This paper uses a variety of in vivo and in vitro assays to address the functional phenotypes of mutations in U4atac that cause the developmental disorder, MOPD I/TALS. Four such mutations inhibit the binding of essential proteins of the U4atac/U6atac di-snRNA and, in so doing, inhibit tri-snRNP assembly. Accordingly, inefficient splicing of U12-dependent introns because of tri-snRNP deficiency is the mechanistic basis for MOPD I disease.
Keywords: splicing, U4atac snRNA, snRNP function, disease
Abstract
Biallelic mutations of the human RNU4ATAC gene, which codes for the minor spliceosomal U4atac snRNA, cause the developmental disorder, MOPD I/TALS. To date, nine separate mutations in RNU4ATAC have been identified in MOPD I patients. Evidence suggests that all of these mutations lead to abrogation of U4atac snRNA function and impaired minor intron splicing. However, the molecular basis of these effects is unknown. Here, we use a variety of in vitro and in vivo assays to address this question. We find that only one mutation, 124G>A, leads to significantly reduced expression of U4atac snRNA, whereas four mutations, 30G>A, 50G>A, 50G>C and 51G>A, show impaired binding of essential protein components of the U4atac/U6atac di-snRNP in vitro and in vivo. Analysis of MOPD I patient fibroblasts and iPS cells homozygous for the most common mutation, 51G>A, shows reduced levels of the U4atac/U6atac.U5 tri-snRNP complex as determined by glycerol gradient sedimentation and immunoprecipitation. In this report, we establish a mechanistic basis for MOPD I disease and show that the inefficient splicing of genes containing U12-dependent introns in patient cells is due to defects in minor tri-snRNP formation, and the MOPD I-associated RNU4ATAC mutations can affect multiple facets of minor snRNA function.
INTRODUCTION
Spliceosomes are complex, dynamic, ribonucleoprotein molecular machines responsible for the catalysis of pre-mRNA splicing (Wahl et al. 2009; Hoskins and Moore 2012; Turunen et al. 2013). In mammals, two types of spliceosomes of differing composition have been characterized (Russell et al. 2006): the major (U2-dependent) type of spliceosome removes >99% of human introns and requires U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins (snRNPs), whereas the minor (U12-dependent) type of spliceosome removes about 800 human introns and requires the snRNPs U11, U12, U4atac, U5, and U6atac (Levine and Durbin 2001; Alioto 2007). The protein composition of the minor spliceosome overlaps significantly with that of the major spliceosome (Turunen et al. 2013).
The formation of the catalytically active spliceosome follows a similar pathway in major and minor spliceosomes (Wahl et al. 2009). During minor intron splicing, after the initial recognition of the splice sites by the U11/U12 di-snRNP, a tri-snRNP complex composed of U4atac, U6atac, and U5 snRNPs associates with the pre-mRNA to form a precatalytic complex. Subsequently, U11 and U4atac snRNPs are destabilized, and a catalytic complex containing U12, U6atac, and U5 snRNPs is formed.
The in vivo kinetic assembly pathway of tri-snRNPs in Cajal bodies has been previously established (Novotný et al. 2011). The tri-snRNP itself is formed from a U4atac/U6atac di-snRNP particle, in which the two snRNAs are tightly held together by extensive base-pairing interactions, plus a 35S U5 snRNP particle. Protein–protein interactions are believed to play a major role in U5 snRNP association with the di-snRNP (Nottrott et al. 2002; Schultz et al. 2006b).
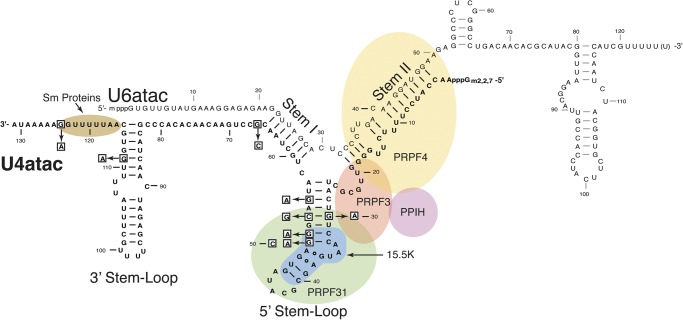
Within the minor spliceosomal di-snRNP and tri-snRNP structures, the U4atac and U6atac snRNAs form a phylogenetically conserved, base-paired, Y-shaped structure consisting of stem I and stem II separated by a U4atac snRNA secondary structure called the 5′ stem–loop (Fig. 1; Shukla et al. 2002; Liu et al. 2011). An evolutionarily conserved 15.5 kD protein (the human NHP2L1 protein, here called 15.5K) binds directly to the kink-turn (K-turn) motif located in the 5′ stem–loop. This RNA–protein complex forms a platform to recruit the PRPF31 protein to the di-snRNP. This ternary complex leads to further assembly of the di-snRNP and its association with the U5 snRNP to yield the tri-snRNP complex (Makarova et al. 2002; Liu et al. 2007).
FIGURE 1.
Diagram of the human U4atac/U6atac di-snRNP. U4atac snRNA (black) is shown base-paired with U6atac snRNA (gray), forming a Y-shaped functional structure that interacts with several essential spliceosomal proteins. The intermolecular stem II domain interacts with three proteins: PRPF4 (yellow), PPIH (purple), and PRPF3 (orange). The U4atac intramolecular 5′ stem–loop domain interacts with the 15.5K (blue) and PRPF31 (green) proteins. PRPF31 directly interacts with PRPF3. A specific sequence located at the 3′ end of U4atac snRNA binds the Sm protein complex (brown). The nucleotides that are mutated in MOPD I patients are boxed.
Recently, mutations in the single copy RNU4ATAC gene coding for U4atac snRNA have been shown to cause the rare, autosomal recessive developmental disorder microcephalic osteodysplastic primordial dwarfism type I (MOPD I), also known as Taybi-Linder syndrome (Edery et al. 2011; He et al. 2011; Nagy et al. 2012). This disorder is characterized by growth retardation, neurological defects, and malformations of the face, long bones, and joints (Abdel-Salam et al. 2011, 2012, 2013; Klingseisen and Jackson 2011; Nagy et al. 2012). Previous data showed that cells from MOPD I patients carrying the RNU4ATAC 51G>A mutation (Fig. 1), had defects in splicing of minor introns that could be rescued by expression of the wild-type U4atac snRNA (He et al. 2011). However, the biochemical defects in these cells were not addressed. Furthermore, there are now nine different mutations in U4atac snRNA that have been found in MOPD I patients, but nothing is known about the functional ramifications of these mutations.
In this report, we describe in vitro and in vivo studies designed to determine the consequences of several MOPD I mutations on U4atac snRNA stability and binding of specific proteins. Using both MOPD I patient fibroblast cells and MOPD I-derived induced pluripotent stem (iPS) cell lines homozygous for the 51G>A mutation, we also demonstrate defects in the formation of U4atac-containing tri-snRNPs.
RESULTS
Distribution of MOPD I mutations in U4atac snRNA
Nine single nucleotide mutations have been described to date in the RNU4ATAC (U4atac snRNA) genes of MOPD I patients in either a homozygous or compound heterozygous configuration (Fig. 1; Abdel-Salam et al. 2011, 2012; Edery et al. 2011; He et al. 2011; Nagy et al. 2012). U4atac and U6atac snRNAs form a base-paired di-snRNP complex that is essential for pre-mRNA splicing of minor-class introns (Shukla et al. 2002; Liu et al. 2007). Six of these mutations, including 30G>A, 50G>A, 50G>C, 51G>A, 53C>G, and 55G>A, are located in the U4atac intramolecular 5′ stem–loop of the U4atac/U6atac duplex (Fig. 1). This RNA structural domain contains a K-turn motif that is recognized by the spliceosomal protein 15.5K. The 15.5K protein belongs to a family of RNA-binding proteins that bind to K-turn motifs through an induced fit interaction (Nottrott et al. 1999, 2002; Turner et al. 2005; Woźniak et al. 2005).
The K-turn motif of U4atac snRNA is comprised of a canonical stem (C-stem) that has two Watson-Crick G-C base pairs, an internal purine-rich loop containing two tandem-sheared G-A base pairs, and a noncanonical stem (NC-stem) that has one G-U and one Watson-Crick G-C base pair attached to an external pyrimidine-rich pentaloop (Cojocaru et al. 2005; Woźniak et al. 2005; Schultz et al. 2006a). The 50G>A, 50G>C and 51G>A MOPD I mutations occur in the K-turn motif, whereas the 30G>A and 53C>G mutations occur in close proximity (Fig. 1).
Binding of the 15.5K protein to the K-turn motif forms a transient binary complex, which recruits the PRPF31 protein followed by the addition of the PPIH/PRPF3/PRPF4 protein complex to form the U4atac/U6atac di-snRNP (Nottrott et al. 2002; Schultz et al. 2006b). The PRPF31 protein simultaneously recognizes the 15.5K protein and the RNA component of the transient binary complex, including the pentaloop, NC stem, and four base pairs of the stem of the 5′ stem–loop domain encompassing the U4atac snRNA positions 30–53 (Schultz et al. 2006a).
Three additional MOPD I mutations lie outside the 5′ stem–loop (Fig. 1). The 124G>A mutation is located immediately adjacent to the U4atac Sm protein binding site. In other spliceosomal snRNAs, binding of the Sm protein complex has been shown to be an essential step in the snRNP assembly process and is required for stability of the snRNA (Seraphin 1995; Urlaub et al. 2001). The 111G>A mutation is located in the apical stem of the intramolecular 3′ stem–loop. This region has been previously shown to be required for U4atac snRNA function (Shukla et al. 2002). Finally, the 66G>C mutation is located at the 3′ end of stem I formed by U4atac/U6atac base-pairing and extends this helix by one additional G-C base pair. In other work, we show that this mutation causes a splicing defect due to the lengthening or hyperstabilization of stem I (R Dietrich, G Shukla, and R Padgett, in prep.).
Expression levels of MOPD I mutant U4atac snRNAs
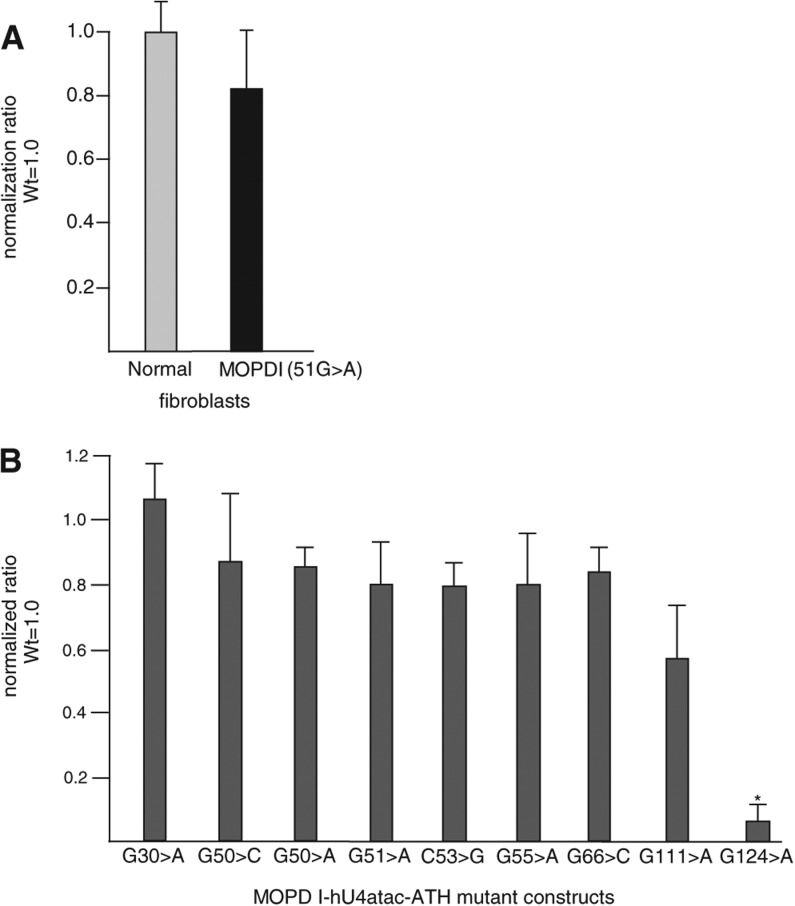
Previous work has shown that several MOPD I mutations led to substantial reductions in U4atac snRNA function when tested in a heterologous in vivo system (He et al. 2011). These functional defects could be related to defects in the splicing mechanism or to reduced levels of the mutant snRNAs. One group has reported that the U4atac 51G>A mutant snRNA is expressed at similar levels to wild type in patient fibroblast cells (Edery et al. 2011). Since MOPD I mutations occur in important structural domains of U4atac snRNA, we analyzed whether they affected steady-state snRNA levels.
First, we confirmed the findings of Edery et al. (2011) in cells homozygous for the 51G>A mutation. Umbilical cord fibroblast cells from two Ohio Amish MOPD I patients (He et al. 2011) were available for us to study the amount of U4atac snRNA directly in patient cells. We used quantitative RT-PCR and the comparative threshold cycle (▵CT) method to measure the relative amounts of U4atac snRNA in patient and control umbilical cord fibroblast cells. Our data showed that, as expected, the 51G>A mutation does not affect the intracellular level of U4atac snRNA (Fig. 2A).
FIGURE 2.
Steady-state levels of mutant U4atac snRNAs. (A) The amounts of U4atac snRNA in normal and homozygous 51G>A MOPD I umbilical cord fibroblast cells were determined by real-time quantitative RT-PCR after isolation of total RNAs. U6 snRNA was used for normalization. The amount of U4atac snRNA in normal cells was set to unity. (B) The amounts of MOPD I-associated mutant U4atac snRNA were determined by real-time quantitative RT-PCR after isolation of total RNAs from CHO cells cotransfected with plasmids expressing either modified wild-type human U4atac snRNA or MOPD I mutant U4atac snRNA as well as modified U6atac snRNA, which is able to base-pair with the modified U4atac snRNA. Primers were designed to amplify only the transfected snRNAs. The level of cotransfected human U12 snRNA was used for normalization. The amount of transfected wild-type human U4atac snRNA was set to unity. Results are expressed as mean and one standard deviation from three independent experiments. (*) A significant difference, P < 0.05, using a two-tailed t-test.
Since patient cell samples carrying other U4atac snRNA mutations were not available, we took advantage of a heterologous test system that has been previously established in our laboratory (Shukla and Padgett 1999, 2001; Shukla et al. 2002; He et al. 2011). We cotransfected human U4atac snRNA expression plasmids with each of the MOPD I mutations embedded in a modified U4atac snRNA in addition to a modified human U6atac snRNA expression plasmid that base-pairs with the modified U4atac snRNA into CHO cells (Shukla et al. 2002). Using primers specific for the modified region allowed us to amplify only the introduced U4atac snRNA in a quantitative RT-PCR assay without interference from the endogenously expressed CHO U4atac snRNA. The amount of each U4atac snRNA mutant was measured by the comparative threshold cycle (▵CT) method and normalized to the level of a cotransfected wild-type human U12 snRNA. These levels were compared with the level seen with transfected wild-type U4atac snRNA (Fig. 2B). The results confirm that the 51G>A mutation yields wild-type levels of U4atac snRNA. Furthermore, although each of these MOPD I mutants reduce splicing activity by at least 90% (He et al. 2011), only the 124G>A mutation had a significant effect on snRNA abundance, which decreased by >90%. Thus, for the majority of the MOPD I mutants, the functional defect(s) must lie elsewhere.
Protein binding by mutant U4atac snRNAs
In U4atac/U6atac (as well as U4/U6) snRNA duplexes, an essential splicing factor, the 15.5K protein, binds through an induced-fit mechanism to the asymmetric internal loop of the 5′ stem–loop and stabilizes a K-turn structure. The 15.5K protein is a nucleating factor that creates a platform for binding of the PRPF31 protein that is required for binding of additional proteins and formation of the U4atac/U6atac.U5 (as well as the U4/U6.U5) tri-snRNP. In the current model of sequential spliceosome assembly, tri-snRNP addition to spliceosomal complex A leads to the formation of complex B.
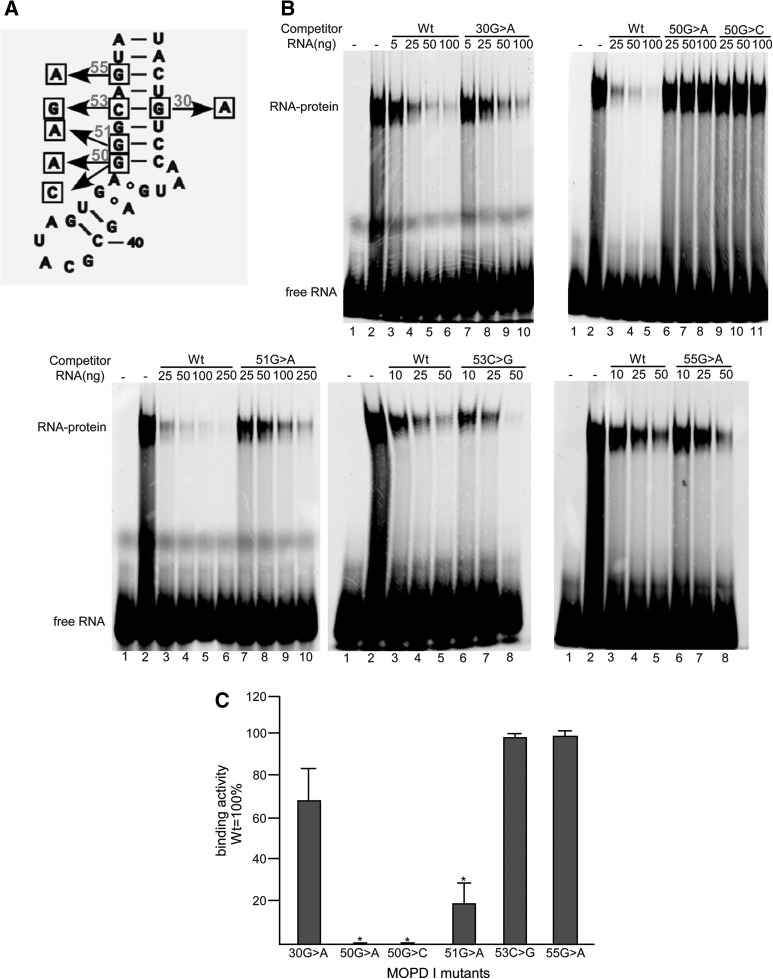
Since binding of the 15.5K and PRPF31 proteins to the 5′ stem–loop of U4atac snRNA is a prerequisite for tri-snRNP formation and six MOPD I-associated mutations are located in this region, we tested whether these mutations interfere with the binding of these proteins to this region. In order to compare the binding affinities of U4atac 5′ stem–loop mutants to wild type, recombinant human 15.5K protein was incubated with a fluorescently 5′ end-labeled 32-nucleotide RNA of the wild-type 5′ stem–loop (nucleotides 26–57). The binding affinities of the mutant RNAs were analyzed by competition experiments using unlabeled 32-nucleotide RNA oligos containing each point mutation in an electrophoretic mobility shift assay. Figure 3B shows the results of adding increasing amounts of each unlabeled RNA to the labeled wild-type RNA in the presence of the 15.5K protein. Figure 3C charts the binding activity of each of the MOPD I mutants.
FIGURE 3.
Binding activity of the 15.5K protein to the 5′ stem–loop of MOPD I U4atac snRNAs. (A) Positions of MOPD I-associated mutations in the intramolecular 5′ stem–loop of human U4atac snRNA. (B) Electrophoretic gel shift analysis of 15.5K protein binding to labeled wild-type RNA in the presence of various amounts of unlabeled competitor RNA. In each panel, lane 1 is labeled wild-type RNA only and lane 2 is labeled wild-type RNA with 15.5K protein. Additional lanes contain the amounts of the various unlabeled competitor RNAs as listed: (Wt) wild type. (C) Quantitation of competition activity compared to wild-type RNA, which was set to 100%. Results are expressed as mean and one standard deviation for three experiments. (*) Indicates a significant difference, P < 0.05, using a two-tailed t-test.
The results show that mutations located in the canonical stem of the K-turn, including 50G>A, 50G>C, and 51G>A, have significantly reduced binding to the 15.5K protein. In fact, the 50G>A and 50G>C mutants show no competition, whereas the 51G>A mutant reduced the competition by >80%. In contrast, the 53C>G and 55G>A mutants that are located in the stem farther from the asymmetric internal loop competed for 15.5K binding similarly to the wild-type sequence. The 30G>A mutation, which disrupts the same G-C base pair as the 53C>G mutation, showed somewhat reduced competition compared to wild type or the 53C>G mutant. These results are consistent with the previous structural and mutational data, indicating that the asymmetric loop is essential for RNA binding to the 15.5K protein (Nottrott et al. 1999; Vidovic et al. 2000; Cojocaru et al. 2005; Woźniak et al. 2005). Mutations that map near this loop and alter the RNA structure disrupt protein binding. Therefore, the molecular basis of the MOPD I-associated 50G>A, 50G>C, and 51G>A U4atac snRNA mutants is likely to be largely due to defective 15.5K protein binding.
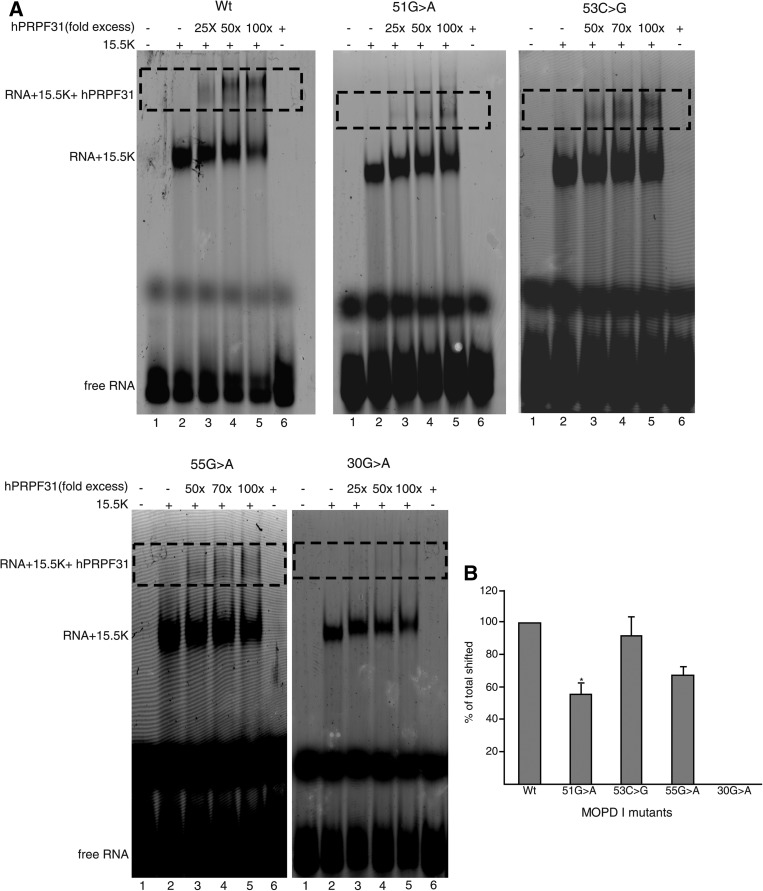
We would expect that, since binding of the PRPF31 protein requires prior 15.5K protein binding to U4atac snRNA (Makarova et al. 2002; Schultz et al. 2006a; Liu et al. 2007, 2011), defects in binding to the 15.5K protein would also lead to defects in binding of the PRPF31 protein. In order to confirm this hypothesis, we tested the binding of recombinant human PRPF31 protein to RNA-15.5K complexes using an electrophoretic super shift assay (Schultz et al. 2006a). The PRPF31 protein was incubated over a range of concentrations with 15.5K protein bound to the labeled wild-type RNA oligo or to labeled oligos containing the various MOPD I mutations (Fig. 4A). As we expected, when labeled G51>A RNA was used, the amount of RNA-15.5K complex shifted by PRPF31 protein was reduced almost by half, which was statistically significant compared to labeled wild-type RNA (Fig. 4B). There were no statistically significant differences between the amounts shifted by PRPF31 protein when either labeled 53C>G or 55G>A RNAs were used compared to wild-type RNA. Interestingly, PRPF31 protein was not able to shift labeled 30G>A RNA (Fig. 4B). This result is in agreement with the previously published results of hydroxyl radical footprinting studies of U4atac snRNA counterpart (U4 snRNA), indicating that nucleotide 30G is protected directly by PRPF31 protein (hPrp31), indicating the possible direct binding of the protein to this region (Schultz et al. 2006a).
FIGURE 4.
Binding of the PRPF31 protein to 5′ stem–loop RNA-15.5K protein complexes. (A) Labeled wild type (Wt) or mutant U4atac 5′ stem–loop RNAs were incubated with 15.5K protein and different amounts of PRPF31 protein. In each panel, lane 1 is RNA only; lane 2 is RNA plus 15.5K protein; lane 6 is RNA plus PRPF31 protein, and lanes 3–5 are reactions containing the listed fold excess of PRPF31 protein to the RNA-15.5K complex. The positions of free RNA, RNA-15.5K complex, and PRPF31-15.5K-RNA complex (dotted regions), are indicated in the figure. (B) Plot showing the percentage amounts of supershifted RNA-15.5K complex. Using ImageJ, the amounts of RNA shifted by 15.5K protein and the amounts of RNA supershifted (dotted region) by 15.5K + PRPF31 proteins were measured for each titration. The percentage of supershifted RNAs was calculated for each titration and normalized to wild type, which was set to 100%.
Derivation of iPS cell lines from MOPD I patient fibroblasts
In the initial descriptions of splicing defects in MOPD I patient cells, only fibroblasts carrying the 51G>A mutation were examined (Edery et al. 2011; He et al. 2011). Due to the rarity of MOPD I, patient samples of other cell types or different mutations are quite difficult to obtain. In addition, human fibroblast cells have a limited life span in culture. To overcome some of these problems, we have derived induced pluripotent stem cells (iPS cells) from fibroblast cell lines from two 51G>A MOPD I patients using viral transduction of reprogramming factors (Takahashi et al. 2007; Yu et al. 2007; Zhou and Freed 2009). PCR amplification and Sanger sequencing of the U4atac snRNA locus showed that the iPS cell lines retained the homozygous 51G>A mutation of the parent cells (data not shown). Comparative genomic hybridization analysis confirmed that the iPS cells had no major chromosomal deletions or duplications (data not shown). We also obtained control normal iPS cell lines prepared using the same reprogramming vector.
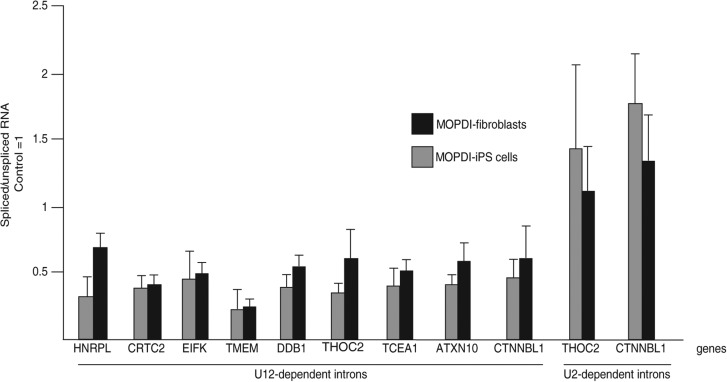
In order to confirm that these MOPD I iPS cells show similar splicing defects as seen in the patient fibroblast cells, we performed quantitative RT-PCR to measure minor intron retention of a set of introns in normal and 51G>A mutant iPS cells as described previously (Fig. 5; He et al. 2011). Both an MOPD I iPS cell line and an MOPD I fibroblast cell line showed increased retention of nine U12-dependent introns while showing robust splicing of two U2-dependent introns. Thus, these MOPD I iPS cells appear to fully recapitulate the minor intron retention observed in patient fibroblasts.
FIGURE 5.
Analysis of 51G>A MOPD I-derived iPS cells. Endogenous U12-dependent introns are inefficiently spliced in iPS cells to similar extents as in MOPD I fibroblast cells. The ratio of spliced to unspliced pre-mRNA for U12- and U2-dependent introns was determined by real-time quantitative RT-PCR as described (He et al. 2011). Two MOPD I-derived iPS cell lines were compared to two MOPD I fibroblast cell lines. For each intron, the ratio seen in control fibroblasts or control iPS cells is set to 1.0. Although U12-dependent introns show increased retention in both MOPD I-iPS and MOPD I fibroblast cell lines, U2-dependent introns are unaffected in both cell lines.
Analysis of snRNP profiles of patient and MOPD I-derived iPS cells by glycerol gradient sedimentation
The aforementioned in vitro studies showed that the MOPD I 51G>A mutation in the 5′ stem–loop of U4atac snRNA is defective in binding to the essential proteins 15.5K and PRPF31. However, these data do not address the functional state of the mutant U4atac snRNA inside cells. Since we confirmed that the MOPD I-derived iPS cells have the same pattern of splicing deficiency as MOPD I fibroblast cells, we used these cells to investigate the distribution of U4atac snRNA-containing snRNP complexes. U4atac snRNPs can exist as mono-snRNPs, as a di-snRNP complex base-paired with U6atac snRNA, and as a tri-snRNP complex with the addition of the U5 snRNP (Tardiff and Rosbash 2006; van der Feltz et al. 2012). The tri-snRNP appears to be the form that is essential for catalytic activation of spliceosome (Wahl et al. 2009; Turunen et al. 2013).
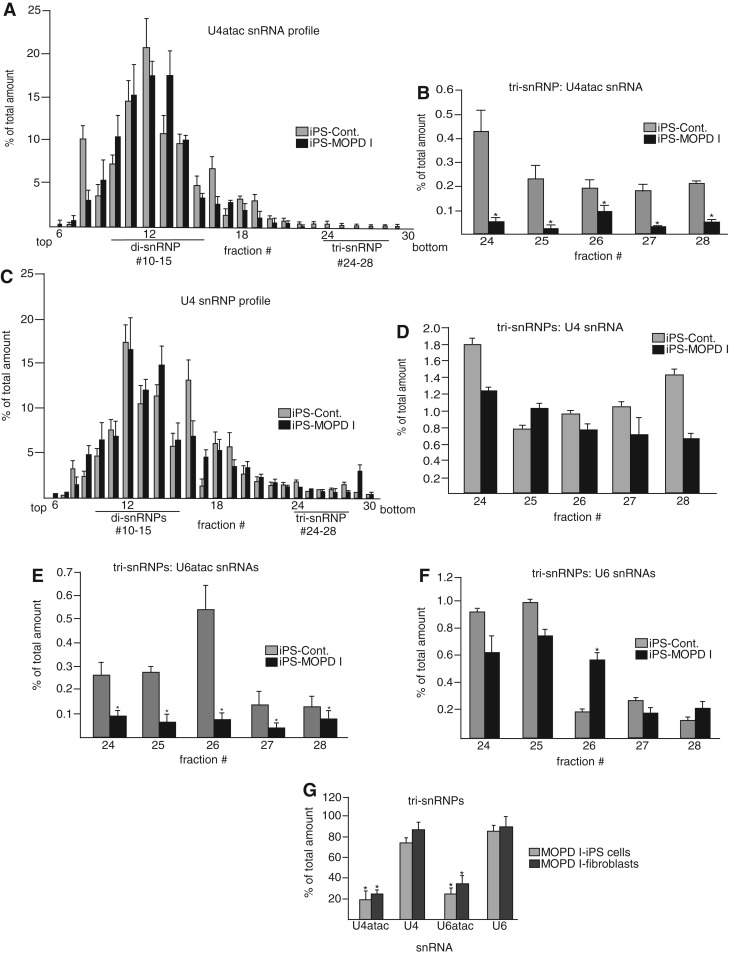
We thus set out to measure the snRNP assembly state as a means of monitoring U4atac snRNA functionality. To examine these processes, whole-cell extracts from normal and 51G>A patient-derived iPS cells were prepared and applied to a 10%–30% glycerol gradient to separate the snRNP species. RNA was prepared from the gradient fractions, and the levels of various snRNAs were determined using quantitative RT-PCR. Figure 6A and C show the overall distribution of U4atac and U4 snRNAs, respectively, in normal and MOPD I iPS cells.
FIGURE 6.
Glycerol gradient sedimentation and characterization of snRNP profiles in 51G>A MOPD I fibroblasts and iPS cell lines. (A) Overall distribution of U4atac snRNA in control and MOPD I iPS cell lines. Whole-cell extracts from control and 51G>A MOPD I-derived iPS cell lines were applied to a 10%–30% glycerol gradient, and snRNP species were separated in different fractions. Total RNA was extracted from each fraction, and U4atac snRNA levels were examined with quantitative RT-PCR by using specific primers for U4atac snRNA. The abundance of U4atac snRNA in each fraction (percentage of total) was calculated as the amount of U4atac snRNA in each fraction divided by the sum of the amount in all fractions. The fraction numbers and the positions of di- and tri-snRNP complexes are shown at the bottom. The direction of sedimentation is from top (left) to bottom (right). (B) Detail of the tri-snRNP fractions shown in A. Results are expressed as mean and one standard deviation from three independent experiments. (C) The overall distribution of U4 snRNA in control and MOPD I iPS cell lines was measured in the same fractions as in A. (D) Detail of U4 snRNA in tri-snRNP fractions. (E) Detail of U6atac snRNA in tri-snRNP fractions. (F) Detail of U6 snRNA in tri-snRNP fractions. (G) An identical analysis was performed using extracts from control and patient fibroblast cells, and the amounts of various snRNAs in the tri-snRNP fractions were compared with the results using control and MOPD I iPS cells. The amounts of the various snRNAs in control cell tri-snRNPs were set to 100%.
The results show that the distribution in both normal and patient samples is dominated by di-snRNPs as seen in other similar analyses (Schneider et al. 2002; Boulisfane et al. 2011). When the gradient fractions containing the tri-snRNPs were plotted separately, it was seen that the level of U4atac snRNA in tri-snRNPs is significantly reduced in the MOPD I samples compared to control samples. In contrast, the levels of U4 snRNA in tri-snRNPs were similar in both cell types (Fig. 6B,D). The same tri-snRNP fractions were assayed for U6atac and U6 snRNAs as shown in Figure 6E (U6atac snRNAs) and Figure 6F (U6 snRNAs). Since U6atac snRNA exists as a base-paired complex with U4atac snRNA in the tri-snRNPs, the levels of U6atac snRNA were also reduced in MOPD I tri-snRNPs, whereas the levels of U6 snRNA were similar in both cell types. The results show that the MOPD I cells contain substantially less U4atac and U6atac snRNAs in the tri-snRNP fractions compared to normal cells, whereas the distribution of U4 and U6 snRNAs is unaffected.
Thus, the 51G>A U4atac snRNA mutant appears to be defective in the process of tri-snRNP formation, which may explain the defects observed in minor intron splicing. The observation of comparable levels of the major spliceosomal tri-snRNPs in both MOPD I and control cells is consistent with the observed comparable levels of major intron splicing in these cells (Fig. 5; Edery et al. 2011; He et al. 2011). The same analyses were also performed with extracts obtained from 51G>A MOPD I fibroblast cells as well as control fibroblast cells with similar results (Fig. 6G).
Levels of tri-snRNP complexes determined by immunoprecipitation
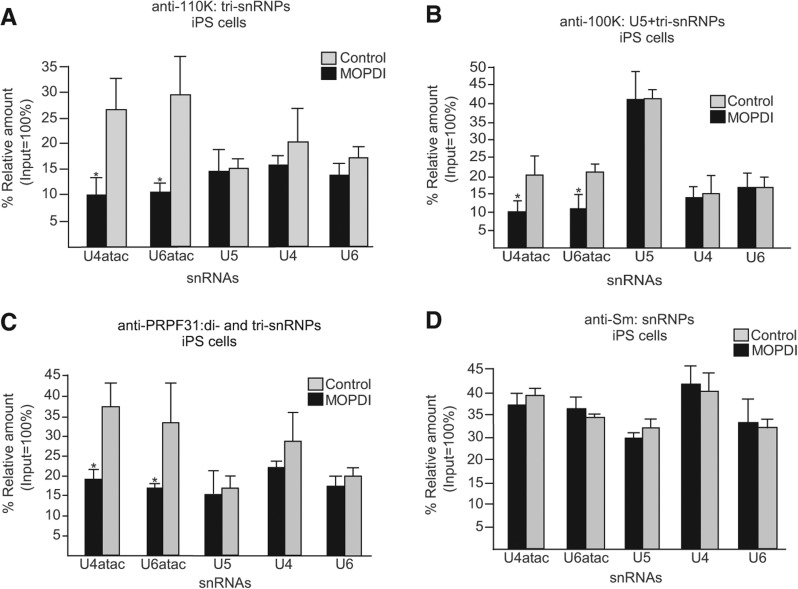
To further confirm the defects in minor tri-snRNP formation observed by glycerol gradient sedimentation, we analyzed the snRNP RNA composition in cell extracts using immunoprecipitation (IP) with antibodies against components of the U5 snRNP as well as of the minor and major spliceosomal tri-snRNPs (Will and Lührmann 2005; Boulisfane et al. 2011). The amount of each immunoprecipitated snRNA was determined by quantitative RT-PCR (Fig. 7). Using antibodies against the 110K tri-snRNP-specific protein (SART1), the amounts of the major spliceosomal snRNAs (U4, U5, and U6) that were precipitated from MOPD I-derived iPS cells were similar when compared to control cells (Fig. 7A). In contrast, the levels of the minor spliceosomal snRNAs (U4atac and U6atac) were reduced by two- to threefold in the immunoprecipitates from MOPD I mutant cells (Fig. 7A). A similar result was seen using antibodies against the U5 snRNP-specific 100K protein (DDX23) that is found in both major and minor spliceosomal tri-snRNP complexes (Fig. 7B). These results are not due to differences in the relative amounts of snRNPs in the extracts since precipitation using anti-Sm protein antibodies recovered equal amounts of snRNAs in both MOPD I and control iPS cells (Fig. 7D).
FIGURE 7.
Immunoprecipitation analysis of snRNPs. (A) Antibody against the 110K tri-snRNP-specific protein was used to immunoprecipitate snRNPs from extracts prepared from control and MOPD I-derived iPS cells. The amounts of immunoprecipitated snRNAs were then measured by quantitative RT-PCR using specific primers for each minor and major snRNA. The amount of input RNA was set to 100%. (B) Immunoprecipitation using antibodies against the 100K U5-specific protein, which is also a component of tri-snRNPs. (C) Immunoprecipitation using antibodies against the PRPF31 di- and tri-snRNP-specific protein. (D) Immunoprecipitation using antibodies against the Sm proteins. Results are expressed as mean and one standard deviation from three independent experiments. (*) Indicates a significant difference, P < 0.05, two-tailed t-test.
As shown above, the in vitro binding of the U4atac/U6atac and U4/U6 di-snRNP-specific proteins are impaired by the 51G>A MOPD I mutation. To confirm this finding in a whole cell environment, a precipitation analysis was performed with antibodies against the PRPF31 protein (Fig. 7C). Again, association with the major spliceosomal snRNAs was unaffected, whereas association with the minor spliceosomal snRNAs was reduced in the MOPD I iPS cells. Similar results were also observed in extracts of MOPD I fibroblast cells (data not shown). Together, these results confirm the defects in the in vivo formation of minor spliceosomal tri-snRNPs, potentially accounting for the observed inefficient minor splicing in 51G>A MOPD I patient cells.
DISCUSSION
An unexpectedly high fraction of human diseases are caused by mutations that alter splicing (Singh and Cooper 2012). A subset of these diseases, such as retinitis pigmentosa (RP), spinal muscular atrophy (SMA), leukemia, and microcephalic osteodysplastic primordial dwarfism type I (MOPD I), are associated with mutations of spliceosomal components (Faustino and Cooper 2003; Padgett 2012; Singh and Cooper 2012). MOPD I is unique in that the causal mutations occur in one of the RNA components of the spliceosome (Edery et al. 2011; He et al. 2011). These mutations occur in the single copy RNU4ATAC gene that encodes U4atac snRNA, an essential component of the minor spliceosome. To date, nine distinct mutations in RNU4ATAC from nearly 30 patients have been characterized as MOPD I causative mutations (Abdel-Salam et al. 2011, 2012; Edery et al. 2011; He et al. 2011; Nagy et al. 2012).
Clinically, MOPD I has a wide spectrum of phenotypes that differ in type and severity. Although the overall syndrome can be clinically recognized, the extent of brain and skeletal abnormalities, the degree of developmental delay, and the overall survival time is quite variable. At the molecular level, MOPD I mutations in RNU4ATAC have been shown to cause impaired splicing of many minor class introns—a phenotype that can be reversed by the expression of normal U4atac snRNA (He et al. 2011). Given that U4atac snRNA forms part of the U4atac/U6atac.U5 tri-snRNP complex, it is likely that the dynamics of the protein–RNA and protein–protein interactions required for the formation and stability of the tri-snRNP is disturbed by the mutations. In this study, we attempted to understand the biochemical and mechanistic basis for the splicing defects observed in MOPD I cells.
The general assembly pathway for U4 or U4atac snRNPs proceeds from a mono-snRNP particle containing the Sm protein complex that is then base-paired to U6 or U6atac snRNA, respectively, to form a Y-shaped U4/U6 or U4atac/U6atac di-snRNP. The base-paired U4/U6 or U4atac/U6atac snRNAs contain several functional and structural domains (Fig. 1). Among the nine point mutations found in U4atac snRNA of MOPD I patients, six are located in the 5′ stem–loop domain indicating, perhaps, the importance of this structural domain in function. The 5′ stem–loop domain is the binding site of several essential proteins, most importantly 15.5K and PRPF31 (Nottrott et al. 2002). The 15.5K protein binds the kink-turn motif in the 5′ stem–loop of U4atac snRNA, inducing changes in the conformations of both RNA and protein (Vidovic et al. 2000; Cojocaru et al. 2005; Woźniak et al. 2005). This binding appears to be essential for the subsequent recruitment of the PRPF31 protein as well as the PRPF4/PPIH/PRPF3 protein complex. The binding of these proteins is crucial for the transition to the next step of the assembly process: the formation of U4atac/U6atac.U5 tri-snRNP (Nottrott et al. 2002; Schultz et al. 2006a).
In the studies reported here, we showed that mutations near the center of the kink-turn, such as 50G>A, 50G>C, and 51G>A, dramatically reduced or abolished the binding of the RNA to 15.5K. Mutations further away from the kink-turn only slightly reduced the affinity of the RNA to 15.5K protein (30G>A) or had no effect (53C>G and 55G>A) (Fig. 3). Interestingly, there is a possible correlation between the severity of the disease based on the age of survival and our in vitro binding studies. For example, patients with homozygous genotype of 55G>A (the same binding activity as wild type) survived much longer than 1 year compared to the majority of patients with homozygous genotype of 51G>A (20% binding activity) with a survival rate <1 year (Nagy et al. 2012).
The PRPF31 binding closely mirrored that of 15.5K binding (Fig. 4). Thus, defects in binding of these two proteins are likely to account for the reduction of U4atac snRNA function in four of the six mutations located in the 5′ stem–loop. We do not have a biochemical explanation for the defects in the 53C>G or 55G>A mutants. However, it is reasonable to suggest that these mutations could cause similar problems with binding of the PRPF4/PPIH/PRPF3 protein complex leading to defects in tri-snRNP formation.
In our in vivo studies, the effects of U4atac snRNA mutation on the higher order formation of macromolecular splicing complexes in patient (iPS) cells with homozygous 51G>A mutation (the most frequently reported MOPD I allele associated with a short postnatal survival period) gave rise to defects in the formation of the U4tac/U6atac.U5 tri-snRNPs. These data are consistent with our in vitro protein binding data, showing impaired interaction of the 51G>A U4atac snRNA mutant with both 15.5K and PRPF31 proteins. It is likely that the other MOPD I mutations in the K-turn region with defective binding to the 15.5K and/or PRPF31 proteins will also show defects in tri-snRNP assembly and splicing.
An impairment of tri-snRNP assembly and, especially minor tri-snRNP assembly, leading to defective splicing and disease is not limited to MOPD I. There are several reports showing defects in tri-snRNP formation in both RP and SMA patient cells. In RP, results from glycerol gradient fractionation, immunoprecipitations and in vitro spliceosome assembly showed that mutations in several core spliceosomal protein components, including the PRPF31 protein, directly or indirectly affect both major and minor tri-snRNP formation and splicing activity in cells (Makarova et al. 2002; Tanackovic et al. 2011). The reduced levels of tri-snRNPs and splicing activity were also observed in patient cells (Makarova et al. 2002; Tanackovic et al. 2011). Although these patients show mild splicing defects in different tissues, the pathogenic effects of the RP mutations are retina-specific; patients do not suffer from syndromic symptoms and have a normal lifespan.
In SMA patients, reduced levels of SMN protein, one of the components necessary for spliceosomal snRNP assembly, leads to lower levels of tri-snRNPs and, particularly, of the minor spliceosomal tri-snRNPs. SMA patient cells displayed differential splicing defects of some but not all minor introns (Zhang et al. 2008; Boulisfane et al. 2011). Although the splicing defects occur in different tissues, SMA is a largely tissue-specific disease limited to motor neurons.
The pathologies seen in MOPD I show some similarities and differences to these two diseases. All three diseases are associated with impaired tri-snRNP formation, but unlike SMA and RP, MOPD I appears to be limited to the minor spliceosomal tri-snRNP. All these diseases show mild global splicing defects with some specific genes being more affected. It is likely that reduced or aberrant splicing of one or a few genes in specific cell types leads to the tissue specificity of each disease. In the case of MOPD I, many, but not all, tissues are affected, perhaps reflecting the wide expression of genes containing minor class introns. Unlike SMA and RP, MOPD I is caused by mutations in an RNA component of the spliceosomes. Although MOPD I is the only reported human disease that is caused by spliceosomal snRNA mutations, a recent report showed that mutations in one of the many genes that code for U2 snRNA in mice causes neurodegeneration and is associated with tissue-specific alternative splicing defects, thus emphasizing the important role of spliceosomal snRNAs in the regulation of mammalian gene expression (Jia et al. 2012).
In summary, our data strongly support the hypothesis that impairment of minor tri-snRNP formation is a primary cause of U12-dependent splicing defects observed in several cases of MOPD I. Of the nine known MOPD I alleles of U4atac snRNA, we have shown that one produces low levels of RNA (124G>A), whereas four others are defective in binding to essential proteins (50G>C, 50G>A, 51G>A, and 30G>A). Further work will be required to define the biochemical defects caused by the other four mutations.
Our study reveals the important role of the U12-dependent splicing machinery in human health and disease. It will be important to further characterize the tissue specificity of MOPD I and make specific connections between affected genes and the disease phenotypes. Since we only have a limited source of patient cells, differentiation of our MOPD I-derived iPS cells to generate different cell types will allow us to address these questions. In the meantime, the results presented here explain how the majority of MOPD I mutations can cause splicing defects and begin to establish a molecular basis for MOPD I.
MATERIALS AND METHODS
Cell culture and extract preparation
The umbilical cord fibroblasts from two individual MOPD I patients with 51G>A mutation were gifts from Dr. A. de la Chapelle's laboratory (He et al. 2011). The MOPD I fibroblasts were cultured in DMEM with high glucose, pyruvate, and 10% heat inactivated FBS. Control umbilical cord fibroblasts (AG14412 and AG14486; Coriell Institute) were cultured in MEM with Earle's salts and nonessential amino acids with 15% FBS. The control and MOPD I fibroblast-derived iPS cell lines were cultured in feeder-free matrigel coated dishes in mTeSR medium (Stemcell Technologies). The CHO cells (CCL-61, ATCC) were cultured in DMEM medium with 10% FBS. Whole-cell extracts were prepared in HNTG buffer (20 mM HEPES, pH 7.9, 150 mM NaCl, 1% Triton X-100, 10% Glycerol, 1 mM MgCl2, 1 mM EGTA, 1 mM protease inhibitor mixture) as described (Boulisfane et al. 2011) and used for glycerol gradient sedimentation and immunoprecipitation experiments.
Derivation of iPS cell lines
Induced pluripotent stem (iPS) cell lines were derived from a 51G>A MOPD I patient (He et al. 2011) and control umbilical cord fibroblasts at the pluripotent stem cell facility at Case Western Reserve University using retroviral transduction of OCT4, KLF4, SOX2, and C-MYC as described (Zhou and Freed 2009; Di Stefano et al. 2010). Briefly, the cells were infected with a single retroviral vector expressing the reprogramming factors. Upon further culture, clones of cells arose with the characteristics of iPS cells. The chromosomal integrity was then confirmed by comparative genomic hybridization (CGH array) (Vermeesch et al. 2005) and karyotyping (data not shown).
Plasmid constructs and transfections
The plasmids expressing each MOPD I-associated U4atac snRNA mutant were generated in the U4atac-Ath background as described (Shukla et al. 2002; He et al. 2011). Plasmids containing the U4atac-Ath constructs were cotransfected with U6atac-Ath and human U12 plasmids into CHO cells as described (He et al. 2011). The levels of snRNAs were measured after 48 h of transfection with SYBR advantage quantitative PCR premix (Clontech) using the 2−▵CT method. The sequences of primers used in this experiment are available upon request.
Electrophoretic gel shift assay
Plasmids expressing the fusion proteins, GST-15.5K and MBP-PRPF31, were gifts from Dr. S. Liu. Proteins were expressed and purified as described (Nottrott et al. 1999; Makarova et al. 2002; Schultz et al. 2006a). For the electrophoretic gel shift analysis, 5′-hexynyl-RNA oligos of wild-type and mutant 5′ stem–loop human U4atac snRNA (nucleotides 26–57) were obtained from IDT. The RNAs were fluorescently labeled with Alexa Fluor 488 using click chemistry as described by the baseclick GmbH click chemistry manual (www.baseclick.eu). For gel shift analysis, 50 ng Alexa Fluor 488 fluorescently labeled U4atac RNA oligonucleotide was incubated with 300 ng recombinant 15.5K protein in a 20-µL reaction mixture in buffer A, and the reaction mixture was incubated for 1 h at 4°C in the presence of 10 µg Escherichia coli tRNA (Roche) as described (Nottrott et al. 1999). For competition studies, increasing amounts of unlabeled wild-type and mutant RNAs were preincubated with the 15.5K protein before addition of the fluorescently labeled wild-type RNA. RNA and RNA-protein complexes were resolved on a native 6% (80:1) polyacrylamide gel containing 0.5× TBE and visualized using a Typhoon Imager. The amount of shifted RNA was quantified using ImageJ. For supershift studies, increasing amounts of recombinant PRPF31 protein (25-, 50-, 70-, and 100-fold excess) were added to the reaction mixture described above and analyzed identically.
Glycerol gradient sedimentation analysis
Whole-cell extracts (500 µL) were prepared as above from four T-75 culture flasks of either fibroblasts or iPS cells. Extracts were then diluted two times with buffer A (50 mM Tris-Cl, pH 7.4, 25 mM NaCl, 5 mM MgCl2) and layered on 11 mL 10%–30% (wt./vol.) glycerol gradients in buffer A. After ultracentrifugation at 35,000 rpm using an SW41 rotor for 16 h at 4°C, 400-µL fractions were collected starting from the top of each tube into which 2 µg of exogenous control RNA (pAW109, Applied Biosystems) was added for normalization (Huggett et al. 2005; Gilsbach et al. 2006). Total RNA was extracted from each fraction using phenol-chloroform and ethanol precipitated (Boulisfane et al. 2011). The RNA samples were then subjected to quantitative real-time PCR in which the relative amount of each snRNA was measured with specific primers using SYBR advantage quantitative PCR premix from Clontech and quantitated by the 2−▵CT method. The sequences of primers used in this experiment are available upon request.
Immunoprecipitation
Antibodies against SART1/110K Ab (Bethyl-Laboratories), DDX23/100K Ab (Bethyl-Laboratories), PRPF31/61K Ab (Lifespan Biosciences), and Sm (Y12) (Abcam) were incubated with Protein G Dynabeads (Invitrogen) in PBS with 0.02% Tween 20 for 2 h at 4°C. Beads were washed three times with the same buffer. Cell extracts prepared in HNTG buffer (Boulisfane et al. 2011) were added and incubated overnight at 4°C. Beads were washed five times using PBS with 0.02% Tween 20. Total RNA was extracted from the beads using phenol-chloroform and ethanol precipitation; 2 µg exogenous control RNA, pAW109 (Applied Biosystems), was added for normalization. The relative amount of each snRNA was measured with quantitative RT-PCR as above.
ACKNOWLEDGMENTS
We thank Dr. Albert de la Chapelle for the MOPD I patient fibroblast cells, Dr. Paul Tesar and the pluripotent stem cell core at Case Western Reserve University for derivation of iPS cells, and Dr. Sunbin Liu for the 15.5K and PRPF31 expression plasmids. This work was supported by grants R01GM093074 and R01GM104059 from the National Institutes of Health to R.A.P. J.M.H. was supported by grant T32GM088088 from the National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.045187.114.
REFERENCES
- Abdel-Salam GMH, Miyake N, Eid MM, Abdel-Hamid MS, Hassan NA, Eid OM, Effat LK, El-Badry TH, El-Kamah GY, El-Darouti M, et al. 2011. A homozygous mutation in RNU4ATAC as a cause of microcephalic osteodysplastic primordial dwarfism type I (MOPD I) with associated pigmentary disorder. Am J Med Genet A 155A: 2885–2896 [DOI] [PubMed] [Google Scholar]
- Abdel-Salam GM, Abdel-Hamid MS, Issa M, Magdy A, El-Kotoury A, Amr K 2012. Expanding the phenotypic and mutational spectrum in microcephalic osteodysplastic primordial dwarfism type I. Am J Med Genet A 158A: 1455–1461 [DOI] [PubMed] [Google Scholar]
- Abdel-Salam GM, Abdel-Hamid MS, Hassan NA, Issa MY, Effat L, Ismail S, Aglan MS, Zaki MS 2013. Further delineation of the clinical spectrum in RNU4ATAC related microcephalic osteodysplastic primordial dwarfism type I. Am J Med Genet A 161A: 1875–1881 [DOI] [PubMed] [Google Scholar]
- Alioto TS 2007. U12DB: A database of orthologous U12-type spliceosomal introns. Nucleic Acids Res 35: D110–D115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonné R 2011. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet 20: 641–648 [DOI] [PubMed] [Google Scholar]
- Cojocaru V, Nottrott S, Klement R, Jovin TM 2005. The snRNP 15.5K protein folds its cognate K-turn RNA: a combined theoretical and biochemical study. RNA 11: 197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B, Buecker C, Ungaro F, Prigione A, Chen H-H, Welling M, Eijpe M, Mostoslavsky G, Tesar P, Adjaye J, et al. 2010. An ES-like pluripotent state in FGF-dependent murine iPS cells. PLoS One 5: e16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery P, Marcaillou C, Sahbatou M, Labalme A, Chastang J, Touraine R, Tubacher E, Senni F, Bober MB, Nampoothiri S, et al. 2011. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 332: 240–243 [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA 2003. Pre-mRNA splicing and human disease. Genes Dev 17: 419–437 [DOI] [PubMed] [Google Scholar]
- Gilsbach R, Kouta M, Bönisch H, Brüss M 2006. Comparison of in vitro and in vivo reference genes for internal standardization of real-time PCR data. Biotechniques 40: 173–177 [DOI] [PubMed] [Google Scholar]
- He H, Liyanarachchi S, Akagi K, Nagy R, Li J, Dietrich RC, Li W, Sebastian N, Wen B, Xin B, et al. 2011. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science 332: 238–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Moore MJ 2012. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci 37: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett J, Dheda K, Bustin S, Zumla A 2005. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6: 279–284 [DOI] [PubMed] [Google Scholar]
- Jia Y, Mu JC, Ackerman SL 2012. Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration. Cell 148: 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingseisen A, Jackson AP 2011. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev 25: 2011–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Durbin R 2001. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res 29: 4006–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Lührmann R, Carlomagno T, Wahl MC 2007. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science 316: 115–120 [DOI] [PubMed] [Google Scholar]
- Liu S, Ghalei H, Lührmann R, Wahl MC 2011. Structural basis for the dual U4 and U4atac snRNA-binding specificity of spliceosomal protein hPrp31. RNA 17: 1655–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Liu S, Vornlocher HP, Lührmann R 2002. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6·U5 tri-snRNP formation and pre-mRNA splicing. EMBO J 21: 1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy R, Wang H, Albrecht B, Wieczorek D, Gillessen-Kaesbach G, Haan E, Meinecke P, de la Chapelle A, Westman JA 2012. Microcephalic osteodysplastic primordial dwarfism type I with biallelic mutations in the RNU4ATAC gene. Clin Genet 82: 140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Hartmuth K, Fabrizio P, Urlaub H, Vidovic I, Ficner R, Lührmann R 1999. Functional interaction of a novel 15.5kD [U4/U6·U5] tri-snRNP protein with the 5′ stem–loop of U4 snRNA. EMBO J 18: 6119–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Urlaub H, Lührmann R 2002. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J 21: 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotný I, Blažíková M, Staněk D, Herman P, Malinsky J 2011. In vivo kinetics of U4/U6·U5 tri-snRNP formation in Cajal bodies. Mol Biol Cell 22: 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett RA 2012. New connections between splicing and human disease. Trends Genet 28: 147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AG, Charette JM, Spencer DF, Gray MW 2006. An early evolutionary origin for the minor spliceosome. Nature 443: 863–866 [DOI] [PubMed] [Google Scholar]
- Schneider C, Will CL, Makarova OV, Makarov EM, Lührmann R 2002. Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol Cell Biol 22: 3219–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz A, Nottrott S, Hartmuth K, Lührmann R 2006a. RNA structural requirements for the association of the spliceosomal hPrp31 protein with the U4 and U4atac small nuclear ribonucleoproteins. J Biol Chem 281: 28278–28286 [DOI] [PubMed] [Google Scholar]
- Schultz A, Nottrott S, Watkins NJ, Lührmann R 2006b. Protein-protein and protein-RNA contacts both contribute to the 15.5K-mediated assembly of the U4/U6 snRNP and the box C/D snoRNPs. Mol Cell Biol 26: 5146–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B 1995. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J 14: 2089–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla GC, Padgett RA 1999. Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants. RNA 5: 525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla GC, Padgett RA 2001. The intramolecular stem-loop structure of U6 snRNA can functionally replace the U6atac snRNA stem-loop. RNA 7: 94–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla GC, Cole AJ, Dietrich RC, Padgett RA 2002. Domains of human U4atac snRNA required for U12-dependent splicing in vivo. Nucleic Acids Res 30: 4650–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Cooper TA 2012. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 18: 472–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Okita K, Nakagawa M, Yamanaka S 2007. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2: 3081–3089 [DOI] [PubMed] [Google Scholar]
- Tanackovic G, Ransijn A, Thibault P, Abou Elela S, Klinck R, Berson EL, Chabot B, Rivolta C 2011. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum Mol Genet 20: 2116–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Rosbash M 2006. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA 12: 968–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DM 2005. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA 11: 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen JJ, Niemelä EH, Verma B, Frilander MJ 2013. The significant other: splicing by the minor spliceosome. Wiley Interdiscip Rev RNA 4: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub H, Raker VA, Kostka S, Lührmann R 2001. Sm protein–Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J 20: 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Feltz C, Anthony K, Brilot A, Pomeranz Krummel DA 2012. Architecture of the spliceosome. Biochemistry 51: 3321–3333 [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Melotte C, Froyen G, Van Vooren S, Dutta B, Maas N, Vermeulen S, Menten B, Speleman F, De Moor B, et al. 2005. Molecular karyotyping: array CGH quality criteria for constitutional genetic diagnosis. J Histochem Cytochem 53: 413–422 [DOI] [PubMed] [Google Scholar]
- Vidovic I, Nottrott S, Hartmuth K, Lührmann R, Ficner R 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell 6: 1331–1342 [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R 2005. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem 386: 713–724 [DOI] [PubMed] [Google Scholar]
- Woźniak AK, Nottrott S, Kühn-Hölsken E, Schröder GF, Grubmüller H, Lührmann R, Seidel CA, Oesterhelt F 2005. Detecting protein-induced folding of the U4 snRNA kink-turn by single-molecule multiparameter FRET measurements. RNA 11: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Freed CR 2009. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 27: 2667–2674 [DOI] [PubMed] [Google Scholar]