Abstract
Microbial pathogens are able to modulate host cells and evade the immune system by multiple mechanisms. For example, Salmonella injects effector proteins into host cells and evades the host immune system in part by inhibiting dendritic cell (DC) migration. The identification of microbial factors that modulate normal host functions should lead to the development of new classes of therapeutics that target these pathways. Current screening methods to identify either host or pathogen genes involved in modulating migration towards a chemical signal are limited because they do not employ stable, precisely controlled chemical gradients. Here, we develop a positive selection microfluidic-based genetic screen that allows us to identify Salmonella virulence factors that manipulate DC migration within stable, linear chemokine gradients. Our screen identified 7 Salmonella effectors (SseF, SifA, SspH2, SlrP, PipB2, SpiC and SseI) that inhibit DC chemotaxis toward CCL19. This method is widely applicable for identifying novel microbial factors that influence normal host cell chemotaxis as well as revealing new mammalian genes involved in directed cell migration.
Inroduction
Salmonella enterica serovars are facultative intracellular pathogens that can cause acute gastroenteritis or systemic typhoid fever in humans, a disease that still burdens millions of people per year world-wide1. Salmonella injects effector proteins into host cells through type-III secretion systems (T3SS), which are required for virulence2. For example, Salmonella enterica serovar Typhimurium (S. typhimurium) translocates an effector protein, SseI, that plays a role in modulating DC migration toward the chemokine CCL193, 4. Normally DCs chemotax toward the T cell zones of secondary lymphoid tissue primarily following the chemokine gradients of CCL19 and CCL215 and help control infection by activating the adaptive immune response6. Previously we have identified the Salmonella factor SseI as a modulator of DC chemotaxis, however, our findings also suggested that Salmonella injects additional factors that influence DC chemotaxis using the Salmonella pathogenicity island 2 (SPI2) T3SS3. Approximately 30 Salmonella effectors have been documented to be translocated by the SPI2 T3SS2 and therefore a screen-based approach would be the most efficient method of determining which of these effectors interfere with DC chemotaxis. The goal of this study was to identify these additional Salmonella factors involved in manipulating DC directed migration using novel engineered genetic screening tools.
Microfluidic techniques provide a vital addition to traditional tissue culture plate-based cell assays by offering more precise temporal and spatial control over the extracellular microenvironment by promoting fast diffusion and efficient mixing on a sub-microliter scale. Because only nano to microliter sample/reagent volumes are necessary, low cost high-throughput testing with expensive or rare clinical samples is achievable. A variety of physical, electrochemical, mechanical or optical detection schemes have been integrated with microfluidic devices for genetic analysis, phenotype screening, cell sorting or cell behavior manipulation at sub-cellular, single-cell and whole organism levels7-10. Microfluidic-based sorting and screens of cell populations can be extremely versatile, using detection schemes that are physical (e.g., microfilters, flow based or acoustic focusing), electrochemical (e.g., impedance or potentiometry), mechanical and optical (e.g., fluorophore conjugation of affinity markers, chemiluminescence, bioluminescence, surface plasmon resonance/refractive index)7, 9, 11-21. A variety of microfluidic devices have been developed for gradient generation based on flow and mixing, microjet or restricted passive diffusion through membrane, microcapillaries and hydrogels22-34. Particularly, microfluidic gradient generators have become more readily accepted in addition to traditional chemotactic assays, which have several inherent limitations. The most significant drawback is that the concentration gradient profiles are transient, unstable, and hard to reproduce in many traditional assays (e.g. micropipette assay,35 Boyden chamber,36-38 Zigmond chamber,39 Dunn chamber40). Secondly, most conventional chemotaxis assays do not support direct cell migration monitoring and detailed single cell analysis, which are required for quantification of important metrics (such as cell speed and directionality of movement)38, 41. In contrast, modern microfluidic chemotaxis devices provide long-term stable and defined concentration gradients with the possibility of real-time imaging. For shear-sensitive or semi-adherent cell cultures (including DCs), soluble25-34, 42 chemical gradients may be generated by passive Fickian chemical diffusion through microcapillaries7, 25-27, 42-44. This device design is also advantageous because specific populations of cells that successfully migrate through the capillaries toward a given chemoattractant may be captured for end-point analysis26. The application of these devices has significantly advanced our understanding of cell migration from neutrophil chemotaxis25, 27 to cancer progression26.
Despite the fast development of novel chemotaxis tools, there is still a lack of high-throughput chemotaxis screening methods. Current protocols focus on the analysis of one cell type per assay compartment that may be arranged in a parallel testing format to form miniature assay arrays (such as micro dots45 or microchannels46). To maximize high-throughput potential, simultaneous monitoring of multiple cell populations with the ability to isolate and capture a targeted population within one single device is needed. Previously, motility based screening has been successfully applied to separate mobile sperm cells from stationary ones in a microfluidic system 47. However, chemotaxis based screening has been more challenging and very limited reports are available. In a study by Tai and colleagues, parallel collection channels were built to collect breast cancer cells that migrated across different distances towards a source of epidermal growth factor 48. However, operation of such a device is challenging and requires a delicate balance of the fluidic streams. There is a great need to develop novel protocols to isolate target cells from heterogeneous tissue cultures or mixed cell populations to support further molecular, genetic or proteomic phenotype identification.
In this study we develop a positive-selection screen for S. typhimurium virulence factors that inhibit DC chemotaxis in a diffusion-based microfluidic device with the potential for high-throughput capacity 7, 44. We had demonstrated previously that wild type (WT) Salmonella modulates the migration of DCs in transwell assays and in infected mice3. Because the chamber and microcapillary design of this device allows multiple cell populations to be monitored and separated on the basis of chemotactic abilities, we developed a chemotaxis-based screen using a library of Salmonella mutants. Our screen led to the identification of 7 Salmonella virulence factors that are involved in modulating DC chemotaxis towards CCL19, 5 of which were previously not implicated in this process. To our knowledge this study is the first reported use of a microfluidic device to conduct a genetic screen for bacterial effectors that influence the chemotaxis of mammalian cells.
Materials and Methods
Bacterial strains and culture conditions
S. typhimurium SL1344 was transformed with the constitutive GFP-expression plasmid pFPV25.149 and this strain was used as the parent strain for all other S. typhimurium strains presented in this work. All S. typhimurium strains were cultured in Luria broth (LB) with 100 μg/ml carbenicillin at 37°C with constant agitation. All S. typhimurium gene deletion mutants (Table 1) were generated using the Datsenko and Wanner method50. Briefly, 70 nucleotide primers were generated (IDT, San Diego, CA) such that the first 50 bases were complementary to the 3’ or 5’ end of the targeted gene and the last 20 bases were complementary to the P1 or P2 sites of pKD4 (flanking the kanamycin resistance gene, kan). These primers were used to PCR amplify the kan locus of pKD4, and the amplified DNA was purified and transformed in to S. typhimurium LT2 expressing λ Red recombinase. Recombinants were selected on 40 μg/ml Kan LB agar plate, and the resulting gene deletion mutations were moved to the GFP-expressing S. typhimurium background by P22 bacteriophage transduction. Deletion of the desired gene was confirmed by PCR amplification (Table 2) and DNA sequencing of the targeted gene (Mclab, South San Francisco, CA). For complementation of the ΔsifA mutant, the SifA expression vector psifA was transformed into the deletion mutant and transformants were selected on LB agar supplemented with 25 μg/ml chloramphenicol.
Table 1.
| SPI2 mutant | Primers used for gene deletion (5’-3’) |
|---|---|
| Δ ssaV | 52 * |
| Δ gogB | ctaggttctaaatcttgcctgaatgaaaataaatgtaataatgatagcttgtgtaggctggagctgcttc caatagggctgctctatatataaatatattaattgcatatttttttaaagcatatgaatatcctccttag |
| Δ pipB | ttgactcacacctataaggagtcggctcacttccataagaaggaatcaaagtgtaggctggagctgcttc agggggcctctgtttgaatacttcttgtttataaaatccctttatctcgacatatgaatatcctccttag |
| Δ pipB2 | atatgataaattttatcatgcactgtgttgctgtctctgggagaaaatatgtgtaggctggagctgcttc cagattgttattcttacattgcttttatttcagatttacgtcaaaaagggcatatgaatatcctccttag |
| Δ sopD2 | aatgagagcaacggattggatcttgctttcgcggtaaataatcaagggaggtgtaggctggagctgcttc aaaagcgtacaaaaaaggctccatatcagtggggcctttttaatgactttcatatgaatatcctccttag |
| Δ spvB | caccgtaaaggaataacgctcatgcatattgatatgtccgcactgtaatggtgtaggctggagctgcttc gacctgactgaccgtaacaatgacattatcctcgagtaccgtaagcaggacatatgaatatcctccttag |
| Δ spvC | ggcggtcagggcgtgcgggttcttctgccgctcatccttcccgaacaggcgtgtaggctggagctgcttc gtactccccggcgtggacggggtgtacctgggccacattgtggatgcaggcatatgaatatcctccttag |
| Δ steA | cggctcagcgagtctgatttctaacaaaactggctaaacataaacgctttcatatgaatatcctccttag agaggagtaggacatataaagctattgagcaaaatttgaaggagtaggatgtgtaggctggagctgcttc |
| Δ steC | ctgtagcgaatgtgcccccggcgattcgcagaaaagaacggaactaaatgcatatgaatatcctccttag ttttatatgtttgcatgtgtattataataaattttcagaggatgagacatgtgtaggctggagctgcttc |
| Δ sspH2 | 52 * |
| Δ sifA | 52 * |
| Δ sifB | 52 * |
| Δ spiC | 52 * |
| Δ sseF | 52 * |
| Δ sseG | 52 * |
| Δ sseI | 3 |
| Δ sseJ | 52 * |
| Δ sseK1 | ctaataatcatcatgtaaaatatgtaatgaagtaagtatggagcatttaattgttgtgtaggctggagctgcttc cgaaacttgatagtttatgccaatattttatgtattcaatagcatgattattgccatttccgcatatgaatatcctccttag |
| Δ sseK2 | 52 * |
| Δ slrP | 52 * |
| Δ sseL | 53 * |
The gene deletion mutation constructed in the referenced work was transduced into S. typhimurium SL1344 (pFPV25.1) for the experiments presented here.
Table 2.
| SPI2 mutant | Primers used for detection by PCR (5’-3’) |
|---|---|
| Δ ssaV | tagcaggattagctgaacgg aagccatcaataactcgccc |
| Δ gogB | cgtcagcatacgatctgctg gaagcagctccagcctacac |
| Δ pipB | tgaagcgtaagctatgctgg gaagcagctccagcctacac |
| Δ pipB2 | aaggcggcaagcgagcgaat gaagcagctccagcctacac |
| Δ sopD2 | gaatggttggcgccgttttg gaagcagctccagcctacac |
| Δ spvB | caggctttacgtgaggaacc gaagcagctccagcctacac |
| Δ spvC | cggtcagtcaggtcgtagcg gaagcagctccagcctacac |
| Δ steA | tcagggttaatccctgcagg gaagcagctccagcctacac |
| Δ steC | cgatgcgctttccaccagac gaagcagctccagcctacac |
| Δ sspH2 | tccgggaatatctttgtcgc atgtctgccggacagatact |
| Δ sifA | gaacgtgacgtctgagaaa ccgcagttgagataaaaaggg |
| Δ sifB | cgatgggcaacatgggataa gttttggtattgccagggga |
| Δ spiC | ggatgtggttgtgagcgaat ctggcataaagggtgaagtc |
| Δ sseF | agattcgccagaatgcgcaa tgagcatttgggctaacagg |
| Δ sseG | ggctgaaaatattgcggcct tgatttccagcagcaaccgt |
| Δ sseI | gcgttgataccggatgatct tgttgttgtcgatctccacc |
| Δ sseJ | ggcattaacctcacgttgttg gagctgtgttttgctcaagg |
| Δ sseK1 | gctcactggtagggtatttatg agcactgcgattttaaagtggc |
| Δ sseK2 | ctcaggacttagcattgtgac gaagggtgagtaaaacaaggc |
| Δ slrP | ctctctcctcggctatgaaa agtcattgacgcagtagtgg |
| Δ sseL | acggtggcatgacagataac gtggttgaatcattgacggc |
Cell culture
Dendritic cells (DCs) were derived from mouse bone marrow based on the method of Lutz et al. 51. Bone marrow was harvested from the femurs and tibia of 129X1/SvJ mice (Jax Mice, Bar Harbor, Me) between the ages of 6 and 12 weeks. Bone marrow cells were resuspended in Dulbecco's modified Eagle medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS, Gibco), 50 mM β-mercaptoethanol, 1 mM HEPES, and 20 ng/ml granulocyte-macrophage colony-stimulating factor (GMCSF, Peprotech, Rocky Hill, NJ). Approximately 20 million bone marrow cells were then seeded into a petri dish and incubated in a humidified chamber at 37°C and 5% CO2. Two days later, 5 ml of DC culture medium was added to each plate and left to incubate for another 2 days. Finally, cells in suspension and loosely adhered to the plate were harvested and used for S. typhimurium infections 24 h later.
S. typhimurium-infection of DCs
For time-lapse video microscopy experiments, the indicated S. typhimurium strain was grown overnight at 37°C with agitation, and approximately 2.5 billion bacteria were harvested, washed once with phosphate buffered saline (PBS), and then added to DCs at a multiplicity of infection (MOI) of 25:1. DCs were incubated with the S. typhimurium bacteria at 37°C for 30 min, and then the medium was changed to DC medium containing 100 μg/ml gentamicin to kill extracellular bacteria. After 1.5 h incubation the infected DCs were harvested by scraping in cold PBS and resuspended at 10 million cells/ml in DC medium containing 10 μg/ml gentamicin. Three μl of this cell suspension was injected into the cell-viewing chamber of the microfluidic device using a P20 micropipette, and the sink and source channels of the device were immediately flushed with 20 μl of DC medium.
Microfluidic Device Fabrication
The microfluidic devices were fabricated using standard soft lithography protocols as previously described45. Briefly, a two-layer SU8 (MicroChem Corp., Newton, MA) master consisting of a 10-μm height capillary layer and a 100-μm height chamber layer was fabricated on a silicon wafer (Fig. 1A), then treated with (3-aminopropyl) trimethoxysilane (Sigma, St. Louis, MO) as a release layer. To make polydimethylsiloxane (PDMS) devices, a 10:1 w/w mixture of Sylgard 184 monomer and hardener (Dow Corning, Corning, NY) was degassed under vacuum over the SU8 master and baked at 65°C for 1 h to cure. Inlet and outlet fluidic ports were punched out manually. Permanent bonding between PDMS chips and the cover glass was achieved by a 40-sec oxygen plasma treatment at 80 watts (Branson IPC oxygen plasma Asher, Hayward, CA).
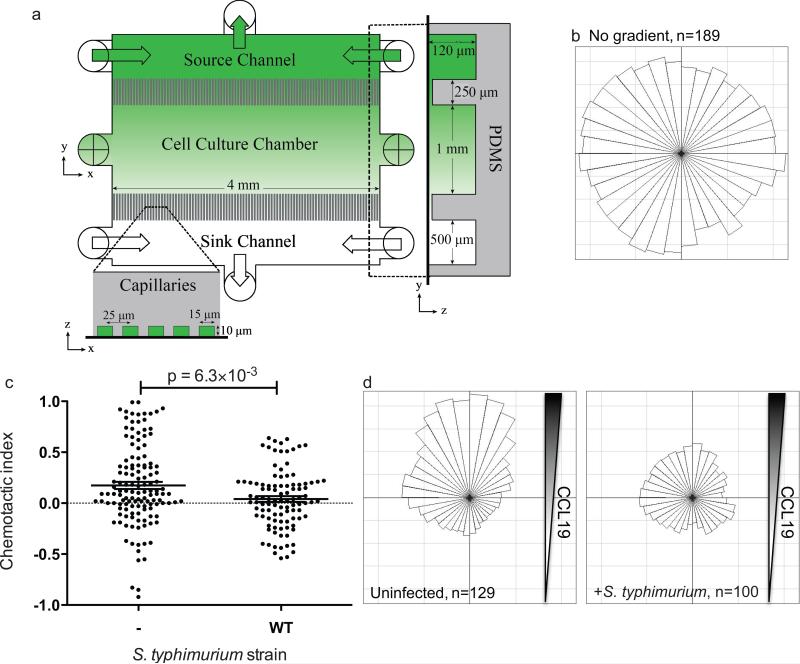
Figure 1.
S. typhimurium inhibits DC migration along a gradient of CCL19 established inside a microfluidic device. A) The microfluidic chemotaxis device includes a cell culture chamber connected by microcapillaries to chemokine source and sink channels, which enable formation of a linear, stable chemokine gradient. Primary DCs were cultured, infected with GFP-expressing S. typhimurium and then injected into the microfluidic chemotaxis device. B) The angular histogram of uninfected mature DC tracks under a uniform distribution of CCL19 is shown. Angular histograms indicate the proportion of cells traveling in each direction when comparing a cell's final position to its initial position and subdivided into 10° bins. C and D) DCs infected with GFP-expressing WT S. typhimurium were cultured in CCL19 gradients and imaged by fluorescence and DIC microscopy every 7 min over the course of 5 h. The cell migratory tracks of infected DCs and their uninfected neighbors were used to calculate their chemotactic indices (C) and angular histograms (D). N is the number of individual cells tracked. Line and error bars represent the mean and SEM, respectively, and all P-values were determined using rank-based Mann-Whitney test.
Preparation of the microfluidic device
On the day the microfluidic device was to be injected with DCs, the microfluidic device was first wetted using 70% ethanol and then washed with deionized sterile water. Next, the inside of the device was coated with 1 μg/ml of mouse fibronectin for 3 h at room temperature. Just before injection of cells, the device was washed with DC medium containing 10 μg/ml gentamicin.
Time-lapse video microscopy
Twenty hours after injection into the microfluidic device, the infected DCs were subjected to a gradient of CCL19 or CXCL12 (Peptrotech, Rocky Hill, NJ) by applying 200 ng/ml of the chemokine and 25 μg/ml of 10 kDa dextran Texas Red (Invitrogen, Grand Island, NY) to the source channel at 3 μl/h for the duration of the time-lapse video microscopy using a syringe pump (WPI, Sarasota, FL) and 100 μl Hamilton gas-tight syringes (Hamilton, Reno, NV). Under similar conditions, Shamloo et al. measured the chemoattractant gradient concentration in the cell-viewing chamber to be linear, with maximum and minimum concentrations equal to 62% and 37%, respectively, of the source channel concentration44. DC medium alone was supplied to the sink channel at an identical flow rate. The syringes were connected to the microfluidic device by PEEK tubing (IDEX Health & Science, Oak Harbor, WA). The DCs were imaged by DIC and fluorescence microscopy at 40X magnification on a Nikon Eclipse TE2000-E microscope with EMCCD camera. The device was placed on an automated stage and phase contrast and fluorescence images were taken and stored every 7 min for 5 h using Openlab software (PerkinElmer, Waltham, MA). Images were compiled into short videos using Volocity (PerkinElmer, Waltham, MA) and Quicktime (Apple, Cupertino, CA) softwares. The phase contrast videos were analyzed using ImageJ software (open source) with the plugins “Manual Tracking” and “Chemotaxis Tool” (Ibidi, Martinsreid, Germany). The Chemotaxis Tool was used to generate a chemotactic index and an average cell speed for every cell track and to make angular histogram plots for the cell track endpoints. Red fluorescence images taken of the 10 kD dextran Texas Red indicated that the chemical gradient was linear and stable throughout the 5 h duration of the time-lapse video microscopy.
Microfluidic-based screen
A library of SPI2 effector gene deletion mutants was generated in the GFP-expressing S. typhimurium SL1344 (pFPV25.1) background using the method of Datsenko and Wanner described above 50. Each mutant was grown overnight at 37°C with agitation, and equal amounts of bacteria from each culture was combined into one tube and used to infect DC at a total MOI of 10:1. These DCs were resuspended at 100 million cells/ml and then injected into the device, the sink and source channels were flushed (in that order) to clear of cells, and the DCs were allowed to recover for 20 h in the incubator (as above). Next, the DCs were subjected to a gradient of CCL19 (as above) for 24 h. The source channel then was flushed with 200 μl of 1% Triton X-100 in plain DMEM to retrieve DCs that had migrated through the capillaries and that remained associated with any part of the source channel. Five min incubation of these DCs in 1% Triton X-100 DMEM with agitation resulted in DC lysis, and the freed intracellular bacteria were spread onto LB agar plates containing 200 μg/ml streptomycin, 40 μg/ml kanamycin, and 100 μg/ml carbenicillin. Colonies that grew overnight were cultured and used for genomic DNA isolation (Qiagen, Valencia, CA). The identities of the clones were determined by PCR and confirmed by DNA sequencing (as above). This screen was carried out 4 times and mutants that were isolated at least 3 out of the 4 screens were identified as positive mutants.
Statistics
The threshold for positive selection in the screen presented here was isolation of the mutant in 3 out of 4 screens. For both cell speed and chemotactic index data, infected cells were compared to their uninfected neighbors within the same microfluidic device using the non-parametric rank-based Mann-Whitney test. The distribution of chemotactic indices and cell speed were tested for normality using the D’Agostino-Pearson test. The sample size (N) for the number of cells tracked for each treatment group was based on our initial experiments measuring chemotactic indices of uninfected DCs toward CCL19 (Fig. 1C). This sample group passed the test for normality and has a mean chemotactic index of 0.1742 (standard deviation = 0.3942) that is significantly different from 0 in both the non-parametric rank-based Wilcoxon signed rank test (p<0.0001) and a one-sample two-tailed Student’s t test (p<0.0001). Initially we presumed that 0.1742 represented the true mean chemotactic index of this cell population (and 0.3942 was the true variance) and calculated the minimum N necessary for a given power (1-β) and a type I error rate (α) of 0.05. For a power level of at least 0.8, N should be at least 43, and therefore more than 43 cells were tracked in every treatment group.
Results
Migration of infected DCs in a microfluidic device
Previous studies aimed at elucidating mechanisms of how pathogens influence immune cell migration have utilized transwells3, 54, 55. For example, we previously demonstrated that S. typhimurium inhibits DC migration in transwells towards the chemokine CCL193, and we identified a SPI2 effector, SseI that hinders DC migration3. We validated the use of the microfluidic device that maintains a stable gradient of CCL19 (Fig. 1A)44 by comparing chemotaxis of uninfected DCs with Salmonella-infected DCs. Primary murine bone marrow-derived DCs were infected with WT S. typhimurium expressing green fluorescent protein (GFP) at a multiplicity of infection (MOI) of 25:1, conditions that lead to approximately 50% of the DCs being infected with Salmonella. The DCs were added to the fibronectin-coated cell culture chamber of the microfluidic device and allowed to recover for 20 h. After this time, the DCs were exposed to a stable CCL19 gradient by flowing DC medium with 200 ng/ml CCL19 through the source channel and DC medium alone through the sink channel at 3 μl/h using a syringe pump44. DIC and fluorescent images were taken every seven minutes for a total of five hours. DIC images were used to detect all DCs, and the fluorescent images were used to detect GFP-Salmonella and Dextran Texas Red, which was used as a tracer molecule to monitor the CCL19 gradient. We tracked DCs over a 5-h period and recorded each cell's speed, location, and overall angular direction of movement with respect to the CCL19 gradient. In the absence of a CCL19 gradient, uninfected mature DCs did not preferentially move in any one angular direction (Fig. 1B). The chemotactic index of each cell's track was calculated as the ratio of the distance traveled in the direction of increasing CCL19 divided by the overall length of the cell's track. We compared the chemotactic indices (Fig. 1C) and angular directions (Fig. 1D) of infected DCs to uninfected cells from the same device, thereby providing an internal control for each individual assay. While uninfected DCs had a clear chemotactic response to the CCL19 gradient, DCs infected with WT S. typhimurium did not chemotax toward CCL19 (Fig. 1C and 1D). Importantly, the degree of chemotaxis inhibition did not correlate with the number of intracellular bacteria per DC (data not shown). These results are in agreement with our previous work demonstrating that WT S. typhimurium modulates DC migration3. Taken together, our results validate the use of this microfluidic device to measure the chemotactic behavior of Salmonella-infected DCs. A representative video of DCs infected with WT S. typhimurium are available as supplementary material online (Supplementary Video). To test the robustness of the microfluidic device in these migration experiments, we also elucidated if S. typhimurium manipulates DC migration toward another chemokine, CXCL12. Salmonella also prevents the chemotaxis of infected DC toward CXCL12, confirming that this system can be used to study a range of host-pathogen interactions (Supplementary Fig. 1).
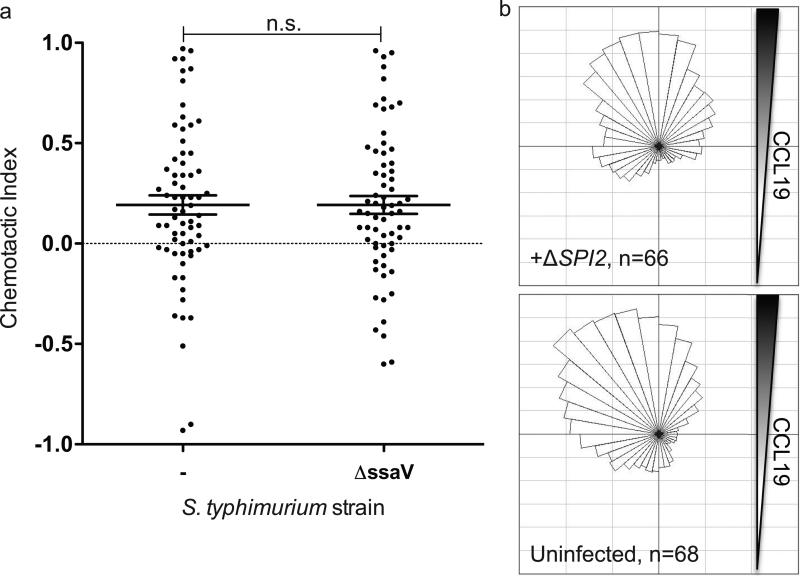
Since CCL19 is very important for migration to secondary lymphoid tissue for mature DCs6, 56,57, we focused on understanding what other Salmonella effectors modulate DC chemotaxis toward this chemokine. Our previous work identified the Salmonella effector SseI, which is translocated into host cells by the SPI2 T3SS, as partially preventing DC migration. Therefore we reasoned that additional SPI2 translocated effectors may contribute to manipulating DC directed migration toward CCL19. To test this hypothesis, we infected DCs with an S. typhimurium strain that does not translocate any SPI2 effectors because it lacks a critical T3SS structural component (ΔssaV)58. In contrast to DCs infected with WT Salmonella, the DCs infected with this SPI2 T3SS-deficient mutant did chemotax toward CCL19 and were indistinguishable from uninfected DCs (Fig. 2A and 2B). The chemotactic indices of DCs infected with this SPI2 T3SS-deficient mutant were significantly higher than DCs infected with WT Salmonella (p < 4.4 × 10−3 in a Mann-Whitney test). These results demonstrate that the translocation of SPI2 effectors are responsible for the inhibition of DC chemotaxis toward CCL19. We next set out to identify these additional Salmonella effectors that are translocated into host cells by the SPI2 T3SS and influence DC chemotaxis towards CCL19.
Figure 2.
Salmonella pathogenicity island 2 (SPI2) is required to inhibit DC chemotaxis toward CCL19. DCs infected with GFP-expressing WT or ΔssaV (SPI2 mutant) S. typhimurium were cultured in a gradient of CCL19 and imaged every 7 min over 5 h. The chemotactic indices (A) and angular histograms (B and C) are presented. Line and error bars represent the mean and SEM, respectively, and n.s. (not significant) indicates P-value > 0.05 in a Mann-Whitney test.
Microfluidic-based genetic screen
Individually testing each SPI2 effector mutant in a microfluidic migration assay would be laborious and time-consuming. Thus, we developed a positive-selection genetic screen, with the potential for high-throughput capacity, for Salmonella mutants that cannot block DC chemotaxis. We adapted the microfluidic device for this screen by enabling collection of cells that migrated through the microcapillaries into the source channel. We took advantage of the precise environment of the microfluidic device to screen a mutant library that consisted of a mixture of 20 different SPI2 effector gene deletion strains simultaneously (Fig. 3A). One of these strains contained a deletion of spiC, a gene that encodes a SPI2-encoded protein that is critical for the translocation of SPI2 effectors in vivo59. Similar to ΔssaV, SPI2 effector translocation into DCs is eliminated, enabling ΔspiC to function as a positive control for the screen. To conduct the screen, DCs were infected with the SPI2 effector mutant library at a MOI of 10:1 such that it would be highly unlikely for a single DC to be infected with two different mutants. The infected DCs were injected into the microfluidic device and allowed to recover for 20 h before exposure to a CCL19 gradient for 24 h. DCs that migrate towards CCL19 should either be uninfected or be infected with a S. typhimurium mutant lacking a SPI2 effector involved in inhibiting DC chemotaxis. To test this idea, DCs that migrated toward CCL19 through the capillaries and into the source channel after 24 h were harvested and lysed to free any intracellular bacteria. The lysate was plated on selective agar plates, and each Salmonella clone was subjected to conventional PCR techniques followed by sequencing for identification of individual SPI2 effector mutants (Fig. 3A).
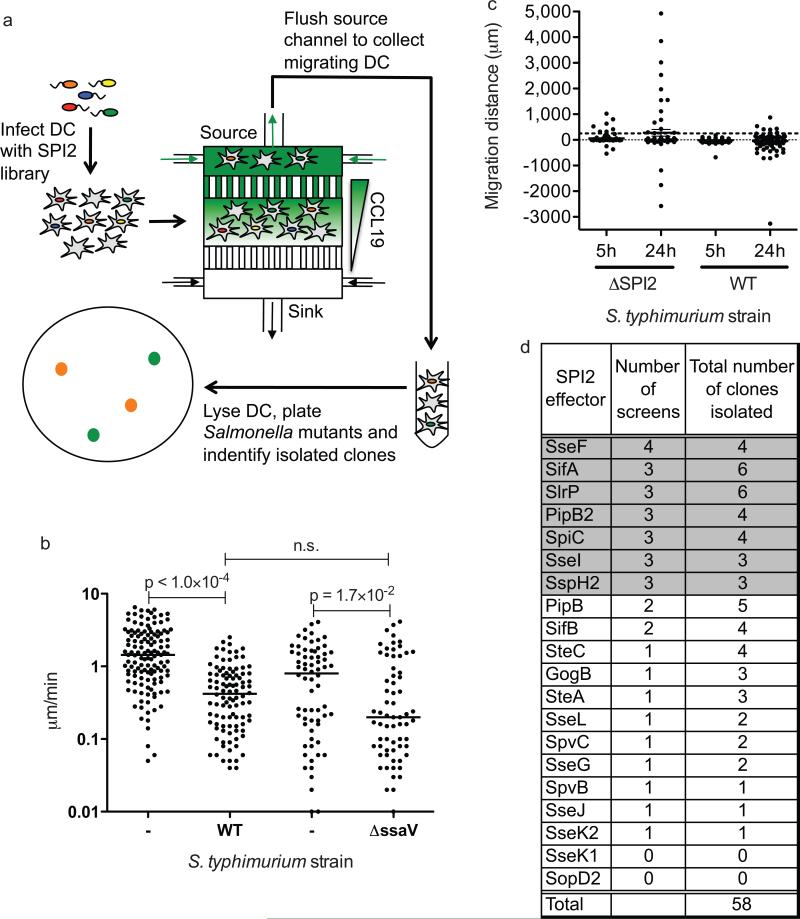
Figure 3.
Positive selection microfluidic-based genetic screen for SPI2 effector mutants unable to inhibit DC migration toward CCL19. A) A diagram of the microfluidic-based screen is presented. DCs were infected with a library of SPI2 effector mutants and allowed to migrate in a CCL19 gradient for 24 hours. DCs that migrated into the source channel were collected and lysed, and the S. typhimurium mutants were isolated as colonies. Mutants were identified by PCR and sequencing. B) DCs infected with either GFP-expressing WT or ΔssaV S. typhimurium were cultured in CCL19 gradients and imaged by fluorescence and DIC microscopy every 7 min over the course of 5 h. The cell migratory tracks of both infected DC and their uninfected neighbors were recorded and used to calculate their migration speed. The line represents the median, and all P-values were determined using the Mann-Whitney test. C) Predicted average distances traveled in the direction of the source channel by each DC infected with either ΔssaV mutant or WT Salmonella were calculated by taking the product of each DC's chemotactic index and speed and the given amount of time (5 h or 24 h). Line and error bars represent the mean and SEM, respectively. Dashed black line drawn at y = 250 μm (length of capillaries). D) The screen was repeated a total of 4 times and the number of times a given mutant was positively selected is indicated as “number of screens” and the total number of times a given mutant was isolated from the source channel over all the screens is indicated as “total number of clones isolated”. The threshold chosen for positive selection was identification in 3 out of the 4 screen repetitions. Positively selected genes are shown in grey. The number of mutants collected ranged between 6 and 24 per screen.
To be considered a positive hit in this screen, a DC must migrate with sufficient speed and sufficient directionality to reach the sink channel within 24 h. Therefore, if DCs infected with WT or ΔssaV mutant Salmonella migrated at different speeds in a CCL19 gradient, our screen could be biased towards detecting DCs infected with Salmonella that migrate faster, unrelated to their chemotactic abilities. We determined that while DCs infected with either WT or ΔssaV migrated at a slower speed than uninfected DCs, their migration speeds were not significantly different from each other (Fig. 3B). While it is biologically intriguing that Salmonella inhibits DC chemotaxis by a different mechanism than it decreases DC migratory speed, it is also beneficial because it allowed us to specifically screen for Salmonella mutants unable to inhibit DC chemotaxis.
To be sure that these target DCs had enough time to migrate into the source channel, we calculated the average distance a population of DCs infected with WT or ΔssaV mutant Salmonella could travel in the direction of the CCL19 source channel after 5 h and 24 h (Fig. 3C). Salmonella-infected DCs must migrate at least the length of the capillaries (250 μm) connecting the cell-viewing chamber to the source channel in order for the Salmonella bacteria to be captured inside of the source channel. While the average distance traveled in 5 h by DCs infected with ΔssaV mutant Salmonella was only 57.5 μm, in 24 h the DCs are predicted to travel an average distance of 275.9 μm, which is sufficient to traverse the capillaries. Therefore, we selected 24 h as the time period for our chemotaxis screen. The average distance travelled by WT Salmonella-infected DCs was not significantly different from zero, but the distribution of these distances varied widely (Fig. 3C) such that a small fraction (0.08) of this DC population would still be able to reach the CCL19 source channel by 24 h. Based on these data, we reasoned that a DC would have an 8% chance of migrating into the source channel when infected with a Salmonella mutant still capable of inhibiting DC chemotaxis (similar to WT Salmonella). The probability that a DC infected with this same Salmonella mutant would migrate repeatedly into the source channel in at least 3 out of 4 repetitions of the screen may be calculated as (0.08)4 + 4(1-30 0.08)(0.08)3 = 0.00193, yielding an acceptably low false-positive error rate. Therefore, SPI2 effectors involved in inhibiting DC chemotaxis were only considered “positively selected” if they were detected in at least 3 out of the 4 screens (Fig. 3D, shaded).
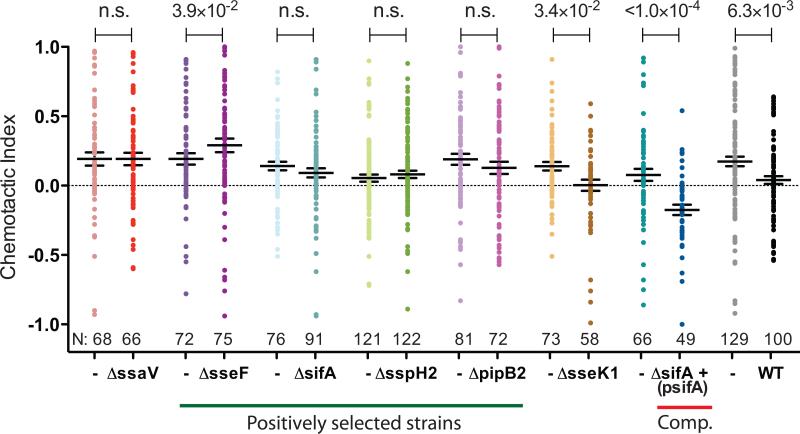
We conducted the microfluidic chemotaxis screen four times and identified 58 individual SPI2 effector mutant clones (Fig. 3D). As expected, the ΔsseI mutant bacterial strain was isolated in 3 of the 4 screens, thus, validating the technique (Fig. 3D). In addition to ΔsseI, the S. typhimurium mutants ΔsseF, ΔsifA, ΔsspH2, ΔslrP, ΔpipB2 and ΔspiC also were positively selected in the screen (Fig. 3D). While SpiC may be translocated into the DC cytoplasm60, SpiC is also required for the formation of the translocon apparatus itself61 and ΔspiC mutants do not translocate SPI2 effectors in vivo59. Thus, the ΔspiC mutant is phenotypically similar to ΔssaV and its positive selection validates the screen. To further confirm the results of our high-throughput screening method, we infected DC separately with different SPI2 effector deletion mutants that were identified in the screen. In a series of separate microfluidic chemotaxis assays using only a single mutant strain per experiment, we confirmed that the mutants ΔsseF, ΔsifA, ΔsspH2 and ΔpipB2 did not inhibit DC chemotaxis toward CCL19 (Fig. 4)., Importantly, the sifA-deletion strain containing a wild type copy of sifA on a plasmid (ΔsifA(psifA)) interfered with DC chemotaxis similar to wild type bacteria, verifying that the positive selection of the ΔsifA mutant in our screen was due to the specific lack of SifA (Fig. 4). Similar to our previously published results3, DC infected with the sseI-deletion strain containing a wild type sseI gene on a plasmid did not chemotax towards CCL19 (data not shown). Consistent with our previous results, DC infected with the SPI2 effector mutants had reduced cell speed (Supplementary Fig. 2). In addition, as a negative control, we confirmed that DCs infected with a SPI2 effector mutant, ΔsseK1, which was not selected in the screen, had a chemotactic index similar to DCs infected with WT Salmonella (Fig. 4). Interestingly, DCs infected with the ΔsseF mutant bacterial strain chemotax towards CCL19 slightly better than their uninfected neighbors (Fig. 4), indicating our screen is sensitive enough to identify DCs with a range of chemotaxis phenotypes. It is intriguing that most of the effectors that were identified in the screen have been shown to interact, either directly or indirectly, with elements of the host cytoskeleton, which is essential for mammalian cell migration3, 62-67.
Figure 4.
SPI2 mutants positively selected by the microfluidic screen do not inhibit DC chemotaxis toward CCL19. DC infected with the indicated S. typhimurium strain were cultured in a gradient of CCL19 and imaged every 7 min over 5 h, and the chemotactic indices are shown here. ΔsseK1 was not positively selected by the screen and serves as a control. ΔsifA+(psifA) refers to the complemented ΔsifA mutant. N is the number of individual cells tracked. Line and error bars represent the mean and SEM, respectively, and P-values were determined using the Mann-Whitney test and are indicated above the graph.
Discussion
Microfluidic devices are now widely used in cell migration assays, because they provide precise, long-term control over the cellular microenvironment while still allowing direct observation41, 68. However, despite the advantages these devices provide, they have not been adopted as screening platforms to identify genes from either the host or the pathogen that influence host cell migration. Here we present a method for performing a positive selection genetic screen for Salmonella translocated effectors that inhibit DC chemotaxis.
While microfluidic devices are commonly used in mammalian cell migration studies44, 69, 70, to our knowledge they have not been used to study how pathogens influence immune cell migration. Using microfluidic devices allowed us to more precisely determine how S. typhimurium inhibits DC migration. Utilizing transwells, we previously demonstrated that Salmonella can hinder DC migration3, however, transwell data can be difficult to interpret because the effects on migratory speed and chemotaxis (directional migration) cannot be differentiated. Because chemokine gradients across a transwell membrane are only present for a short amount of time, any cellular perturbation that decreases migration speed potentially could be misinterpreted as a decrease in chemotaxis. This may be a particularly important consideration for studies of host-pathogen interactions, as all Salmonella infected DCs were found to have significantly decreased migration rates (Fig. 4 and Supplementary Fig. 1).
Through microfluidic migration assays, we confirmed that Salmonella attenuates DC chemotaxis toward CCL19 (Fig. 1) and CXCL12 (Supplementary Fig. 1). Recently, CCL19 was shown to have a much greater chemotactic potency than CXCL12 toward DCs71, and we have shown that SPI2 translocated effectors are completely responsible for Salmonella-dependent inhibition of chemotaxis toward CCL19 (Fig. 2).
To determine which SPI2 effectors target DC chemotaxis, we developed a high-throughput positive-selection genetic screen in our microfluidic device. We exploited the geometric design of the microfluidic device to screen for those DCs that were capable of efficient chemotaxis (Fig. 3). The chemotaxing cells can migrate through the microcapillaries to the source channel where they can be collected for genetic analysis. Our screen successfully identified two previously known and five new SPI2 effectors that are necessary for inhibiting DC chemotaxis toward CCL19 (Fig. 4).
It is striking that we identified several effectors involved in modulating host cell chemotaxis, and that all the single mutants have strong phenotypes. Our results indicate that the manipulation of DC directed migration by Salmonella is quite complex. It will be important to determine if the effectors that block DC chemotaxis work individually or together in a complex. It is likely that these factors, either separately or together, disrupt the ability of DCs to either sense or respond to the chemokine gradient. DCs sense CCL19 using the chemokine receptor CCR772. Since Salmonella-infection does not compromise CCR7 expression by DCs72, it is likely that these SPI2 effectors target host factors downstream of CCR7 that help the cell respond to the CCL19 gradient. Stimulation of CCR7 by CCL19 leads to chemotaxis toward CCL19 and an increase in migratory speed73. Chemotaxis toward a CCR7 ligand is controlled through a signaling module made up of heterotrimeric G proteins of the Gi subfamily and the MAPK protein family, whereas speed is governed by the Rho/Pyk2/cofilin module73. Chemotaxis requires both actin filaments and microtubules, as well as a wide array of cytoskeletal regulatory proteins, to extend the plasma membrane forward and ultimately migrate in the direction of a chemical gradient74, 75. Because SPI2 effectors mainly target DC chemotaxis and not migration speed, it is likely that they interfere with either the Gi/MAPK signaling module and/or the host cytoskeleton.
SseF, SifA, SspH2, PipB2, and SseI can interact directly or indirectly with the cytoskeleton3, 62-67, 76. Both SseI and SspH2 interact with filamin, a F-actin cross-linking protein66. SseI also binds to the cytoskeletal regulator and scaffolding protein, IQGAP13, 77. SseF and PipB2 both interact with the microtubule-associated molecular motors, dynein and kinesin-1, respectively62, 64, 65. SifA also interacts with kinesin-1 indirectly by binding SKIP and is required for maintenance of the Salmonella containing vacuole (SCV) in DCs63, 78. Except for SseI, all of these factors are required for Salmonella-dependent inhibition of antigen presentation in DC79, an activity that also requires a functional cytoskeleton80. Understanding the molecular mechanism for how these Salmonella effectors (either separately or together) inhibit chemotaxis and whether it involves interactions with components of the host cytoskeleton is the subject of further study.
The current understanding of the main role of SPI2 effectors is to maintain the integrity and correct positioning of the SCV inside of the host cell during infection, partially through acting on the host cell cytoskeleton62-65, 67. Here, we expand the role of SPI2 beyond the SCV to include the regulation of DC chemotaxis, potentially having important consequences for the host's immune response. In order to clear a Salmonella infection, the host must mount an adaptive immune response against this bacterial pathogen81, and immune cell migration along CCL19 gradients is essential to establishing an adaptive immune response82. However, a fraction of hosts fail to completely clear systemic Salmonella infections and become persistently infected83. The results of our screen revealed that Salmonella targets the ability of DCs to chemotax toward CCL19 using a specific set of SPI2 effectors, and these results provide a potential mechanism for how these bacteria avoid clearance.
In addition to Salmonella, many other bacterial pathogens are able to evade the host's immune response, often through translocating bacterial effectors directly into host cells84. The microfluidic positive-selection screening method presented here is a novel, useful technique to identify bacterial effectors from many different pathogens that impact directed host cell migration toward a given chemokine. Additionally, this method could be adapted for increased capacity of high-throughput screening techniques. For example, by using DNA sequencing methods instead of conventional PCR techniques to identify mutants, much larger mutant libraries could be screened quickly and efficiently. These methods will allow researchers to determine which bacterial effectors are responsible for modulating host cell migration, which likely helps the bacteria regulate the host immune response. These effectors of immune cell migration may serve as excellent targets for future pharmaceutical development.
Beyond use in investigating host-pathogen interactions affecting host cell migration, this microfluidic-based screen can be adapted for general studies of mammalian cell chemotaxis. For example, mammalian cells could be screened for genes influencing chemotaxis using a retroviral insertion library and subjecting them to a gradient in the microfluidic device. Using negative selection, mutants that are unable to migrate towards the gradient could be identified by sequencing of the insertion site. This would provide a high-throughput mammalian cell chemotaxis screen that takes advantage of the precise control over gradient shape and stability provided by microfluidic devices. This genetic screening method will allow researchers to genetically probe increasingly complex questions concerning host cell chemotaxis and how pathogens manipulate that chemotaxis.
Conclusions
We have developed and validated a microfluidic-based positive selection genetic screening method and we have used this new method to investigate how a pathogen such as Salmonella alters host cell migration. Our screen successfully identified Salmonella effectors that are necessary for inhibiting DC chemotaxis toward CCL19 (Fig. 4), thereby expanding our understanding of how this pathogen manipulates the host cell and potentially the immune response. However, we believe this new tool is not limited to the study of host-pathogen interactions, but instead is broadly applicable to the study of cell migration in potentially any biological system.
Supplementary Material
Supplemental Figure 1: S. typhimurium inhibits DC migration toward CXCL12 independently of SseI. A-D) DCs infected with GFP-expressing WT or ΔsseI S. typhimurium were cultured in a gradient of CXCL12 and imaged every 7 min over 5 h. The chemotactic indices (A) and angular histograms (B and C) are presented. A) Line and error bars represent the mean and SEM, respectively, and all P-values were determined using the Mann-Whitney.
Supplemental Figure 2: DCs infected with SPI2 mutants positively selected by the microfluidic screen have lower migratory speed compared to uninfected DCs. DCs infected with the indicated S. typhimurium strains were cultured in a gradient of CCL19 and imaged every 7 min over 5 h. The cell migratory tracks of both infected DC and their uninfected neighbors were recorded and used to calculate their speeds. N is the number of individual cells tracked. Lines represent the median, and P-values were determined using the Mann-Whitney test and are indicated above the graph.
Supplemental Figure 3: DCs located inside of the capillaries during time-lapse experiments do not affect the chemical gradient. A) Time-lapse images were taken every 84 min of the capillaries leading to the source channel in a chemical gradient containing CCL19 and 10 kD dextran Texas Red; DIC, red fluorescence images and plot profiles of the red fluorescence are shown (yellow outline denotes plot profile range, taken using imageJ). B) The number of DCs per capillary averaged over 3 separate videos (error bars indicate SEM). C) The speed of DCs infected with ΔsifA(psifA) Salmonella inside the capillaries (N =6) compared to infected DCs in the cell-viewing chamber (bar indicates median speed).
Acknowledgements
We thank Manuel Amieva for his help and expertise with live-cell time-lapse video microscopy, data analysis and very helpful discussions. We thank Amir Shamloo for his help with computational analysis of time-lapse video microscopy data. We also thank Nicole Romano for her preparation of the detailed schematic of the microfluidic device, as well as Andrea Almaguer for her assistance with DC infection and migration measurements. We thank Sam Miller for providing psifA for complementation studies. Denise Monack, Ph.D. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. Additional support was provided by NIH 1-T32-HL098049 to Stanford Cardiovascular Institute (H.X.), and NIH DP2-OD006477 (S.C.H.). Sarah Carden is supported by the Lucille P. Markey Stanford Biomedical Research Graduate Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contribution
Conceived and designed the experiments: LMM, HX, SCH, DMM. Performed the experiments: LMM, SEC, HX. Analyzed the data: LMM, SEC, HX, SF, MR, SCH, DMM. Contributed reagents/materials/analysis tools: LMM, SEC, HX, SCH, DMM. Wrote the paper: LMM, SEC, DMM, HX.
References
- 1.Wang MM, Tu E, Raymond DE, Yang JM, Zhang H, Hagen N, Dees B, Mercer EM, Forster AH, Kariv I, Marchand PJ, Butler WF. Nat Biotechnol. 2005;23:83–87. doi: 10.1038/nbt1050. [DOI] [PubMed] [Google Scholar]
- 2.Figueira R, Holden DW. Microbiology. 2012;158:1147–1161. doi: 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 3.McLaughlin LM, Govoni GR, Gerke C, Gopinath S, Peng K, Laidlaw G, Chien YH, Jeong HW, Li Z, Brown MD, Sacks DB, Monack D. PLoS Pathog. 2009;5:e1000671. doi: 10.1371/journal.ppat.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Worley MJ, Nieman GS, Geddes K, Heffron F. Proc Natl Acad Sci U S A. 2006;103:17915–17920. doi: 10.1073/pnas.0604054103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schumann K, Lammermann T, Bruckner M, Legler DF, Polleux J, Spatz JP, Schuler G, Forster R, Lutz MB, Sorokin L, Sixt M. Immunity. 2010;32:703–713. doi: 10.1016/j.immuni.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 6.Jang MH, Sougawa N, Tanaka T, Hirata T, Hiroi T, Tohya K, Guo Z, Umemoto E, Ebisuno Y, Yang BG, Seoh JY, Lipp M, Kiyono H, Miyasaka M. J Immunol. 2006;176:803–810. doi: 10.4049/jimmunol.176.2.803. [DOI] [PubMed] [Google Scholar]
- 7.Xu H, Heilshorn SC. Small. 2013;9:585–595. doi: 10.1002/smll.201202208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung K, Crane MM, Lu H. Nature methods. 2008;5:637–643. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- 9.Rohde CB, Zeng F, Gonzalez-Rubio R, Angel M, Yanik MF. Proc Natl Acad Sci U S A. 2007;104:13891–13895. doi: 10.1073/pnas.0706513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takayama S, Ostuni E, LeDuc P, Naruse K, Ingber DE, Whitesides GM. Nature. 2001;411:1016. doi: 10.1038/35082637. [DOI] [PubMed] [Google Scholar]
- 11.Applegate RW, Jr., Squier J, Vestad T, Oakey J, Marr DW, Bado P, Dugan MA, Said AA. Lab Chip. 2006;6:422–426. doi: 10.1039/b512576f. [DOI] [PubMed] [Google Scholar]
- 12.MacDonald MP, Spalding GC, Dholakia K. Nature. 2003;426:421–424. doi: 10.1038/nature02144. [DOI] [PubMed] [Google Scholar]
- 13.Saliba AE, Saias L, Psychari E, Minc N, Simon D, Bidard FC, Mathiot C, Pierga JY, Fraisier V, Salamero J, Saada V, Farace F, Vielh P, Malaquin L, Viovy JL. Proc Natl Acad Sci U S A. 2010;107:14524–14529. doi: 10.1073/pnas.1001515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugino H, Ozaki K, Shirasaki Y, Arakawa T, Shoji S, Funatsu T. Lab Chip. 2009;9:1254–1260. doi: 10.1039/b815765k. [DOI] [PubMed] [Google Scholar]
- 15.Lee C, Lee J, Kim HH, Teh SY, Lee A, Chung IY, Park JY, Shung KK. Lab Chip. 2012;12:2736–2742. doi: 10.1039/c2lc21123h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan YC, Fisher JS, Lee AI, Cristini V, Lee AP. Lab Chip. 2004;4:292–298. doi: 10.1039/b403280m. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Chen S, Kong M, Wang Z, Costa KD, Li RA, Sun D. Lab Chip. 2011;11:3656–3662. doi: 10.1039/c1lc20653b. [DOI] [PubMed] [Google Scholar]
- 18.Wei H, Chueh BH, Wu H, Hall EW, Li CW, Schirhagl R, Lin JM, Zare RN. Lab Chip. 2011;11:238–245. doi: 10.1039/c0lc00121j. [DOI] [PubMed] [Google Scholar]
- 19.Yao B, Luo GA, Feng X, Wang W, Chen LX, Wang YM. Lab Chip. 2004;4:603–607. doi: 10.1039/b408422e. [DOI] [PubMed] [Google Scholar]
- 20.Chabert M, Viovy JL. Proc Natl Acad Sci U S A. 2008;105:3191–3196. doi: 10.1073/pnas.0708321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, Bessette PH, Qian J, Meinhart CD, Daugherty PS, Soh HT. Proc Natl Acad Sci U S A. 2005;102:15757–15761. doi: 10.1073/pnas.0507719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keenan TM, Hsu CH, Folch A. Applied Physics Letters. 2006:89. [Google Scholar]
- 23.Bhattacharjee N, Li N, Keenan TM, Folch A. Integr Biol (Camb) 2010;2:669–679. doi: 10.1039/c0ib00038h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung BG, Lin F, Jeon NL. Lab Chip. 2006;6:764–768. doi: 10.1039/b512667c. [DOI] [PubMed] [Google Scholar]
- 25.Saadi W, Rhee S, Lin F, Vahidi B, Chung B, Jeon N. Biomedical Microdevices. 2007;9:627–635. doi: 10.1007/s10544-007-9051-9. [DOI] [PubMed] [Google Scholar]
- 26.Breckenridge MT, Egelhoff TT, Baskaran H. Biomed Microdevices. 2010;12:543–553. doi: 10.1007/s10544-010-9411-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Irimia D, Charras G, Agrawal N, Mitchison T, Toner M. Lab on a Chip. 2007;7:1783–1790. doi: 10.1039/b710524j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haddox JL, Knowles IW, Sommers CI, Pfister RR. Journal of Immunological Methods. 1994;171:1–14. doi: 10.1016/0022-1759(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 29.Cao X, Shoichet MS. Neuroscience. 2001;103:831–840. doi: 10.1016/s0306-4522(01)00029-x. [DOI] [PubMed] [Google Scholar]
- 30.Wong AP, Perez-Castillejos R, Christopher Love J, Whitesides GM. Biomaterials. 2008;29:1853–1861. doi: 10.1016/j.biomaterials.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng S-Y, Heilman S, Wasserman M, Archer S, Shuler ML, Wu M. Lab on a Chip. 2007;7:763–769. doi: 10.1039/b618463d. [DOI] [PubMed] [Google Scholar]
- 32.Wu H, Huang B, Zare RN. Journal of the American Chemical Society. 2006;128:4194–4195. doi: 10.1021/ja058530o. [DOI] [PubMed] [Google Scholar]
- 33.Abhyankar VV, Lokuta MA, Huttenlocher A, Beebe DJ. Lab on a Chip. 2006;6:389–393. doi: 10.1039/b514133h. [DOI] [PubMed] [Google Scholar]
- 34.Diao J, Young L, Kim S, Fogarty EA, Heilman SM, Zhou P, Shuler ML, Wu M, DeLisa MP. Lab on a Chip. 2006;6:381–388. doi: 10.1039/b511958h. [DOI] [PubMed] [Google Scholar]
- 35.Adler J. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 36.Colvin RA, Means TK, Diefenbach TJ, Moita LF, Friday RP, Sever S, Campanella GS, Abrazinski T, Manice LA, Moita C, Andrews NW, Wu D, Hacohen N, Luster AD. Nat Immunol. 2010;11:495–502. doi: 10.1038/ni.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Kumamoto Y, Wang P, Gan X, Lehmann D, Smrcka AV, Cohn L, Iwasaki A, Li L, Wu D. J Biol Chem. 2009;284:28599–28606. doi: 10.1074/jbc.M109.047282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyden S. J Exp Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zigmond SH. Methods Enzymol. 1988;162:65–72. doi: 10.1016/0076-6879(88)62064-7. [DOI] [PubMed] [Google Scholar]
- 40.Zicha D, Dunn G, ones G. Methods Mol Biol. 1997;75:449–457. doi: 10.1385/0-89603-441-0:449. [DOI] [PubMed] [Google Scholar]
- 41.Irimia D. Annu Rev Biomed Eng. 2010;12:259–284. doi: 10.1146/annurev-bioeng-070909-105241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paliwal S, Iglesias PA, Campbell K, Hilioti Z, Groisman A, Levchenko A. Nature. 2007;446:46–51. doi: 10.1038/nature05561. [DOI] [PubMed] [Google Scholar]
- 43.Shamloo A, Xu H, Heilshorn S. Tissue Eng Part A. 2012;18:320–330. doi: 10.1089/ten.tea.2011.0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shamloo A, Ma N, Poo MM, Sohn LL, Heilshorn SC. Lab Chip. 2008;8:1292–1299. doi: 10.1039/b719788h. [DOI] [PubMed] [Google Scholar]
- 45.Onuki-Nagasaki R, Nagasaki A, Hakamada K, Uyeda TQ, Fujita S, Miyake M, Miyake J. Lab Chip. 2008;8:1502–1506. doi: 10.1039/b803879a. [DOI] [PubMed] [Google Scholar]
- 46.Hattori H, Subramanian KK, Sakai J, Jia Y, Li Y, Porter TF, Loison F, Sarraj B, Kasorn A, Jo H, Blanchard C, Zirkle D, McDonald D, Pai SY, Serhan CN, Luo HR. Proc Natl Acad Sci U S A. 2010;107:3546–3551. doi: 10.1073/pnas.0914351107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cho BS, Schuster TG, Zhu X, Chang D, Smith GD, Takayama S. Analytical Chemistry. 2003;75:1671–1675. doi: 10.1021/ac020579e. [DOI] [PubMed] [Google Scholar]
- 48.Tai W, Yeh CF, Wu CC, Hsu CH. 15th International Conference on Miniaturized Systems for Chemistry and Life Sciences Seattle. Washington, USA: 2011. [Google Scholar]
- 49.Valdivia RH, Hromockyj AE, Monack D, Ramakrishnan L, Falkow S. Gene. 1996;173:47–52. doi: 10.1016/0378-1119(95)00706-7. [DOI] [PubMed] [Google Scholar]
- 50.Datsenko KA, Wanner BL. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lutz MB, Kukutsch N, Ogilvie AL, Rossner S, Koch F, Romani N, Schuler G. J Immunol Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 52.Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. PLoS Pathog. 2006;2:e11. doi: 10.1371/journal.ppat.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rytkonen A, Poh J, Garmendia J, Boyle C, Thompson A, Liu M, Freemont P, Hinton JC, Holden DW. Proc Natl Acad Sci U S A. 2007;104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konradt C, Frigimelica E, Nothelfer K, Puhar A, Salgado-Pabon W, di Bartolo V, Scott-Algara D, Rodrigues CD, Sansonetti PJ, Phalipon A. Cell Host Microbe. 2011;9:263–272. doi: 10.1016/j.chom.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Rajashree P, Supriya P, Das SD. Immunobiology. 2008;213:567–575. doi: 10.1016/j.imbio.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 56.Humrich JY, Humrich JH, Averbeck M, Thumann P, Termeer C, Kampgen E, Schuler G, Jenne L. Immunology. 2006;117:238–247. doi: 10.1111/j.1365-2567.2005.02292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ouwehand K, Santegoets SJ, Bruynzeel DP, Scheper RJ, de Gruijl TD, Gibbs S. Eur J Immunol. 2008;38:3050–3059. doi: 10.1002/eji.200838384. [DOI] [PubMed] [Google Scholar]
- 58.Hensel M, Shea JE, Raupach B, Monack D, Falkow S, Gleeson C, Kubo T, Holden DW. Mol Microbiol. 1997;24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 59.Freeman JA, Rappl C, Kuhle V, Hensel M, Miller SI. J Bacteriol. 2002;184:4971–4980. doi: 10.1128/JB.184.18.4971-4980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shotland Y, Kramer H, Groisman EA. Mol Microbiol. 2003;49:1565–1576. doi: 10.1046/j.1365-2958.2003.03668.x. [DOI] [PubMed] [Google Scholar]
- 61.Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW. Science. 2010;328:1040–1043. doi: 10.1126/science.1189000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abrahams GL, Muller P, Hensel M. Traffic. 2006;7:950–965. doi: 10.1111/j.1600-0854.2006.00454.x. [DOI] [PubMed] [Google Scholar]
- 63.Diacovich L, Dumont A, Lafitte D, Soprano E, Guilhon AA, Bignon C, Gorvel JP, Bourne Y, Meresse S. J Biol Chem. 2009;284:33151–33160. doi: 10.1074/jbc.M109.034975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henry T, Couillault C, Rockenfeller P, Boucrot E, Dumont A, Schroeder N, Hermant A, Knodler LA, Lecine P, Steele-Mortimer O, Borg JP, Gorvel JP, Meresse S. Proc Natl Acad Sci U S A. 2006;103:13497–13502. doi: 10.1073/pnas.0605443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuhle V, Jackel D, Hensel M. Traffic. 2004;5:356–370. doi: 10.1111/j.1398-9219.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 66.Miao EA, Brittnacher M, Haraga A, Jeng RL, Welch MD, Miller SI. Mol Microbiol. 2003;48:401–415. doi: 10.1046/j.1365-2958.2003.t01-1-03456.x. [DOI] [PubMed] [Google Scholar]
- 67.Ohlson MB, Huang Z, Alto NM, Blanc MP, Dixon JE, Chai J, Miller SI. Cell Host Microbe. 2008;4:434–446. doi: 10.1016/j.chom.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim S, Kim HJ, Jeon NL. Integr Biol (Camb) 2010;2:584–603. doi: 10.1039/c0ib00055h. [DOI] [PubMed] [Google Scholar]
- 69.Haessler U, Pisano M, Wu M, Swartz MA. Proc Natl Acad Sci U S A. 2011;108:5614–5619. doi: 10.1073/pnas.1014920108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yan J, Mihaylov V, Xu X, Brzostowski JA, Li H, Liu L, Veenstra TD, Parent CA, Jin T. Dev Cell. 2012;22:92–103. doi: 10.1016/j.devcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ricart BG, John B, Lee D, Hunter CA, Hammer DA. J Immunol. 2011;186:53–61. doi: 10.4049/jimmunol.1002358. [DOI] [PubMed] [Google Scholar]
- 72.Cheminay C, Schoen M, Hensel M, Wandersee-Steinhauser A, Ritter U, Korner H, Rollinghoff M, Hein J. Microb Pathog. 2002;32:207–218. doi: 10.1006/mpat.2002.0497. [DOI] [PubMed] [Google Scholar]
- 73.Riol-Blanco L, Sanchez-Sanchez N, Torres A, Tejedor A, Narumiya S, Corbi AL, Sanchez-Mateos P, Rodriguez-Fernandez JL. J Immunol. 2005;174:4070–4080. doi: 10.4049/jimmunol.174.7.4070. [DOI] [PubMed] [Google Scholar]
- 74.Mattila PK, Lappalainen P. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 75.Ridley AJ. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 76.Beuzon CR, Meresse S, Unsworth KE, Ruiz-Albert J, Garvis S, Waterman SR, Ryder TA, Boucrot E, Holden DW. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim H, White CD, Sacks DB. FEBS Lett. 2011 doi: 10.1016/j.febslet.2011.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Petrovska L, Aspinall RJ, Barber L, Clare S, Simmons CP, Stratford R, Khan SA, Lemoine NR, Frankel G, Holden DW, Dougan G. Cell Microbiol. 2004;6:1071–1084. doi: 10.1111/j.1462-5822.2004.00419.x. [DOI] [PubMed] [Google Scholar]
- 79.Halici S, Zenk SF, Jantsch J, Hensel M. Infect Immun. 2008;76:4924–4933. doi: 10.1128/IAI.00531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peachman KK, Rao M, Palmer DR, Zidanic M, Sun W, Alving CR, Rothwell SW. Immunol Lett. 2004;95:13–24. doi: 10.1016/j.imlet.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 81.Hess J, Ladel C, Miko D, Kaufmann SH. J Immunol. 1996;156:3321–3326. [PubMed] [Google Scholar]
- 82.Ziegler E, Gueler F, Rong S, Mengel M, Witzke O, Kribben A, Haller H, Kunzendorf U, Krautwald S. J Am Soc Nephrol. 2006;17:2521–2532. doi: 10.1681/ASN.2005070782. [DOI] [PubMed] [Google Scholar]
- 83.Monack DM, Bouley DM, Falkow S. J Exp Med. 2004;199:231–241. doi: 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bedoui S, Kupz A, Wijburg OL, Walduck AK, Rescigno M, Strugnell RA. J Immunol. 2010;184:2237–2242. doi: 10.4049/jimmunol.0902871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: S. typhimurium inhibits DC migration toward CXCL12 independently of SseI. A-D) DCs infected with GFP-expressing WT or ΔsseI S. typhimurium were cultured in a gradient of CXCL12 and imaged every 7 min over 5 h. The chemotactic indices (A) and angular histograms (B and C) are presented. A) Line and error bars represent the mean and SEM, respectively, and all P-values were determined using the Mann-Whitney.
Supplemental Figure 2: DCs infected with SPI2 mutants positively selected by the microfluidic screen have lower migratory speed compared to uninfected DCs. DCs infected with the indicated S. typhimurium strains were cultured in a gradient of CCL19 and imaged every 7 min over 5 h. The cell migratory tracks of both infected DC and their uninfected neighbors were recorded and used to calculate their speeds. N is the number of individual cells tracked. Lines represent the median, and P-values were determined using the Mann-Whitney test and are indicated above the graph.
Supplemental Figure 3: DCs located inside of the capillaries during time-lapse experiments do not affect the chemical gradient. A) Time-lapse images were taken every 84 min of the capillaries leading to the source channel in a chemical gradient containing CCL19 and 10 kD dextran Texas Red; DIC, red fluorescence images and plot profiles of the red fluorescence are shown (yellow outline denotes plot profile range, taken using imageJ). B) The number of DCs per capillary averaged over 3 separate videos (error bars indicate SEM). C) The speed of DCs infected with ΔsifA(psifA) Salmonella inside the capillaries (N =6) compared to infected DCs in the cell-viewing chamber (bar indicates median speed).