Abstract
Hydrolysis of the phosphodiester bonds of the transcript by bacterial RNA polymerase is assisted by 3′NMP of the RNA. Here we provide evidence that this mechanism is also involved in RNA cleavage by eukaryotic RNA polymerase II, suggesting that transcript assisted hydrolysis has emerged before divergence of bacteria and archaea/eukaryotes.
Keywords: RNA polymerase II, transcript assisted proofreading, phosphodiester bond hydrolysis, elongation complex, backtracking
Multisubunit RNA polymerase (RNAP), the enzyme accomplishing transcription in all living organisms, has emerged before the divergence of bacteria and archaea/eukaryotes.1 Accordingly, the molecular mechanisms involved in RNA synthesis are highly conserved in evolution. Besides the synthesis of phosphodiester bonds (and pyrophosphorolysis, which is a direct reversal of this reaction) during transcript elongation, RNAP active center can catalyze the hydrolysis of the phosphodiester bonds of the nascent RNA. As well as the synthesis, the hydrolysis is catalyzed by the two metal (Mg2+) ion mechanism. In addition, in bacterial RNAP, a flexible domain of the active center, the Trigger Loop, plays a critical role in hydrolysis by participating in the reaction as a general base.2 This appears to be different from eukaryotic RNA polymerase II (RNAP II), whose Trigger Loop fails to adapt a catalytically active conformation during hydrolysis,3,4 explaining a much slower intrinsic hydrolysis by RNAP II as compared with bacterial RNAPs.
Earlier, we have discovered that an NMP at the 3′ end of the RNA transcript in the bacterial elongation complex also participates in catalysis of phosphodiester bond hydrolysis.5 For this to occur, RNAP has to backtrack by 1 base pair, thus positioning the penultimate (second from the 3′ end of the RNA) phosphodiester bond in the active center, making it ready for cleavage. The 3′NMP disengages from the template strand and flips backward to approach the site of the reaction, and helps to chelate the second Mg2+ ion, Mg2+II (which otherwise is bound weakly), to position the attacking water molecule and, possibly, to participate in the catalysis as a general acid/base.5 Such transcript-assisted hydrolysis of the second phosphodiester bond becomes even more prominent when RNAP is stabilized in the 1 base pair backtracked state via misincorporation of NMP that is not complementary to the base in the template strand. Chemical groups of 3′AMP, CMP, GMP and UMP contribute differently during the hydrolysis.5 Furthermore, the nature of the misincorporation event, i.e., the base in the template strand, may also influence the involvement of the chemical groups of the 3′NMP in the reaction. The erroneously incorporated NMP can be imagined to help to excise itself from the transcript, thus contributing to the proofreading of misincorporated events, i.e., overall fidelity of transcription.5,6
The “self-correcting” function of the transcript led to the proposition that transcript assisted second phosphodiester bond hydrolysis could be an ancient feature of transcription by multisubunit RNAPs, which may have emerged before the divergence of bacterial and archaeal/eukaryotic lineages. A possible involvement of the 3′NMP in the RNA hydrolysis by eukaryotic RNAP became evident from the crystal structure of the backtracked elongation complex of RNAP II3 (Fig. 1A). As was proposed for bacterial RNAP,5 the 3′NMP (GMP in this case) flips backward coming closer to the second phosphodiester bond and Mg2+I, possibly making itself available for assisting the reaction. This was also consistent with the faster cleavage by RNAP II of the second phosphodiester bond as compared with the ultimate one, even in the complexes that were not stabilized in the backtracked state.7 However, no biochemical evidence for the involvement of the 3′NMP, i.e., transcript assistance, in the hydrolysis of the second phosphodiester bond exists so far.
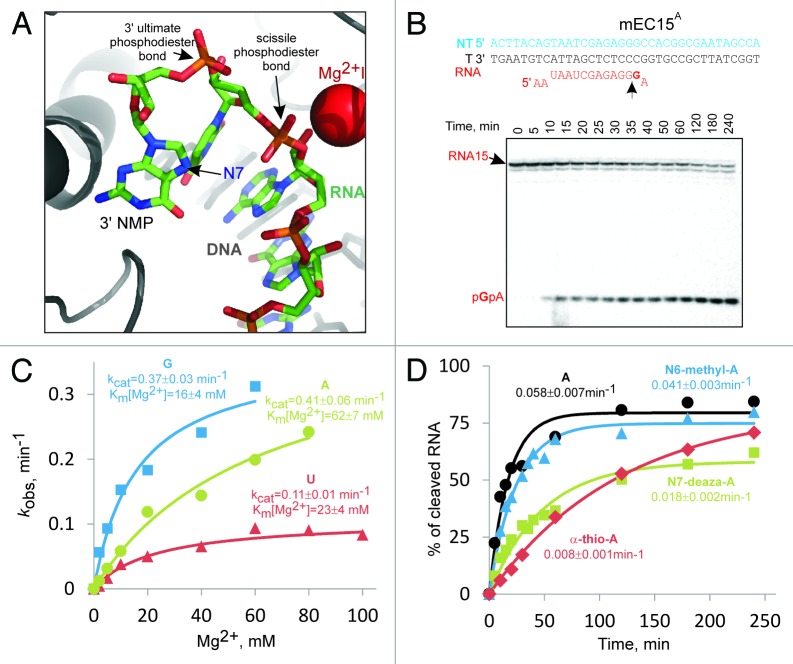
Figure 1. Transcript assisted proofreading by RNAP II. (A) In the 1 base pair backtracked elongation complex of RNAP II3 (pdbid: 3GTJ), RNA’s 3′NMP flips away from the template base and approaches the second phosphodiester bond. Protein elements and DNA are shown in gray. (B) Representative kinetics of the second phosphodiester bond cleavage in RNAP II 1 base pair backtracked complex, mEC15A. The nucleic acids scaffold of the complex is shown above the gel; T – template strand, NT – non-template strand. 32P-labeled GMP is in bold, the scissile phosphodiester bond is marked with an arrow. Note that the mismatched AMP was misincorporated by RNAP II prior to the start of the cleavage kinetics. (C) Mg2+ dependence of second phosphodiester bond hydrolysis in mEC15A, mEC15G and mEC15C. The data were fitted into a hyperbolic equation using SigmaPlot software. The Km[Mg2+] and the rate of the reaction in the saturating Mg2+ concentration are shown next to the plots. (D) The kinetics of second phosphodiester bond cleavage in elongation complexes carrying misincorporated AMP, N6-methyl-AMP, N7-deaza-AMP and α-thio-AMP in 10 mM Mg2+. The data were fitted in a single exponential equation using SigmaPlot software. The rates of reactions are shown next to the plots.
We decided to analyze the possibility of the assistance from the transcript’s 3′NMP during second phosphodiester bond hydrolysis by RNAP II active center. To do so, we used artificial elongation complexes assembled with S. cerevisiae RNAP II, fully complementary synthetic template and non-template DNA strands and synthetic RNA transcript (see scheme in Figure 1B). These complexes are indistinguishable from “native” stalled elongation complexes obtained by transcription on the double-stranded DNA from the promoter, but allow omitting the complicated step of transcription initiation and easy changing of the transcribed sequences.2,8-12 The complexes were assembled and immobilized on Ni-NTA agarose beads through 6-histidine tag of the RNAP II as described.2 To exclude the effect of translocation equilibrium on the rate of hydrolysis, we prepared complexes stabilized in the 1 base pair backtracked state due to a mismatch at the 3′ end of RNA. Instead of assembling complexes with mismatched RNA (which, in the case of RNAP II, may lead to incorrectly assembled elongation complexes; unpublished), we forced RNAP II in the “correct” elongation complex to misincorporate an NMP non-complementary to the template base (Fig. 1B). The RNA was 32P-labeled at the penultimate phosphodiester bond by incorporation of α[32P]GMP prior to the misincorporation, thus allowing us to monitor the excision of the dinucleotide upon second phosphodiester bond cleavage (Fig. 1B). All substrates were washed away before the start of the reaction, which was started, stopped and analyzed as described2 (see also legend of Fig. 1).
We misincorporated GMP, UMP and AMP opposite to dTMP, dTMP and dGMP in the template strand, respectively, and analyzed RNA hydrolysis in the formed misincorporated elongation complexes (mEC15G, mEC15U and mEC15A) (Fig. 1B). We determined the affinity to Mg2+II (Km[Mg2+]) and the rate of the reaction in the saturating Mg2+ (kcat). As seen from Figure 1C, the maximal rate of the second phosphodiester bond hydrolysis in mEC15U was ~4 times slower than that in mEC15G or mEC15A, suggesting that 3′purine increases the rate of the reaction as compared with pyrimidines. Km[Mg2+] in mEC15A was 3–4 times higher than that in mEC15G or mEC15U. This result suggests that the 3′ UMP and GMP may participate in the chelation of Mg2+II, while 3′ AMP may not provide additional coordination bonds.
To analyze if chemical groups of 3′purine are involved in the accelerated second phosphodiester bond hydrolysis, we used chemically modified 3′AMP. We misincorporated α-thio-AMP, 7-deaza-AMP and N6-methyl-AMP at the 3′ end of the transcript and measured the rate of second phosphodiester bond hydrolysis in the resultant complexes. Given that 3′AMP (i.e., its chemical groups) unlikely participates in the Mg2+ binding, we measured the rate of the reaction only in 10 mM Mg2+. As seen from Figure 1D, N6-methyl group had little effect on the rate of the reaction. However, substitution of nitrogen in the position 7 of the purine rings with carbon reduced the rate of the reaction ~3-fold, while the thio-group in the ultimate phosphodiester bond slowed down the reaction ~7-fold.
The above result suggests that the nitrogen in position 7 of the adenine ring may participate in the penultimate phosphodiester bond hydrolysis as an acid/base and/or by coordinating the attacking water molecule. This hypothesis is supported by the crystal structure of the backtracked RNAP II elongation complex3 (pdbid: 3GTJ), in which the N7 of the 3′guanine moiety is turned toward the penultimate phosphodiester bond where it can assist the reaction (Fig. 1A). The phosphate of the 3′GMP in this structure is also positioned so that it can directly or through the network of hydrogen bonding interact with the attacking water molecule (Fig. 1A), which is consistent with our results.
All together, our results suggest that, like bacterial RNAP, RNAP II also may use the transcript-assisted hydrolysis of the phosphodiester bond. As mentioned above, the Trigger Loop of RNAP II seems not to fold fully in the backtracked elongation complex, and thus cannot participate in the transcript assisted hydrolysis, as it does in the case of bacterial RNAP.2 This deviation and some differences in the amino acid content of the active centers of bacterial RNAP and RNAP II likely influence the way the chemical groups of 3′NMPs are involved in the reaction. For example, N7 of the 3′AMP of bacterial elongation complex was proposed to participate in the chelation of Mg2+II.5 In the case of RNAP II, however, 3′AMP does not contribute strongly to binding of Mg2+II. Instead, N7 of 3′AMP seems to accelerate the reaction rate. In contrast, however, for example, the phosphate group of 3′AMP seems to be involved in the reaction similarly for both, bacterial RNAP5 and RNAP II.
The 3′AMP assisted hydrolysis is ~4 times faster than 3′UMP assisted hydrolysis in both bacterial and RNAP II elongation complexes. It is also ~4 times faster than cleavage assisted by 3′CMP in bacterial RNAP (we did not analyze assistance from 3′CMP for RNAP II). Though cleavage assisted by 3′GMP in bacterial elongation complex is 4–5 times slower than with 3′AMP,5 the rates of cleavage by RNAP II assisted by 3′AMP or 3′GMP are close. Together, these data may indicate that the 3′purine accelerates phosphodiester bond hydrolysis more efficiently than 3′pyrimidines, possibly due to the presence of the imidazole ring. It is possible, that the participation of 3′purines in the reaction has been similar earlier in evolution but diverged later, with N7 of 3′AMP losing its ability to chelate Mg2+ during cleavage by RNAP II, and N7 of 3′GMP losing the ability to accelerate the reaction by bacterial RNAP.
Deeper biochemical analysis, such as determining pH profiles of reactions and analysis of a more diverse panel of chemically modified 3′NMPs are required to build the full picture of similarities/differences of transcript assisted RNA cleavage between bacterial and eukaryotic RNAPs. However, the above data together with our earlier results5 support an intriguing hypothesis that transcript-assisted phosphodiester bond hydrolysis may have emerged before the divergence of bacteria and archaea/eukaryotes, at the stage of the Last Universal Common Ancestor (LUCA). This mechanism may have been critical in the absence of elongation cleavage factors (evolutionary unrelated factor S of archaea/eukaryotes and Gre factor of bacteria, which must have emerged after divergence of domains of life) to rescue backtracked and/or misincorporated transcription elongation complexes.
Acknowledgments
This work was supported by UK Biotechnology and Biological Sciences Research Council and the European Research Council [ERC-2007-StG 202994-MTP].
Glossary
Abbreviations:
- RNAP
RNA polymerase
- RNAP II
RNA polymerase II
- NMP
nucleotide monophosphate
- NTA
nitrilotriacetate
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Werner F, Grohmann D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 2.Yuzenkova Y, Zenkin N. Central role of the RNA polymerase trigger loop in intrinsic RNA hydrolysis. Proc Natl Acad Sci U S A. 2010;107:10878–83. doi: 10.1073/pnas.0914424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science. 2009;324:1203–6. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–53. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 5.Zenkin N, Yuzenkova Y, Severinov K. Transcript-assisted transcriptional proofreading. Science. 2006;313:518–20. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 6.Cramer P. Molecular biology. Self-correcting messages. Science. 2006;313:447–8. doi: 10.1126/science.1131205. [DOI] [PubMed] [Google Scholar]
- 7.Thomas MJ, Platas AA, Hawley DK. Transcriptional fidelity and proofreading by RNA polymerase II. Cell. 1998;93:627–37. doi: 10.1016/S0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 8.Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA:DNA hybrid in the processivity of transcription. Mol Cell. 1998;2:55–64. doi: 10.1016/S1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 9.Yuzenkova Y, Bochkareva A, Tadigotla VR, Roghanian M, Zorov S, Severinov K, Zenkin N. Stepwise mechanism for transcription fidelity. BMC Biol. 2010;8:54. doi: 10.1186/1741-7007-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuzenkova Y, Roghanian M, Bochkareva A, Zenkin N. Tagetitoxin inhibits transcription by stabilizing pre-translocated state of the elongation complex. Nucleic Acids Res. 2013;41:9257–65. doi: 10.1093/nar/gkt708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochkareva A, Yuzenkova Y, Tadigotla VR, Zenkin N. Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. EMBO J. 2012;31:630–9. doi: 10.1038/emboj.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen S, Yuzenkova Y, Zenkin N. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 2013;340:1577–80. doi: 10.1126/science.1237934. [DOI] [PMC free article] [PubMed] [Google Scholar]