This paper addresses the mechanism of recognition and cleavage of primary microRNAs (pri-miRNAs) by the Microprocessor (DGCR8/Drosha) complex. The authors use an extensive panel of closely related miRNA variants to analyze the role of pri-miRNA structures on Drosha cleavage. They conclude that a combination of multiple structural elements coordinates Drosha cleavage position and efficiency. They show that not only the junction with the terminal loop, but also the basal junction, are important in determining the cleavage site. This confirms a recent observation by Ma and colleagues, but importantly also adds to it since it shows that internal bulges strongly affect the Drosha cleavage site.
Keywords: Drosha, DGCR8, pri-miRNA, miRNA, shRNA, RNAi
Abstract
Processing of primary microRNA (pri-miRNA) stem–loops by the Drosha–DGCR8 complex is the initial step in miRNA maturation and crucial for miRNA function. Nonetheless, the underlying mechanism that determines the Drosha cleavage site of pri-miRNAs has remained unclear. Two prevalent but seemingly conflicting models propose that Drosha–DGCR8 anchors to and directs cleavage a fixed distance from either the basal single-stranded (ssRNA) or the terminal loop. However, recent studies suggest that the basal ssRNA and/or the terminal loop may influence the Drosha cleavage site dependent upon the sequence/structure of individual pri-miRNAs. Here, using a panel of closely related pri-miRNA variants, we further examine the role of pri-miRNA structures on Drosha cleavage site selection in cells. Our data reveal that both the basal ssRNA and terminal loop influence the Drosha cleavage site within three pri-miRNAs, the Simian Virus 40 (SV40) pri-miRNA, pri-miR-30a, and pri-miR-16. In addition to the flanking ssRNA regions, we show that an internal loop within the SV40 pri-miRNA stem strongly influences Drosha cleavage position and efficiency. We further demonstrate that the positions of the internal loop, basal ssRNA, and the terminal loop of the SV40 pri-miRNA cooperatively coordinate Drosha cleavage position and efficiency. Based on these observations, we propose that the pri-miRNA stem, defined by internal and flanking structural elements, guides the binding position of Drosha–DGCR8, which consequently determines the cleavage site. This study provides mechanistic insight into pri-miRNA processing in cells that has numerous biological implications and will assist in refining Drosha-dependent shRNA design.
INTRODUCTION
MicroRNAs (miRNAs) are small RNAs (∼22 nt) that direct post-transcriptional repression of gene expression (Bartel 2004; Kim 2005). The majority of miRNAs are generated via an established biogenesis pathway. The RNase III enzyme Drosha cleaves stem–loop structures embedded in longer primary miRNA (pri-miRNA) transcripts to liberate pre-miRNAs (Lee et al. 2003; Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). The RNase III enzyme Dicer subsequently processes pre-miRNAs (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001) into duplex miRNA strands (Lee et al. 2003). One strand is then incorporated into the RNA-induced silencing complex (RISC) and guides mRNA transcript association (Khvorova et al. 2003; Schwarz et al. 2003; Liu et al. 2004). While the mechanisms of pre-miRNA recognition and cleavage by Dicer are well characterized (Zhang et al. 2002, 2004; Vermeulen et al. 2005; MacRae et al. 2006, 2007; Park et al. 2011), an understanding of how Drosha and its binding partner DGCR8 (DiGeorge syndrome critical region gene 8) (Landthaler et al. 2004) selectively recognize and precisely cleave pri-miRNAs remains unclear. This is evident in the high false-positive rates of ab initio pri-miRNA prediction algorithms (Bentwich et al. 2005; Nam et al. 2005; Chiang et al. 2010) and the lack of methods to precisely predict the position of pri-miRNA cleavage in cells.
As the initial processing step in the miRNA biogenesis pathway, Drosha–DGCR8 accomplishes two important functions: (1) pri-miRNA recognition by Drosha–DGCR8 distinguishes RNAs intended for miRNA production from other structured RNAs, and (2) Drosha cleavage of pri-miRNAs directly establishes the sequence and, therefore, indirectly the targetome of the derivative miRNAs. Primary sequences in the basal ssRNA and terminal loop of human pri-miRNAs enhance pri-miRNA recognition by Drosha–DGCR8 (Auyeung et al. 2013). In addition, common structural features, such as the basal ssRNA, stem, and terminal loop, have also been shown to be important for recognition and processing (Zeng and Cullen 2005; Zeng et al. 2005; Han et al. 2006; Zhang and Zeng 2010; Auyeung et al. 2013). Two prevalent models, using two different model pri-miRNAs, ascribe conflicting mechanisms for pri-miRNA recognition and processing by Drosha–DGCR8. The “loop-anchor” model proposes that Drosha binds the stem–loop and directs cleavage ∼22 bp from the terminal loop of pri-miR-30a (Zeng et al. 2005). The “base-anchor” model proposes that DGCR8 anchors to the basal ssRNA and directs Drosha cleavage ∼11 bp up the stem of pri-miR-16 (Han et al. 2006). However, recent studies suggest that the basal ssRNA and/or the terminal loop, dependent upon sequence/structural motifs of a particular pri-miRNA, may be important for Drosha cleavage cite selection (Auyeung et al. 2013; Ma et al. 2013). Thus, the mechanism that guides the Drosha cleavage position of pri-miRNAs remains indeterminate.
Here, we demonstrate that an internal loop, the basal ssRNA, and the terminal loop of the SV40 pri-miRNA cooperatively coordinate cleavage position, specificity, and efficiency by Drosha–DGCR8 in cells. We propose an alternative mechanism for pri-miRNA processing whereby structural elements within the stem and the flanking ssRNA structures cooperatively position Drosha–DGCR8 on the basal and apical double-stranded RNA (dsRNA) regions of the pri-miRNA stem, and in turn, this determines the Drosha cleavage site.
RESULTS
Experimental design: systematic mutagenesis of the Simian Virus 40 pri-miRNA
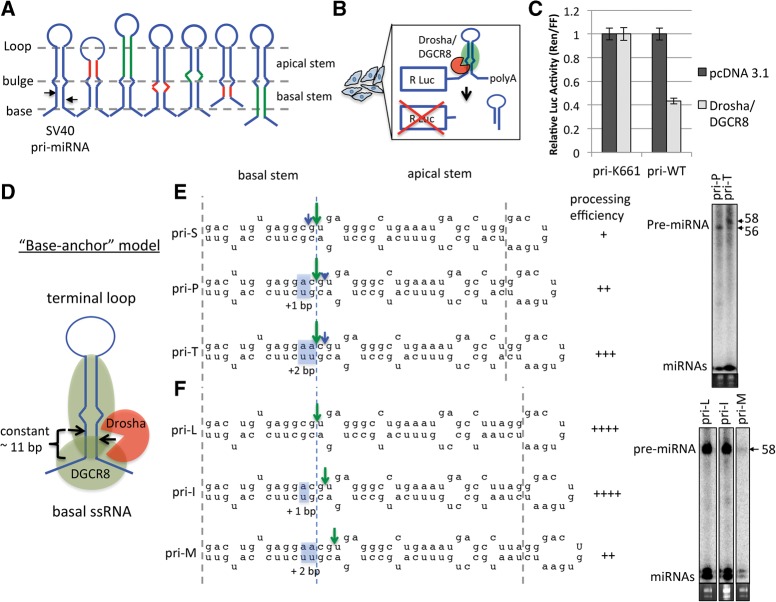
To explore the structural determinants that mediate pri-miRNA cleavage in cells, we focused on a mammalian viral pri-miRNA for which numerous natural variants have been identified (Chen et al. 2013). Analysis of the pri-miRNA encoded by strains of Simian Virus 40 (SV40) revealed variants in which the positions of the basal ssRNA, internal loops/bulges within the stem, and/or the terminal loop seemingly altered the Drosha cleavage site (Chen et al. 2013; Supplemental Fig. S1A,B). To systematically test the contributions of these structural elements, we generated a panel of 26 SV40 (strain 776) pri-miRNA variants encompassing permutations in which the positions of the basal ssRNA, internal loop, and/or terminal loop were altered (Fig. 1A; detailed in Supplemental Fig. S1C). Primary sequences in or near the terminal loop and the basal ssRNA known to regulate Drosha processing were not altered in our mutagenesis strategy (García-Mayoral et al. 2008; Trabucchi et al. 2009; Auyeung et al. 2013). To determine the Drosha cleavage site of each pri-miRNA variant, the pri-miRNAs were expressed in HEK293T cells and small RNA sequencing (RNA seq) was performed to map the pre-miRNAs, which are direct products of Drosha cleavage (Supplemental Table S1). For many variants, Northern blot analysis was performed to independently verify the RNA seq data (Supplemental Fig. S1D). The predicted secondary structures of the pri-miRNA stem–loops with indicated nomenclature and mapped cleavage sites for all the SV40 pri-miRNA variants are shown in Supplemental Figure S2. To determine the Drosha-DGCR8 processing efficiency of the pri-miRNA variants in cells, we performed a Drosha cleavage efficiency assay that we previously developed (Fig. 1B; Kincaid et al. 2012). pri-miRNA variants were inserted into the 3′ UTR of Renilla luciferase. Drosha processing efficiency was quantified as the reduction in Renilla luciferase activity with Drosha/DGCR8 coexpression and relative to the SV40 K661 pri-miRNA (pri-K661), a wild variant that we have previously demonstrated to be a defective Drosha–DGCR8 substrate (Fig. 1C; Chen et al. 2013). As this assay measures direct cleavage of the Renilla transcript by Drosha–DGCR8, possible biases in downstream effectors that alter pre-miRNA levels, such as Dicer, were avoided. The processing efficiencies for all the SV40 pri-miRNA variants are shown in Supplemental Figure S3.
FIGURE 1.
The distance from basal ssRNA of the SV40 pri-miRNA is only a partial determinant of Drosha cleavage. (A) Schematic diagram illustrating the mutagenesis strategy to generate the SV40 pri-miRNA variants. The basal ssRNA, internal loop, and/or the terminal loop positions of the SV40 pri-miRNA were increased (green), unchanged (blue), or decreased (red) in all combinations (detailed in Supplemental Fig. 1C). (B) Schematic diagram of the cellular-based Drosha–DGCR8 processing efficiency assay. (C) Renilla luciferase constructs containing either the K661 (pri-K661) or 776 (pri-WT) SV40 pri-miRNA were cotransfected with either pcDNA3.1+ or Drosha/DGCR8 expression vectors. The average luciferase activity ratio (Ren/FF) is graphed and normalized to pri-K661; error bars indicate SE (n = 6). (D) Schematic diagram of the base-anchor model (Han et al. 2006). DGCR8 (green) binds the basal ssRNA and directs Drosha (red) to cleave ∼11 bp up the stem. (E) Illustration of the Drosha cleavage sites of the indicated SV40 pri-miRNA variants determined by RNA seq (Supplemental Table S1) and Northern blot analysis. The large/green arrows and small/blue arrows represent the dominant and minor cleavage sites, respectively. The length of the arrows roughly correlates to the ratio of the derivative pre-miRNAs. The gray dotted lines indicate the basal and apical stem-ssRNA junctions of pri-S. The dotted blue line indicates the dominant cleavage site of pri-S. Alteration to the basal stems of pri-P and pri-T relative to pri-S are highlighted in blue. Each (+) represents a 20% increase in cleavage efficiency relative to pri-K661 (Supplemental Fig. S3). (F) Illustration of the cleavage site and efficiency of the indicated SV40 pri-miRNA variants as the basal stem was extended (highlighted in blue).
The basal ssRNA of the SV40 pri-miRNA influences the Drosha cleavage site
To test the influence of the basal ssRNA on cleavage site selection, we analyzed the cleavage site of SV40 pri-miRNA variants that differ only in the basal stem length. According to the base-anchor model (Han et al. 2006), as the basal stem is extended, the Drosha cleavage site should remain a constant distance from the basal ssRNA (Fig. 1D). Indeed, as the basal stems of some variants were sequentially extended +1 bp, such as from pri-S to pri-P to pri-T, the dominant cleavage site remained a constant distance from the basal ssRNA, while the processing efficiency increased (Fig. 1E; Supplemental Fig. S4A). These results are consistent with the base-anchor model and reiterate the importance of the basal stem for efficient Drosha processing (Zeng et al. 2005; Han et al. 2006; Auyeung et al. 2013). In contrast, extension of the basal stem in many variants, such as from pri-L to pri-I to pri-M, which only differ from the pri-miRNAs in Figure 1E by the apical stem length, resulted in a shift of the cleavage site up the stem (toward the loop) (Fig. 1F; Supplemental Fig. S4B,C). These results are consistent with the loop-anchor model. Thus, the basal ssRNA can influence cleavage site selection in some SV40 pri-miRNA variants, while the loop-anchor model better predicts the cleavage site in other closely related SV40 pri-miRNA variants.
The terminal loop position of the SV40 pri-miRNA influences Drosha cleavage site selection
The above data (Fig. 1F) suggest that structures in the upper stem influence cleavage site selection. To examine the influence of the terminal loop, we analyzed pri-miRNA variants in which only the apical stem length was altered. According to the loop-anchor model (Zeng et al. 2005), the Drosha cleavage position should remain a constant distance from the terminal loop (Fig. 2A). Consistent with this model, as the apical stem of some pri-miRNA variants was sequentially extended +1 bp, such as from pri-P to pri-B to pri-I, the dominant cleavage site shifted up the stem (toward the loop) (Fig. 2B; Supplemental Fig. S4D,E), indicating that the terminal loop position influences cleavage site selection. Extension of the apical stem also increased processing efficiency, suggesting that, in addition to the basal stem, the apical stem is important for pri-miRNA recognition and processing by Drosha–DGCR8. In contrast, when the apical stem length was sequentially extended +1 bp in many variants, such as from pri-Y to pri-U to pri-W, the dominant cleavage site did not shift up the stem but remained a constant distance from the basal ssRNA, consistent with the base-anchor model (Fig. 2C; Supplemental Fig. S4F). These results indicate that the terminal loop position influences cleavage site selection in some SV40 pri-miRNA variants, while the base-anchor model better accounts for the cleavage position in others.
FIGURE 2.
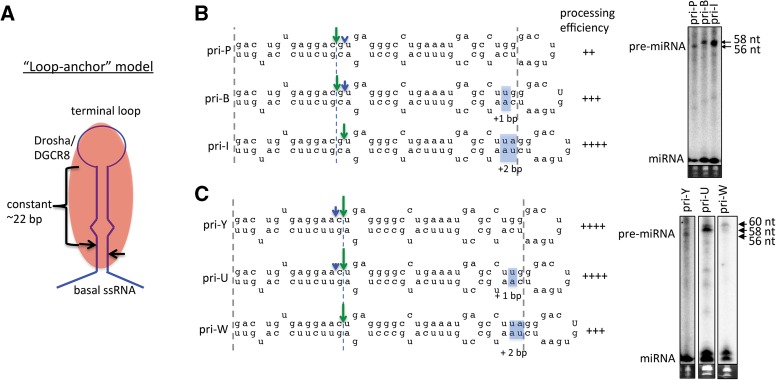
The terminal loop position of the SV40 pri-miRNA influences the Drosha cleavage position. (A) Schematic diagram of the loop-anchor model (Zeng et al. 2005). Drosha–DGCR8 (red) binds and directs cleavage ∼22 bp from the terminal loop. (B,C) Illustration of the cleavage sites and processing efficiency of indicated SV40 pri-miRNA variants when the apical stem was extended by 1 or 2 bp (highlighted).
Combined, the above data (Figs. 1, 2) demonstrate that while both the basal ssRNA and the terminal loop of the SV40 pri-miRNA variants are able to influence Drosha cleavage site selection, the Drosha cleavage site is not precisely measured from either the basal ssRNA or terminal loop.
Structural elements within the SV40 pri-miRNA stem influence cleavage position
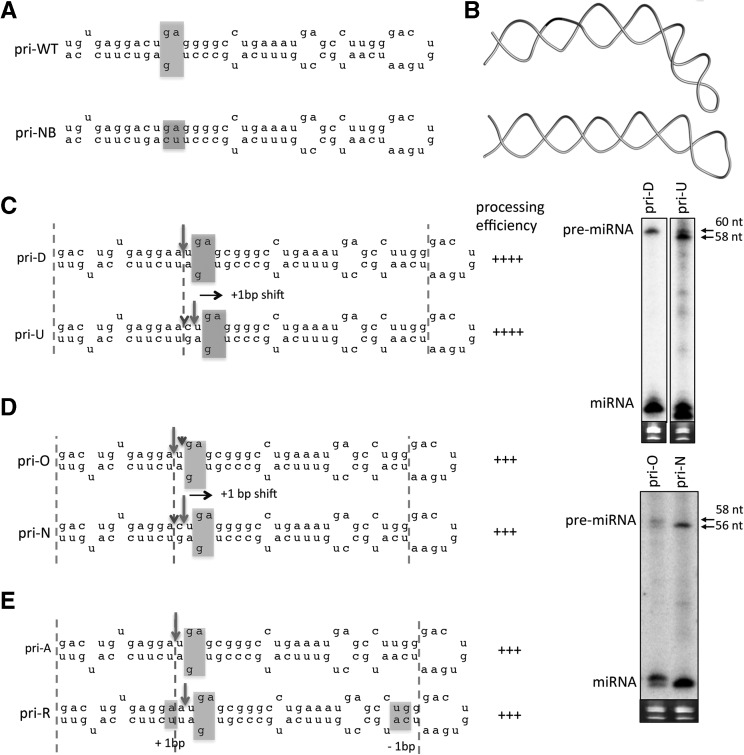
Next, we examined the influence of structural elements within the SV40 pri-miRNA stem on Drosha cleavage site selection. Three-dimensional modeling of the SV40 pri-miRNA revealed that an internal loop proximal to the cleavage site might induce a bend in the pri-miRNA stem (Fig. 3A,B). To test whether the position of the internal loop placement could influence Drosha cleavage site selection, we analyzed the cleavage site of SV40 pri-miRNA variants in which only the internal loop position was altered. Surprisingly, as the internal loop was shifted +1 bp up the stem in many variants, such as from pri-D to pri-U and from pri-O to pri-N, the cleavage site remained a constant distance from the internal loop while shifting up the stem relative to both the basal ssRNA and terminal loop (Fig. 3C,D; Supplemental Fig. S4G–J). This is inconsistent with both the base- and the loop-anchor models. Similarly, simultaneous removal of 1 bp in the apical stem and insertion of 1 bp in the basal stem, such as from pri-A to pri-R, which neither the base- nor the loop-anchor models would predict to alter cleavage position, shifted cleavage +1 bp up the stem (Fig. 3E; Supplemental Fig. S4K,L). Thus, in addition to the flanking ssRNA regions, these results demonstrate that the internal loop within the SV40 pri-miRNA stem contributes to cleavage site selection by Drosha–DGCR8.
FIGURE 3.
The internal loop within the SV40 pri-miRNA stem influences cleavage site selection. (A) Illustration of the predicted secondary structure of pri-WT and a hypothetical variant (pri-NB) in which the internal loop was removed by pairing the bulged nucleotides in pri-WT (highlighted). (B) Three-dimensional model of pri-WT and pri-NB. (C,D) Illustration of the cleavage sites and processing efficiency for the indicated SV40 pri-miRNA variants as the internal loop (highlighted) is shifted +1 bp (arrow) up the stem (toward the loop). (E) Illustration of the cleavage sites and processing efficiency for the indicated SV40 pri-miRNA variants when 1 bp is removed from the apical stem while 1 bp is simultaneously inserted in the basal stem (highlighted).
The basal ssRNA, internal loop, and terminal loop cooperatively coordinate the Drosha cleavage position
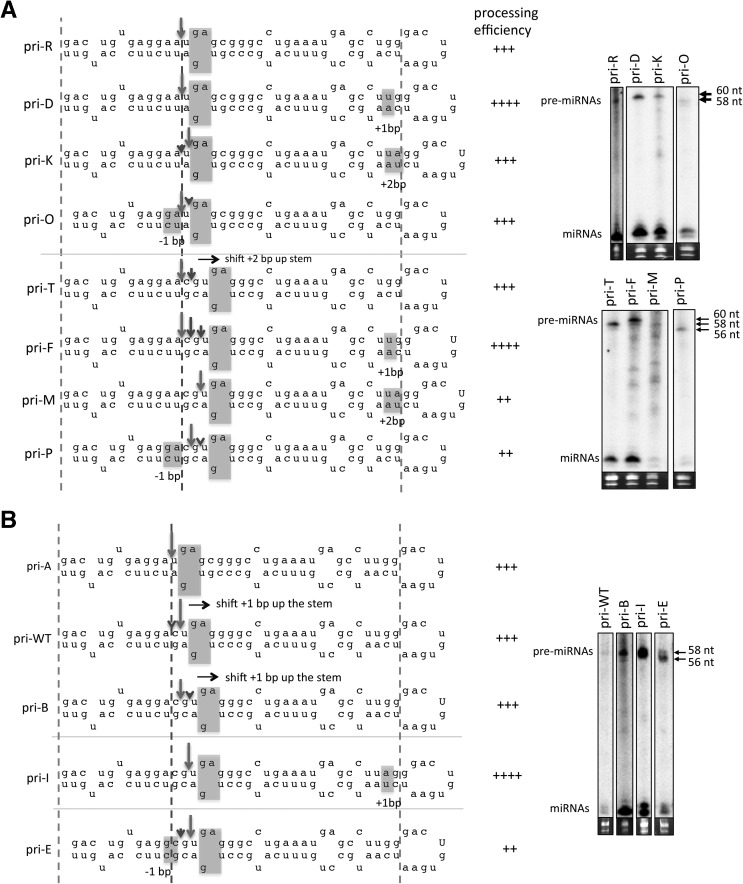
Because we show that basal ssRNA, internal loop, and terminal loop positions can individually influence the cleavage site in some SV40 pri-miRNA variants but not others (Figs. 1–3), we hypothesized that these structures may cooperate to coordinate the Drosha cleavage site. To test this, we examined whether the internal loop position could alter the ability of the flanking ssRNA regions to direct the cleavage site. We first identified variants in which the basal ssRNA and terminal loop did not robustly influence cleavage site selection. For example, sequential +1 bp extensions of the apical stem from pri-R to pri-D to pri-K only partially shifted cleavage +1 bp up the stem, while shortening of the basal stem by −1 bp from pri-R to pri-O did not alter the cleavage position (Fig. 4A). Thus, the flanking ssRNA regions did not alter the cleavage site as predicted (up the stem) by the respective base- or loop-anchor models. To test whether the internal loop position restricted the cleavage site, we examined identical basal- and apical-stem alterations to a pri-miRNA (pri-T) in which the internal loop was positioned +2 bp up the stem as compared with pri-R. In this context, we now observed that sequential +1-bp extensions of the apical stem from pri-T to pri-F to pri-M shifted the cleavage site +2 bp up the stem, consistent with the loop-anchor model. Similarly, shortening of the basal stem −1 bp from pri-T to pri-P shifted the cleavage site +1 bp up the stem, consistent with the base-anchor model. Similar phenomena were observed for other SV40 pri-miRNA variants (Supplemental Fig. S5A). These results demonstrate that the placement of the internal loop within the SV40 pri-miRNA stem can modulate the ability of the flanking ssRNA-regions to define the cleavage site.
FIGURE 4.
The basal ssRNA, internal loop, and terminal loop cooperatively coordinate Drosha cleavage of the SV40 pri-miRNA. Illustration of the cleavage sites and processing efficiency for the indicated SV40 pri-miRNAs variants. (A) The apical and basal stem alterations relative to pri-R are indicated in pri-D, pri-K, and pri-O. The gray line indicates that the internal loop position is shifted +2 bp up the stem in the following pri-miRNA variants: pri-T, pri-F, pri-M, and pri-P. The basal and apical alterations relative to pri-T are indicated in pri-F, pri-M, and pri-P. (B) The internal loop position of pri-A is shifted up the stem +1 bp (pri-WT) and +2 bp (pri-B). The gray lines indicate 1-bp extension of the apical stem (pri-I) or a 1-bp deletion in the basal stem (pri-E) relative to pri-B.
To further examine coordination of the cleavage site, we determined whether the flanking ssRNA regions could restrict the ability of the internal loop to direct cleavage. To test this, we first identified variants in which the internal loop position did not direct the cleavage site. For example, while positioning of the internal loop +1 bp upward from pri-A to pri-WT shifted cleavage +1 bp up the stem, a further positioning of the internal loop an additional +1 bp upward from pri-WT to pri-B did not further shift the cleavage site (Fig. 4B). Rather, the cleavage site remained constant relative to the flanking ssRNA regions. To test whether the basal ssRNA and/or terminal loop restricted the ability of the internal loop to direct cleavage up the stem from pri-WT to pri-B, we then analyzed variants of pri-B that had either a lengthened apical stem or shortened basal stem. Indeed, extension of the apical stem (relative to the internal loop) from pri-B to pri-I resulted in cleavage +1 bp up the stem, suggesting that the terminal loop position restricted the ability of the internal loop to direct cleavage up the stem from pri-WT to pri-B. Similarly, shortening of the basal stem (relative to the internal loop) from pri-B to pri-E also shifted cleavage +1 bp up the stem, which suggests that the repositioning of the basal ssRNA overrode the restriction imposed by the terminal loop in pri-B. This indicates coordination of the cleavage site between the basal ssRNA and terminal loop. Similar results were observed for other SV40 pri-miRNA variants (Supplemental Fig. S5B,C). Combined, these data indicate that the basal ssRNA, internal loop, and the terminal loop cooperatively coordinate the position at which Drosha cleaves the SV40 pri-miRNA in cells.
Coordination between the basal ssRNA and the terminal loop of two prototypic human pri-miRNAs guides the Drosha cleavage site
Next, we examined whether the basal ssRNA and terminal loop coordinate the cleavage position of human pri-miRNAs. To test this, we generated variants of pri-miR-30a, which was the model pri-miRNA for establishing the loop-anchor model. In accordance with the loop-anchor model, insertion of 2 bp in the apical stem of pri-miR-30a (pri-miR-30a L+2) resulted in cleavage sites shifted toward the loop (Fig. 5). However, when 2 bp were removed in the apical stem of pri-miR-30a (pri-miR-30a L−2), cleavage did not shift down the stem but remained a constant distance from the basal ssRNA, consistent with the base-anchor model. Interestingly, insertion of 2 bp in the basal stem (pri-miR-30a B+2) resulted in two pre-miRNA bands observed via Northern blot analysis, one of equal and one of greater length than the miR-30a pre-miRNA. This indicates that Drosha cleaved at two sites, one shifted +2 bp up the stem, as predicted by the loop-anchor model, and one closer to the basal ssRNA, consistent with the base-anchor model (Fig. 5). Similar results were obtained for analogous variants of pri-miR-16, which was the model pri-miRNA for establishing the base-anchor model (Supplemental Fig. S5D). These results demonstrate that, similar to the SV40 pri-miRNA variants, both the basal ssRNA and the terminal-loop positions influence Drosha cleavage site selection of the two model pri-miRNAs, miR-30a and miR-16, used to develop the respective anchor models.
FIGURE 5.
The basal ssRNA and terminal loop coordinate the Drosha cleavage site of human pri-miRNAs. Illustration of the Drosha cleavage sites of pri-miR-30a and the indicated variants, as determined via a combination of RNA seq of the pre- and mature-miRNAs and Northern blot analysis of the pre-miRNAs. Alterations to the basal and apical stems are highlighted. (*) Only the pre-miRNA lengths, as determined by Northern blot analysis, were used to map the indicated cleavage site(s).
The SV40 pri-miRNA stem influences recognition and processing by Drosha–DGCR8
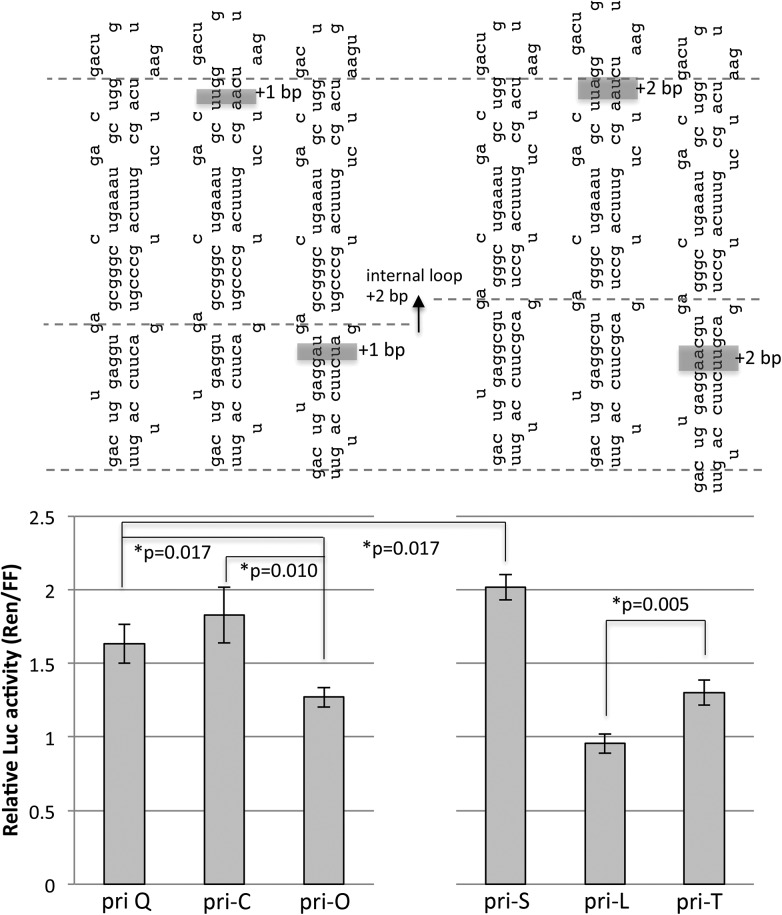
In addition to cleavage site selection, our above data (Figs. 1–4) indicate that altering the basal and the apical stem dimensions influences processing efficiency, suggesting that both regions are important for pri-miRNA recognition and processing by Drosha–DGCR8. However, equivalent alterations to either the basal or the apical stem similarly affected processing efficiency in many variants (Supplemental Fig. S3), which suggests that these effects may have resulted from altering the total stem length. To determine whether the basal and apical stem regions distinctly influence recognition and processing by Drosha–DGCR8, we analyzed the change in processing efficiency of pri-miRNA variants when the basal stem or the apical stem lengths were equally altered to control for stem length. Extension (+1 bp) of the apical stem from pri-Q to pri-C did not alter Drosha processing efficiency, while extension (+1 bp) of the basal stem from pri-Q to pri-O increased the relative processing efficiency ∼25% (Fig. 6). This result suggests that the basal stem influences pri-miRNA recognition and/or processing by Drosha–DGCR8. Interestingly, this indicates that the dsRNA in the pri-miRNA stem is divided into basal-dsRNA and apical-dsRNA. We hypothesized that structures within the stem, such as the internal loop, might differentiate the basal-dsRNA from the apical-dsRNA. Indeed, when the internal loop was positioned +2 bp up the stem (from pri-Q to pri-S), which lengthens the basal stem and shortens the apical stem (relative to the internal loop), processing efficiency decreased ∼20%. This suggests that the dimensions of the basal stem and apical stem relative to the internal loop influence processing efficiency. Further, extension (+2 bp) of the apical stem from pri-S to pri-L now increased relative processing efficiency ∼30% more than extension (+2 bp) of the basal stem from pri-S to pri-T (Fig. 6). Thus, equivalent alterations to either the apical stem or the basal stem had differential effects on Drosha processing dependent upon the internal loop position. Combined, these data suggest that the basal stem and apical stem of the SV40 pri-miRNA are distinct dsRNA-regions, differentiated by the internal loop, which are important for pri-miRNA recognition and/or processing by Drosha–DGCR8.
FIGURE 6.
The apical- and basal-stem dimensions distinctly influence Drosha processing efficiency of the SV40 pri-miRNA. The secondary pri-miRNA structures for the indicated SV40 pri-miRNA variants (differences highlighted) are shown above a graphical representation of the processing efficiency for each pri-miRNA. Gray dashed lines represent the basal ssRNA-, internal loop-, and terminal loop-junctions. Bar graphs depict the mean relative luciferase activity (Ren/FF); error bars represent the SE (n = 6).
DISCUSSION
The underlying mechanism of how Drosha–DGCR8 interacts with pri-miRNAs to facilitate precise cleavage of pri-miRNAs has remained unclear and controversial. Two prevalent models propose that Drosha–DGCR8 anchors to and directs cleavage at a fixed distance from either the basal ssRNA or the terminal loop (Zeng et al. 2005; Han et al. 2006), dependent upon the pri-miRNA. However, we have shown that neither model alone is sufficient to account for the precise cleavage position of pri-miRNAs in cells. Instead, our work reveals that both the basal ssRNA and terminal loop are able to influence the cleavage position of pri-miRNAs (Figs. 1, 2). Consistent with our findings, Ma et al. (2013) reported that the lower and upper stem–ssRNA junctions influence Drosha cleavage precision. These observations suggest an alternative mechanism whereby both the basal ssRNA and terminal loop contribute to cleavage site.
Our mutagenesis of the SV40 pri-miRNA reveals three main findings that provide further insight into the mechanism of pri-miRNA processing by Drosha–DGCR8: (1) Structural elements within the pri-miRNA stem can influence the Drosha cleavage site (Fig. 3). (2) The basal ssRNA, structural elements within the stem, and the terminal loop can cooperatively coordinate the position of pri-miRNA cleavage by Drosha–DGCR8 in cells (Figs. 4, 5). (3) In addition to stem length, both the basal and the apical dsRNA-regions of the SV40 pri-miRNA stem, which can be differentiated by structural elements within the stem, are important for pri-miRNA recognition and/or processing by Drosha–DGCR8 (Fig. 6). Thus, in contrast to Drosha–DGCR8 anchoring to and cleaving a fixed distance from either flanking ssRNA region, our data suggest that Drosha–DGCR8 can be “slid” bidirectionally along the dsRNA of the pri-miRNA stem as the basal and/or apical stem dimensions are altered. Thus, we propose that the flanking ssRNA regions and structural elements within the stem cooperatively position/restrict Drosha–DGCR8 on the basal- and apical-dsRNA of pri-miRNA stems, and this in turn directs the cleavage position by Drosha–DGCR8 (Fig. 7). As our data suggest that multiple structural elements cooperatively coordinate the Drosha cleavage position, the degree of influence of a particular structure on cleavage site selection for individual pri-miRNAs may differ due to sequence/structural composition.
FIGURE 7.
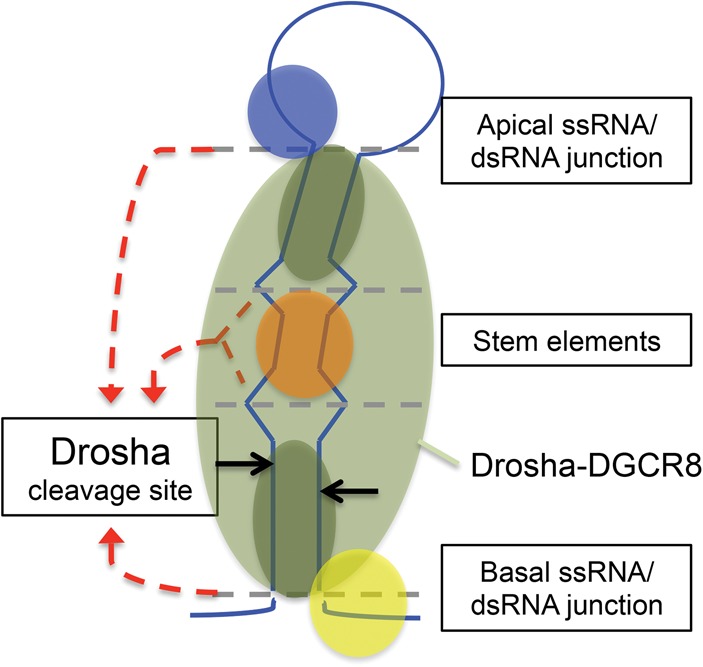
Model of pri-miRNA recognition and processing by Drosha–DGCR8. Illustration of the ssRNA-cooperation model for Drosha–DGCR8 recognition and processing of pri-miRNAs. The dashed gray lines represent ssRNA/dsRNA junctions. The dashed red arrows represent the influence from the indicated structures on Drosha cleavage site selection (black arrows). Drosha/DGCR8 (light green); DGCR8 dsRBDs (dark green); hypothetical loop recognition factor (blue); hypothetical stem cofactor (orange); hypothetical basal ssRNA recognition factor (yellow).
Drosha–DGCR8 does not cleave long dsRNA (Han et al. 2006). Accordingly, both the basal ssRNA and the terminal-loop structures have been shown to be important for Drosha processing (Zeng and Cullen 2005; Zeng et al. 2005; Han et al. 2006; Zhang and Zeng 2010; Auyeung et al. 2013), and our data show that both ssRNA regions coordinate cleavage site selection. However, neither Drosha nor DGCR8 has been shown to have strong binding affinity for ssRNA (Han et al. 2006; Sohn et al. 2007). Thus, how the flanking ssRNA regions of pri-miRNAs interact with Drosha–DGCR8 to guide pri-miRNA recognition and processing has remained unclear. One possibility consistent with our results is that the flanking ssRNA regions confine the binding position of DGCR8, which primarily binds dsRNA (Sohn et al. 2007), on the pri-miRNA stem. This effect may be necessary for efficient processing by Drosha and contribute to cleavage site selection. Solving Drosha–DGCR8/pri-miRNA structures will provide further insight into how the flanking ssRNA regions of pri-miRNAs interact with Drosha–DGCR8 to influence cleavage site selection. As we performed our experiments in cells and our mutagenesis strategy altered the sequence/structure of the stem, we cannot rule out that alterations to the stem may have changed the binding affinity for known or unknown Drosha–DGCR8 cofactors that recognize sequences/structures in the basal ssRNA, the stem, and/or the terminal loop (Auyeung et al. 2013). Thus, it is possible that cofactor recognition of sequence/structural motifs in multiple regions of the pri-miRNA influences Drosha–DGCR8/pri-miRNA interactions. This may permit the coordination of Drosha cleavage position and efficiency by multiple pri-miRNA structural elements in cells (Fig. 7).
In addition to the flanking ssRNA, our data demonstrate that the internal loop within the SV40 pri-miRNA stem influences the Drosha cleavage site and contributes to the variable cleavage position relative to the flanking ssRNA regions. Whether the internal loop within the SV40 pri-miRNA influences Drosha cleavage or the binding position of Drosha–DGCR8 on the pri-miRNA is unclear. However, two dsRNA-binding domains (dsRBDs) in the core of DGCR8 have been modeled to bind the basal and apical stem of pri-miRNAs (Sohn et al. 2007). Accordingly, our data indicate that both the basal- and the apical-dsRNA stem regions influence pri-miRNA recognition and/or processing by Drosha–DGCR8. Thus, the internal loop within the SV40 pri-miRNA stem may function as a structural barrier in the dsRNA that governs the binding positions of the DGCR8 dsRBDs on the basal-dsRNA and/or apical-dsRNA. Alternatively/additionally, the internal loop within the SV40 pri-miRNA may bend the pri-miRNA stem (Fig. 3B). As bulged-structures that bend pri-miRNA stems have been shown to enhance processing efficiency (Quarles et al. 2013), the position of the internal loop within the SV40 pri-miRNA and resulting bend in the pri-miRNA stem could facilitate an optimal Drosha–DGCR8/pri-miRNA interaction, dictating the Drosha–DGCR8-binding position and cleavage site. Although many mammalian pri-miRNAs contain internal loops/bulges within the pri-miRNA stems, whether these structures similarly influence Drosha cleavage site selection and processing efficiency remains to be tested.
Our results suggest that multiple structural elements contribute to the processing efficiency of the SV40 pri-miRNA. This can have important biological consequences, such as suboptimal processing of the SV40 pri-miRNA, which may be necessary to avoid over-regulation of the SV40 miRNA targets—the viral early transcripts (Sullivan et al. 2005). Furthermore, coordination of pri-miRNA processing by multiple structural elements may minimize nonspecific cleavage of transcripts by Drosha–DGCR8. This is made evident by the significant decrease in processing efficiency observed with minimal alterations to the stem of the SV40 pri-miRNA, highlighting the difficulty in predicting cellular Drosha–DGCR8 substrates. Additionally, as small variations in the SV40 pri-miRNA induce heterogeneous miRNA products, structural coordination of the cleavage site can allow for heterogeneous cleavage of pri-miRNAs. This may be advantageous for the evolution of new miRNA seed sequences. Thus, pri-miRNA structures can be “fine-tuned” to regulate Drosha cleavage precision and efficiency, adding another level of regulation to miRNA activity.
In summary, our data demonstrate that multiple structural elements of three model pri-miRNAs cooperatively influence the position and efficiency of pri-miRNA cleavage by Drosha–DGCR8. This work has implications for better understanding host and viral miRNA biology, improving pri-miRNA prediction models, and refining the design of Drosha-dependent shRNAs.
MATERIALS AND METHODS
Plasmids and cell culture
SV40 (776), miR-16, and miR-30a pri-miRNAs and variants were produced by fill-in PCR of overlapping oligos (Supplemental Table S2) using Phusion polymerase (NEB). The pri-miRNA sequences encoding the stem–loop and ∼25 bp flanking the stem were then cloned into the Kpn1/Xho1 site of pcDNA3.1+ and the Xho1/Xba1 site of Renilla luciferase (pcDNA3.1dsRluc). HEK293T cells were obtained from American Type Culture Collection (ATCC) and maintained in DMEM supplemented with 10% (vol/vol) FBS (Cellgro).
Small RNA sequencing
HEK293T cells seeded (70% confluency) in a 6-well format were transfected with 2 µg of pcDNA 3.1 pri-miRNA expression vectors using Lipofectamine 2000 (Life Technologies). Total RNA was harvested 48 h post-transfection with PIG-B (Weber et al. 1998). RNA was fractionated on a 15% UREA-PAGE gel. The pre-miRNA-sized RNAs were isolated for the SV40 pri-miRNA variants, and the pre- and mature-miRNA-sized RNAs and were isolated for the miR-16 and miR-30a pri-miRNA variants as previously described (Chen et al. 2011). The RNAs derived from pri-miRNAs that harbor the same basal-stem modifications were then pooled. Small RNA libraries were then prepared (GSAF, UT Austin) for Illumina small RNA sequencing and sequenced on a Illumina HiSeq 250. Adapter sequences were trimmed from the reads using customPython scripts. The preprocessed reads were then mapped to the respective pri-miRNA variant sequences using the SHRiMP2 software package (Rumble et al. 2009). 5′ start site counts were calculated using customPython scripts and visualized using the gnuplot software package. pre-miRNAs were screened for via sequence abundance, size (∼60 nt), and the characteristic 2-nt 3′ overhang (RNAs with 1- and 3-nt 3′ overhangs were observed and included in the data set).
Northern Blot analysis
HEK293T cells seeded (70% confluency) in 6-well plates were transfected with 2 µg of pri-miRNA expression vector using Lipofectamine 2000. Total RNA was harvested 48 h post-transfection with PIG-B. RNA (15 µg) was fractionated on 10.5% (vol/vol) UREA-PAGE gels. Northern blot analysis was performed as previously described (McClure et al. 2011). Briefly, RNA was transferred to Amersham Hybond –N+ membrane (GE Healthcare) and probed with indicated DNA oligos: SV40 3p probe (5′-GGCATGAAACAGGCA-3′); miR 30a loop probe (5′-ATCTGTGGCTTCACAG-3′); miR 16 loop probe (5′-GATAATTTTAGAATCTTAAC-3′).
Drosha processing efficiency assay
HEK293T cells seeded (70% confluency) in 24-well plates were cotransfected with 0.5 ng Renilla (pcDNA3.1dsRluc) 3′ UTR reporter, 0.5 ng firefly reporter (pcDNA3.1dsLuc2CP), and either 0.5 µg of pcDNA3.1+ or 0.25 μg of Drosha and 0.25 μg of DGCR8 expression vectors (Han et al. 2006) using Lipofectamine 2000. Forty hours after transfection cell lysates were collected and processed with the Dual-Glo Luciferase Assay System (Promega). Luciferase activity was measured with a Luminoskan Ascent luminometer (Thermo Electronic).
Structural predictions of pri-miRNAs
Secondary structures of pri-miRNAs were generated using the RNA-folding forum on the Mfold web server (Zuker 2003). Three-dimensional structural modeling of the pri-miRNA stem–loops were generated using the MC-Fold|MC-Sym pipeline (Parisien and Major 2008). Primary sequences for secondary structure prediction were submitted to MC-Fold with parameters to explore the best 15% of suboptimal structures and return the 20 best scoring. The best-scoring (lowest energy) MC-Fold prediction was submitted to the MC-Sym pipeline and 1000 structural models were generated for each RNA. The structures were scored and sorted by the MC-Fold P-Score function. The lowest P-Score structures were selected for visualization and further analysis. Structure visualization and image generation was performed with the UCSF Chimera package (Pettersen et al. 2004).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. V. Narry Kim for plasmid reagents; Dr. Stephen Trent for use of equipment; and the C.S.S. laboratory for comments regarding this manuscript. This work was supported by Grant R01AI077746 from the National Institutes of Health, a Burroughs Wellcome Investigators in Pathogenesis Award, Grant RP110098 from the Cancer Prevention and Research Institute of Texas, and a University of Texas at Austin Institute for Cellular and Molecular Biology fellowship.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.044537.114.
REFERENCES
- Auyeung VC, Ulitsky I, McGeary SE, Bartel DP 2013. Beyond secondary structure: Primary-sequence determinants license pri-miRNA hairpins for processing. Cell 152: 844–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37: 766–770 [DOI] [PubMed] [Google Scholar]
- Chen CJ, Kincaid RP, Seo GJ, Bennett MD, Sullivan CS 2011. Insights into Polyomaviridae microRNA function derived from study of the bandicoot papillomatosis carcinomatosisviruses. J Virol 85: 4487–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Cox JE, Kincaid RP, Martinez A, Sullivan CS 2013. Divergent microRNA targetomes of closely related circulating strains of a polyomavirus. J Virol 87: 11135–11147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev 24: 992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- García-Mayoral MF, Díaz-Moreno I, Hollingworth D, Ramos A 2008. The sequence selectivity of KSRP explains its flexibility in the recognition of the RNA targets. Nucleic Acids Res 36: 5290–5296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN 2004. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN 2006. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125: 887–901 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim VN 2005. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 6: 376–385 [DOI] [PubMed] [Google Scholar]
- Kincaid RP, Burke JM, Sullivan CS 2012. RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci 109: 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T 2004. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167 [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Ma H, Wu Y, Jang-Gi C, Wu H 2013. Lower and upper stem–single-stranded RNA junctions together determine the Drosha cleavage site. Proc Natl Acad Sci 110: 20687–20692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA 2006. Structural basis for double-stranded RNA processing by Dicer. Science 311: 195–198 [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA 2007. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol 14: 934–940 [DOI] [PubMed] [Google Scholar]
- McClure LV, Lin YT, Sullivan CS 2011. Detection of viral microRNAs by Northern blot analysis. Methods Mol Biol 721: 153–171 [DOI] [PubMed] [Google Scholar]
- Nam J, Shin K, Han J, Lee Y, Kim VN 2005. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Res 33: 3570–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M, Major F 2008. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452: 51–55 [DOI] [PubMed] [Google Scholar]
- Park J, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN 2011. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475: 201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 13: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Quarles KA, Sahu D, Havens MA, Forsyth ER, Wostenberg C, Hastings ML, Showalter SA 2013. Ensemble analysis of primary microRNA structure reveals an extensive capacity to deform near the Drosha cleavage site. Biochemistry 52: 795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M 2009. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol 5: e1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Sohn SY, Bae WJ, Kim JJ, Yeom K, Kim VN, Cho Y 2007. Crystal structure of human DGCR8 core. Nat Struct Mol Biol 14: 847–853 [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435: 682–686 [DOI] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral MF, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG 2009. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459: 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A 2005. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA 11: 674–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Bolander ME, Sarkar G 1998. PIG-B: a homemade monophasic cocktail for the extraction of RNA. Mol Biotechnol 9: 73–77 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Cullen BR 2005. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J Biol Chem 280: 27595–27603 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 24: 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zeng Y 2010. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res 38: 7689–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118: 57–68 [DOI] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]