Abstract
Hepatitis C Virus (HCV) infection occurs frequently in patients with preexisting mental illness. Treatment for chronic hepatitis C using interferon formulations often increases risk for neuropsychiatric symptoms. Pegylated-Interferon-α (PegIFN-α) remains crucial for attaining sustained virologic response (SVR); however, PegIFN-α based treatment is associated with psychiatric adverse effects, which require dose reduction and/or interruption. This study's main objective was to identify genes induced by PegIFN-α and expressed in the central nervous system and immune system, which could mediate the development of psychiatric toxicity in association with antiviral outcome. Using peripheral blood mononuclear cells from Human Immunodeficiency Virus (HIV)/HCV co-infected donors (N=28), DNA microarray analysis was performed and 21 differentially regulated genes were identified in patients with psychiatric toxicity vs. those without. Using these 21 expression profiles a two-way-ANOVA was performed to select genes based on antiviral outcome and occurrence of neuropsychiatric adverse events. Microarray analysis demonstrated that Interferon-stimulated-exonuclease-gene 20kDa (ISG20) and Interferon-alpha-inducible-protein 27 (IFI27) were the most regulated genes (P<0.05) between three groups that were built by combining antiviral outcome and neuropsychiatric toxicity. Validation by bDNA assay confirmed that ISG20 expression levels were significantly associated with these outcomes (P<0.035). Baseline levels and induction of ISG20 correlated independently with no occurrence of psychiatric adverse events and non-response to therapy (P<0.001). Among the 21 genes that were associated with psychiatric adverse events and 20 Interferon-inducible genes (IFIGs) used as controls, only ISG20 expression was able to link PegIFN-α related neuropsychiatric toxicity to distinct HCV-responses in patients co-infected with HIV and HCV in vivo.
Keywords: HIV/HCV co-infection, Interferon-α, Neuropsychiatric Toxicity, ISG20
Introduction
Among 40 million people worldwide infected with the Human Immunodeficiency Virus (HIV), approximately 5 million are also co-infected chronically with the hepatitis C Virus (HCV) [Alter, 2006]. To date, treatment with pegylated Interferon-α (PegIFN-α) in combination with ribavirin (RBV) has been proven to achieve sustained virologic response (SVR) in up to 40% of all treated patients. However, PegIFN-α/RBV triggers the development of neuropsychiatric adverse effects including anxiety, irritability, mood lability, depression and suicidal behavior [Laguno et al., 2004; Weiss and Gorman, 2006]. Psychiatric toxicity occurs within the first few months of treatment and may affect up to 50% of all patients receiving PegIFN-α based therapy [Sulkowski and Thomas, 2005]. Such severe adverse events represent a frequent reason for dose reduction or treatment discontinuation [Zdilar et al., 2000], especially in patients co-infected with HIV and HCV, who exhibit higher rates of neuropsychiatric illness relative to HIV mono-infected persons or the general population [Goulet et al., 2005; Weiss and Gorman, 2006].
Previously, a diagnostic biomarker panel has been described as able to predict emergent psychiatric toxicity in patients co-infected with HIV and HCV undergoing PegIFN-α/RBV treatment [Rasimas et al., 2012]. This study was aimed to further explore the molecular basis of the clinical outcome observed in patients who experience a psychiatric toxicity and achieve sustained virologic response) [Rasimas et al., 2012], similar to findings published using HCV mono-infected patients [Loftis et al., 2004]. To date, it is unclear if psychiatric adverse events are more commonly associated with a favorable therapeutic outcome. If this is correct, then proper management of these adverse events and continued therapy is critical to obtain optimal sustained virologic response rates in these patients. Furthermore, it is also not clear whether the molecular mechanisms involved in pathogenesis of PegIFN-α associated mood disorders and those involved in permanent viral clearance are necessarily the same. Although direct acting antiviral therapies have been developed to offer a viable Interferon-free regimen, PegIFN-α remains the backbone of present treatment for chronic hepatitis C at the moment. In this regard, this study was aimed to discover novel molecular targets/mechanisms that govern viral response and neuropsychiatric toxicity in patients co-infected with HIV and HCV receiving PegIFN-α based treatment. To this end, emergence of psychiatric symptoms like depression, mood lability, anxiety or psychotic behavior was evaluated as detailed elsewhere [Rasimas et al., 2012].
This study is distinct from previous work [Rasimas et al., 2012] as the primary focus has been put on specific genes/pathways that are enriched in immune cells and CNS cells. Here, DNA microarray analysis has been performed using peripheral blood mononuclear cells (PBMCs) obtained from patients co-infected with HIV and HCV before and after treatment with PegIFN-α/RBV and a four-step annotation algorithm resulted in the identification of two molecular markers significantly involved in the occurrence of sustained virologic response and psychiatric toxicity, i.e., Interferon stimulated exonuclease gene 20kDa (ISG20) and Interferon alpha-inducible protein 27 (IFI27). These findings provide yet another evidence in support of the hypothesis that gene regulation in peripheral immune cells might be connected causally to neuropsychiatric events in the brain, since it has been shown that HIV infection is associated with disruption of the blood-brain-barrier and increased leukocyte penetration into the CNS [Lafrenie et al., 1997; Persidsky et al., 2000].
Materials and Methods
Study subjects
Patients (N=32) with stable HIV disease and chronic HCV infection were enrolled in Institutional Review Board approved studies at the National Institute of Allergy and Infectious Diseases in Bethesda, Maryland for treatment of HCV infection with pegylated Interferon-α2b (1.5μg/Kg/wk) and ribavirin 1000-1200mg/day. Four patients were taken off study: three were lost to follow up and one dropped out for social reasons after only one dose of PegIFN-α.
Ethics statement
All patients signed an Institutional Review Board approved protocol informed consent document.
Study inclusion and exclusion criteria
Patients co-infected with HIV and HCV were eligible if they were older than 18 years, had a CD4 count >100 cells/mm3, HCV viral load >2000 copies/ml, had histological evidence of chronic HCV infection and stable HIV disease being managed according to current HIV treatment guidelines. Participants with active psychiatric illness, i.e. mood lability, anxiety, or psychotic symptoms, had been treated and stabilized prior to enrollment; 3 patients got new prescriptions for citalopram, 1 was prescribed a higher dose of escitalopram, 1 was given risperidone and 1 maintained mirtazapine throughout the anti-HCV treatment as reported previously [Rasimas et al., 2012]. Four patients who participated in this study (including two of those given citalopram) increased their frequency of psychotherapy visits before study entry [Rasimas et al., 2012].
Psychiatric evaluations
All patients underwent standard pretreatment psychiatric evaluations conducted by board-certified psychiatrists from the National Institute of Mental Health as noted elsewhere [Rasimas et al., 2012]. The clinical assessment of mood stability was corroborated by Beck Depression Inventory (BDI)-II scores of less than or equal to 9 at the time of PegIFN-α/RBV treatment initiation [Rasimas et al., 2012]. Patients (N=28) who received at least 4 weeks of PegIFN-α/RBV treatment were classified into two groups, i.e., those with psychiatric toxicity and those without, after initiation of therapy in a blinded fashion prior to gene expression analysis as stated previously [Rasimas et al., 2012].
Microarray analysis
PBMCs from patients co-infected with HIV and HCV (N=28) were obtained at baseline and post treatment (within five days after last administration of PegIFN-α/RBV), and were subjected to DNA microarray analysis (Affymetrix HG-U133A) as described previously [Lempicki et al., 2006]. Post treatment, 3 out of 28 samples were excluded from the analysis as they failed chip quality control tests. Data analysis was carried out in four steps. STEP ONE: one-way-ANOVA (PARTEK Genomics Suite) was performed based on two outcome parameters: presence or absence of psychiatric toxicity. A Mean Fold Difference (MFD) representing the absolute mean difference in gene expression (log2) between both groups (patients with or without psychiatric toxicity) was calculated for selected genes at two different time points, i.e, pre and post treatment. Furthermore, a Mean Fold Change (MFC) was calculated for every gene within each group as the absolute mean difference in gene expression (log2) between two different time points (pre versus post treatment). This statistical approach is unique from the one that was employed in previous work [Rasimas et al., 2012], which was aimed exclusively to identify the genes that predict emergence of neuropsychiatric toxicity. For all analyses in the present study, a P-value of <0.05, an absolute log2MFD of >0.38 and an absolute log2MFC of >0.38 were considered significant. Subsequently, sixteen gene subsets were identified that passed these strict cutoffs for significance. STEP TWO: Venn diagrams were assembled between these sixteen gene subsets in order to identify expression profiles with consistently significant modulation in at least two of the categories (psychiatric toxicity, sustained virologic response and or induction by therapy). STEP THREE: Further selection was based on gene characterization using the functional annotation tool DAVID [Huang da et al., 2009]. Finally, four gene subsets (including twenty two probe IDs) involving 21 unique genes reached statistical significance based on the above criteria and exhibited significant enrichment in brain tissue or were associated with neuropsychiatric disorders based on DAVID [Huang da et al., 2009]. There were five genes that were common to previous work [Rasimas et al., 2012] (Supplementary Object 1). STEP FOUR: Using expression data of those 21 unique genes, two-way-ANOVA was performed based on pair-wise comparisons between three groups: patients with psychiatric toxicity who reached sustained virological response (N=8, 28%) vs. patients with psychiatric toxicity who did not (N=10, 36%) vs. patients who remained uneventful non-responders (N=10, 36%). There were no patients who achieved sustained virologic response but did not experience a psychiatric adverse event in this study. This could be a random epidemiological outcome and lacks any other reasonable explanation for this observation. Selected results were validated by a custom bDNA assay 1.0 for IFIGs.
QuantiGene-Plex Assay (QGP 1.0)
Validation of DNA microarray data was performed using bDNA assay capable of detecting expression levels of 20 genes. This 20-plex IFIGs panel was custom-ordered from Panomics, Inc. (Fremont, CA; http://www.panomics.com/index.php?id=qgpsc&page=5). PBMCs from all patients at both time points were pelleted and lysed in 2:1 PBS and Panomics' Lysis Mixture. The assay was performed following the manufactures' protocol as described elsewhere [Kottilil et al., 2009].
Results
Overall, 18 patients co-infected with HIV and HCV developed psychiatric adverse events during treatment with PegIFN-α/RBV, while 10 patients did not. Baseline characteristics of patients who developed toxicity and those who did not were similar with regards to age, sex, race, Body Mass Index, HCV genotype, HCV viral load, CD4+ T-cell counts, HIV viral load, highly active antiretroviral therapy (HAART), underlying psychiatric history and psychiatric medication prior to enrollment. (For details please see under Materials and Methods → Study inclusion and exclusion criteria and elsewhere [Rasimas et al., 2012]). In this study, the incidence of PegIFN-α induced depression (29%) was lower than reported by other investigators [Laguno et al., 2004].
Using a combined statistical and functional characterization algorithm as described in the Materials and Materials section a total of 21 unique genes were selected as regulated significantly in at least two outcome based analyses. A supervised clustering (heat map I-IV) was performed after mean-shift normalization of the data in order to highlight expression differences between these genes pre and post therapy (Figure 1). Based on DAVID, all genes included in heat map I [Anikster et al., 2002; Iwamoto et al., 2004; Millar et al., 2005], heat map III [Gardner and Ghorpade, 2003; Labrada et al., 2002; Maisel et al., 2007; Sakamoto et al., 2007] and heat map IV [Beasley et al., 2005; Cicin-Sain et al., 2008; Han et al., 2007; Jentsch, 1992; Le-Niculescu et al., 2009; Simmen et al., 2008; Wegner et al., 2008; Wu, 2006; Zhang et al., 2004] were enriched significantly in various brain tissues (PI<0.03; PIII<0.02; PIV<0.01), while all genes included in heat map II [Ainiala et al., 2004; Georgieva et al., 2008; Hamilton et al., 2004; Millar et al., 2005; Skibinski et al., 2005; Spleiss et al., 1998] were associated significantly with a number of neuropsychiatric symptoms or syndromes (PII<0.01) (Figure 1). Interestingly, 11 (52%) out of 21 selected genes were enriched significantly in CD4+ T-cells (P<0.025) according to DAVID (Supplementary Object 2).
Figure 1. Expression differences of 21 genes between patients with psychiatric events (PsAE) and without (NoPsAE), pre and post therapy.
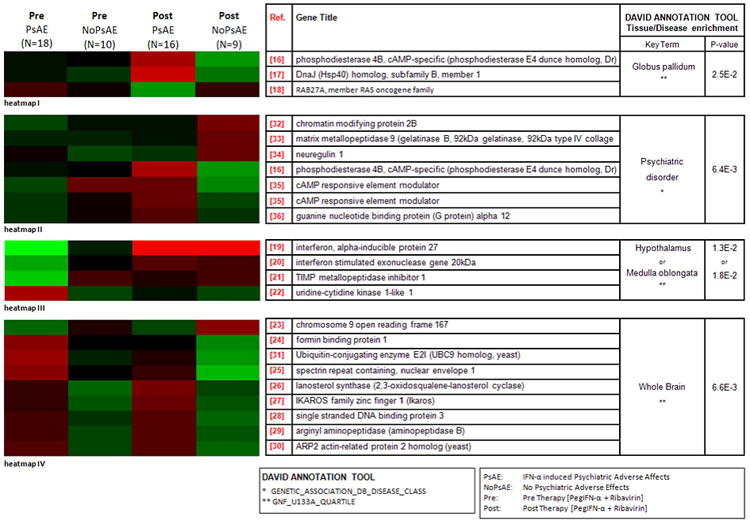
In patients (N=28) co-infected with the Human Immunodeficiency Virus/Hepatitis C Virus (HIV/HCV) DNA microarray analysis was performed using peripheral blood mononuclear cells (PBMCs). Serial overlapping of selected genes based on multi-contrast statistical and functional characterization algorithms led to identification of three gene subsets (heat map I-III) representing a panel of 12 unique molecular factors that reflect different biological states with respect to the development of Interferon-α (IFN-α) associated psychiatric adverse effects (PsAE). Furthermore, a 9-gene signature (heat map IV) characterized by sustained expression differences between eventful and uneventful patients pre and post therapy with pegylated Interferon-α and ribavirin (PegIFN-α/RBV) has been identified as containing genetic imprints associated likely with HIV and/or HCV related neurobehavioral disorders.
The objective of this study was to extend previous observations of a significant correlation between a sustained virologic response and the occurrence of psychiatric adverse events (P<0.025) and further elucidate the molecular mechanisms involved in the in vivo interaction between PegIFN-α induced viral response and neuropsychiatric toxicity. In this regard, two-way-ANOVA was performed after assigning patients into three groups, those who achieved sustained virologic response and had psychiatric toxicity (N=8) vs. those who did not achieve sustained virologic response but had psychiatric toxicity (N=10) vs. those who did not achieve sustained virologic response nor had toxicity (N=10), using expression values of 21 unique genes in three different settings: pre (baseline), post (end of treatment), and pre-to-post (induced) PegIFN-α/RBV therapy. Pre treatment: Looking for expression profiles that had an ANOVA P-value (PA) of <0.05 and were regulated significantly (P<0.05 and absolute log2MFD>0.38) in at least two contrasts, only 2 genes could be selected: ISG20 (Figure 2A, Figure 2C) and IFI27 (Supplementary Object 3A). Pre-to-post treatment: Interestingly, ISG20 and IFI27 were the only genes that showed a PA of <0.05 and were induced significantly (P<0.05 and absolute log2MFC>0.38) in at least two groups: patients who had psychiatric toxicity with or without sustained virologic response. In those who had neither sustained virologic response nor toxicity, only IFI27 was induced significantly (P<0.0001; absolute log2MFC=4.6) (Supplementary Object 3A). Post treatment: Utilizing identical criteria and cut-offs as above, only the gene “lanosterol synthase (LSS)” was found as regulated significantly in the same inter-group contrasts (data not shown). Based on these findings, genes with known significant expression in the CNS such as ISG20 and IFI27 might represent novel potential biomarkers and/or regulatory molecules for the in vivo interaction between behavioral toxicity and virologic response during PegIFN-α/RBV therapy in HIV/HCV co-infection.
Figure 2. Expression levels of ISG20, pre and post therapy.
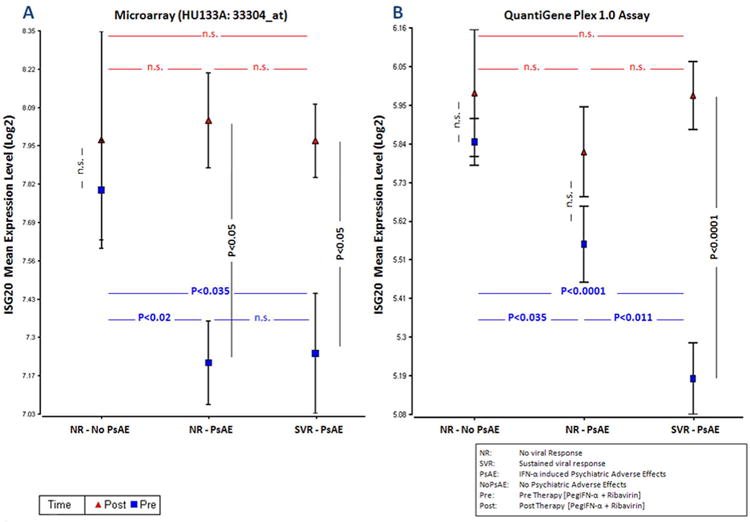
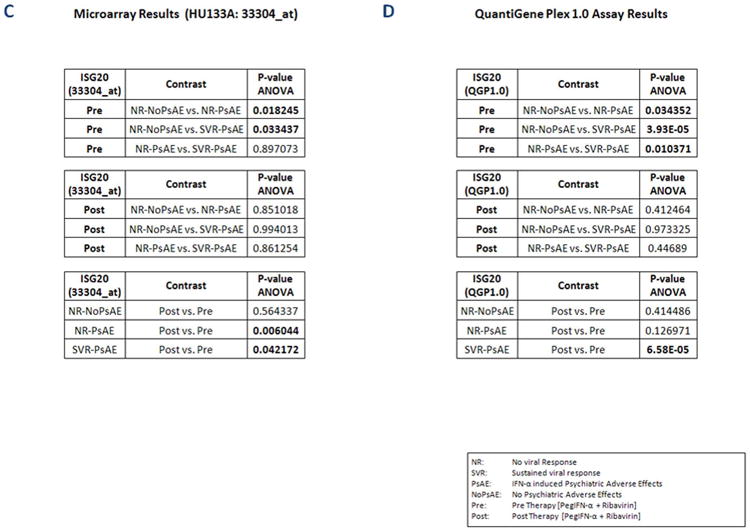
Microarray analysis yielded expression of Interferon stimulated exonuclease gene 20kDa (ISG20) at highest level in the group of uneventful non-responders (NR-NoPsAE) at baseline (Figure 2A). Once validated, eventful responders (SVR-PsAE) showed the lowest ISG20 expression at baseline (Figure 2B). Statistical results for all contrasts pre (baseline), pre-to-post (induction) or post therapy are shown in Figure 2C and 2D.
Microarray data were confirmed using the QGP 1.0 assay that detected overall significant differences in gene expression of ISG20 (PA<0.0002) at baseline (P<0.05 and absolute log2MFD>0.38) along with a lack of induction in patients who did not achieve sustained virologic response regardless of toxicity (Figure 2B, Figure 2D). Of note, linear regression analysis showed overall consistent detection performance of ISG20 expression by two different probe sets (33304_at and 204698_at) (P<5.8E-11), pre and post treatment. In a further effort to delineate its functional specificity regarding CNS toxicity, gene expression of ISG20 was compared to that of nineteen other IFIGs that also had been evaluated by the QGP 1.0 assay, at baseline; of those, ISG20 was the only gene regulated significantly among non-responders who experienced a psychiatric toxicity when compared to the ones who did not (Table 1). In addition, validation of microarray data using the QGP1.0 Assay confirmed independently a significant up-regulation of ISG20 at baseline in patients who did not develop a psychiatric adverse event (P<0.001) and those who did not achieve sustained virological response (P<0.0003) (Table 2A). However, these were not observed with IFI27 expression at baseline (Supplementary Object 3B).
Table 1. Correlation between baseline expression levels of IFIGs (N=20) and response-toxicity outcomes.
For control reasons, evaluation of gene expression of Interferon inducible genes (IFIGs, N=20) was performed using a bDNA multiplex assay (QGP 1.0, Panomics®). Only expression of Interferon stimulated exonuclease gene 20kDa (ISG20) differed significantly in all comparisons (P<0.035) before initiation of therapy with pegylated Interferon-α and ribavirin (PegIFN-α/RBV). Gene names and symbols listed in this table correspond to terms explained in EntrezGene: http://www.ncbi.nlm.nih.gov/gene
| overall ANOVA P-value | NR-NoPsAE vs. NR-PsAE | NR-NoPsAE vs. SVR-PsAE | NR-PsAE vs. SVR-PsAE | |
|---|---|---|---|---|
| Gene Symbol | P-value | P-value | P-value | P-value |
| TRIM5 | 9.64E-05 | 0.618378 | 5.35E-05 | 0.000242357 |
| APOBEC3G | 0.000130875 | 0.416413 | 5.37E-05 | 0.000526096 |
| OAS2 | 0.00118548 | 0.481903 | 0.000462119 | 0.00316296 |
| IFIT1 | 0.00125874 | 0.268183 | 0.000371204 | 0.00669527 |
| ISG20 | 0.000180288 | 0.0343525 | 3.93E-05 | 0.0103719 |
| OAS1 | 0.00708266 | 0.579752 | 0.00281471 | 0.0120099 |
| IFITM1 | 0.00399497 | 0.380528 | 0.00129636 | 0.0122214 |
| IFIT3 | 0.0784446 | 0.497076 | 0.0278317 | 0.115344 |
| MX1 | 0.0556843 | 0.335928 | 0.0177016 | 0.132397 |
| MX2 | 0.0628115 | 0.341076 | 0.0201968 | 0.144085 |
| STAT1 | 0.282406 | 0.930856 | 0.149474 | 0.183131 |
| EIF2AK2 | 0.0879966 | 0.301756 | 0.0290891 | 0.216966 |
| ISG15 | 0.0760249 | 0.260831 | 0.0246772 | 0.223032 |
| IFI44 | 0.20454 | 0.428514 | 0.0779284 | 0.31044 |
| PLSCR1 | 0.725129 | 0.812797 | 0.570623 | 0.438556 |
| LY6E | 0.208519 | 0.206418 | 0.0943669 | 0.640991 |
| APOBEC3A | 0.80747 | 0.532544 | 0.893426 | 0.644969 |
| SP110 | 0.832107 | 0.841938 | 0.552333 | 0.694313 |
| IFI27 | 0.803717 | 0.752103 | 0.513094 | 0.732396 |
| IRF7 | 0.704648 | 0.412934 | 0.624917 | 0.763441 |
[SVR: sustained viral response; NR: viral non-response; PsAE: IFN-α induced psychiatric adverse effects; NoPsAE: No psychiatric adverse effects]
Table 2. Association between unique clinical outcomes (response or toxicity) and gene expression levels of ISG20, pre and post therapy.
Association between unique clinical outcomes, i.e. viral response (SVR vs. NR) or neuropsychiatric toxicity (PsAE vs. NoPsAE), and gene expression of Interferon stimulated exonuclease gene 20kDa (ISG20) determined by microarray analysis and validated by bDNA assay at baseline (N=28 patients / Table 2A) and post therapy with pegylated Interferon-α and ribavirin (PegIFN-α/RBV) (N=25 patients / Table 2B).
| A | Pre therapy (PegIFN-α/RBV) | ISG20 gene expression | P-value (ANOVA) |
|---|---|---|---|
| Response | (microarray; HU133A/A_2, Affymetrix®) | Up-regulated in NR | 0.2016 |
| Toxicity | (microarray; HU133A/A_2, Affymetrix®) | Up-regulated in NoPsAE | 0.0084 |
| Response | (bDNA assay; QGP 1.0, Panomics®) | Up-regulated in NR | 0.0003 |
| Toxicity | (bDNA assay; QGP 1.0, Panomics®) | Up-regulated in NoPsAE | 0.0010 |
| B | Post therapy (PegIFN-α/RBV) | ISG20 gene expression | P-value (ANOVA) |
|---|---|---|---|
| Response | (microarray; HU133A/A_2, Affymetrix®) | Up-regulated in NR | 0.7604 |
| Toxicity | (microarray; HU133A/A_2, Affymetrix®) | Up-regulated in PsAE | 0.8975 |
| Response | (bDNA assay; QGP 1.0, Panomics®) | Up-regulated in SVR | 0.5887 |
| Toxicity | (bDNA assay; QGP 1.0, Panomics®) | Up-regulated in NoPsAE | 0.6180 |
[SVR: sustained viral response; NR: viral non-response; PsAE: IFN-α induced psychiatric adverse effects; NoPsAE: No psychiatric adverse effects]
Discussion
This study demonstrates a significant association between occurrence of a psychiatric adverse event and a sustained virologic response in subjects co-infected with HIV and HCV and treated with PegIFN-α/RBV. Further, a novel molecular marker, ISG20, has been identified whose baseline expression is able to discriminate patients co-infected with HIV and HCV based on virologic response and psychiatric toxicity.
As previously reported, occurrence of toxicity to PegIFN-α based therapy in subjects co-infected with HIV and HCV is closely associated with achieving virologic suppression and effectiveness of therapy [Rasimas et al., 2012]. As consistent observations have been reported by other investigators in this regard [Loftis et al., 2004] a DNA microarray analysis was utilized to shed light on molecular mechanisms that could govern this crucial clinical interaction outcome. Hereby, a novel molecular marker, ISG20, could be identified and validated as capable to distinguish clearly between three different “response-toxicity” interaction outcomes.
Thus far, cumulative research data suggested that development of toxicity might serve as clinical marker for a well functioning IFN-α pathway and therefore also for effective responses to antiviral treatment in patients co-infected with HIV and HCV [Osinusi et al.]. In addition, it has been shown previously that up-regulation of IFIGs expression at baseline is associated with a lack of response to therapy in this difficult to treat population [Lempicki et al., 2006]. This study demonstrated that ISG20 expression levels were clearly associated with occurrence of a sustained virologic response as well as occurrence of a psychiatric toxicity. In this regard, the lowest ISG20 expression levels at baseline and the strongest ISG20 induction by PegIFN-α were associated with occurrence of sustained virologic response along with psychiatric toxicity. These important observations provide a molecular basis for PegIFN-α induced neuro-psychiatric toxicity in association with sustained vilological responses in patients co-infected with HIV and HCV.
When exploring further the relevance of ISG20 regulation on CNS toxicity, evidence appeared to support the association of a robust ISG20 expression with the survival of motor neuron (SMN)-containing macromolecular nuclear complexes. While these complexes are required for biogenesis of various small nuclear ribonucleoproteins [Espert et al., 2006], a few candidates have been reported as deregulated significantly in association with major depression in microarray based studies using post mortem prefrontal cortices [Tochigi et al., 2008]. These results suggest that the loss of SMN is incompatible with life while decreased levels lead to serious inherited motor neuron diseases [Monani, 2005]. Also, accelerated SMN protein expression has been described during differentiation of human mesenchymal stem cells into functional neuron-like cells [Alexanian et al., 2008]. Hence, it is not surprising that ISG20 is enriched and expressed at high levels in neural progenitor cells (NPC) in several regions of the adult human brain such as the cortex, the hippocampus and the subventricular zone of the lateral ventricles [Maisel et al., 2007]. NPC are pluripotent stem cells that are capable to self-renew and evolve into three cell types of the central nervous system (neurons, oligodendrocytes and astrocytes) [Maisel et al., 2007]. Cellular integrity in these zones has been shown to be relevant in the pathogenesis of several mental illnesses. Specifically, decreased cell volume in the hippocampus and the subventricular zone have been documented in depression using animal models [Lau et al., 2007]. Furthermore, these anatomic changes can be reversed with serotonergic antidepressant treatment [Paizanis et al., 2007], which is effective for helping patients to complete HCV therapy when neuropsychiatric adverse events occur [Osinusi et al.]. Thus, findings from this study indicate that increased ISG20 expression levels in PBMCs from IFN-α naïve HIV/HCV co-infected individuals may serve as a molecular marker for protection against the occurrence of psychiatric adverse events. Although, the authors did not study CNS tissues, a peripheral Interferon stimulated gene (ISG) expression may be associated with high CNS expression of ISG20 and could lead to a reduced susceptibility to PegIFN-α induced neuropsychiatric toxicity. Most interestingly, ISG20 was the only gene among twenty different Interferon stimulated genes (used as controls) confirmedly capable of discriminating between the occurrence of a neuropsychiatric event among those who did not achieve an sustained virologic response, suggesting a mechanistic role for ISG20 regulation on the development of neuropsychiatric toxicity in patients co-infected with HIV and HCV.
In support of the hypothesis that PegIFN therapy can lead to induction of ISGs in the CNS, it has been shown that type I IFNs can promote directly NPC survival in mice [Hirsch et al., 2009]. This study reiterates the fact that systemic IFN-α can gain access to and act directly within the CNS in mice and in humans [Raison et al., 2009; Wang et al., 2008].
Furthermore, there is evidence to suggest that both ISG20 and SMN proteins are expressed (inside the cell nucleus) in proximity of or overlap with coiled bodies also known as Cajal bodies [Young et al., 2001]. Although the human SMN protein is present in virtually all tissues, it is expressed at highest levels in the CNS and liver [Young et al., 2001]. An accumulation of ISG20 in Cajal bodies after IFN treatment [Espert et al., 2006], may interact with SMN proteins mediating neuropsychiatric event in the CNS along with hepatic viral clearance in patients co-infected with HIV and HCV receiving treatment with PegIFN-α/RBV.
Recent studies revealed that ISG20 encodes a 3′ to 5′ exoribonuclease member of the DEDD superfamily of exonucleases that degrades specifically single-stranded RNA [Espert et al., 2006; Zhou et al.]. When expressed at high levels in vitro, ISG20 was capable of restricting infections by HIV [Espert et al., 2005] and also inhibiting HCV replication [Jiang et al., 2008]. In this regard, PegIFN-α induced up-regulation of ISG20 has been shown to be associated with favorable therapeutic outcomes in several studies [Lempicki et al., 2006].
In summary, ISG20 expression is associated with IFN-α response culminating in HCV clearance (in the liver) as well as regulation of neuropsychiatric signals (in the CNS). These data may contribute to the development of individualized prevention strategies against psychiatric toxicities in patients likely to achieve sustained virologic response and could be particularly beneficial for patients co-infected with HIV and HCV as these individuals are considered especially vulnerable to depression and other mood disorders [Goulet et al., 2005; Weiss and Gorman, 2006]. Finally, these results could also be of benefit for the management of patients receiving type I IFN based therapy for other diseases such as melanoma or multiple sclerosis [Zdilar et al., 2000].
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH (National Institute of Allergy and Infectious Diseases, and the Clinical Research Center; Grant number: HHSN261200800001E). The content of this publication does not necessarily reflect the views orpolicies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Disclaimer: The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government.
Conflict of interest statement: None of the authors have any conflicts of interest to report.
References
- Ainiala H, Hietaharju A, Dastidar P, Loukkola J, Lehtimaki T, Peltola J, Korpela M, Heinonen T, Nikkari ST. Increased serum matrix metalloproteinase 9 levels in systemic lupus erythematosus patients with neuropsychiatric manifestations and brain magnetic resonance imaging abnormalities. Arthritis Rheum. 2004;50:858–865. doi: 10.1002/art.20045. [DOI] [PubMed] [Google Scholar]
- Alexanian AR, Maiman DJ, Kurpad SN, Gennarelli TA. In vitro and in vivo characterization of neurally modified mesenchymal stem cells induced by epigenetic modifiers and neural stem cell environment. Stem Cells Dev. 2008;17:1123–1130. doi: 10.1089/scd.2007.0212. [DOI] [PubMed] [Google Scholar]
- Alter MJ. Epidemiology of viral hepatitis and HIV co-infection. J Hepatol. 2006;44(1 Suppl):S6–9. doi: 10.1016/j.jhep.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Anikster Y, Huizing M, Anderson PD, Fitzpatrick DL, Klar A, Gross-Kieselstein E, Berkun Y, Shazberg G, Gahl WA, Hurvitz H. Evidence that Griscelli syndrome with neurological involvement is caused by mutations in RAB27A, not MYO5A. Am J Hum Genet. 2002;71:407–414. doi: 10.1086/341606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley CL, Honer WG, Bergmann K, Falkai P, Lutjohann D, Bayer TA. Reductions in cholesterol and synaptic markers in association cortex in mood disorders. Bipolar Disord. 2005;7:449–455. doi: 10.1111/j.1399-5618.2005.00239.x. [DOI] [PubMed] [Google Scholar]
- Cicin-Sain L, Simaga S, Froebe A, Abramic M. Central aminopeptidase and serotonin system activities: possible relationship. Neuropeptides. 2008;42:435–440. doi: 10.1016/j.npep.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Espert L, Degols G, Lin YL, Vincent T, Benkirane M, Mechti N. Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J Gen Virol. 2005;86:2221–2229. doi: 10.1099/vir.0.81074-0. [DOI] [PubMed] [Google Scholar]
- Espert L, Eldin P, Gongora C, Bayard B, Harper F, Chelbi-Alix MK, Bertrand E, Degols G, Mechti N. The exonuclease ISG20 mainly localizes in the nucleolus and the Cajal (Coiled) bodies and is associated with nuclear SMN protein-containing complexes. J Cell Biochem. 2006;98:1320–1333. doi: 10.1002/jcb.20869. [DOI] [PubMed] [Google Scholar]
- Gardner J, Ghorpade A. Tissue inhibitor of metalloproteinase (TIMP)-1: the TIMPed balance of matrix metalloproteinases in the central nervous system. J Neurosci Res. 2003;74:801–806. doi: 10.1002/jnr.10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva L, Dimitrova A, Ivanov D, Nikolov I, Williams NM, Grozeva D, Zaharieva I, Toncheva D, Owen MJ, Kirov G, O'Donovan MC. Support for neuregulin 1 as a susceptibility gene for bipolar disorder and schizophrenia. Biol Psychiatry. 2008;64:419–427. doi: 10.1016/j.biopsych.2008.03.025. [DOI] [PubMed] [Google Scholar]
- Goulet JL, Fultz SL, McGinnis KA, Justice AC. Relative prevalence of comorbidities and treatment contraindications in HIV-mono-infected and HIV/HCV-co-infected veterans. AIDS. 2005;19 Suppl 3:S99–105. doi: 10.1097/01.aids.0000192077.11067.e5. [DOI] [PubMed] [Google Scholar]
- Hamilton SP, Slager SL, Mayo D, Heiman GA, Klein DF, Hodge SE, Fyer AJ, Weissman MM, Knowles JA. Investigation of polymorphisms in the CREM gene in panic disorder. Am J Med Genet B Neuropsychiatr Genet. 2004;126B:111–115. doi: 10.1002/ajmg.b.20121. [DOI] [PubMed] [Google Scholar]
- Han L, Wong DL, Tsai G, Jiang Z, Coyle JT. Promoter analysis of human glutamate carboxypeptidase II. Brain Res. 2007;1170:1–12. doi: 10.1016/j.brainres.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch M, Knight J, Tobita M, Soltys J, Panitch H, Mao-Draayer Y. The effect of interferon-beta on mouse neural progenitor cell survival and differentiation. Biochem Biophys Res Commun. 2009;388:181–186. doi: 10.1016/j.bbrc.2009.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Iwamoto K, Kakiuchi C, Bundo M, Ikeda K, Kato T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol Psychiatry. 2004;9:406–416. doi: 10.1038/sj.mp.4001437. [DOI] [PubMed] [Google Scholar]
- Jentsch S. The ubiquitin-conjugation system. Annu Rev Genet. 1992;26:179–207. doi: 10.1146/annurev.ge.26.120192.001143. [DOI] [PubMed] [Google Scholar]
- Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, Guo JT. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol. 2008;82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottilil S, Yan MY, Reitano KN, Zhang X, Lempicki R, Roby G, Daucher M, Yang J, Cortez KJ, Ghany M, Polis MA, Fauci AS. Human immunodeficiency virus and hepatitis C infections induce distinct immunologic imprints in peripheral mononuclear cells. Hepatology. 2009;50:34–45. doi: 10.1002/hep.23055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrada L, Liang XH, Zheng W, Johnston C, Levine B. Age-dependent resistance to lethal alphavirus encephalitis in mice: analysis of gene expression in the central nervous system and identification of a novel interferon-inducible protective gene, mouse ISG12. J Virol. 2002;76:11688–11703. doi: 10.1128/JVI.76.22.11688-11703.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafrenie RM, Wahl LM, Epstein JS, Yamada KM, Dhawan S. Activation of monocytes by HIV-Tat treatment is mediated by cytokine expression. J Immunol. 1997;159:4077–4083. [PubMed] [Google Scholar]
- Laguno M, Blanch J, Murillas J, Blanco JL, Leon A, Lonca M, Larrousse M, Biglia A, Martinez E, Garcia F, Miro JM, de Pablo J, Gatell JM, Mallolas J. Depressive symptoms after initiation of interferon therapy in human immunodeficiency virus-infected patients with chronic hepatitis C. Antivir Ther. 2004;9:905–909. [PubMed] [Google Scholar]
- Lau WM, Qiu G, Helmeste DM, Lee TM, Tang SW, So KF. Corticosteroid decreases subventricular zone cell proliferation, which could be reversed by paroxetine. Restor Neurol Neurosci. 2007;25:17–23. [PubMed] [Google Scholar]
- Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, McMahon FJ, Schork NJ, Nurnberger JI, Jr, Niculescu AB., 3rd Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:155–181. doi: 10.1002/ajmg.b.30887. [DOI] [PubMed] [Google Scholar]
- Lempicki RA, Polis MA, Yang J, McLaughlin M, Koratich C, Huang DW, Fullmer B, Wu L, Rehm CA, Masur H, Lane HC, Sherman KE, Fauci AS, Kottilil S. Gene expression profiles in hepatitis C virus (HCV) and HIV coinfection: class prediction analyses before treatment predict the outcome of anti-HCV therapy among HIV-coinfected persons. J Infect Dis. 2006;193:1172–1177. doi: 10.1086/501365. [DOI] [PubMed] [Google Scholar]
- Loftis JM, Socherman RE, Howell CD, Whitehead AJ, Hill JA, Dominitz JA, Hauser P. Association of interferon-alpha-induced depression and improved treatment response in patients with hepatitis C. Neurosci Lett. 2004;365:87–91. doi: 10.1016/j.neulet.2004.04.058. [DOI] [PubMed] [Google Scholar]
- Maisel M, Herr A, Milosevic J, Hermann A, Habisch HJ, Schwarz S, Kirsch M, Antoniadis G, Brenner R, Hallmeyer-Elgner S, Lerche H, Schwarz J, Storch A. Transcription profiling of adult and fetal human neuroprogenitors identifies divergent paths to maintain the neuroprogenitor cell state. Stem Cells. 2007;25:1231–1240. doi: 10.1634/stemcells.2006-0617. [DOI] [PubMed] [Google Scholar]
- Millar JK, Pickard BS, Mackie S, James R, Christie S, Buchanan SR, Malloy MP, Chubb JE, Huston E, Baillie GS, Thomson PA, Hill EV, Brandon NJ, Rain JC, Camargo LM, Whiting PJ, Houslay MD, Blackwood DH, Muir WJ, Porteous DJ. DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science. 2005;310:1187–1191. doi: 10.1126/science.1112915. [DOI] [PubMed] [Google Scholar]
- Monani UR. Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Osinusi A, Rasimas JJ, Bishop R, Proschan M, McLaughlin M, Murphy A, Cortez KJ, Polis MA, Masur H, Rosenstein D, Kottilil S. HIV/Hepatitis C virus-coinfected virologic responders to pegylated interferon and ribavirin therapy more frequently incur interferon-related adverse events than nonresponders do. J Acquir Immune Defic Syndr. 53:357–363. doi: 10.1097/QAI.0b013e3181c7a29d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paizanis E, Hamon M, Lanfumey L. Hippocampal neurogenesis, depressive disorders, and antidepressant therapy. Neural Plast. 2007;2007:73754. doi: 10.1155/2007/73754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persidsky Y, Zheng J, Miller D, Gendelman HE. Mononuclear phagocytes mediate blood-brain barrier compromise and neuronal injury during HIV-1-associated dementia. J Leukoc Biol. 2000;68:413–422. [PubMed] [Google Scholar]
- Raison CL, Borisov AS, Majer M, Drake DF, Pagnoni G, Woolwine BJ, Vogt GJ, Massung B, Miller AH. Activation of central nervous system inflammatory pathways by interferon-alpha: relationship to monoamines and depression. Biol Psychiatry. 2009;65:296–303. doi: 10.1016/j.biopsych.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasimas J, Katsounas A, Raza H, Murphy AA, Yang J, Lempicki RA, Osinusi A, Masur H, Polis M, Kottilil S, Rosenstein D. Gene expression profiles predict emergence of psychiatric adverse events in HIV/HCV-coinfected patients on interferon-based HCV therapy. J Acquir Immune Defic Syndr. 2012;60:273–281. doi: 10.1097/QAI.0b013e31824c17c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Cansev M, Wurtman RJ. Oral supplementation with docosahexaenoic acid and uridine-5′-monophosphate increases dendritic spine density in adult gerbil hippocampus. Brain Res. 2007;1182:50–59. doi: 10.1016/j.brainres.2007.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen FA, Su Y, Xiao R, Zeng Z, Simmen RC. The Kruppel-like factor 9 (KLF9) network in HEC-1-A endometrial carcinoma cells suggests the carcinogenic potential of dys-regulated KLF9 expression. Reprod Biol Endocrinol. 2008;6:41. doi: 10.1186/1477-7827-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski G, Parkinson NJ, Brown JM, Chakrabarti L, Lloyd SL, Hummerich H, Nielsen JE, Hodges JR, Spillantini MG, Thusgaard T, Brandner S, Brun A, Rossor MN, Gade A, Johannsen P, Sorensen SA, Gydesen S, Fisher EM, Collinge J. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- Spleiss O, van Calker D, Scharer L, Adamovic K, Berger M, Gebicke-Haerter PJ. Abnormal G protein alpha(s) - and alpha(i2)-subunit mRNA expression in bipolar affective disorder. Mol Psychiatry. 1998;3:512–520. doi: 10.1038/sj.mp.4000393. [DOI] [PubMed] [Google Scholar]
- Sulkowski MS, Thomas DL. Perspectives on HIV/hepatitis C virus co-infection, illicit drug use and mental illness. AIDS (London, England) 2005;19 Suppl 3:S8–12. doi: 10.1097/01.aids.0000192064.09281.48. [DOI] [PubMed] [Google Scholar]
- Tochigi M, Iwamoto K, Bundo M, Sasaki T, Kato N, Kato T. Gene expression profiling of major depression and suicide in the prefrontal cortex of postmortem brains. Neurosci Res. 2008;60:184–191. doi: 10.1016/j.neures.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Wang J, Campbell IL, Zhang H. Systemic interferon-alpha regulates interferon-stimulated genes in the central nervous system. Mol Psychiatry. 2008;13:293–301. doi: 10.1038/sj.mp.4002013. [DOI] [PubMed] [Google Scholar]
- Wegner AM, Nebhan CA, Hu L, Majumdar D, Meier KM, Weaver AM, Webb DJ. N-wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J Biol Chem. 2008;283:15912–15920. doi: 10.1074/jbc.M801555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss JJ, Gorman JM. Psychiatric behavioral aspects of comanagement of hepatitis C virus and HIV. Curr HIV/AIDS Rep. 2006;3:176–181. doi: 10.1007/s11904-006-0013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. Structure and functional characterization of single-strand DNA binding protein SSDP1: carboxyl-terminal of SSDP1 has transcription activity. Biochem Biophys Res Commun. 2006;339:977–984. doi: 10.1016/j.bbrc.2005.11.098. [DOI] [PubMed] [Google Scholar]
- Young PJ, Le TT, Dunckley M, Nguyen TM, Burghes AH, Morris GE. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res. 2001;265:252–261. doi: 10.1006/excr.2001.5186. [DOI] [PubMed] [Google Scholar]
- Zdilar D, Franco-Bronson K, Buchler N, Locala JA, Younossi ZM. Hepatitis C, interferon alfa, and depression. Hepatology (Baltimore, Md. 2000;31:1207–1211. doi: 10.1053/jhep.2000.7880. [DOI] [PubMed] [Google Scholar]
- Zhang J, Moseley A, Jegga AG, Gupta A, Witte DP, Sartor M, Medvedovic M, Williams SS, Ley-Ebert C, Coolen LM, Egnaczyk G, Genter MB, Lehman M, Lingrel J, Maggio J, Parysek L, Walsh R, Xu M, Aronow BJ. Neural system-enriched gene expression: relationship to biological pathways and neurological diseases. Physiol Genomics. 2004;18:167–183. doi: 10.1152/physiolgenomics.00220.2003. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Wang N, Woodson SE, Dong Q, Wang J, Liang Y, Rijnbrand R, Wei L, Nichols JE, Guo JT, Holbrook MR, Lemon SM, Li K. Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology. 409:175–188. doi: 10.1016/j.virol.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


