This paper describes an analysis of the selenocysteine incorporation machinery in nematodes. Some nematodes, including C. elegans, encode only one selenoprotein, while others, including plant parasitic nematodes, have lost the ability to incorporate selenocysteine despite retaining parts of the incorporation machinery.
Keywords: selenocysteine, SECIS-binding protein, SBP2, Caenorhabditis elegans, selenoprotein, translation initiation
Abstract
Selenocysteine (Sec) is encoded by an UGA codon with the help of a SECIS element present in selenoprotein mRNAs. SECIS-binding protein (SBP2/SCBP-2) mediates Sec insertion, but the roles of its domains and the impact of its deficiency on Sec insertion are not fully understood. We used Caenorhabditis elegans to examine SBP2 function since it possesses a single selenoprotein, thioredoxin reductase-1 (TRXR-1). All SBP2 described so far have an RNA-binding domain (RBD) and a Sec-incorporation domain (SID). Surprisingly, C. elegans SBP2 lacks SID and consists only of an RBD. An sbp2 deletion mutant strain ablated Sec incorporation demonstrating SBP2 essentiality for Sec incorporation. Further in silico analyses of nematode genomes revealed conservation of SBP2 lacking SID and maintenance of Sec incorporation linked to TRXR-1. Remarkably, parasitic plant nematodes lost the ability to incorporate Sec, but retained SecP43, a gene associated with Sec incorporation. Interestingly, both selenophosphate synthetase (SPS) genes are absent in plant parasitic nematodes, while only Cys-containing SPS2 is present in Sec-incorporating nematodes. Our results indicate that C. elegans and the nematode lineage provide key insights into Sec incorporation and the evolution of Sec utilization trait, selenoproteomes, selenoproteins, and Sec residues. Finally, our study provides evidence of noncanonical translation initiation in C. elegans, not previously known for this well-established animal model.
INTRODUCTION
Selenocysteine (Sec) is the twenty-first amino acid and is inserted into selenoproteins at in-frame UGA codons by a specific tRNASec (Leinfelder et al. 1988; Hatfield and Gladyshev 2002). Insertion of Sec requires translational recoding, which requires a Sec insertion sequence (SECIS) element, a cis-acting stem–loop structure present in selenoprotein mRNAs (Berry and Larsen 1993). In eukaryotes, SECIS element binds SECIS-binding protein 2 (SBP2) and then recruits the Sec-specific elongation factor (EfSec), which binds tRNASec. After association with the ribosome, SBP2 exchanges for the ribosomal protein L30. This leads to a conformational change in the SECIS element that triggers the release of tRNASec and the hydrolysis of GTP (Chavatte et al. 2005; Allmang et al. 2009). In mammals, nucleolin and eIF4a3 also bind the SECIS element of some selenoprotein mRNAs and regulate Sec incorporation (Budiman et al. 2009; Miniard et al. 2010). SBP2 is likely an essential and limiting factor for Sec incorporation in mammals (Mehta et al. 2004). Invertebrate and vertebrate SBP2 differ significantly in length (∼850 and 350 amino acids long, respectively), however the N-terminal half of mammalian SBP2 is not required for Sec incorporation (Donovan and Copeland 2009). Invertebrate SBP2 and the C-terminal half of mammalian SBP2 possess an RNA-binding domain (RBD) and a Sec-incorporation domain (SID), which are presumed to be essential for decoding Sec (Takeuchi et al. 2009; Donovan and Copeland 2010). Sec synthesis is the additional part of the pathway required for biosynthesis of selenoproteins. It takes place on tRNASec, which is first aminoacylated with serine by a canonical seryl-tRNA synthetase (SerS) (Leinfelder et al. 1988; Lee et al. 1989). The seryl-tRNASec is phosphorylated to form O-phosphoseryl-tRNASec by O-phosphoseryl-transfer tRNASec kinase (PSTK) (Carlson et al. 2004), and then Sec synthase (SecS) catalyzes the formation of selenocysteinyl-tRNASec from O-phosphoseryl-tRNASec and selenophosphate (Xu et al. 2007b). In mammals, selenophosphate synthetase 2 (SPS2) is responsible for the synthesis of selenophosphate from selenide and ATP (Bock et al. 1991), while SPS1 has been recently postulated to function in a pathway unrelated to Sec biosynthesis (Xu et al. 2007a). Finally, other genes are present only in Sec-incorporating species but their precise function is not known; one of these genes is SecP43, thought to participate in the nucleus-cytosol transport of tRNASec (Small-Howard et al. 2006).
The evolution of Sec incorporation has not been fully explored across eukaryotes. Evolution of Sec utilization in vertebrates has been characterized (Mariotti et al. 2012). In addition, Sec incorporation evolution has been examined in detail in arthropods, a lineage that contains numerous species with sequenced genomes. These studies have led to the identification of the only known animal lineage in which some representatives lack selenoproteins (Chapple and Guigó 2008; Lobanov et al. 2008). However, the Sec incorporation and biosynthesis machinery has not been fully examined in Nematoda, a diverse phylum that includes free-living as well as plant and animal parasitic species with sequenced genomes. Interestingly, the nematode Caenorhabditis elegans possesses the entire Sec insertion system dedicated to a single Sec residue in its proteome, located in the active site of thioredoxin reductase-1 (TRXR-1) (Buettner et al. 1999; Gladyshev et al. 1999). trxr-1 is not an essential gene under standard laboratory conditions, but it is required for molting when glutathione reductase (GSR-1) function is compromised (Stenvall et al. 2011). These facts along with its powerful genetics make C. elegans an excellent experimental model to study Sec incorporation and utilization. Furthermore, several Caenorhabditis species and additional nematodes from other clades inhabiting very diverse environments have been completely sequenced. Therefore, Nematoda is an exceptional lineage to study the evolution of Sec incorporation and biosynthesis, selenoproteomes, and Sec residues.
In this study, we identified the genes involved or putatively involved in Sec biosynthesis and incorporation in C. elegans and silenced their expression, generated an SBP2 deletion mutant strain, and examined the nematode lineage for the genes involved in Sec utilization. We found that (i) nematodes, including C. elegans, possess the shortest eukaryote SBP2 that lacks SID, (ii) C. elegans SBP2 is essential for inserting Sec in TRXR-1, (iii) plant nematode parasites have lost the ability to incorporate Sec, lack both SPS genes, but retained SecP43, a gene previously associated exclusively with selenium incorporation.
RESULTS
Identification and silencing of genes involved in Sec incorporation in C. elegans
C. elegans and C. briggsae code for only one selenoprotein that contains a single Sec (Buettner et al. 1999; Gladyshev et al. 1999; Taskov et al. 2005). Therefore, we hypothesized that adjustments in Sec incorporation machinery may have occurred in representatives of this lineage (e.g., due to relaxed SECIS-binding constraints). To examine this possibility, we first searched and analyzed the nematode genes involved in Sec incorporation. For this purpose, we used human amino acid sequences as queries except for SBP2, as homology analyses revealed no clear ortholog of the human SBP2. Using a multiple query consisting of the C-terminal half of human SBP2, human SBP2-like, and invertebrate SBP2, we identified the gene scbp-2 (hereafter referred to as sbp2) as the putative C. elegans SBP2, which is significantly shorter than any known eukaryotic SBP2. We further identified 10 out of 11 genes involved in Sec decoding in humans: (i) the essential and presumptively essential genes for Sec incorporation: tRNASec (K11H12.t1), SPS (seld-1), seryl-tRNA synthetase (sars-1), phosphoseryl-tRNA (Ser/Sec) kinase (pstk-1), Sec-tRNA synthase (secs-1), EfSec (efsc-1, formerly named in WormBase as selb-1) a gene previously identified by Fagegaltier et al. (2000), and ribosomal protein L30 (rpl-30); (ii) involved in Sec insertion, but not essential for this pathway: SecP43 (T07F10.3); and (iii) two of the three genes present in organisms that incorporate Sec, but are absent in those that do not incorporate Sec (i.e., putatively involved only in Sec incorporation): R13A5.9 and tag-321 (sequences are shown in Supplemental Fig. 1). Interestingly, in contrast to other metazoa, C. elegans possesses only a Cys-containing sps gene.
In order to identify a phenotype associated with Sec incorporation, we knocked down the expression of genes involved in this process by RNA interference (RNAi). These experiments were carried out with three C. elegans strains: N2 (wild type), rrf-3(pk1426), and eri-1(mg366). The latter two strains are more sensitive to RNAi (Simmer et al. 2002; Kennedy et al. 2004). These experiments showed the same outcome in all strains assayed: (i) Interference of the expression of any gene involved in both canonical translation and Sec decoding (rpl-30 and sars-1) showed a lethal phenotype, (ii) interference of the expression of genes involved or putatively involved only in Sec decoding did not exhibit any apparent phenotype. These results were consistent with the known model of Sec synthesis and insertion and the fact that trxr-1 is not essential in this model under laboratory conditions. Indeed, both trxr-1 RNAi down-regulation and the null alleles trxr-1(sv47) and efsc-1(sv36) did not exhibit any apparent phenotype (Stenvall et al. 2011). Interference of the expression of genes associated with Sec synthesis but without a known function (R13A5.9 and tag-321) did not generate any evident phenotype, indicating that their functions are not essential for C. elegans under laboratory conditions.
C. elegans SBP2 is the first C. elegans protein with a non-AUG initiation of translation codon and the first SBP2 without SID
As described above, we identified scbp-2 as the putative C. elegans SBP2. The annotated gene was significantly shorter than any known SBP2 whether from vertebrates or invertebrates. Mammalian SBP2 are ∼800 amino acids long, while insect SBP2 are ∼400 amino acids long (Donovan and Copeland 2009). Surprisingly, the predicted C. elegans gene codes for an 85-residue-long protein, corresponding to the C-terminal end of eukaryote SBP2.
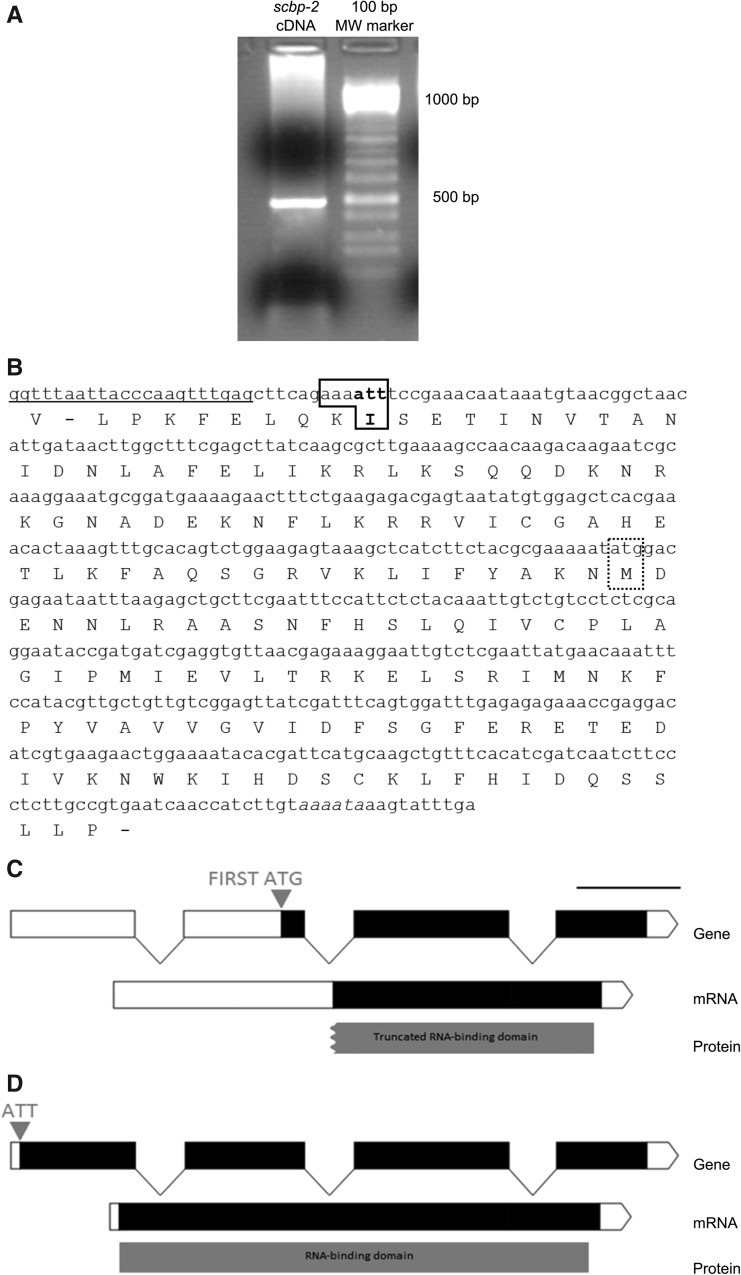
In order to determine if the identified gene was correctly annotated, we amplified the full-length cDNA from embryos using a reverse gene specific primer and the splice leader 1, a mini exon trans-spliced at the 5′ end of certain mRNAs, as a forward primer (Blumenthal and Steward 1997; Hastings 2005). We obtained a 493-bp product (Fig. 1A,B) identical to the transcript sequence annotated in WormBase. The first in-frame ATG codon was 231 nucleotides (nt) downstream from the beginning of the amplified sequence and it would encode an 85-amino-acid protein with a truncated RBD (Fig. 1C). Further analysis of the sequence showed that upstream of the first ATG, the reading frame continued for an additional 228 nt (Fig. 1B). We found that near the beginning of the full-length cDNA sequence there is an ATT codon preceded by AAA, which provides a favorable Kozak context for potential noncanonical translation initiation in C. elegans (Fig. 1B; Blumenthal and Steward 1997). Importantly, translation from this first AUU would complete the functional RBD (Fig. 1D). Also consistent with a non-AUG initiator codon is the fact that the amino acid homology with other eukaryotic SBP2 extends upstream of the first AUG to the first AUU (Fig. 2). In addition, C. elegans Ribo-seq data indicate translation upstream of the first AUG (Michel et al. 2014). Translation initiation from a non-AUG codon is a striking and conspicuous feature of SBP2 mRNA since it has not been previously described for any C. elegans mRNA. Our results indicate that for sbp2 there is noncanonical initiation of translation and that an AUU codon in a favorable translation initiation context likely signals the beginning of translation.
FIGURE 1.
cDNA, protein, and genomic organization of C. elegans sbp2 gene coding for a SECIS-binding protein. (A) Amplification of the full-length sbp2 mRNA with SL1 as forward primer and a reverse specific primer. A 493-bp product corresponding to the full-length cDNA was obtained (lane 1), 100-bp gene ruler (lane 2). (B) The conceptual translation of the amplified mRNA shows that the reading frame remains open upstream of the first in-frame ATG codon (dotted frame) and that there is an ATT codon at the beginning of the cDNA (bold frame) in a favorable Caenorhabditis Kozak context. (C) The gene model proposed by WormBase (black and white rectangles correspond to coding and noncoding exons, respectively), codes for an 85-amino-acid-long protein and has a long 5′ UTR and a truncated RBD (gray). The first in-frame ATG codon is indicated in gray. (D) The gene model put forward in the present work codes for a 153-amino-acid-long protein with a short 5′ UTR and a complete RBD. The ATT codon that serves for translation initiation is indicated in gray.
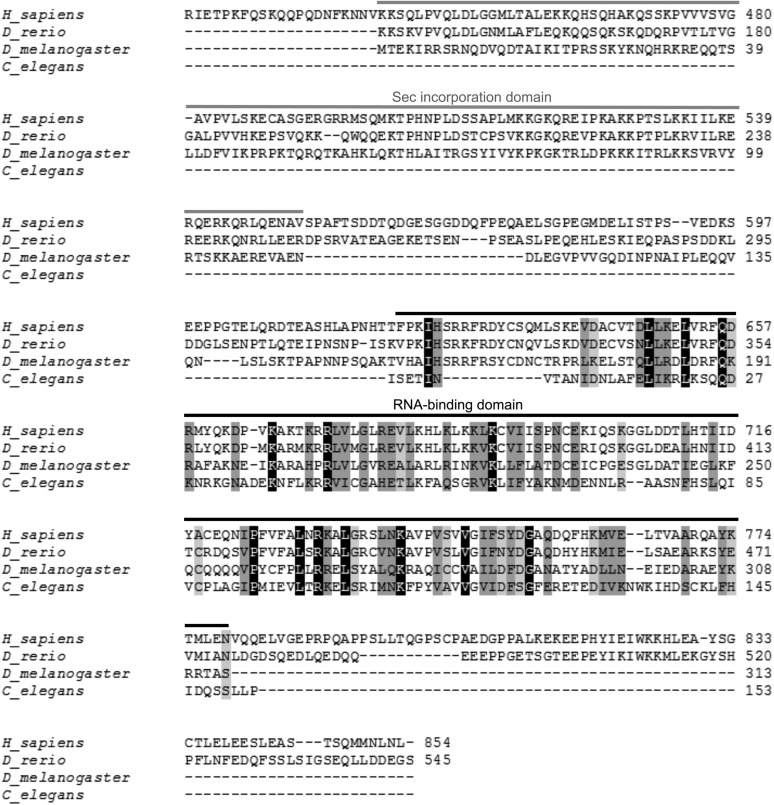
FIGURE 2.
Sequence alignment of SBP2 from Homo sapiens, Danio rerio, Drosophila melanogaster, and the identified C. elegans SBP2. The reading frame of C. elegans SBP2 remains open upstream of the first Met, and translation from the first Ile completes the RBD (overlined in black). C. elegans SBP2 lacks the Sec incorporation domain (overlined in gray). The full-length D. melanogaster and C. elegans SBP2s are shown; in the case of vertebrate SBP2s, the first 400 amino acids (absent in invertebrate SBP2s) are not shown. Black blocks indicate positions which have a single, fully conserved residue. Dark gray and light gray blocks indicate conservation between groups of strongly or weakly similar properties, respectively. Numbers indicate the amino acid residue in the polypeptide chain.
Another remarkable feature of C. elegans SBP2 is the absence of the SID. SBP2-flanking genes have neither a related function nor a SID. Even more, SBP2 is not a “split gene” in C. elegans, since the SID is absent in the entire genome. No hits were found in BLAST searches using several eukaryotic SIDs (independent or concatenated) or conserved regions of the SID as queries. This finding suggests that the C. elegans Sec-incorporating machinery differs from that in other eukaryotes.
Sec biosynthesis and incorporation in the Caenorhabditis lineage
The peculiar C. elegans SBP2 prompted us to analyze the Caenorhabditis lineage taking advantage of several Caenorhabditis species that have been completely sequenced. We analyzed C. briggsae, C. brenneri, C. remanei, C. japonica, C. sp5, and C. sp11 genomes annotated in WormBase as of July 2013 (WS238). We found that all these species possess the machinery to incorporate Sec. All of them encode a single Sec residue in the entire proteome: the Sec residue of trxr-1. Interestingly, unlike other metazoa, all Caenorhabditis species encode a single SPS. Even though SPS displays slightly higher identity to SPS1 than to SPS2, the C. elegans gene is phylogenetically an SPS2 (M Mariotti, pers. comm.) with a Cys residue at the active site.
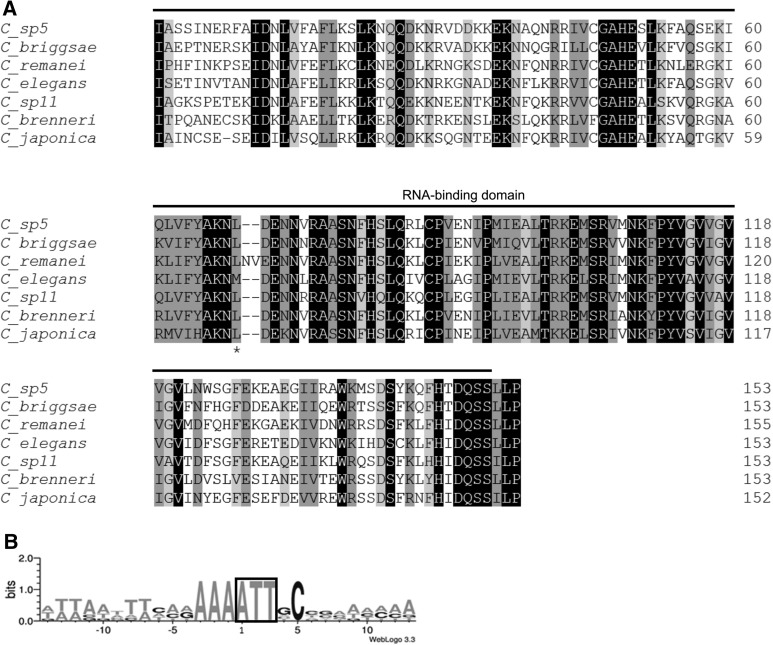
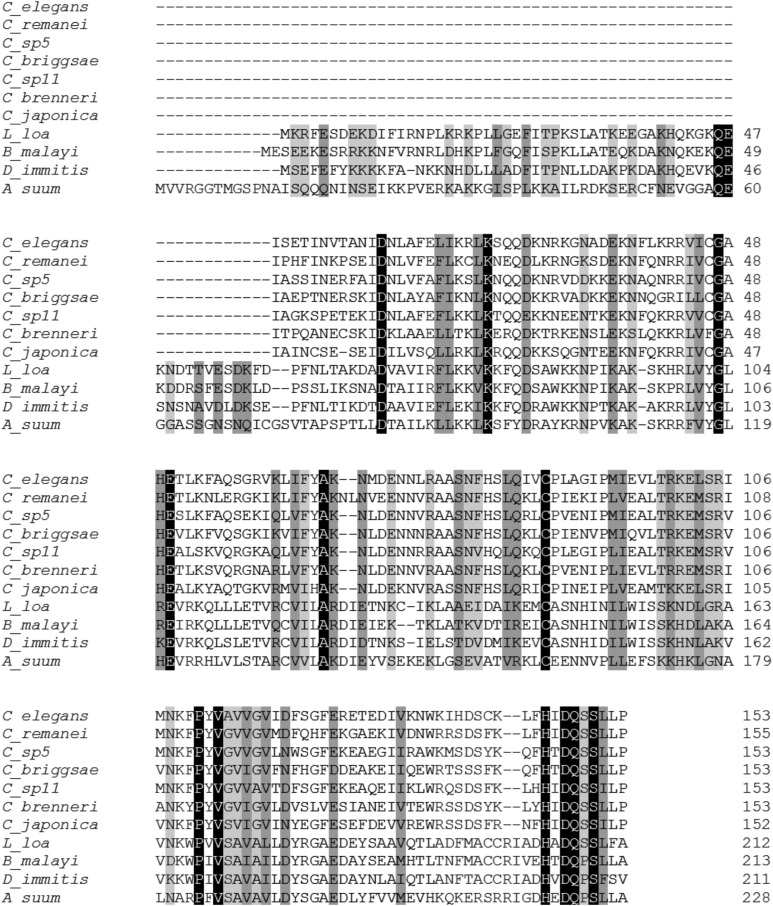
We further identified and analyzed Caenorhabditis sbp2. All analyzed Caenorhabditis species lacked the SID and coded for a short sbp2, i.e., 152–155 amino acid long (Fig. 3A), with an identical gene structure: four exons and three introns. We then examined signals for translation initiation in Caenorhabditis sbp2 and found that they all shared the putative AUU initiation codon, preceded by a highly conserved Kozak sequence −4AAA−1 (Fig. 3B). They also shared a conserved cytosine in the +5 position, suggesting that this position may also be important for non-AUG translation initiation. In addition, we found that all Caenorhabditis SBP2 displayed high levels of similarity at the amino acid sequences from the first isoleucine residue (encoded by the proposed AUU initiator codon) including conservation of basic residues at the beginning of the RBD (Fig. 3A). Strikingly, at the homologous position of the first in-frame methionine of C. elegans SBP2, a leucine residue was found in all other Caenorhabditis spp., providing further evidence that C. elegans sbp2 translation initiation is not at this AUG codon (Fig. 3A). It is also important to mention that, of all analyzed Caenorhabditis SBP2, only the C. japonica protein has a methionine upstream of the homologous position of the first methionine of C. elegans SBP2. Together, these results strongly support that noncanonical translation initiation occurs in Caenorhabditis SBP2. Although described in other species, noncanonical translation initiation has not been previously reported for any Caenorhabditis (Rhoads et al. 2006).
FIGURE 3.
Caenorhabditis SBP2s. (A) Alignment of SBP2 proteins in all sequenced Caenorhabditis species. The RBD is overlined; the first methionine encoded in C. elegans SBP2 and amino acids corresponding to the homologous position in the other Caenorhabditis SBP2 are marked with an asterisk. Black blocks indicate positions which have a single, fully conserved residue. Dark gray and light gray blocks indicate conservation between groups of strongly or weakly similar properties, respectively. Numbers indicate the amino acid residue in the polypeptide chain. (B) Frequency of nucleotides found at the beginning of the cDNA of Caenorhabditis sbp2. Adenine +1 corresponds to the first base of the coding sequence. A complete conservation of a favorable Caenorhabditis Kozak context is observed at positions −1 to −3.
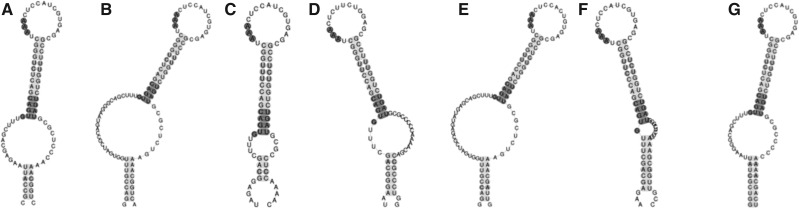
With regard to Sec incorporation, the results of our analysis strongly indicate that Caenorhabditis SBP2 lack SID. The absence of a SID may relate to the number and type of SECIS elements encoded by Caenorhabditis. All Caenorhabditis genomes encode a single selenoprotein (TRXR-1). The analysis of trxr-1 SECIS elements using the SECISearch3 program (Mariotti et al. 2013) revealed the presence of the unusual type I SECIS element typical of nematodes with a guanine immediately upstream of the non-Watson-Crick base-pairing quartet (Fig. 4; Grundner-Culemann et al. 1999; Taskov et al. 2005).
FIGURE 4.
SECIS elements of Caenorhabditis Sec-containing thioredoxin reductase. (A) C. brenneri, (B) C. briggsae, (C) C. elegans, (D) C. japonica, (E) C. remanei, (F) C. sp5, (G) C. sp11. All Caenorhabditis encode a type I SECIS element with a guanine immediately upstream of the non-Watson-Crick base-pairing quartet. The non-Watson-Crick base-pairing quartet (SBP2-binding site) and the AAR triplet are highlighted.
Finally, it is worth mentioning that the Caenorhabditis SBP2 contain a short C-terminal tripeptide which is highly conserved in this lineage but absent in vertebrates.
C. elegans SBP2 is a functional and essential gene for Sec incorporation
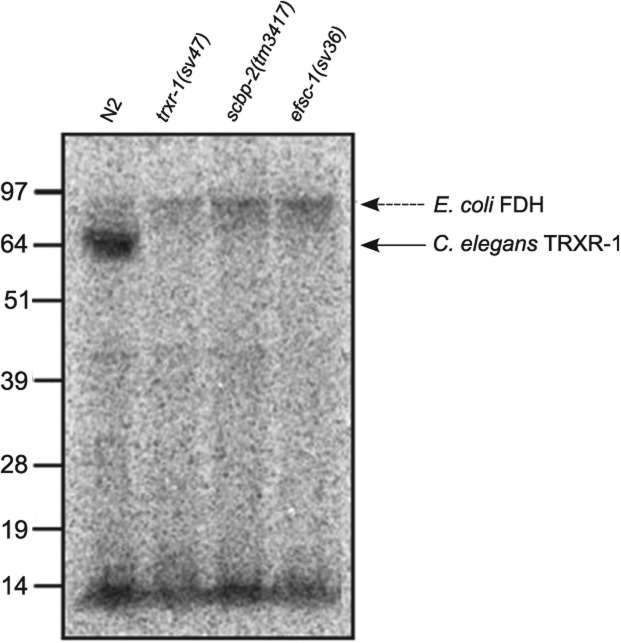
We performed metabolic incorporation of radioactive 75Se in the scbp-2(tm3417) mutant to analyze the relevance of sbp2 for Sec incorporation. We used the N2 wild-type strain as a Sec incorporation positive control and both trxr-1(sv47) and efsc-1(sv36) mutants as Sec incorporation negative controls. This experiment showed that the scbp-2(tm3417) was unable to incorporate Sec (Fig. 5). In a previous study, it was shown that the trxr-1 and efsc-1 deletion mutant strains showed larval arrest phenotype when the expression of gsr-1 gene was silenced (Stenvall et al. 2011). Importantly, gsr-1 RNAi in scbp-2(tm3417) mutant also reproduced the larval arrest phenotype.
FIGURE 5.
C. elegans SBP2 deletion mutant strain is unable to incorporate Sec. Metabolic incorporation of radioactive 75Se showed that the deletion mutant strain scbp-2(tm3417) was unable to incorporate Se in TRXR-1 (lane 3) as well as the deletion mutant strains trxr-1(sv47) and efsc-1(sv36) (lanes 2 and 4, respectively). The wild-type strain N2 incorporates Se in the only selenoprotein expressed in C. elegans: TRXR-1 (lane 1), shown by the bold arrow. The dashed arrow shows E. coli formate dehydrogenase. The bands at the low part of the gel show unbound Se and Se-labeled tRNA in E. coli.
These results indicate that the identified sbp2 is an essential gene for Sec incorporation in C. elegans and therefore provides support to the concept that lineage-specific adjustments may have occurred in the Sec insertion machinery particularly regarding requirements for SECIS element decoding.
Analysis of transgenic SBP2 strain provides further evidence for a noncanonical start of translation and function of the identified gene
To gain further information, we immobilized the trxr-1 SECIS element (wtSECIS) in order to pull down the native SBP2 from a total wild-type worm aqueous extract (see Materials and Methods). The proteins retained by the wtSECIS column were analyzed by SDS-page and MS/MS. We isolated the eukaryotic translation elongation factor 1α (eef-1A.2) and several ribosomal proteins: rpl-3, rpl-4, rpl-22, and Y37E3.8; however, SBP2 was not identified. This result can be explained considering that ribosomal proteins and general translation factors are abundant, whereas SBP2 expression in C. elegans is expected to be low considering that it is used for insertion of just one Sec residue. Furthermore, SBP2 is thought to be the limiting factor for Sec incorporation (Mehta et al. 2004).
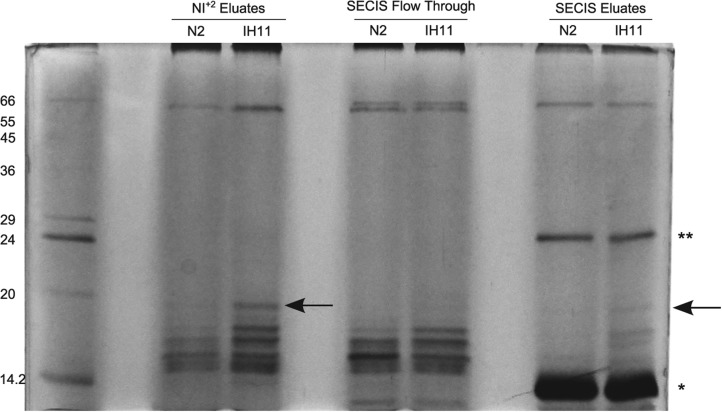
To overcome the low endogenous levels of SBP2, we generated the transgenic strain IH11, ciuEx3[pLO109(Psur-5::scbp-2::polyHis); pRF4(rol-6(su1006))] that overexpresses SBP2 with a C-terminal poly-histidine extension under the control of the sur-5 promoter to assure constitutive expression. Aqueous extracts of adult ciuEx3 and wild-type control animals obtained from liquid culture were chromatographed on Ni-NTA magnetic agarose beads. Two differential bands, present in the ciuEx3 extract, but absent in the wild-type extract, were retained by the metal affinity magnetic beads (Fig. 6). One of them corresponded to the expected size of the full-length SBP2, starting from the AUU codon in the appropriate Kozak context. The other poly-His-enriched protein present in the transgenic strain may be associated with SBP2 and copurify with it or be a truncated form of SBP2 that retained the poly-His tail. Soluble mutated trxr-1 SECIS element (mutSECIS, having mutations at the non-Watson-Crick quartet, required for SBP2 binding [Fletcher et al. 2001; Allmang et al. 2002], and at the conspicuous AAR triplet present at the apical loop; see Materials and Methods for sequence) was added to the proteins eluted from the Ni-NTA magnetic beads, in order to decrease binding of RNA-binding proteins to wtSECIS in the following chromatographic step. The mixes were applied to a wtSECIS column as previously described, and retained proteins were analyzed by SDS-PAGE (Fig. 6). The enriched 18- and 16-kDa bands present in the transgenic strain, but absent in the N2 strain, were bound to wtSECIS. These results support the noncanonical initiation of translation of SBP2.
FIGURE 6.
Isolation of an 18-kDa band by SECIS affinity chromatography. Sequential metal and SECIS affinity chromatography of aqueous extracts of the wild-type strain N2 (as control) and the transgenic strain IH11, ciuEx3[pLO109(Psur-5::scbp-2::polyHis); pRF4(rol-6 (su1006))] overexpressing sbp2. An 18-kDa protein band is purified from the IH11 extract (arrows), but not from the control. This mass corresponds to the expected to SBP2 if translation starts from the proposed AUU codon. (Lane 1) Molecular weight marker (MWM). (Lanes 3,4) Eluates from the Ni+2 column. (Lanes 6,7) Flow through the wtSECIS element column. (Lanes 9,10) Eluates from the wtSECIS element column from wild type and ciuEx3, respectively (containing soluble mutSECIS). One asterisk shows the streptavidin monomer and two asterisks show the streptavidin dimer.
Selenoprotein extinction in plant nematode parasites
Since Caenorhabditis lineage has a single Sec residue in its proteome and its SBP2 lacks SID, we examined the Sec incorporation machinery in additional nematode lineages with completely sequenced genomes. These lineages corresponded to the clade III (Spirurina, a clade that includes several animal parasitic nematodes) and clade IV (Tylenchina, a clade that includes plant parasitic nematodes). We first searched for the Sec incorporation trait using SecS and PSTK genes as signatures of the trait. If the trait was present, we then searched for their SBP2 and other genes involved in Sec incorporation.
For the Spirurina lineage, we analyzed Ascaris suum, Brugia malayi, Dirofilaria immitis, and Loa loa genomes. These nematodes fully conserved the ability to incorporate Sec. However, they encoded a longer SBP2 than the Caenorhabditis proteins, between 211 and 228 amino acids long, but shorter than other invertebrate SBP2 (Fig. 7). The N-terminal extension of Spirurina SBP2 is rich in basic amino acid residues. Similar to Caenorhabditis SBP2, Spirurina SBP2 consist of an RBD and a C-terminal motif conserved in nematodes but not in other eukaryotes, but they lack the SID. All analyzed spirurina sbp2 had a canonical initiation of translation. Interestingly, the data from transcriptome for additional nematode lineages indicate that they possess more than one selenoprotein (Taskov et al. 2005) (data not shown).
FIGURE 7.
Sequence alignment of the identified nematode SBP2s. Spirurina SBP2 are longer than Caenorhabditis SBP2s; however, all identified nematode SBP2 lack the SID. The N-terminal extension of Spirurina SBP2s is rich in basic residues. Black blocks indicate positions which have a single, fully conserved residue. Dark gray and light gray blocks indicate conservation between groups of strongly or weakly similar properties, respectively. Numbers indicate the amino acid residue in the polypeptide chain.
We then examined Meloidogyne hapla, Meloidogyne incognita, and Globodera pallida genomes of the Tylenchina clade. These plant parasitic nematodes have lost the machinery for Sec incorporation: They lack secs-1 and pstk-1 (Fig. 8). Furthermore, they lacked both Sec and Cys-containing trxr genes; instead, they appear to rely on gsr as the core enzyme for disulfide reduction pathways. Interestingly, plants neither incorporate Sec nor encode large thioredoxin reductases. These results reinforce the idea that thioredoxin reductase is the selenoprotein that maintains the selenium incorporation trait in the nematode lineage and suggest adaptations of parasite metabolism governed by the host metabolism. Finally, the analysis of the other genes involved in Sec incorporation in plant parasitic nematode genomes indicated the loss of all genes involved in Sec incorporation, including SPS, but the presence of SecP43. This finding suggests that SecP43 might be involved in processes other than Sec incorporation, or that in this lineage this gene acquired a new function.
FIGURE 8.
Sec incorporation and SBP2 in the nematode lineage. Nematodes from the clades III, IV, and V were examined for the presence of Sec incorporation machinery. Nematodes from the clade IV have lost the ability to incorporate Sec. Nematode SBP2s are the shortest eukaryote SBP2. For comparison D. melanogaster and human SBP2s are included in the phylogenetic analysis. RBDs are represented in light upward diagonal and Sec incorporation domains in wide upward diagonal.
DISCUSSION
SBP2 is a key player in dictating Sec incorporation; however, its essentiality for Sec incorporation has not been conclusively demonstrated. C. elegans SBP2 had not been previously identified, remaining as an elusive gene. We identified C. elegans SBP2 by sequence comparison analysis and proved that the identified gene is essential for Sec incorporation in TRXR-1, the only Sec-containing protein in C. elegans. Furthermore, the larval arrest phenotype observed when gsr-1 expression was silenced on trxr-1 or efsc-1 mutants was also observed in the SBP2 deletion mutant strain. Caution should be taken to avoid generalizations regarding SBP2 essentiality in Sec incorporation for all selenoproteins in any lineage.
C. elegans SBP2 is the shortest eukaryotic SBP2 described so far: It lacks a SID and comprises exclusively an RBD. Other Caenorhabditis spp. also have a short SBP2 without SID. Remarkably, SID is completely absent from the genomes of these organisms. The absence of SID emphasizes that adjustments in the Sec incorporation machinery occurred in some eukaryotes and questions the proposed minimal requirements for Sec incorporation in eukaryotes. We compared other key proteins involved in Sec decoding to test whether additional and/or compensatory adjustments occurred in these sequences, but found no differences with these proteins in other lineages. Although speculative, the possibility that SBP2 could interact with the SECIS indirectly, via protein–protein interactions with another genuine SECIS-binding protein, cannot be ruled out. The minimal requirement of an RBD for SECIS binding in nematodes provides a potential clue to identify the archaeal missing link in Sec decoding. Indeed, archaeal selenoprotein mRNAs contain the SECIS element in the 3′ UTR, but the SECIS-binding protein has not been identified yet (Rother et al. 2001).
The C. elegans SBP2 mRNA does not begin translation at an AUG codon. This is a striking feature since noncanonical translation initiation has not been previously described for this well-studied animal model. The evidence we have obtained is compelling: Translation from the first AUG codon leads to an incomplete RBD, the ORF and the amino acid sequence homology extends well beyond the first methionine, a different amino acid is present at the homologous position in other Caenorhabditis spp., and the Ribo-seq data available indicate translation upstream of the first AUG. The evidence for translation initiation from an AUU is also compelling: This codon is absolutely conserved together with the appropriate Caenorhabditis Kozak consensus −4AAA−1 sequence, amino acid conservation extends until this codon, and the biochemical evidence obtained from the transgenic strain expressing SBP2 is consistent with the length expected for translation initiation from this codon. As this is the first gene that has been shown to have a non-AUG initiation of translation, we anticipate that examination of full-length mRNAs with long 5′ UTRs will lead to correction of additional gene models and would provide further clues regarding noncanonical initiation of translation in C. elegans.
Analyzing the genome sequences of other nematodes, we found that the Spirurina lineage also possessed short SBP2 that lack SID, reinforcing the idea that the RBD may suffice for Sec incorporation. Interestingly, the RBD of Spirurina SBP2 is preceded by a K/R rich region. This fact may reflect the need for recognizing more than one SECIS element. Another remarkable finding of our study is that nematode plant parasites that belong to the Tylenchina clade have lost the Sec trait. Sec extinction in these nematodes does not appear to be a recent event: All essential genes in Sec incorporation were lost. We could only identify SecP43 as the only gene predicted to be involved in Sec incorporation, suggesting that it might be involved in some other cellular processes in this lineage. Furthermore, rather than a Sec→Cys conversion, we observed selenoprotein gene loss, which may also indicate a distant and irreversible event in selenoprotein extinction. In this context, it is interesting to note that plants do not incorporate Sec, and thus selenoproteinless nematodes likely evolved as an adaptation of this lineage to its selenium-incompatible niche. So far, the only reported Metazoa unable to incorporate Sec were a few insect species (Chapple and Guigó 2008; Lobanov et al. 2008). This finding enlarges the group of selenoproteinless animals, stressing a highly dynamic evolutionary process and highlighting that in the nematode lineage Sec incorporation is maintained by thioredoxin reductase.
Interestingly, in some selenoproteinless insects SBP2, SecP43, and SPS1 were still conserved (Chapple and Guigó 2008). Although SPS are responsible for the synthesis of selenophosphate from selenide and ATP, it has been suggested that SPS1 functions in a pathway unrelated to Sec biosynthesis (Lobanov et al. 2008). Our results portray a slightly different scenario in nematodes: Both sps genes are absent in plant parasite nematodes, and only one sps gene is present in Sec-incorporating nematodes. This SPS is the ortholog to metazoan SPS2 (M Mariotti, pers. comm.) but contains Cys instead of Sec at its active site.
Our results indicate that SID is not required for Sec decoding in nematodes and that nematodes provide an interesting scenario to study Sec incorporation, selenoproteomes, selenoproteins, and Sec/Cys evolution. Finally, the demonstration that a non-AUG translation initiation occurs in C. elegans is an important observation that should lead to revision in protein sequences, beyond SBP2, in this organism.
MATERIALS AND METHODS
C. elegans strains and culture conditions
Bristol-N2 strain was obtained from the CGC Center; VB1414 trxr-1(sv47) IV, VB0740 efsc-1(sv36) I, VB1646 rrf-3(pk1426) II; trxr-1(sv47) IV, and VB1286 rrf-3(pk1426) II; efsc-1(sv36) I strains were kindly provided by Dr. Simon Tuck (Umeå University, Sweden); TM3417 scbp-2(tm3417) I was obtained from the C. elegans National Bioresource Project of Japan. Escherichia coli strains OP50 and HT115 were obtained from the CGC. RNAi clones were kindly provided by Dr. Peter Askjaer (Universidad Pablo de Olavide, Spain). The standard methods for culturing and maintaining C. elegans were used as described in Brenner (1974). All experiments were performed at 20°C. To obtain IH2 scbp-2(tm3417) I strain, TM3417 scbp-2(tm3417) I was backcrossed six times with N2 strain.
Amplification of C. elegans sbp2 cDNA
RNA from N2 embryos was kindly provided by Dr. Peter Askjaer (Universidad Pablo de Olavide, Spain). Reverse transcription was performed using the gene-specific reverse primer (TCACGGCAAGAGGGAAG). Amplification of sbp2 cDNA was carried out using the same specific reverse primer and a forward primer (GGTTTAATTACCCAAGTTTGAG) derived from the SL1 exon trans- spliced to the 5′ end of the full-length mRNAs. The PCR product was analyzed by electrophoresis, purified from 1.5% agarose gel, cloned into pGEM-T Easy (PROMEGA), and sequenced.
Identification of proteins involved in Sec decoding in C. elegans and nematodes
Searches for genes and proteins involved in Sec decoding in C. elegans were performed using as queries human tRNASec, EfSec, PSTK, L30, SecS, SecP43, SerS, SPS1, SPS2, and SBP2. Additionally, D. melanogaster sequences were used as queries for some genes and proteins: SPS1, SPS2, SBP2-like and the protein sequences encoded by genes present in Sec-incorporating organisms and absent in nonincorporating ones: Q7KUF9, Q8T3V7, and Q9VEH4. These sequences were used in blastn, tblastn, or blastp searches against WormBase (WS238). In all cases, the hit sequence was retroblasted to obtain the best reciprocal hit. For identifying C. elegans SBP2, additional searches were carried out using several invertebrate SBP2 sequences independently and concatenated.
SecS and PSTK genes were used as signature for maintenance of the Sec incorporation trait in other members of the Caenorhabditis genus and in other nematodes. C. elegans genes were used as queries to identify nematode orthologs searching by blastp and tblastn using WormBase (WS238). To confirm the absence of the trait in plant nematode parasites, the following databases were also examined: M. hapla 10× (Plant Nematode Genomics Group, North Carolina State University), M. incognita genome, proteins and dbEST (Meloidogyne incognita resources, INRA), and G. pallida predicted genes and contigs (Wellcome Trust Sanger Institute).
C. brenneri, C. briggsae, C. japonica, C. remanei, C. sp5, and C. sp11 SBP2 were obtained by tblastn using C. elegans SBP2 as query and WS238 databases. Other nematode SBP2 were obtained by tblastn using C. elegans SBP2 as query and WormBase (WS238), nematode.net v3.0 (http://nematode.net/NN3_frontpage.cgi), UniProtKB (http://www.uniprot.org/), 959 Nematode Genomes (http://www.nematodes.org/nematodegenomes/index.php/Main_Page) databases. Once a new nematode SBP2 sequence was obtained, it was also used as query for searching for SBP2 of closely related nematodes.
Multiple sequence alignments
Multiple sequence alignments were performed using clustalw program and its default parameters at the European Bioinformatics Institute website: http://www.ebi.ac.uk/Tools/msa/clustalw2/. All alignments were manually refined.
SECISearch analyses of nematode sequences
The 3′ UTR sequences of the Sec-containing trxr-1 of Caenorhabditis species were obtained from WormBase (WS238) via blastn tool, using C. elegans TRXR-1 sequence as query and C. brenneri, C. briggsae, C. japonica, C. remanei, C. sp5, and C. sp11 genome databases. The obtained 3′ UTR sequences were then analyzed using SECISearch3 tool for prediction of SECIS elements (Mariotti et al. 2013) at http://seblastian.crg.es/ using infernal method and threshold 10.
Identification of homologs of known selenoprotein genes
By blastp and tblastn similarity searches, we identified homologs to the following eukaryotic selenoproteins in Caenorhabditis spp.: glutathione peroxidases (Gpx), selenoprotein 15 (Sel15), selenophosphate synthetase (SPS), selenoprotein K (SelK), selenoprotein R (SelR), selenoprotein T (SelT), selenoprotein U (SelU), thioredoxin reductases (TRXR-1 and TRXR-2). No homologs were found to other eukaryotic selenoproteins. In all cases, except TRXR-1, these homologs contained Cys at the homologous Sec position in mammals.
Metabolic labeling with 75Se
E. coli OP50 strain was grown overnight at 37°C in 5 mL Luria Broth (LB) medium supplemented with cysteine and Na2SeO3 radiolabeled with 75Se (20 μCi of 75Se) . Two hundred microliters of the overnight culture was spread onto a Nematode Growth Media (NGM) agar plate and allowed to grow for 24 h at room temperature (∼20°C). Then, the plate was seeded with 10 L4 hermaphrodite worms of the appropriate strain. The worms were harvested shortly before they had consumed all of the bacteria (∼7 d). Worms were then collected, washed, and boiled in sample buffer. The total protein extracts were analyzed by SDS-PAGE and then transferred to a PVDF membrane. Radioactive signal was detected with a PhosphorImager.
RNAi assays
HT115 E. coli strain was transformed with the plasmid pL4440 containing the sequence of the gene to be down-regulated or empty vector as a control. The bacteria were first grown overnight at 37°C in LB medium containing 100 µg/mL ampicillin. Then, they were seeded in plates containing NMG agar with 100 µg/mL ampicillin and 1 mM IPTG and incubated overnight at 37°C to induce the expression of dsRNA. Worms were then seeded onto plates at 20°C and phenotypes scored from the first generation onward by allowing interfered gravid hermaphrodites to lay eggs during 2 h on fresh RNAi plates.
Sequential Ni+2 SECIS chromatography
Adult N2 and IH11 ciuEx3[pLO109(Psur-5::scbp-2::polyHis); pRF4(rol-6 (su1006))] worms were harvested from liquid culture and extensively washed. Worms were ground to a powder under liquid nitrogen using a mortar and a pestle, resuspended in buffer containing 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 0.05% Tween and 1 mM PMSF, sonicated, and the suspension centrifuged at 20,000g for 20 min. Six milliliters of the aqueous protein extract of 15.71 mg/mL of N2 and 16.60 mg/mL of IH11 were added to 120 µL Ni-NTA Magnetic Agarose Beads (Qiagen), washed, and eluted in 150 µL following manufacturer's recommendations and protocol.
The buffer of the eluates obtained from the Ni+2 column was exchanged to PBS using an Amicon Ultra-0.5 (Millipore) cartridge in order to decrease imidazole concentration (<10 µM). To 85 µL of the resulting protein solution we added 2 µL Ribolock RNase Inhibitor (Thermo) to avoid RNA degradation and 0.5 µL of 100 mM mutSECIS: trxr-1 SECIS element that has the critical SBP2-binding site (the non-Watson-Crick quartet), and the typical AAR triplet mutated (uucacacgaccuuuggcuccacuccaucgugagcgccucuggucugaugc) to minimize unspecific binding to the wild-type SECIS element (wtSECIS) (uugugacgaccuuuggcuaaacuccaucgugagcgccucuggucugaugc). The mixes were incubated with biotinylated wtSECIS immobilized on streptavidin magnetic beads (Thermo), following manufacturer's recommendations and protocol. After binding and washing, bound proteins were eluted adding SDS-PAGE sample buffer and boiled for 5 min. The N2 and IH11 eluates and the flow through fractions were analyzed by 15% SDS-PAGE and silver stained according to the protocol described in Shevchenko et al. (1996).
Generation of transgenic C. elegans strain
The sbp2 sequence was amplified using genomic DNA (extracted from N2 adults), a specific forward primer (ACGTGCTAGCCTCTTCCTGTTTTATGAAATC), and a specific reverse primer that adds a C-terminal poly-His tag (TCGGTACCATGGTTAGTGATGATGGTGATGATGCGGCAAGAGGGAAGATTG). The amplified sequence was analyzed and purified from 1% agarose gel and then cloned into pPD49.26 vector that has already cloned the sur-5 promoter (kindly provided by Dr. Ana Bratanich, UBA) and sequenced. This construct was microinjected into N2 L4 larvae gonads along with pRF4 plasmid—that carries the rol-6(su1006) dominant transformation marker—to generate stable transgenic lines (50 ng/µL each construct).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. Peter Askjaer (Universidad Pablo de Olavide, Spain) for provision of C. elegans embryos mRNA and helpful discussions, Dr. Simon Tuck (Umeå University, Sweden) for kindly providing stains, Dr. Ana Bratanich (UBA, Argentina) for providing the pPD49.26 vector, and Dr. Briseida Cacho-Valadez for experimental assistance. We are grateful to the Caenorhabditis Genetics Center (CGC) and the Japanese National Bioresource Project for the Experimental Animal Nematode C. elegans for provision of E. coli and C. elegans strains. We also thank the instructors and assistants of the CSHL C. elegans course attended by G.S. This work was supported by Universidad de la República, Uruguay (Grant Number 418 to G.S., PhD fellowship to L.O.); Asociación Española de Cooperación Internacional (C/7646/07 to A.M.-V. and G.S.; A/016083/08 to A.M.-V. and G.S.); Asociación Universitaria Iberoamericana de Posgrado and Agencia Nacional de Innovación e Investigación (BE_POS_2009_183 and BE_POS_2010_2160 to L.O.), and was partially funded by FOCEM (MERCOSUR Structural Convergence Fund), [COF 03/11].
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.043877.113.
REFERENCES
- Allmang C, Carbon P, Krol A 2002. The SBP2 and 15.5 kD/Snu13p proteins share the same RNA binding domain: identification of SBP2 amino acids important to SECIS RNA binding. RNA 8: 1308–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Wurth L, Krol A 2009. The selenium to selenoprotein pathway in eukaryotes: more molecular partners than anticipated. Biochim Biophys Acta 1790: 1415–1423 [DOI] [PubMed] [Google Scholar]
- Berry MJ, Larsen PR 1993. Recognition of UGA as a selenocysteine codon in eukaryotes: a review of recent progress. Biochem Soc Trans 21: 827–832 [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Steward K 1997. RNA processing and gene structure. In C. elegans II (ed. Riddle DL, et al. ), pp. 117–145 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Bock A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F 1991. Selenocysteine: the 21st amino acid. Mol Microbiol 5: 515–520 [DOI] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budiman ME, Bubenik JL, Miniard AC, Middleton LM, Gerber CA, Cash A, Driscoll DM 2009. Eukaryotic initiation factor 4a3 is a selenium-regulated RNA-binding protein that selectively inhibits selenocysteine incorporation. Mol Cell 35: 479–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner C, Harney JW, Berry MJ 1999. The Caenorhabditis elegans homologue of thioredoxin reductase contains a selenocysteine insertion sequence (SECIS) element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J Biol Chem 274: 21598–21602 [DOI] [PubMed] [Google Scholar]
- Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL 2004. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci 101: 12848–12853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple CE, Guigó R 2008. Relaxation of selective constraints causes independent selenoprotein extinction in insect genomes. PLoS One 3: e2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Brown BA, Driscoll DM 2005. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat Struct Mol Biol 12: 408–416 [DOI] [PubMed] [Google Scholar]
- Donovan J, Copeland PR 2009. Evolutionary history of selenocysteine incorporation from the perspective of SECIS binding proteins. BMC Evol Biol 9: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan J, Copeland PR 2010. Threading the needle: getting selenocysteine into proteins. Antioxid Redox Signal 12: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A 2000. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J 19: 4796–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JE, Copeland PR, Driscoll DM, Krol A 2001. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA 7: 1442–1453 [PMC free article] [PubMed] [Google Scholar]
- Gladyshev VN, Krause M, Xu XM, Korotkov KV, Kryukov GV, Sun QA, Lee BJ, Wootton JC, Hatfield DL 1999. Selenocysteine-containing thioredoxin reductase in C. elegans. Biochem Biophys Res Commun 259: 244–249 [DOI] [PubMed] [Google Scholar]
- Grundner-Culemann E, Martin GW III, Harney JW, Berry MJ 1999. Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA 5: 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings KE 2005. SL trans-splicing: easy come or easy go? Trends Genet 21: 240–247 [DOI] [PubMed] [Google Scholar]
- Hatfield DL, Gladyshev VN 2002. How selenium has altered our understanding of the genetic code. Mol Cell Biol 22: 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun G 2004. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature 427: 645–649 [DOI] [PubMed] [Google Scholar]
- Lee BJ, Worland PJ, Davis JN, Stadtman TC, Hatfield DL 1989. Identification of a selenocysteyl-tRNASer in mammalian cells that recognizes the nonsense codon, UGA. J Biol Chem 264: 9724–9727 [PubMed] [Google Scholar]
- Leinfelder W, Zehelein E, Mandrand-Berthelot MA, Bock A 1988. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature 331: 723–725 [DOI] [PubMed] [Google Scholar]
- Lobanov AV, Hatfield DL, Gladyshev VN 2008. Selenoproteinless animals: selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein Sci 17: 176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti M, Ridge PG, Zhang Y, Lobanov AV, Pringle TH, Guigo R, Hatfield DL, Gladyshev VN 2012. Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS One 7: e33066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti M, Lobanov AV, Guigo R, Gladyshev VN 2013. SECISearch3 and Seblastian: new tools for prediction of SECIS elements and selenoproteins. Nucleic Acids Res 41: e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Rebsch CM, Kinzy SA, Fletcher JE, Copeland PR 2004. Efficiency of mammalian selenocysteine incorporation. J Biol Chem 279: 37852–37859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Fox G, M Kiran A, De Bo C, O'Connor PBF, Heaphy SM, Mullan JPA, Donohue CA, Higgins DG, Baranov PV 2014. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res 42(Database issue): D859–D864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miniard AC, Middleton LM, Budiman ME, Gerber CA, Driscoll DM 2010. Nucleolin binds to a subset of selenoprotein mRNAs and regulates their expression. Nucleic Acids Res 38: 4807–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads RE, Dinkova TD, Korneeva NL 2006. Mechanism and regulation of translation in C. elegans. In WormBook (ed. The C. elegans Research Community), pp. 1–18. 10.1895/wormbook.1.63.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rother M, Resch A, Gardner WL, Whitman WB, Bock A 2001. Heterologous expression of archaeal selenoprotein genes directed by the SECIS element located in the 3′ non-translated region. Mol Microbiol 40: 900–908 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12: 1317–1319 [DOI] [PubMed] [Google Scholar]
- Small-Howard A, Morozova N, Stoytcheva Z, Forry EP, Mansell JB, Harney JW, Carlson BA, Xu XM, Hatfield DL, Berry MJ 2006. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol 26: 2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvall J, Fierro-Gonzalez JC, Swoboda P, Saamarthy K, Cheng Q, Cacho-Valadez B, Arner ES, Persson OP, Miranda-Vizuete A, Tuck S 2011. Selenoprotein TRXR-1 and GSR-1 are essential for removal of old cuticle during molting in Caenorhabditis elegans. Proc Natl Acad Sci 108: 1064–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A, Schmitt D, Chapple C, Babaylova E, Karpova G, Guigo R, Krol A, Allmang C 2009. A short motif in Drosophila SECIS Binding Protein 2 provides differential binding affinity to SECIS RNA hairpins. Nucleic Acids Res 37: 2126–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taskov K, Chapple C, Kryukov GV, Castellano S, Lobanov AV, Korotkov KV, Guigo R, Gladyshev VN 2005. Nematode selenoproteome: the use of the selenocysteine insertion system to decode one codon in an animal genome? Nucleic Acids Res 33: 2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Carlson BA, Irons R, Mix H, Zhong N, Gladyshev VN, Hatfield DL 2007a. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem J 404: 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL 2007b. Biosynthesis of selenocysteine on its tRNA in eukaryotes. PLoS Biol 5: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]