OHC in hepatoma cells. Hepatoma Huh 7 cells were treated with 0.1% ETOH, 10 μM 22(R)-OHC, 0.1% DMSO, 10 μM CDCA, and RIF for 30 h. Total cellular RNAs were isolated and subjected to RT-PCR using BSEP-specific primers with 35 cycles. B, the up-regulation of BSEP mRNA expression was attenuated by actinomycin D. Human primary hepatocytes and Huh 7 cells were treated with 0.1% ETOH, 10 μM 22(R)-OHC, 5 μM actinomycin D (ACTD), and a combination of 10 μM 22(R)-OHC and 5 μM ACTD. Total cellular RNAs were isolated and subjected to RT-PCR. The PCR cycles were 30 for both hepatocytes and Huh 7-derived samples.
Oxysterol 22(R)-OHC Regulates BSEP Expression at Transcriptional Level
To determine whether the increase in BSEP mRNA upon 22(R)-OHC treatment is due to transactivation or decreased degradation of BSEP mRNA, human primary hepatocytes and hepatoma Huh 7 cells were treated with a combination of 10 μM 22(R)-OHC and 5 μM actinomycin D, a potent transcription inhibitor, for 24 h, and the levels of BSEP mRNA were determined by RT-PCR. As shown in Fig. 2B, treatment with actinomycin D completely abolished the induction of BSEP mRNA expression by oxysterol 22(R)-OHC in both hepatocytes and Huh 7 cells, indicating that oxysterol 22(R)-OHC transcriptionally up-regulates BSEP expression.
The Up-Regulation of BSEP Expression by Oxysterol 22(R)-OHC Is Achieved through Nuclear Receptor FXR but Not LXR
Studies have demonstrated that oxysterol 22(R)-OHC is a potent endogenous agonist for nuclear receptor LXR. Activation of LXR by 22(R)-OHC transactivates a group of LXR targets (Janowski et al., 1996; Peet et al., 1998a). On the other hand, CDCA-mediated transactivation of BSEP expression is through activation of nuclear receptor FXR (Ananthanarayanan et al., 2001; Plass et al., 2002). We hypothesized that transcriptional up-regulation of BSEP expression by 22(R)-OHC is through activation of LXR.
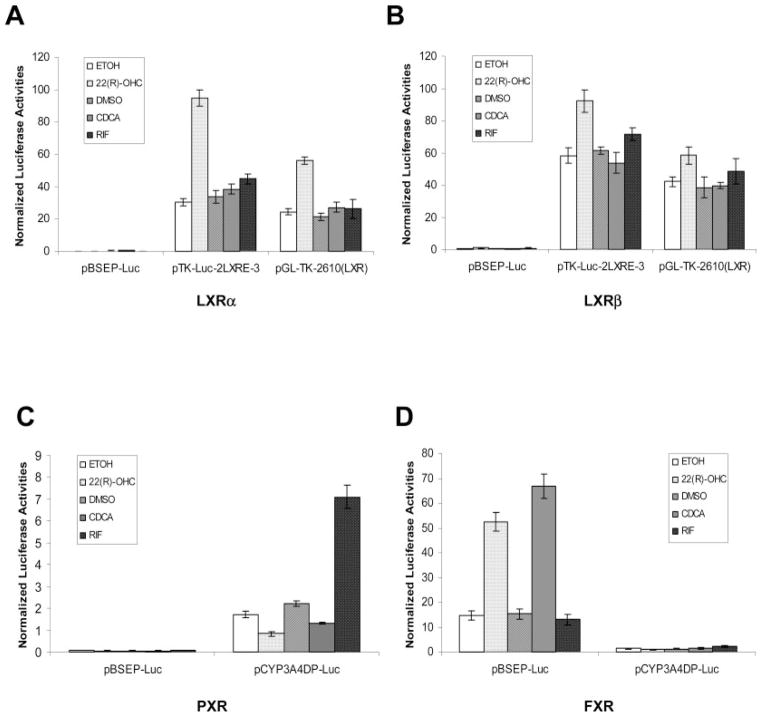
To test the hypothesis, a BSEP promoter reporter was constructed by cloning a 2.6-kb genomic fragment upstream of the BSEP transcription start site into the luciferase vector pGL4.10. The resulting pBSEP-Luc construct was cotransfected with nuclear receptor LXRα or LXRβ, followed by treatment of the cotransfected cells with 22(R)-OHC, CDCA, or rifampicin. In contrast to our hypothesis, BSEP promoter was not activated at all by either LXRα or LXRβ in the presence of oxysterol 22(R)-OHC (Fig. 3, A and B). On the other hand, as positive controls, the two LXR element (LXRE)-containing reporters pTK-Luc-2LXRE-3 and pGL-TK-2610 (LXR) were strongly activated by both LXRα and LXRβ, and an additional induction was observed by the treatment with 22(R)-OHC (Fig. 3, A and B). The results demonstrated that 22(R)-OHC-induced activation of BSEP was not through nuclear receptor LXRα or LXRβ.
Fig. 3.
Oxysterol 22(R)-OHC-mediated up-regulation of BSEP promoter activity was achieved through nuclear receptor FXR but not LXR or PXR. Huh 7 cells were transiently transfected with 100 ng of BSEP promoter reporter pBSEP-Luc or other nuclear receptor target reporters, 100 ng of expression plasmid for either nuclear receptor LXRα or LXRβ, PXR, and FXR, and 10 ng of the null-Renilla luciferase plasmid as an internal control. Sixteen hours post-transfection, cells were treated with 0.1% ETOH, 10 μM 22(R)-OHC, 0.1% DMSO, 10 μM CDCA, and RIF for 30 h. Luciferase activities were measured by the Dual-Luciferase Reporter assay system. The firefly luminescence was normalized based on the Renilla luminescence signal. Data are presented as mean ± S.D. of at least three separate experiments. A, cells were cotransfected with LXRα and pBSEP-Luc or pTK-Luc-2LXRE-3 or pGL-TK-2610 (LXR). B, cells were cotransfected with LXRβ and pBSEP-Luc or pTK-Luc-2LXRE-3 or pGL-TK-2610 (LXR). C, cells were cotransfected with PXR and pBSEP-Luc or pCYP3A4DP-Luc. D, cells were cotransfected with FXR and pBSEP-Luc or pCYP3A4DP-Luc.
To test whether nuclear receptor pregnane X receptor (PXR) or FXR was involved in the activation of BSEP by 22(R)-OHC, Huh 7 cells were cotransfected with pBSEP-Luc and PXR or FXR expression plasmid, followed by treatment with 22(R)-OHC, CDCA, and rifampicin. As a positive control for PXR and negative control for FXR, a PXR target gene reporter, pCYP3A4DP-Luc, was included in the experiment. As shown in Fig. 3C, no induction of BSEP promoter activity was detected in cells cotransfected with PXR and pBSEP-Luc in the presence of 22(R)-OHC or PXR agonist rifampicin, whereas strong induction was observed in cells cotransfected with PXR and pCYP3A4DP-Luc, and an additional induction was detected upon treatment with rifampicin. However, as shown in Fig. 3D, marked activation of BSEP promoter was achieved by cotransfection with nuclear receptor FXR (Fig. 3B). More importantly, an additional more than 3-fold induction was detected upon treatment with 22(R)-OHC. The level of induction of BSEP promoter activity by 22(R)-OHC was comparable with that mediated by CDCA (Fig. 3D), consistent with the previous observation for BSEP mRNA induction in hepatocytes by 22(R)-OHC and CDCA (Fig. 1). The data demonstrated that 22(R)-OHC-induced expression of BSEP was achieved through FXR. It should be mentioned that activation of BSEP promoter activity upon FXR transfection without 22(R)-OHC or CDCA treatment may result from the presence of endogenous FXR agonists, such as bile acids and oxysterols. Taken together, those data demonstrated that oxysterol 22(R)-OHC mediated transcriptional up-regulation of BSEP was achieved through nuclear receptor FXR but not LXR or PXR.
Oxysterol/FXR-Mediated Up-Regulation of BSEP Promoter Activity Is Time- and Dose-Dependent
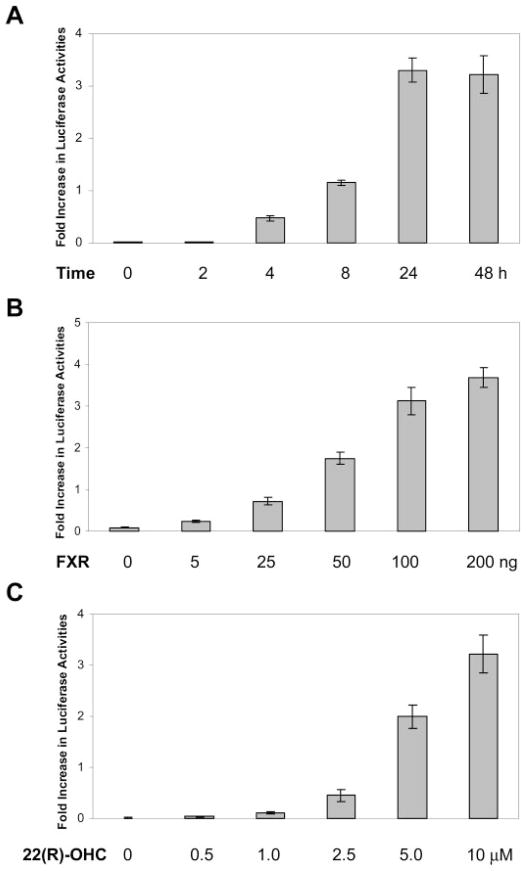
To gain insight into the molecular mechanism on oxysterol/FXR-mediated regulation of BSEP expression, time and dose-response studies were performed. For the time course study, Huh 7 cells were cotransfected with pBSEP-Luc plasmid DNA and FXR expression construct, followed by treatment with 22(R)-OHC for 0, 2, 4, 8, 24, and 48 h. As shown in Fig. 4A, an increase in luciferase activity was first detected at 4 h post-treatment, elevated to approximately 1-fold at 8 h, and reached the plateau at 24 and 48 h.
Fig. 4.
A, time-dependent activation of BSEP promoter by 22(R)-OHC. Huh 7 cells were cotransfected with 100 ng of BSEP promoter reporter pBSEP-Luc, 100 ng of FXR expression construct, and 10 ng of null-Renilla luciferase plasmid. Sixteen hours post-transfection, cells were treated with 10 μM 22(R)-OHC for 0, 2, 4, 8, 24, and 48 h. Luciferase activities were measured by the Dual-Luciferase Reporter assay system. B, FXR dose-dependent activation of BSEP promoter by 22(R)-OHC. Huh 7 cells were transfected with 100 ng of BSEP promoter reporter pBSEP-Luc, 10 ng of the null-Renilla luciferase plasmid, and increasing amounts of FXR expression construct (0, 5, 25, 50, 100, and 200 ng). Sixteen hours post-transfection, cells were treated with 10 μM 22(R)-OHC for 30 h. Luciferase activities were measured as described above. C, dose-dependent induction of BSEP promoter activity by 22(R)-OHC. Huh 7 cells were cotransfected with 100 ng of BSEP promoter reporter pBSEP-Luc, 100 ng of FXR expression construct, and 10 ng of the null-Renilla luciferase plasmid. Sixteen hours post-transfection, cells were treated with various concentrations of 22(R)-OHC (0, 0.5, 1.0, 2.5, 5, and 10 μM) for 30 h. Luciferase activities were measured by the Dual-Luciferase Reporter assay system.
For the FXR dose-response study, Huh 7 cells were cotransfected with pBSEP-Luc plasmid and increasing amounts of FXR constructs (5, 10, 25, 50, 100, and 200 ng), followed by treatment with 22(R)-OHC. As shown in Fig. 4B, induction of luciferase activities correlated very well with increasing amounts of FXR, indicating that the level of FXR expression is one of the limiting factors in the induction of the BSEP expression by 22(R)-OHC.
For the 22(R)-OHC dose-response study, Huh 7 cells were cotransfected with pBSEP-Luc and FXR plasmid DNA, followed by treatment of transfected cells with increasing concentrations of 22(R)-OHC (0, 0.5, 1.0, 2.5, 5, and 10 μM). As shown in Fig. 4C, an increase in luciferase activity was first detected in cells treated with 2.5 μM 22(R)-OHC, followed by strong induction (3-fold) in cells treated with 5 μM 22(R)-OHC, and highest increase (~ 4-fold) in cells treated with 10 μM 22(R)-OHC. The data indicate that the up-regulation of BSEP promoter activity by 22(R)-OHC is dose-dependent. Higher concentration of 22(R)-OHC was not used in the study because of the fact that concentration higher than 10 μM resulted in death of Huh 7 cells, consistent with the report that oxysterols induced cell death at higher concentration (Pettersen et al., 1991). In summary, oxysterol-mediated up-regulation of BSEP expression is time- and dose-dependent on both nuclear receptor FXR and oxysterol 22(R)-OHC.
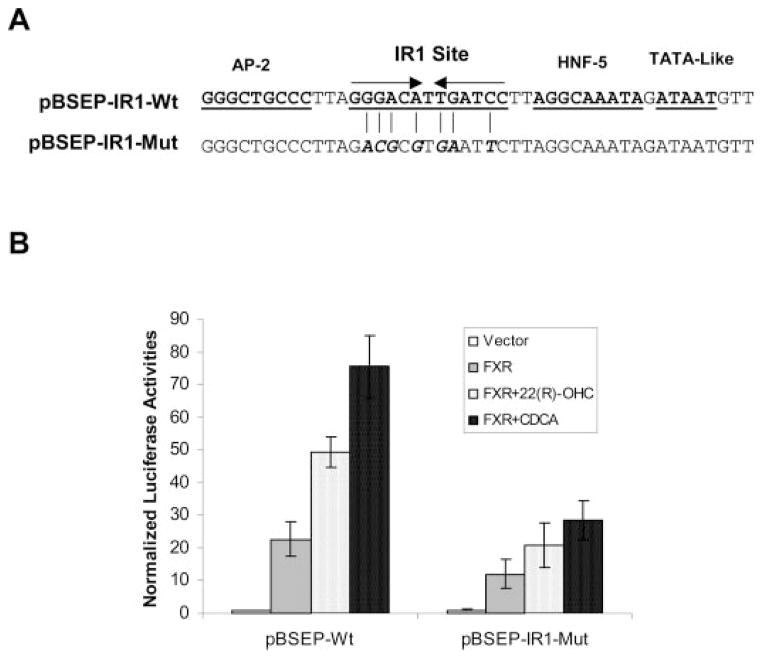
IR1 Element in the BSEP Promoter Is Required for Maximal Induction of BSEP Promoter Activity by 22(R)-OHC
Transactivation of the BSEP promoter by CDCA is through the IR1 element in the BSEP promoter (Ananthanarayanan et al., 2001; Plass et al., 2002). To determine whether the oxysterol/FXR-mediated transcriptional regulation of BSEP expression is through the same IR1 element, site-directed mutagenesis was performed to disrupt the IR1 element, and the resulting mutant was tested for its ability to respond to 22(R)-OHC. As shown in Fig. 5A, the wt IR1 sequence was mutated in multiple positions to ensure complete destruction of the site. Four and three nucleotides were substituted in the first half and the second half, respectively. The resulting mutant, pBSEP-IR1-Mut, was cotransfected into Huh 7 cells with FXR, followed by treatment with 22(R)-OHC for 30 h. As shown in Fig. 5B, destruction of the IR1 element significantly decreased the ability of BSEP promoter to respond to 22(R)-OHC as well as CDCA, although a considerable level of induction was still observed in cells transfected with pBSEP-IR1-Mut compared with vehicle-treated cells. The data demonstrated that the IR1 element was required for maximal induction of BSEP expression by 22(R)-OHC.
Fig. 5.
A, substitution of the IR1 element in the BSEP promoter. Multiple nucleotide substitutions were introduced in the IR1 element by site-directed mutagenesis, resulting in an IR1 mutant, pBSEP-IR1-Mut. Four and three nucleotides were mutated in the first half and the second half, respectively, to ensure complete destruction of the site. B, the IR1 element was required for maximal activation of BSEP promoter by 22(R)-OHC. Huh 7 cells were transfected with 100 ng of either pBSEP-Luc (pBSEP-IR1-wt) or the IR1 mutant (pBSEP-IR1-Mut) with 100 ng of FXR expression plasmid, and 10 ng of the null-Renilla luciferase plasmid. Sixteen hours post-transfection, cells were treated with 10 μM 22(R)-OHC for 30 h. Luciferase activities were measured by the Dual-Luciferase Reporter assay system.
Mutation of Residues L340 and R351 of FXR Differentiates CDCA-and 22(R)-OHC-Mediated Activation
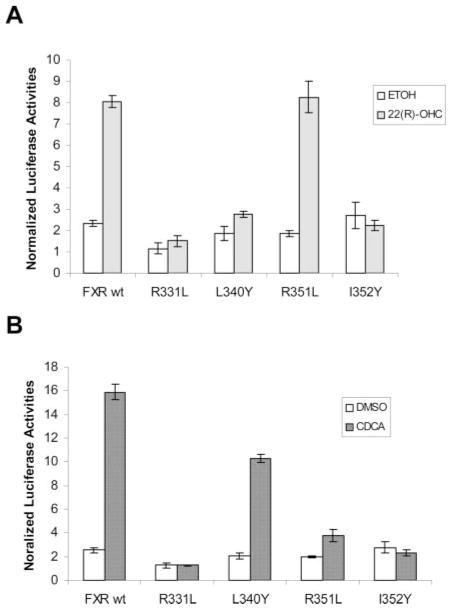
To determine whether 22(R)-OHC and CDCA bind to similar structural features of FXR, a series of FXR mutants were made in the FXR LBD, guided by the crystal structures of rat and human FXR LBD in complex with 6ECDCA or fexaramine (Downes et al., 2003; Mi et al., 2003). The amino acid residues selected for mutation are in the binding pocket with ligand 6ECDCA or fexaramine. Unfavorable substitutions were introduced for all the residues to disrupt its binding capability with respective ligands. The resulting FXR mutants were tested for their ability to activate BSEP promoter in the presence of 22(R)-OHC or CDCA.
As shown in Fig. 6, mutation of residue 331 from arginine to leucine (R331L) or residue 352 from isoleucine to tyrosine (I352Y) abolished the activation mediated by both 22(R)-OHC and CDCA, indicating that the two residues are important in binding with 22(R)-OHC and CDCA. More importantly, substitution of residue 351 from arginine to leucine (R351L) or residue 340 from leucine to tyrosine (L340Y) exhibited differential activation mediated by 22(R)-OHC and CDCA. As shown in Fig. 6B, mutation R351L almost completely abolished CDCA-induced activation; however, the mutant maintained its ability to activate BSEP promoter with 22(R)-OHC at the level similar to that achieved with FXR wt (Fig. 6A). The results suggest that residue R351 is critical for CDCA but not 22(R)-OHC-mediated activation. In contrast to R351L mutant, FXR L340Y mutant exhibited the opposite effect on CDCA- and 22(R)-OHC-induced activation. Substitution of residue 340 from leucine to tyrosine resulted in complete loss of its ability to activate BSEP promoter with 22(R)-OHC (Fig. 6A). However, CDCA-induced activation remained largely intact (Fig. 6B). The data indicate that residue L340 is critical for oxysterol-mediated activation.
Fig. 6.
Activation of FXR mutants by oxysterol 22(R)-OHC (A) and bile acid CDCA (B). Amino acid residues R331, L340, R351, and I352 in the FXR LBD were mutated to leucine, tyrosine, leucine, and tyrosine, respectively. After sequence verification, each of the resultant FXR mutants (100 ng) was cotransfected into Huh 7 cells on 24-well plates with 100 ng of pBSEP-Luc and 10 ng of the null-Renilla luciferase plasmid. Sixteen hours post-transfection, cells were treated with 10 μM 22(R)-OHC and 1% ETOH or 10 μM CDCA and 1% DMSO. The treatments were continued for 30 h, followed by detection of luciferase activities with the Dual-Luciferase Reporter assay system. Data are presented as mean ± S.D. of at least three separate experiments.
Discussion
Oxysterols are cholesterol derivatives acting as sensors for cholesterol levels in the body. It is well established that oxysterols are natural endogenous ligands for nuclear receptor LXR, which regulates a class of target genes important in cholesterol metabolism and transport (Peet et al., 1998b; Chiang et al., 2001; Davis et al., 2002; Edwards et al., 2002; Repa et al., 2002). On the other hand, bile acids are a group of cholesterol metabolites and function as potent endogenous ligands for nuclear receptor FXR, which regulates a set of target genes in bile acid synthesis, transport, and elimination (Rizzo et al., 2005). Therefore, oxysterol/LXR and bile acid/FXR pathways coordinately regulate their target genes to maintain the cholesterol and bile acid homeostasis.
BSEP is responsible for the canalicular secretion of bile acids, a rate-limiting step in the bile enterohepatic circulation (Meier and Stieger, 2002; Fuchs, 2003; Kullak-Ublick et al., 2004). Its expression is up-regulated by its substrate bile acids, such as CDCA, through activation of FXR (Ananthanarayanan et al., 2001; Plass et al., 2002). In this study, we have demonstrated that oxysterol 22(R)-OHC, a potent LXR ligand, markedly induced BSEP expression, and the induction is achieved through activation of FXR but not LXR. The results suggest that 22(R)-OHC functions not only as key regulators for LXR targets but also for FXR target BSEP. Other endogenous intermediates in the synthesis of cholesterol, including farnesol and lanosterol, also function as ligand for nuclear receptor FXR (Forman et al., 1995; Otte et al., 2003). It becomes evident that in addition to LXR, FXR also plays vital role in regulating genes important in the synthesis, transport, and metabolism of cholesterol. Consistent with our finding that 22(R)-OHC functions as a dual ligand for LXR and FXR is a study showing that a potent synthetic LXR ligand, T0901317, activates FXR more potently than CDCA (Houck et al., 2004). Our finding raises a series of not only interesting but also important questions, such as whether other oxysterols serve as FXR ligands to regulate FXR target genes, how oxysterols interplay with bile acids to regulate FXR target genes, and how oxysterols behave as dual ligands in the presence of FXR and LXR in consideration of the fact that FXR, LXR, and oxysterols coexist in many tissues including liver, intestine, kidney, adrenal, and vascular epithelial cells (Bishop-Bailey et al., 2004). Those questions are currently under investigation.
Transactivation of a target gene by FXR or LXR requires formation of FXR or LXR with their obligate partner, retinoid X receptor (RXR). In Huh 7 cells, strong induction of BSEP promoter activity by 22(R)-OHC and CDCA was achieved when Huh 7 cells were cotransfected with FXR, suggesting that endogenous RXR is sufficient to support the activation. Indeed, an abundant RXR-specific band was detected by RT-PCR in Huh 7 cells (data not shown). The fact that exogenously added FXR was required for maximal induction of BSEP promoter activity indicates that FXR rather than RXR is the limiting factor in the formation of FXR/RXR heterodimer in Huh 7 cells. It should be mentioned that BSEP promoter activity was strongly increased in cells cotransfected with FXR with vehicle treatment (Fig. 3). The increase in BSEP promoter activity upon FXR transfection without 22(R)-OHC or CDCA treatment may result from the presence of endogenous FXR agonists, such as bile acids and oxysterols. It cannot be excluded, however, that FXR may directly transactivate BSEP promoter in a ligand-independent mechanism.
Bile acids mediated up-regulation of BSEP expression is achieved by binding of the bile acid-activated FXR to the IR1 element in the BSEP promoter. In this study, we have demonstrated that destruction of the IR1 element significantly attenuates the induction of BSEP promoter activity by 22(R)-OHC and CDCA, indicating that a direct binding of the activated FXR to the IR1 site is required for the 22(R)-OHC and CDCA-mediated induction of BSEP. However, as shown in Fig. 5B, the FXR-dependent induction of BSEP promoter activity was not completely abolished by the destruction of the IR1 element. Approximately 8-fold increases in promoter activity were detected in cells cotransfected with IR1 mutant and FXR compared with vector-cotransfected cells, and furthermore, an additional 71 or 134% induction was detected upon 22(R)-OHC or CDCA treatment, respectively. A similar phenomenon was observed in a study using rat FXR and human BSEP promoter (Plass et al., 2002). The data suggest an additional IR1-independent mechanism to support the transactivation of BSEP promoter by FXR.
Residue R331 of rat FXR is involved in hydrogen bonding with its ligand 6ECDCA (Mi et al., 2003). In this study, substitution of the corresponding residues in human FXR from arginine to leucine completely abolished CDCA- and 22(R)-OHC-mediated activation, indicating that the residue may mediate hydrogen bonding with CDCA and 22(R)-OHC. Residue I352 of human FXR is in contact with the methyl ester chain of fexaramine through van der Waals interaction (Downes et al., 2003). Mutation of the residue resulted in loss of its activation by CDCA and 22(R)-OHC, indicating that the residue may be in direct contact with CDCA and 22(R)-OHC as well. Residue R351 is next to residue E350, which mediates hydrogen bonding with 6ECDCA (Mi et al., 2003). Mutation of the residue abolished CDCA-mediated activation but not that mediated by 22(R)-OHC. The data suggest that residue R351 is important for CDCA-mediated activation through either direct interaction with CDCA or maintaining the structure required for CDCA binding. However, the residue does not play critical role in 22(R)-OHC-mediated activation. In contrast to R351L mutant, L340Y mutant exhibited the opposite effect on CDCA- and 22(R)-OHC-induced activation. Mutation of the residue from leucine to tyrosine dramatically decreased 22(R)-OHC-induced transactivation; however, CDCA-mediated activation was largely intact. Residue L340 of rat FXR is not in direct contact with 6ECDCA (Mi et al., 2003), but the corresponding residue in human FXR makes van der Waals interaction with the hexyl group of fexaramine (Downes et al., 2003). Our results suggest that residue L340 is not directly in contact with CDCA as revealed in the crystal structure of rat FXR in complex with 6ECDCA. However, the residue plays important role in 22(R)-OHC-mediated activation, as shown in the crystal structure of human FXR in complex with fexaramine.
In summary, this study demonstrated that expression of BSEP was induced by oxysterol 22(R)-OHC, and the induction was mediated through activation of nuclear receptor FXR but not LXR. The IR1 element in the BSEP promoter was required for maximal induction of BSEP expression by 22(R)-OHC, and mutation of residues L340 and R351 in the FXR LBD differentiated CDCA- and 22(R)-OHC-mediated activation.
Acknowledgments
We thank Dr. David Mangelsdorf for providing human FXR, LXRα, and LXRβ expression plasmids; Dr. Bryan Goodwin for providing pGL-TK-2610 (LXR) construct; and Dr. Peter Edwards for providing pTK-Luc-2LXRE-3 reporter plasmid. The contributions to the project by Dr. Ma Yuzhong and Song Xiulong are gratefully acknowledged.
This work was partially supported by National Institutes of Health Grants R01GM61988 and R01ES07965, and Rhode Island-IDeA Network of Biomedical Research Excellence Grant P20RR16457 from National Center for Research Resources/National Institutes of Health.
ABBREVIATIONS
- LXR
liver X receptor
- BSEP
bile salt export pump
- CDCA
chenodeoxycholic acid
- FXR
farsenoid X receptor
- IR1
inverted repeat spacing by one nucleotide
- CYP7A1
cholesterol 7α-hydroxylase
- 22(R)-OHC
22(R)-hydroxycholesterol
- LBD
ligand binding domain
- DMSO
dimethyl sulfoxide
- DMEM
Dulbecco’s modified Eagle’s medium
- PCR
polymerase chain reaction
- PXR
pregnane X receptor
- 6ECDCA
6α-ethyl-chenodeoxycholic acid
- RT-PCR
reverse transcription coupled-polymerase chain reaction
- LXRE
LXR element
- wt
wild type
- Mut
mutant
- RXR
retinoid X receptor
- ETOH
ethanol
- RIF
rifampicin
References
- Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001;276:28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- Auchus RJ. Overview of dehydroepiandrosterone biosynthesis. Semin Reprod Med. 2004;22:281–288. doi: 10.1055/s-2004-861545. [DOI] [PubMed] [Google Scholar]
- Bishop-Bailey D, Walsh DT, Warner TD. Expression and activation of the farnesoid X receptor in the vasculature. Proc Natl Acad Sci USA. 2004;101:3668–3673. doi: 10.1073/pnas.0400046101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang JY. Bile acid regulation of gene expression: roles of nuclear hormone receptors. Endocr Rev. 2002;23:443– 463. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- Chiang JY, Kimmel R, Stroup D. Regulation of cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRal-pha) Gene. 2001;262:257–265. doi: 10.1016/s0378-1119(00)00518-7. [DOI] [PubMed] [Google Scholar]
- Chiang JY, Kimmel R, Weinberger C, Stroup D. Farnesoid X receptor responds to bile acids and represses cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J Biol Chem. 2000;275:10918–10924. doi: 10.1074/jbc.275.15.10918. [DOI] [PubMed] [Google Scholar]
- Crocenzi FA, Mottino AD, Sanchez-Pozzi EJ, Pellegrino JM, Rodriguez Garay EA, Milkiewicz P, Vore M, Coleman R, Roma MG. Impaired localisation and transport function of canalicular Bsep in taurolithocholate induced cholestasis in the rat. Gut. 2003;52:1170–1177. doi: 10.1136/gut.52.8.1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Huang L, Zhao A, Lew JL, Yu J, Sahoo S, Meinke PT, Royo I, Pelaez F, Wright SD. Guggulsterone is a farnesoid X receptor antagonist in coactivator association assays but acts to enhance transcription of bile salt export pump. J Biol Chem. 2003;278:10214–10220. doi: 10.1074/jbc.M209323200. [DOI] [PubMed] [Google Scholar]
- Davis RA, Miyake JH, Hui TY, Spann NJ. Regulation of cholesterol-7alpha-hydroxylase: BAREly missing a SHP. J Lipid Res. 2002;43:533–543. [PubMed] [Google Scholar]
- Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer JL, Anisfeld AM, Edwards PA, et al. A chemical, genetic and structural analysis of the nuclear bile acid receptor FXR. Mol Cell. 2003;11:1079–1092. doi: 10.1016/s1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PA, Kennedy MA, Mak PA. LXRs; oxysterol-activated nuclear receptors that regulate genes controlling lipid homeostasis. Vascul Pharmacol. 2002;38:249–256. doi: 10.1016/s1537-1891(02)00175-1. [DOI] [PubMed] [Google Scholar]
- Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687– 693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- Fuchs M. Bile acid regulation of hepatic physiology: III. Regulation of bile acid synthesis: past progress and future challenges. Am J Physiol. 2003;284:G551–G557. doi: 10.1152/ajpgi.00468.2002. [DOI] [PubMed] [Google Scholar]
- Horikawa M, Kato Y, Tyson CA, Sugiyama Y. Potential cholestatic activity of various therapeutic agents assessed by bile canalicular membrane vesicles isolated from rats and humans. Drug Metab Pharmacokinet. 2003;18:16–22. doi: 10.2133/dmpk.18.16. [DOI] [PubMed] [Google Scholar]
- Houck KA, Borchert KM, Hepler CD, Thomas JS, Bramlett KS, Michael LF, Burris TP. T0901317 is a dual LXR/FXR agonist. Mol Genet Metab. 2004;83:184–187. doi: 10.1016/j.ymgme.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature (Lond) 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- Jansen PL, Strautnieks SS, Jacquemin E, Hadchouel M, Sokal EM, Hooiveld GJ, Koning JH, De Jager-Krikken A, Kuipers F, Stellaard F, et al. Hepatocanalicular bile salt export pump deficiency in patients with progressive familial intrahepatic cholestasis. Gastroenterology. 1999;117:1370–1379. doi: 10.1016/s0016-5085(99)70287-8. [DOI] [PubMed] [Google Scholar]
- Kullak-Ublick GA, Stieger B, Meier PJ. Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology. 2004;126:322–342. doi: 10.1053/j.gastro.2003.06.005. [DOI] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Meier PJ, Stieger B. Bile salt transporters. Annu Rev Physiol. 2002;64:635– 661. doi: 10.1146/annurev.physiol.64.082201.100300. [DOI] [PubMed] [Google Scholar]
- Menke JG, Macnaul KL, Hayes NS, Baffic J, Chao YS, Elbrecht A, Kelly LJ, Lam MH, Schmidt A, Sahoo S, et al. A novel liver X receptor agonist establishes species differences in the regulation of cholesterol 7alpha-hydroxylase (CYP7a) Endocrinology. 2002;143:2548–2558. doi: 10.1210/endo.143.7.8907. [DOI] [PubMed] [Google Scholar]
- Mi LZ, Devarakonda S, Harp JM, Han Q, Pellicciari R, Willson TM, Khorasanizadeh S, Rastinejad F. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Mol Cell. 2003;11:1093–1100. doi: 10.1016/s1097-2765(03)00112-6. [DOI] [PubMed] [Google Scholar]
- Noe J, Stieger B, Meier PJ. Functional expression of the canalicular bile salt export pump of human liver. Gastroenterology. 2002;123:1659–1666. doi: 10.1053/gast.2002.36587. [DOI] [PubMed] [Google Scholar]
- Otte K, Kranz H, Kober I, Thompson P, Hoefer M, Haubold B, Remmel B, Voss H, Kaiser C, Albers M, et al. Identification of farnesoid X receptor beta as a novel mammalian nuclear receptor sensing lanosterol. Mol Cell Biol. 2003;23:864– 872. doi: 10.1128/MCB.23.3.864-872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsley E, Chiang JY. Guggulsterone antagonizes farnesoid X receptor induction of bile salt export pump but activates pregnane X receptor to inhibit cholesterol 7alpha-hydroxylase gene. Biochem Biophys Res Commun. 2003;304:191–195. doi: 10.1016/s0006-291x(03)00551-5. [DOI] [PubMed] [Google Scholar]
- Peet DJ, Janowski BA, Mangelsdorf DJ. The LXRs: a new class of oxysterol receptors. Curr Opin Genet Dev. 1998a;8:571–575. doi: 10.1016/s0959-437x(98)80013-0. [DOI] [PubMed] [Google Scholar]
- Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, Man-gelsdorf DJ. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998b;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- Pettersen KS, Boberg KM, Stabursvik A, Prydz H. Toxicity of oxygenated cholesterol derivatives toward cultured human umbilical vein endothelial cells. Arterioscler Thromb. 1991;11:423– 428. doi: 10.1161/01.atv.11.2.423. [DOI] [PubMed] [Google Scholar]
- Plass JR, Mol O, Heegsma J, Geuken M, Faber KN, Jansen PL, Muller M. Farnesoid X receptor and bile salts are involved in transcriptional regulation of the gene encoding the human bile salt export pump. Hepatology. 2002;35:589–596. doi: 10.1053/jhep.2002.31724. [DOI] [PubMed] [Google Scholar]
- Redinger RN. Nuclear receptors in cholesterol catabolism: molecular biology of the enterohepatic circulation of bile salts and its role in cholesterol homeostasis. J Lab Clin Med. 2003;142:7–20. doi: 10.1016/S0022-2143(03)00088-X. [DOI] [PubMed] [Google Scholar]
- Repa JJ, Berge KE, Pomajzl C, Richardson JA, Hobbs H, Mangelsdorf DJ. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J Biol Chem. 2002;277:18793–18800. doi: 10.1074/jbc.M109927200. [DOI] [PubMed] [Google Scholar]
- Rizzo G, Renga B, Mencarelli A, Pellicciari R, Fiorucci S. Role of FXR in regulating bile acid homeostasis and relevance for human diseases. Curr Drug Targets Immune Endocr Metabol Disord. 2005;5:289–303. doi: 10.2174/1568008054863781. [DOI] [PubMed] [Google Scholar]
- Roman ID, Fernandez-Moreno MD, Fueyo JA, Roma MG, Coleman R. Cyclosporin A induced internalization of the bile salt export pump in isolated rat hepatocyte couplets. Toxicol Sci. 2003;71:276–281. doi: 10.1093/toxsci/71.2.276. [DOI] [PubMed] [Google Scholar]
- Russell DW. The enzymes, regulation and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- Schroepfer GJ. Oxysterols: modulators of cholesterol metabolism and other processes. Physiol Rev. 2000;80:361–554. doi: 10.1152/physrev.2000.80.1.361. [DOI] [PubMed] [Google Scholar]
- Song X, Xie M, Zhang H, Li Y, Sachdeva K, Yan B. The pregnane X receptor binds to response elements in a genomic context-dependent manner and PXR activator rifampicin selectively alters the binding among target genes. Drug Metab Dispos. 2004;32:35– 42. doi: 10.1124/dmd.32.1.35. [DOI] [PubMed] [Google Scholar]
- Strautnieks SS, Bull LN, Knisely AS, Kocoshis SA, Dahl N, Arnell H, Sokal E, Dahan K, Childs S, Ling V, et al. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet. 1998;20:233–238. doi: 10.1038/3034. [DOI] [PubMed] [Google Scholar]
- Tiemann M, Han Z, Soccio R, Bollineni J, Shefer S, Sehayek E, Breslow JL. Cholesterol feeding of mice expressing cholesterol 7alpha-hydroxylase increases bile acid pool size despite decreased enzyme activity. Proc Natl Acad Sci USA. 2004;101:1846–1851. doi: 10.1073/pnas.0308426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Lo JL, Huang L, Zhao A, Metzger E, Adams A, Meinke PT, Wright SD, Cui J. Lithocholic acid decreases expression of bile salt export pump through farnesoid X receptor antagonist activity. J Biol Chem. 2002;277:31441–31447. doi: 10.1074/jbc.M200474200. [DOI] [PubMed] [Google Scholar]