This paper describes the development of a database containing known RNA interactions, both with protein and other RNAs.
Keywords: RNA–RNA, RNA–protein, interaction, database
Abstract
Transcriptomic analyses have revealed an unexpected complexity in the eukaryote transcriptome, which includes not only protein-coding transcripts but also an expanding catalog of noncoding RNAs (ncRNAs). Diverse coding and noncoding RNAs (ncRNAs) perform functions through interaction with each other in various cellular processes. In this project, we have developed RAID (http://www.rna-society.org/raid), an RNA-associated (RNA–RNA/RNA–protein) interaction database. RAID intends to provide the scientific community with all-in-one resources for efficient browsing and extraction of the RNA-associated interactions in human. This version of RAID contains more than 6100 RNA-associated interactions obtained by manually reviewing more than 2100 published papers, including 4493 RNA–RNA interactions and 1619 RNA–protein interactions. Each entry contains detailed information on an RNA-associated interaction, including RAID ID, RNA/protein symbol, RNA/protein categories, validated method, expressing tissue, literature references (Pubmed IDs), and detailed functional description. Users can query, browse, analyze, and manipulate RNA-associated (RNA–RNA/RNA–protein) interaction. RAID provides a comprehensive resource of human RNA-associated (RNA–RNA/RNA–protein) interaction network. Furthermore, this resource will help in uncovering the generic organizing principles of cellular function network.
INTRODUCTION
In the past decades, systematic human protein interaction screens have provided a valuable platform to explore the functional organization of the cells (Bossi and Lehner 2009; Vidal et al. 2011). Consequently, text mining-based annotations of this huge number of protein–protein interactions (PPIs) have been established and lead to a more comprehensive understanding of protein function and cellular processes (STRING) (Franceschini et al. 2013), eggnog (Powell et al. 2012). However, recent development has indicated that PPIs are perhaps only half of the story in cells, since an expanding catalog of noncoding RNAs (ncRNAs) are actively involved in multiple biological processes such as cell death, developmental timing, and fat metabolism (Guttman and Rinn 2012; Xu et al. 2012; Li et al. 2013).
The cross-talks within ncRNAs and among RNA–protein are far more intricate and dynamic (Konig et al. 2011; Bernstein et al. 2012; Muller-McNicoll and Neugebauer 2013). For example, experimental evidences indicated that metastasis-associated lung adenocarcinoma transcript 1 (malat1), one of up-regulated long noncoding RNAs (lncRNAs) in many malignant tumors, can stimulate cancer invasion and promote tumorigenicity via binding to several key tumor-suppressor proteins, such as BCL2 and BCLXL1 (Li et al. 2009; Guo et al. 2010). Similarly, recent investigations discovered a novel regulatory RNA circuit, in which RNAs cross-regulate each other by competing for shared ncRNAs (Salmena et al. 2011; Sumazin et al. 2011). For instance, linc-MD1 can sponge miR-133 and miR-135 to modulate the expression of MAML1 and MEF2C, thus act as a competing endogenous RNA (ceRNA) to govern the time of muscle differentiation in mouse and human myoblasts (Cesana et al. 2011). Hence, considerable attention should be focused on the expanding RNA-associated (RNA–RNA/RNA–protein) interaction.
Since the comprehensive regulating cross-talk between diverse RNAs and protein still remains ambiguous, we have developed an RNA-associated interaction database (RAID, http://www.rna-society.org/raid) by integrating experimental evidence from tens of thousands of references. The current version of RAID documents over 6100 human RNA-associated (RNA–RNA/RNA–protein) interactions that are extracted from more than 2100 published papers. RAID provides a valuable resource to manipulate, visualize, and analyze human RNA–RNA/RNA–protein interactions. By integrating the RNA–RNA and RNA–protein interactions into a global network, users can follow RNA-associated (RNA–RNA/RNA–protein) interaction trajectory and determine their functional significance in the whole RNA-associated interaction network.
DATA SOURCES AND IMPLEMENTATION
In order to collect all available RNA and Protein symbols, we have downloaded and integrated all types of RNA and protein symbols including approved symbols, approved names, previous symbols, and names and synonyms in the HGNC database (Gray et al. 2013). Because the research for some ncRNAs is still in its infancy, such as promoter-associated small RNAs (PASRs), PIWI-interacting RNAs (piRNA), promoter upstream transcripts (PROMPTs), transcription initiation RNAs (tiRNA), and TSS-associated RNAs (TSSa-RNAs), etc. (Esteller 2011), and there are not-unified nomenclatures, we instead searched the PubMed database by using these ncRNA category names to replace specific ncRNA symbols. In order to reduce the great challenge of manual curation, we have written scripts to screen in advance all abstracts and full-text articles in the PubMed database for the following keywords combinations: (1) RNA–RNA interactions: (RNA symbols or RNA category names) and/or (RNA symbols or RNA category names) and/or (“interaction” or “binding,” etc.); (2) RNA–protein interactions: (RNA symbols or RNA category names) and/or (protein symbols) and/or (interaction or binding, etc.). The scripts mainly consist of two steps: (1) extract PMC and Pubmed IDs from NCBI through Entrez Programming Utilities (eUtils) based on the combination of keywords; (2) download of the matched abstracts or full articles from NCBI. Then, these screened results were further revised manually. The functional information such as RNA/protein interactions, validated methods, and expressing tissues were extracted. At the same time, the interactions predicted in silico were discarded. This manual checking process ensured the high reliability of data.
In addition, RAID also integrated the miRNA-associated interactions collected in some focused databases such as miRTarBase, miRDeathDB, MNDR (Mammalian ncRNA-disease repository) (Xu and Li 2012; Wang et al. 2013), and other resources (NPInter and PRD) (Wu et al. 2006; Hsu et al. 2011; Fujimori et al. 2012). All of the above third-party data contain a manual collection of RNA regulation interactions usually produced from precise experiments (Xu et al. 2012; Wang et al. 2013; Hsu et al. 2014). On the other hand, some data sets generated by high-throughput techniques or outdated data such as those collected in dorina, Tarbase, and starbase (Yang et al. 2011; Anders et al. 2012; Vergoulis et al. 2012; Li et al. 2014) haven't been integrated into RAID because of the possible higher false-positive targets.
The RAID database is implemented using HTML and PHP language in a window environment connected to the MySQL server, and the interface component consists of the web pages designed and implemented in HTML/CSS. It has been tested in Google Chrome, Safari, Mozilla Firefox, and Internet Explorer web browsers.
CONTENT OF THE DATABASE
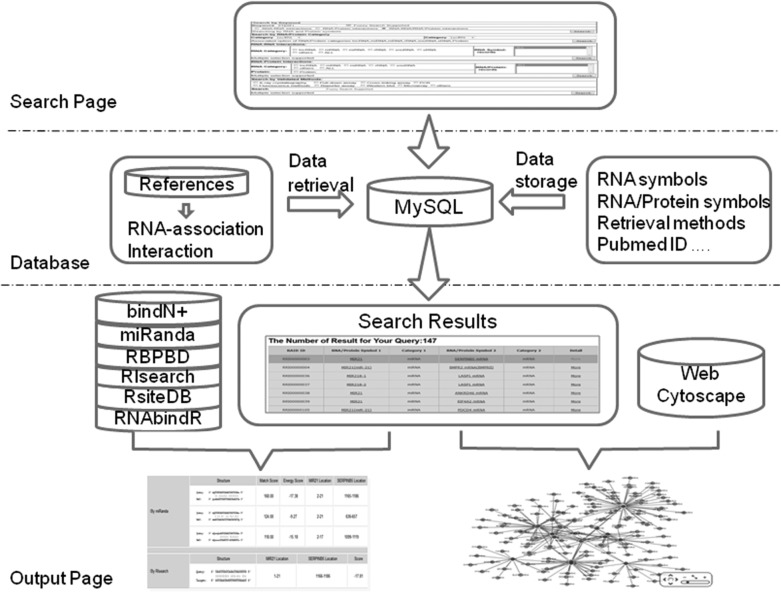
According to the PubMed database, we collected the references published before April 2013. Based on keyword combinations, we have automatically screened tens of thousands of abstracts and full-text articles by in-house scripts. In total, more than 2100 literatures were documented and 4493 RNA–RNA interactions and 1619 RNA–protein interaction entries for a total of 6112 curated entries were documented. Among these RNA-associated (RNA–RNA/RNA–protein) interaction entries, there were 2070 nonredundancy RNA symbols and 395 nonredundancy protein symbols. In the current version of RAID, each entry contains detailed information on an RNA-associated (RNA–RNA/RNA–protein) interaction, including RAID ID, RNA/protein symbol, RNA/protein categories, validated method, expressing tissue, a literature reference (Pubmed ID), and detailed functional description (Fig. 1). To facilitate researchers in accessing information from external resources, we linked RNA and protein symbols to the HGNC database (Gray et al. 2013), which can efficiently retrieve plenty of genomic-associated data from external resources. In addition, RAID also welcomes researchers to submit experimentally identified novel RNA-associated (RNA–RNA/RNA–protein) interaction. All of the RNA-associated interactions can be downloaded directly in the Excel format, and RAID provides a publicly available interface (API) for automatic data retrieval in the Download and API page.
FIGURE 1.
The overview of RAID database.
SEARCHING PATHS AND BROWSING
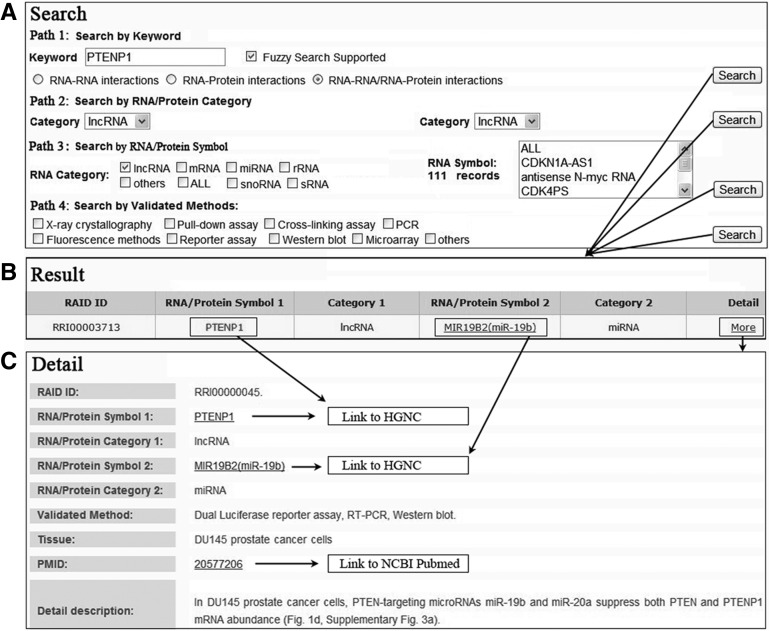
In the search page, RAID provides an interface for convenient retrieval of RNA-associated (RNA–RNA/RNA–protein) interactions. Users can browse and obtain any RNA-associated (RNA–RNA/RNA–protein) interaction through four paths (Fig. 2A). Path 1 (by keyword): browsing the RNA-associated interactions by inputting the keywords (any RNA and protein symbol) with fuzzy search supported. Users can obtain a list of RNA-associated (RNA–RNA/RNA–protein) interactions for any keywords. Path 2 (by RNA/protein category): Users can search all RNA-associated interactions between two defined RNA/protein categories. Similarly, users can retrieve all interactions between two defined RNA/protein symbols in Path 3 (by RNA/protein symbol). Path 4 (by validated method): browsing the RNA-associated interactions by experimental validated methods with multiple selection supported. The main table of results contains RAID ID, RNA/protein symbol 1 and category 1, RNA/protein symbol 2 and category 2, and detail “More” (Fig. 2B). When clicking the “More” link in each record, users can have access to more specific information such as RAID ID, RNA/protein symbol, RNA/protein category, validated method, expression tissue, a literature reference (Pubmed ID), and detailed functional description (Fig. 2C). Similarly, in the browser page, users can also browse any RNA-associated interaction by interaction type (RNA–RNA or RNA–protein), such as lncRNA-associated RNA–RNA interactions (229 entries).
FIGURE 2.
A flowchart for retrieving RNA-associated interaction entry. (A) Four searching paths for retrieving the RNA-associated interaction. (B) The result of a representative database entry. (C) The detailed information for an RNA-associated interaction. In the result and detail pages, RAID linked each RNA/protein symbol and PMID to their corresponding databases.
THE PREDICTED BINDING SITES AND NETWORK VISUALIZATION
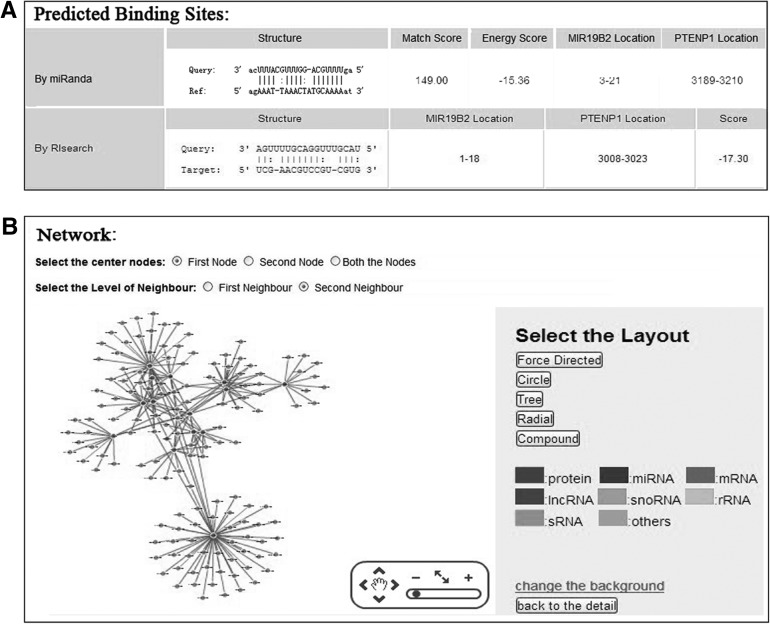
In addition to archive RNA-associated interaction, RAID also intends to integrate a variety of useful tools to analyze these data. Because the identification of RNA–RNA/RNA–protein binding sites can provide valuable insights for underlying the detailed regulating mechanism of the various RNAs, RAID also contains the predicted binding sites for RNA-associated interaction. Specifically, RAID adopts the predicted binding sites and scores by miRanda for a miRNA and its targets (John et al. 2004), while containing the predicted binding sites and score by RIsearch for the RNA–RNA interactions (Fig. 3; Wenzel et al. 2012). For RNA–protein interactions, bindN (Wang and Brown 2006), bindN+ (Wang et al. 2010), Pprint (Kumar et al. 2008), and RNAbindR (Terribilini et al. 2007) are commonly used tools to predict RNA-binding residues in proteins (Puton et al. 2012). Similarly, RAID also merges the predicted RNA-binding residues and scores from these tools. Additionally, RAID also integrates the experimentally verified RNA-binding sites in proteins documented in the RBPBD (Cook et al. 2011) and RsiteDB (Shulman-Peleg et al. 2009) databases. The parameters used by these predictive tools were documented in the Parameter of Help Page.
FIGURE 3.
Representative screenshots of the Binding and Network pages. (A) The Binding page: representing the predicted binding sites and/or constants. (B) The Network page: representing the interaction subnetwork of interacting RNA/protein.
Besides the detailed analysis of RNA interaction sites, RAID also supports the users to globally observe the RNA-associated (RNA–RNA/RNA–protein) interaction network. Cytoscape Web (cytoscapeweb.cytoscape.org/) is a visualization tool that is suitable for displaying small to medium sized networks in a web-based manner (Lopes et al. 2010). In the visualization option at Network Page (Fig. 3B), RNA-associated interaction subnetworks can be rapidly and independently represented by embedding interactive networks with the Cytoscape Web. The “First Node” or “Second Node” option represents the subnetwork of interacting RNA/protein with the first or second interaction RNA/protein, the “Both the Nodes” option represents the subnetwork of interacting RNA/protein with both interaction nodes. The “First Neighbour” represents the subnetwork of direct interacting with the center node, the “Second Neighbour” represents the subnetwork of direct and second-step interacting with the center node. Interaction of a subnetwork based on the two nodes of this interaction may help the researchers represent all interacting partners immediately. Thus, multiple RNA/protein data resources can be combined in a single visualization for each RNA/protein with its interaction partner. Since the compelling visualization architecture is pan-and-zoom, users can observe specific RNA/protein within the RNA-associated interaction network and the “Selection of the Layout” option can provide the different layout types for this subnetwork.
DISCUSSION AND FUTURE DIRECTIONS
High-throughput proteomics and protein–protein interaction screens have enabled rapid progress in mapping the protein interactome (Bossi et al. 2009; Vidal et al. 2011). However, the RNA-associated interactome is likely to be much larger and more complex due to the huge numbers of transcripts identified by global analyses (Konig et al. 2011; Bernstein et al. 2012; Derrien et al. 2012; Frazer 2012; Muller-McNicoll et al. 2013). Recent investigations indicated that there are complex regulations among diverse ncRNAs and protein-coding genes (Konig et al. 2011; Bernstein et al. 2012; Derrien et al. 2012; Frazer 2012; Muller-McNicoll et al. 2013). Consequently, we systematically collect experimentally verified human RNA-associated (RNA–RNA/RNA–protein) interactions and established the first database centering on the interaction network between diverse RNAs and RNAs/Proteins. RAID will be of particular interest to the life-science community and facilitates the biologists to unravel the role of RNAs/proteins in a variety of biological processes. In the future, we will continuously curate and update the reference data. Complemented with the successful PPI databases, RAID will provide a valuable skeleton for a better understanding of the functional organization of the cell.
ACKNOWLEDGMENTS
This work was supported by the National High Technology Research and Development Program of China (863 Program, 2014AA021102), the Major State Basic Research Development Program of China (973 Program, 2014CB910504), the National Natural Science Foundation of China (31100901, U1304311, 91129710, 61170154), China Postdoctoral Science Foundation (2013M531064), Natural Science Foundation of Heilongjiang Province of China (QC2010012 and ZD201114), Heilongjiang Postdoctoral Foundation (LBH-Z12171), Plan of Nature Science Fundamental Research in Henan University of Technology (11JCYJ11), and China Undergraduate Training Programs for Innovation (201210226016).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.044776.114.
REFERENCES
- Anders G, Mackowiak SD, Jens M, Maaskola J, Kuntzagk A, Rajewsky N, Landthaler M, Dieterich C 2012. doRiNA: a database of RNA interactions in post-transcriptional regulation. Nucleic Acids Res 40: D180–D186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi A, Lehner B 2009. Tissue specificity and the human protein interaction network. Mol Syst Biol 5: 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147: 358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR 2011. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res 39: D301–D308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M 2011. Non-coding RNAs in human disease. Nat Rev Genet 12: 861–874 [DOI] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. 2013. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41: D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA 2012. Decoding the human genome. Genome Res 22: 1599–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori S, Hino K, Saito A, Miyano S, Miyamoto-Sato E 2012. PRD: a protein–RNA interaction database. Bioinformation 8: 729–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KA, Daugherty LC, Gordon SM, Seal RL, Wright MW, Bruford EA 2013. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res 41: D545–D552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Li Y, Liu Y, Wang J, Li G 2010. Inhibition of metastasis-associated lung adenocarcinoma transcript 1 in CaSki human cervical cancer cells suppresses cell proliferation and invasion. Acta Biochim Biophys Sin 42: 224–229 [DOI] [PubMed] [Google Scholar]
- Guttman M, Rinn JL 2012. Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. 2011. miRTarBase: A database curates experimentally validated microRNA–target interactions. Nucleic Acids Res 39: D163–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al. 2014. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 42: D78–D85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS 2004. Human microRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Luscombe NM, Ule J 2011. Protein–RNA interactions: new genomic technologies and perspectives. Nat Rev Genet 13: 77–83 [DOI] [PubMed] [Google Scholar]
- Kumar M, Gromiha MM, Raghava GP 2008. Prediction of RNA binding sites in a protein using SVM and PSSM profile. Proteins 71: 189–194 [DOI] [PubMed] [Google Scholar]
- Li L, Feng T, Lian Y, Zhang G, Garen A, Song X 2009. Role of human noncoding RNAs in the control of tumorigenesis. Proc Natl Acad Sci 106: 12956–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhuang L, Wang Y, Hu Y, Wu Y, Wang D, Xu J 2013. Connect the dots: a systems level approach for analyzing the miRNA-mediated cell death network. Autophagy 9: 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JH, Liu S, Zhou H, Qu LH, Yang JH 2014. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42: D92–D97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD 2010. Cytoscape Web: an interactive web-based network browser. Bioinformatics 26: 2347–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-McNicoll M, Neugebauer KM 2013. How cells get the message: dynamic assembly and function of mRNA–protein complexes. Nat Rev Genet 14: 275–287 [DOI] [PubMed] [Google Scholar]
- Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T, et al. 2012. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res 40: D284–D289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puton T, Kozlowski L, Tuszynska I, Rother K, Bujnicki JM 2012. Computational methods for prediction of protein-RNA interactions. J Struct Biol 179: 261–268 [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146: 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman-Peleg A, Nussinov R, Wolfson HJ 2009. RsiteDB: a database of protein binding pockets that interact with RNA nucleotide bases. Nucleic Acids Res 37: D369–D373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, et al. 2011. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147: 370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terribilini M, Sander JD, Lee JH, Zaback P, Jernigan RL, Honavar V, Dobbs D 2007. RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res 35: W578–W584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T, Hatzigeorgiou AG 2012. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res 40: D222–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabasi AL 2011. Interactome networks and human disease. Cell 144: 986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown SJ 2006. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res 34: W243–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Huang C, Yang MQ, Yang JY 2010. BindN+ for accurate prediction of DNA and RNA-binding residues from protein sequence features. BMC Syst Biol 4 Suppl 1: S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen L, Chen B, Li X, Kang J, Fan K, Hu Y, Xu J, Yi L, Yang J, et al. 2013. Mammalian ncRNA-disease repository: a global view of ncRNA-mediated disease network. Cell Death Dis 4: e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A, Akbasli E, Gorodkin J 2012. RIsearch: fast RNA–RNA interaction search using a simplified nearest-neighbor energy model. Bioinformatics 28: 2738–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Wang J, Liu C, Zhang Y, Shi B, Zhu X, Zhang Z, Skogerbo G, Chen L, Lu H, et al. 2006. NPInter: the noncoding RNAs and protein related biomacromolecules interaction database. Nucleic Acids Res 34: D150–D152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li YH 2012. miRDeathDB: a database bridging microRNAs and the programmed cell death. Cell Death Differ 19: 1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang Y, Tan X, Jing H 2012. MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy 8: 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH 2011. starBase: a database for exploring microRNA–mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 39: D202–D209 [DOI] [PMC free article] [PubMed] [Google Scholar]