The stability of PIF3 LIKE1 (PIL1) is regulated through physical interactions with COP1 and phyB, and PIL1 coordinates with HFR1 to inhibit the transcriptional activity of PIFs and promote photomorphogenic development of Arabidopsis.
Abstract
In Arabidopsis thaliana, the cryptochrome and phytochrome photoreceptors act together to promote photomorphogenic development. The cryptochrome and phytochrome signaling mechanisms interact directly with CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1), a RING motif–containing E3 ligase that acts to negatively regulate photomorphogenesis. COP1 interacts with and ubiquitinates the transcription factors that promote photomorphogenesis, such as ELONGATED HYPOCOTYL5 and LONG HYPOCOTYL IN FAR-RED1 (HFR1), to inhibit photomorphogenic development. Here, we show that COP1 physically interacts with PIF3-LIKE1 (PIL1) and promotes PIL1 degradation via the 26S proteasome. We further demonstrate that phyB physically interacts with PIL1 and enhances PIL1 protein accumulation upon red light irradiation, probably through suppressing the COP1–PIL1 association. Biochemical and genetic studies indicate that PIL1 and HFR1 form heterodimers and promote photomorphogenesis cooperatively. Moreover, we demonstrate that PIL1 interacts with PIF1, 3, 4, and 5, resulting in the inhibition of the transcription of PIF direct-target genes. Our results reveal that PIL1 stability is regulated by phyB and COP1, likely through physical interactions, and that PIL1 coordinates with HFR1 to inhibit the transcriptional activity of PIFs, suggesting that PIL1, HFR1, and PIFs constitute a subset of antagonistic basic helix-loop-helix factors acting downstream of phyB and COP1 to regulate photomorphogenic development.
INTRODUCTION
Light serves not only as a source of energy, but also as an informational signal to modulate almost every aspect of plant development throughout the life cycle, such as seed germination, seedling deetiolation, shade avoidance, phototropism, circadian rhythm, and floral transition (Fankhauser and Chory, 1997; Deng and Quail, 1999). Using Arabidopsis thaliana as a model organism has led to substantial advances in understanding the light control of seedling development. Photomorphogenesis is one of the well-studied light responses. Arabidopsis seedlings grown in the dark display an etiolated phenotype such as folded apical hooks, elongated hypocotyls, and absence of chlorophyll synthesis, while those in the light display expanded cotyledons, short hypocotyls, and accumulation of chlorophyll (McNellis and Deng, 1995). Two well-characterized families of photoreceptors are primarily responsible for light promotion of photomorphogenesis: cryptochromes and phytochromes. The cryptochrome family members containing CRY1 and CRY2 absorb blue light (300 to 500 nm), whereas the phytochrome family members (phyA to phyE) perceive red and far-red light (600 to 750 nm) (Kendrick and Kronenberg, 1994; Briggs and Olney, 2001; Lin, 2002). phyA functions predominately in far-red light, while phyB primarily regulates plant growth in red light (Whitelam et al., 1993; Neff et al., 2000). Phytochromes exist in two photoreversible forms: a red light–absorbing Pr form (biologically inactive) and a far-red light–absorbing Pfr form (biologically active) (Rockwell et al., 2006). Most recently, UVR8, a UV-B light photoreceptor, has been found to exclusively act in UV-B signaling (Brown et al., 2005; Heijde and Ulm, 2012).
The CONSTITUTIVELY PHOTOMORPHOGENIC (COP)/DE-ETIOLATED/FUSCA proteins are repressors of photomorphogenesis that act downstream of the multiple photoreceptors. They consist of at least three distinct complexes: the COP1-SPA complex, the COP9 signalosome, and the COP10-DET1-DDB1 (CDD) complex (Deng et al., 1991; Wei et al., 1994; Suzuki et al., 2002; Serino and Deng, 2003; Laubinger et al., 2004). COP1 is a RING-finger E3 ubiquitin ligase, which contains three recognizable structural domains: a Zn2+ binding RING finger motif, a coiled-coil domain, and a WD-40 repeat domain (Deng et al., 1992; McNellis et al., 1994). A myriad of transcription factors that promote photomorphogenesis were found to be suppressed by COP1 through direct interactions, such as ELONGATED HYPOCOTYL5 (HY5), a bZIP transcription factor, and LONG HYPOCOTYL IN FAR-RED1 (HFR1), an atypical basic helix-loop-helix (bHLH) transcription factor (Ang et al., 1998; Osterlund et al., 2000; Jang et al., 2005; Yang et al., 2005). SPAs (SUPPRESSOR OF PHYA-105s; SPA1, SPA2, SPA3, and SPA4), isolated as suppressors of phyA, show high sequence similarity to COP1 (Hoecker et al., 1998, 1999; Laubinger et al., 2004). SPA1 physically interacts with COP1 and enhances its E3 ligase activity (Saijo et al., 2003; Seo et al., 2003). Photoactivated CRY1 and CRY2 physically interact with SPAs, resulting in the dissociation of the COP1-SPA complex and eventually the accumulation of downstream COP1 substrates such as HY5 and HFR1 (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011).
Upon exposure to light, phytochromes translocate from the cytoplasm to the nucleus (Nagy et al., 2000; Nagy and Schäfer, 2000; Nagatani, 2004) and interact with a group of transcription factors called PHYTOCHROME-INTERACTING FACTORS (PIFs) (Castillon et al., 2007; Leivar and Quail, 2011). PIFs belong to the bHLH superfamily and negatively regulate photomorphogenic development (Ni et al., 1998; Huq et al., 2004; Monte et al., 2004; Shin et al., 2007; Leivar et al., 2008). PIFs contain conserved N-terminal sequences, including the active phyA binding (APA) and phyB binding (APB) motifs (Khanna et al., 2004; Leivar and Quail, 2011), which are required for their interactions with phyA and phyB, respectively (Zhu et al., 2000; Khanna et al., 2004; Al-Sady et al., 2006; Shen et al., 2008). It is well established that, upon light exposure, PIF1, PIF3, PIF4, and PIF5 are rapidly phosphorylated in a phytochrome-dependent manner, which induces their degradation via the 26S proteasome (Park et al., 2004; Shen et al., 2005, 2008; Al-Sady et al., 2006; Shen et al., 2007). Chromatin immunoprecipitation studies identified a number of sites in the promoters of genes including PIL1, XTR7, and IAA19, bound by PIFs, which contain a series of G-boxes (CACGTG) (Martinez-Garcia et al., 2000; Shin et al., 2007; de Lucas et al., 2008; Moon et al., 2008; Hornitschek et al., 2009; Oh et al., 2009; Sun et al., 2013; Zhang et al., 2013).
PIF3-LIKE1 (PIL1) shares amino acid sequence similarity to PIF3 and localizes to the nucleus (Yamashino et al., 2003; Khanna et al., 2006). The G-boxes in the PIL1 promoter are responsible for the rapid upregulation of PIL1 by PIFs (Hornitschek et al., 2009; Zhang et al., 2013). The pil1 mutants display elongated hypocotyls and smaller cotyledons, compared with the wild type under continuous red and far-red light (Salter et al., 2003; Khanna et al., 2006; Roig-Villanova et al., 2006), suggesting that PIL1 functions as a positive regulator of photomorphogenesis.
Although previous studies have characterized a number of COP1-interacting proteins, it is unknown whether there are additional COP1-interacting proteins that may act in cryptochrome- or phytochrome-mediated signaling pathways. In this study, we demonstrate that PIL1 interacts with COP1 and is degraded in a COP1-dependent manner. PIL1 physically interacts with phyB, and its abundance is promoted by phyB upon red light exposure. Furthermore, PIL1 interacts with HFR1 and PIFs (PIF1, 3, 4, and 5 studied here) and coordinates with HFR1 to suppress the transcriptional activity of PIFs and promote photomorphogenesis. Taken together, our results suggest a mechanism by which PIL1, HFR1, and PIFs constitute a subset of antagonistic bHLH transcription factors, whose activities are modulated by COP1 and phyB.
RESULTS
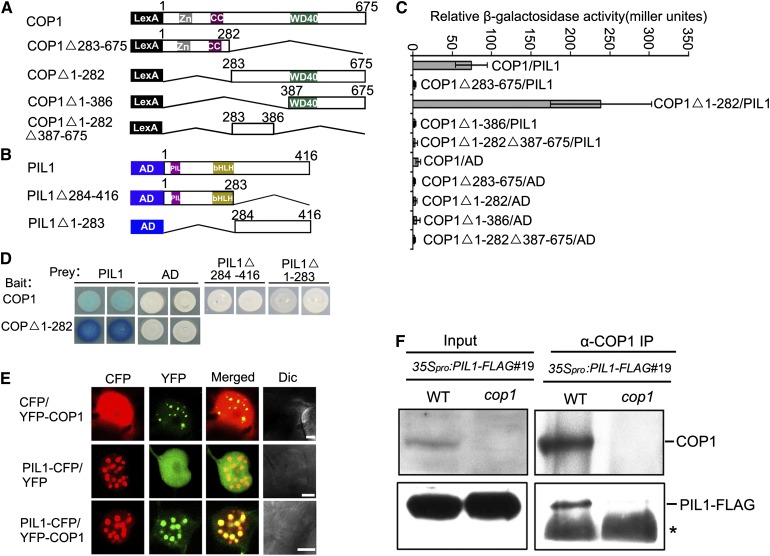
PIL1 Physically Interacts with COP1 in Yeast Cells
The signaling mechanism of Arabidopsis cryptochromes, phytochromes, and UVR8 involves direct interaction with COP1 (Yang et al., 2000, 2001; Wang et al., 2001; Seo et al., 2004; Favory et al., 2009; Jang et al., 2010). A number of transcription factors, such as HY5, HFR1, and CO, have been shown to interact with COP1 and are degraded in a COP1-dependent manner (Ang et al., 1998; Osterlund et al., 2000; Jang et al., 2005; Yang et al., 2005; L.J. Liu et al., 2008). We sought to identify new COP1-interacting proteins and designated PIL1 as a candidate based on the previous demonstrations that PIL1 shares high amino acid sequence similarity to HFR1 and that the pil1 mutant shows reduced light responsiveness, similar to the hfr1 mutant (Duek and Fankhauser, 2003; Salter et al., 2003; Khanna et al., 2006; Roig-Villanova et al., 2006; Zhang et al., 2008). We further analyzed the phenotype of the pil1-2 T-DNA insertion mutant and found that this mutant allele displays taller hypocotyls than the wild type in continuous blue, red, and far-red light, respectively, confirming a positive role of PIL1 in regulating photomorphogenesis (Supplemental Figure 1). We performed a yeast two-hybrid assay to examine whether PIL1 might interact with COP1. To do this, a bait construct expressing the LexA DNA binding domain fused to the full-length COP1 protein and a prey construct expressing the B42 transcriptional activation domain fused to full-length PIL1 were prepared (Figures 1A and 1B). The results show that the full-length COP1 interacts with PIL1 as indicated by the high β-galactosidase activity (Figures 1C and 1D). Moreover, domain mapping assays demonstrate that PIL1 strongly interacts with the C-terminal WD40-containing fragment of COP1 (COP1△1-282) but does not interact with either the N-terminal fragment of COP1 (COP1△283-675), a shorter C-terminal WD40-containing fragment (COP1△1-386), or an internal fragment between the N-terminal and WD40-domain (COP1△1-282△387-675) (Figures 1C and 1D). Immunoblot analysis indicates that the protein levels of the various COP1 fragments are similar to or higher than those of the full-length COP1 in yeast (Supplemental Figure 3A). These results indicate that the C-terminal domain of COP1 may mediate the interaction of COP1 with PIL1 in yeast cells. To determine the domains of PIL1 that mediate the interaction with COP1, we generated two prey constructs expressing an N-terminal 283 amino acid fragment of PIL1, including the PIL and bHLH domains (PIL1△284-416) and a C-terminal fragment of PIL1 (PIL1△1-283), respectively (Figure 1B), and performed a yeast two-hybrid assay. These results show that neither the N-terminal nor the C-terminal fragment of PIL1 interacts with COP1 (Figure 1D). Immunoblot analysis demonstrates that the protein levels of the PIL1△1-283 and PIL1△284-416 are higher and a little lower than those of the full-length PIL1 in yeast, respectively (Supplemental Figure 3B). Therefore, it is likely that the overall structure of PIL1 is required for its interaction with COP1.
Figure 1.
PIL1 Physically Interacts with COP1.
(A) Yeast two-hybrid bait constructs comprising COP1 fragments fused to the LexA DNA binding domain (LexA).
(B) Prey constructs of PIL1 fragments fused to the B42 transcriptional activation domain (AD).
(C) Quantitative yeast two-hybrid assay defines domains of COP1 essential for the interactions with PIL1. All the vector combinations are given as bait/prey. Error bars represent ±sd (n = 10).
(D) Plate assays showing the interaction of PIL1 fragments with COP1 or the C-terminal WD40-containing fragment of COP1 (COP1△1-282). The panels show the corresponding β-galactosidase activities represented by blue precipitates.
(E) PIL1 and COP1 localize together to NBs in tobacco cells. Dic, differential interference contrast. Bars = 5 μm.
(F) COP1 interacts with PIL1 in vivo. Four-day-old blue light–grown 35Spro:PIL1-FLAG#19/WT and 35Spro:PIL1-FLAG#19/cop1 seedlings were transferred to darkness for 16 h and subjected to a Co-IP assay using anti-COP1 antibody, and the immunoprecipitates were detected using anti-COP1 and anti-FLAG antibodies, respectively. Asterisks show the heavy chain of IgG.
PIL1 Physically Interacts with COP1 in Plant Cells
To examine whether PIL1 interacts with COP1 in plant cells, we first transiently expressed PIL1 tagged with cyan fluorescent protein (PIL1-CFP) and COP1 tagged with yellow fluorescent protein (YFP-COP1), either individually or together, in tobacco (Nicotiana tabacum) cells. As anticipated, PIL1 and COP1 are colocalized in the same nuclear bodies (NBs) (Figure 1E), indicating their interaction in plant cells. We then investigated whether PIL1 interacts with COP1 in Arabidopsis. To do this, we prepared a construct expressing PIL1 fused to 3×FLAG driven by the 35S promoter of cauliflower mosaic virus and transformed it into the wild-type background. We found that hypocotyl elongation was inhibited to a greater extent in the transgenic lines than in the wild type under continuous blue, red, and far-red light, respectively, suggesting that the PIL1-FLAG fusion protein is biologically functional (Supplemental Figure 1). One representative line, 35Spro:PIL1-FLAG#19/WT, was introgressed into the cop1 mutant background by genetic crossing. We used the 35Spro:PIL1-FLAG#19/WT and 35Spro:PIL1-FLAG#19/cop1 transgenic plants to perform a coimmunoprecipitation (Co-IP) assay. The results show that immunoprecipitation of endogenous COP1 pulls down PIL1-FLAG in 35Spro:PIL1-FLAG#19/WT seedlings, but not in 35Spro:PIL1-FLAG#19/cop1, demonstrating that PIL1 interacts with COP1 in Arabidopsis (Figure 1F).
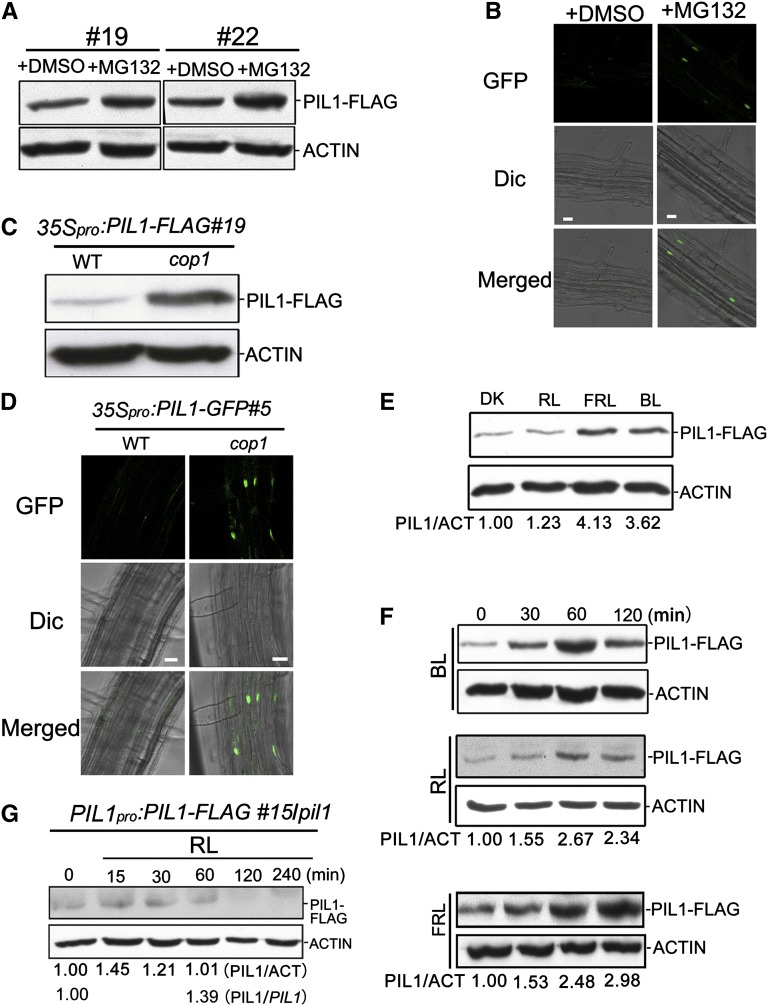
PIL1 Undergoes Degradation in a COP1-Dependent Manner in Darkness
With the demonstration that COP1 physically interacts with PIL1, we asked whether PIL1 stability is regulated by COP1. To test this possibility, we first determined whether PIL1 is degraded via the 26S proteasome. 35Spro:PIL1-FLAG#19/WT and 35Spro:PIL1-FLAG#22/WT seedlings were grown in darkness for 4 d and then treated with MG132 or DMSO for 6 h. Total protein was extracted and equal amounts were subjected to immunoblot analysis. The results reveal that MG132 treatment significantly increases the accumulation of PIL1-FLAG in these two independent transgenic lines, indicating that PIL1 is subjected to 26S proteasome-mediated proteolysis in darkness (Figure 2A). We also introduced a construct expressing PIL1 fused to green fluorescent protein (GFP) driven by the 35S promoter into the wild-type background and found that MG132, but not DMSO promotes the nuclear accumulation of PIL1 in the dark (Figure 2B). We then examined whether COP1 is responsible for PIL1 degradation in the dark by analyzing PIL1 accumulation in dark-grown 35Spro:PIL1-FLAG#19/WT and 35Spro:PIL1-FLAG#19/cop1 seedlings, respectively. The results show that PIL1-FLAG accumulates at much higher levels in the cop1 mutant than in the wild type (Figure 2C). We also introgressed the 35Spro:PIL1-GFP transgene from the wild type into the cop1 mutant background by genetic crossing to generate 35Spro:PIL1-GFP#5/cop1 transgenic plants and analyzed the GFP signal through fluorescence microscopy. The GFP signal is clearly detected in root cells of dark-grown 35Spro:PIL1-GFP#5/cop1 seedlings, but hardly detected in 35Spro:PIL1-GFP#5/WT (Figure 2D). These results indicate that COP1 promotes PIL1 degradation in darkness.
Figure 2.
PIL1 Protein Is Degraded in a COP1-Dependent Manner and Accumulates upon Light Irradiation.
(A) and (B) PIL1 is stabilized by the 26S proteasome inhibitor MG132.
(A) Total protein extracted from two independent lines of the 35Spro:PIL1-FLAG/WT (#19 and #22) transgenic plants grown in darkness for 4 d and then treated with MG132 or DMSO for 6 h was subjected to immunoblot analysis with anti-FLAG and anti-ACTIN antibodies, respectively.
(B) Root cells of 35Spro:PIL1-GFP#5/WT seedlings grown under conditions of (A) were analyzed by fluorescence microscopy. Bars = 20 μm.
(C) and (D) PIL1 accumulated in the cop1 mutant background.
(C) Total protein extracted from 4-d-old dark-grown 35Spro:PIL1-FLAG#19/WT and 35Spro:PIL1-FLAG#19/cop1-4 seedlings was subjected to immunoblot analysis with anti-FLAG and anti-ACTIN antibodies, respectively.
(D) Root cells of the 35Spro:PIL1-GFP#5/WT and 35Spro:PIL1-GFP#5/cop1 seedlings grown under conditions of (C) were analyzed by fluorescence microscopy. Bars = 20 μm.
(E) Immunoblot showing PIL1-FLAG in 35Spro:PIL1-FLAG#19/WT seedlings grown in continuous indicated conditions for 4 d.
(F) PIL1 accumulated by exposure to the indicated light conditions. Total protein extracted from the 35Spro:PIL1-FLAG#19/WT seedlings grown in darkness for 4 d and then transferred to the indicated light conditions for the indicated periods of time was subjected to immunoblot analysis.
(G) Immunoblot analysis of PIL1-FLAG in PIL1pro:PIL1-FLAG#15/pil1 seedlings grown in continuous darkness for 4 d and then exposed to red light for the indicated time periods. PIL1/PIL1 represents the ratio of PIL1 protein to PIL1 mRNA (from Supplemental Figure 4) under the indicated light conditions.
(E) to (G) Total protein was subjected to immunoblot analysis with anti-FLAG and anti-ACTIN antibodies. PIL1/ACT indicates the relative band intensities of PIL1-FLAG normalized to ACTIN and is presented relative to that in darkness set at unity. DK, darkness; BL, blue light; RL, red light; FRL, far-red light.
Light Exposure Promotes PIL1 Accumulation
We examined whether light regulates PIL1 accumulation by immunoblotting analysis of the 35Spro:PIL1-FLAG#19/WT seedlings grown in continuous darkness, blue, red, and far-red light, respectively. The results indicate that blue and far-red light strongly promote PIL1 accumulation, whereas red light has mild promotion effects (Figure 2E). Next, we analyzed the dynamic changes of PIL1 by immunoblot analysis of 35Spro:PIL1-FLAG#19/WT seedlings grown in darkness for 4 d and then exposed to blue, red, and far-red light for 30, 60, and 120 min, respectively. The results show that 30 to 60 min of blue and far-red light irradiation dramatically promotes PIL1-FLAG, whereas 30 to 60 min red light exposure moderately enhances PIL1 accumulation (Figure 2F). Prolonged far-red light irradiation (120 min) still increases PIL1-FLAG accumulation, whereas prolonged blue and red light exposure attenuate PIL1-FLAG accumulation, compared with 60 min exposure (Figure 2F).
Considering that the PIL1 transcript rapidly decreases in response to red light illumination (Khanna et al., 2006), we asked whether light-grown Arabidopsis seedlings accumulate PIL1. To do this, we generated a construct expressing PIL1-FLAG driven by the PIL1 native promoter (−2000 to −1 bp) and transformed it into the pil1-2 mutant. PIL1pro:PIL1-FLAG/pil1 transgenic plants show the wild-type phenotypes (Supplemental Figure 2), indicating that the PIL1pro:PIL1-FLAG transgene is biologically functional. Quantitative RT-PCR (qRT-PCR) analysis indicates that the transcript levels of the PIL1 transgene in all these transgenic plants are reduced upon 60 min red light irradiation, but to a lesser extent than endogenous PIL1 in the wild type (Supplemental Figure 4). It is possible that the selected 2.0-kb promoter fragment of PIL1 lacks some transcriptional regulatory elements present in its native promoter. Immunoblot analysis of PIL1 in PIL1pro:PIL1-FLAG#15/pil1 seedlings grown in continuous blue, red, and far-red light indicates that, consistent with the results obtained for 35Spro:PIL1-FLAG#19/WT, blue and far-red light strongly enhance PIL1 accumulation, whereas red light hardly does (Supplemental Figure 5). We then analyzed the dynamic changes of PIL1-FLAG in PIL1pro:PIL1-FLAG#15/pil1 seedlings grown in darkness for 4 d and then exposed to red light for 15 to 240 min. PIL1 accumulation slightly increases upon 30 min red light illumination and declines to a level similar to that in darkness upon 60 min exposure (Figure 2G). An exposure of longer than 60 min further attenuates PIL1 accumulation. The ratio of PIL1 protein to PIL1 transgene mRNA (PIL1/PIL1) increases by 39% upon a 60 min exposure (Figure 2G; Supplemental Figure 4). Given that the level of endogenous PIL1 transcripts declines much more quickly than the PIL1 transgene upon 60 min red light irradiation (Khanna et al., 2006; Supplemental Figure 4), these results indicate that initial red light illumination promotes PIL1 protein accumulation.
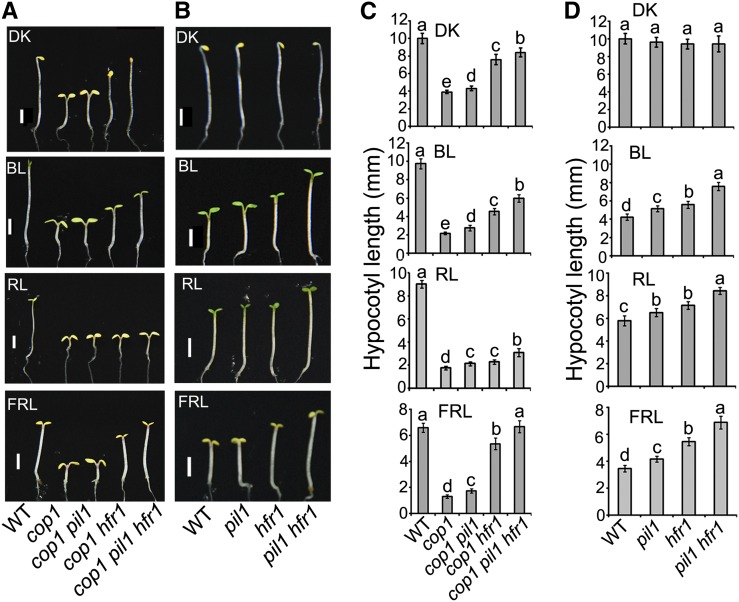
PIL1 and HFR1 Act Genetically Downstream of COP1 to Promote Photomorphogenesis
Since COP1 physically interacts with PIL1 and as the PIL1 protein level is regulated by COP1 (Figures 1, 2C, and 2D), we analyzed the genetic interaction between COP1 and PIL1 by generating a cop1 pil1 double mutant, and we found that this double mutant displays a slightly but statistically significantly (Tukey’s LSD test, P ≤ 0.01) elongated hypocotyl phenotype compared with the cop1 single mutant in the dark, blue, red, and far-red light, respectively (Figures 3A and 3C). Since both PIL1 and HFR1 act to promote photomorphogenesis and undergo protein degradation in a COP1-dependent manner (Jang et al., 2005; Yang et al., 2005; Figures 1, 2C, and 2D; Supplemental Figures 1 and 2), we asked whether PIL1 may act additively with HFR1 to promote photomorphogenesis. To test this possibility, we first generated the pil1 hfr1 double mutant and found that the hypocotyls of the double mutant are significantly taller than those of the pil1 and hfr1 single mutant seedlings under blue, red, and far-red light, respectively (Figures 3B and 3D). We then constructed a cop1 pil1 hfr1 triple mutant and found that, compared with the cop1 pil1 or cop1 hfr1 double mutant, the triple mutant seedlings develop dramatically elongated hypocotyls in darkness and various light conditions and folded or reduced cotyledons in the dark, blue, and far-red light, respectively (Figures 3A and 3C). These results indicate that PIL1 and HFR1 act additively to regulate photomorphogenesis and are situated genetically downstream of COP1.
Figure 3.
PIL1 and HFR1 Act Genetically Downstream of COP1.
(A) and (B) Photographs of representative seedlings of the indicated genotypes. Seedlings grown under continuous light conditions for 4 d. Bars = 2 mm.
(A) DK, darkness; BL, blue light (0.3 μmol/s·m2); RL, red light (0.3 μmol/s·m2); FRL, far-red light (0.5 μmol/s·m2).
(B) DK, darkness; BL, blue light (15 μmol/s·m2); RL, red light (8 μmol/s·m2); FRL, far-red light (2 μmol/s·m2).
(C) and (D) Quantification of hypocotyl lengths of the seedlings shown in (A) and (B). Error bars represent ±sd (n = 30). The letters “a” to “e” indicate statistically significant differences between means for hypocotyl lengths of the indicated genotypes, as determined by Tukey’s LSD test (P ≤ 0.01).
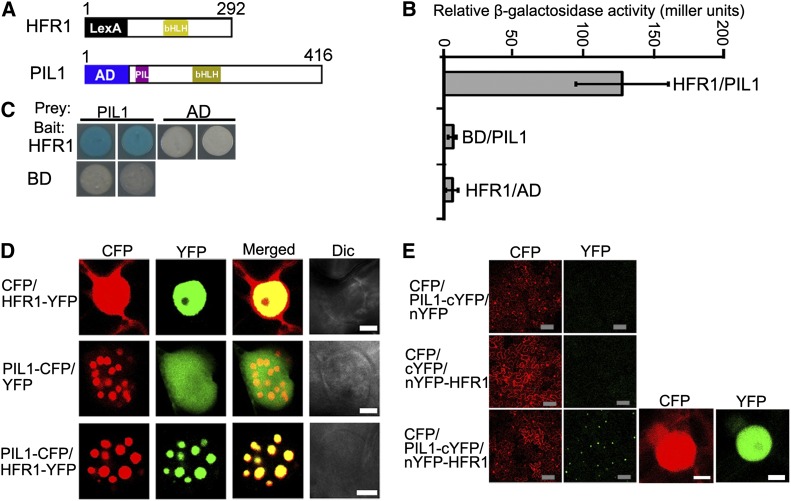
PIL1 Physically Interacts with HFR1
The genetic interaction between PIL1 and HFR1 prompted us to investigate whether PIL1 and HFR1 physically interact. We first performed a yeast two-hybrid assay with constructs expressing the LexA DNA binding domain fused to HFR1 and pB42 AD-PIL1 (Figure 4A). Indeed, a strong interaction between PIL1 and HFR1 was observed in yeast cells (Figures 4B and 4C). The PIL1–HFR1 interaction was then confirmed by in vivo protein colocalization and bimolecular fluorescence complementation (BiFC) assays. For the protein colocalization assay, we transiently expressed PIL1-CFP and HFR1 tagged with YFP (HFR1-YFP) either individually or together in tobacco cells. As shown in Figure 4D, HFR1-YFP diffuses exclusively in the nucleus, while coexpression of PIL1-CFP and HFR1-YFP show the colocalized NBs, indicating that PIL1 and HFR1 interact in tobacco cells. For the BiFC assay, we fused the N- and C-terminal halves of YFP to the N terminus of HFR1 and C terminus of PIL1, respectively. Strong YFP fluorescence was observed when nYFP-HFR1 and PIL1-cYFP were coexpressed, suggesting that PIL1 interacts with HFR1 (Figure 4E). Taken together, these results demonstrate that PIL1 physically interacts with HFR1.
Figure 4.
PIL1 Physically Interacts with HFR1.
(A) Yeast two-hybrid bait construct of HFR1 fused to LexA DNA binding domain (LexA) and a prey construct of PIL1 fused to the B42 transcriptional activation domain (AD).
(B) Quantitative yeast two-hybrid assay showing the PIL1–HFR1 interaction. All vector combinations are given as bait/prey. Error bars represent ±sd (n = 10).
(C) Plate assays showing the PIL1–HFR1 interaction. The plates show the corresponding β-galactosidase activities represented by blue precipitates.
(D) PIL1 and HFR1 localize together to NBs in tobacco cells. Dic, differential interference contrast. Bars = 5 μm.
(E) BiFC assay of the PIL1–HFR1 interaction in tobacco leaf cells. CFP serves as the internal control. The left two panels show the wide field view to indicate the BiFC efficiency. The right two panels show a single nucleus at high magnification. Gray bars = 100 μm; white bars = 5 μm.
PIL1 Preferentially Interacts with Photoactive phyB
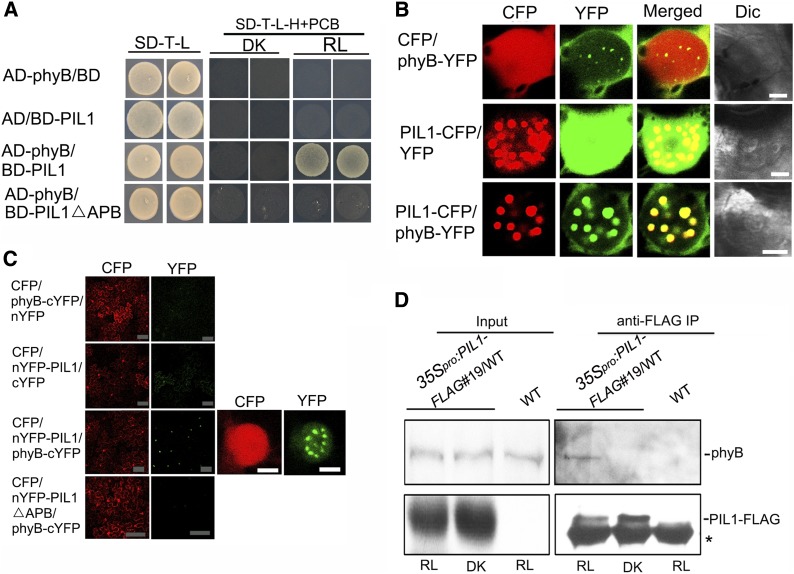
Since PIL1 is closely related to PIFs and possesses a putative APB motif (Yamashino et al., 2003; Khanna et al., 2004; Leivar and Quail, 2011), we asked whether PIL1 might interact with phyB. To test this possibility, the yeast two-hybrid assay was performed with phyB and PIL1 fused to a GAL4 activation (AD) and binding (BD) domain, respectively. Since phycocyanobilin (PCB) has long been used as a phytochromobilin analog to reconstitute Arabidopsis phytochrome holoproteins (Elich and Lagarias, 1989; Wahleithner et al., 1991), the selective medium was supplemented with PCB to reconstitute photoactive phyB (Pfr form). As shown in Figure 5A, yeast cells coexpressing AD-phyB and BD-PIL1 are able to grow on selective media with PCB incubated in red light for 3 d, but not in darkness. When the incubation time was extended to 6 d, yeast cells coexpressing AD-phyB and BD-PIL1 are able to grow in darkness, but not as robustly as in red light, suggesting that PIL1 interacts preferentially with the Pfr form of phyB (Supplemental Figure 6). To investigate whether the putative APB motif in PIL1 is required for the phyB–PIL1 interaction, we generated a bait construct expressing PIL1 lacking the APB motif (BD-PIL1△APB). No interaction was found between PIL1△APB and phyB in either red light or darkness (Figure 5A), indicating that the APB motif of PIL1 is required for phyB–PIL1 interaction in yeast cells.
Figure 5.
PIL1 Preferentially Physically Interacts with Photoactive phyB.
(A) The phyB–PIL1 interaction, as analyzed by a yeast two-hybrid assay. Yeast cells coexpressing the indicated combinations of constructs were grown on nonselective (SD-T-L) or selective media with 25 μM PCB (SD-T-L-H+PCB), in continuous red light (RL = 3 μmol/s·m2) or darkness (DK) for 3 d.
(B) PIL1 and phyB colocalize to NBs in tobacco cells. Dic, differential interference contrast. Bars = 5 μm.
(C) BiFC assay of the phyB–PIL1 interaction in tobacco leaf cells. CFP serves as an internal control. The left two panels show the wide field view to indicate the BiFC efficiency. The right two panels show a single nucleus at high magnification. Gray bars = 100 μm; white bars = 5 μm. The tobacco leaves infiltrated with the combinations shown in (B) and (C) were adapted in darkness for 30 to 36 h and then exposed to red light (15 μmol/s·m2) for 12 h before analysis by confocal microscopy.
(D) phyB interacts with PIL1 in vivo. Four-day-old blue light–grown 35Spro:PIL1-FLAG#19/WT and wild-type seedlings were transferred to darkness for 4 h and then exposed to red light for 12 h (RL) or kept in darkness for another 12 h (DK). Total protein was immunoprecipitated using FLAG-beads, and immunoprecipitates were probed with anti-phyB and anti-FLAG antibodies, respectively. Asterisks show the heavy chain of IgG.
Next, we performed protein colocalization and BiFC assays to confirm the phyB–PIL1 interaction in plant cells. For the colocalization assay, PIL1-CFP and phyB tagged with YFP (phyB-YFP), either individually or together, were transiently coexpressed in tobacco cells. When exposed to red light, phyB-YFP and PIL1-CFP display NBs, but CFP and YFP do not when coexpressed with phyB-YFP and PIL1-CFP, respectively (Figure 5B). Coexpression of PIL1-CFP and phyB-YFP result in the formation of the same colocalized NBs (Figure 5B), indicating their interaction in plant cells. We also transiently coexpressed PIL1-CFP and YFP-tagged phyA (phyA-YFP), either individually or together, in tobacco cells and found that PIL1 and phyA also colocalized in the same NBs (Supplemental Figure 7), indicating that PIL1 might interact with phyA.
For the BiFC assay, we fused the N- and C-terminal halves of YFP to the N terminus of PIL1 and the C terminus of phyB, respectively. When exposed to red light, strong YFP fluorescence was observed when nYFP-PIL1 and phyB-cYFP were coexpressed, suggesting PIL1 interacts with phyB (Figure 5C). To investigate whether the putative APB motif in PIL1 is required for the phyB–PIL1 interaction in plant cells, we generated a construct containing nYFP fused to PIL1 lacking the APB motif (nYFP-PIL1△APB) and performed the BiFC assay. No YFP signal was observed when nYFP-PIL1△APB and phyB-cYFP are coexpressed (Figure 5C), indicating that the APB motif of PIL1 is required for the phyB–PIL1 interaction.
To further determine whether PIL1 interacts with phyB in Arabidopsis, we performed a Co-IP assay using 35Spro:PIL1-FLAG#19/WT and wild-type seedlings. The anti-phyB antibody was prepared and its specificity was analyzed by immunoblotting (Supplemental Figure 8). As shown in Figure 5D, immunoprecipitation of PIL1-FLAG pulls down the endogenous phyB from the 35Spro:PIL1-FLAG#19/WT seedlings exposed to red light, but not from either wild-type control or dark-grown 35Spro:PIL1-FLAG#19/WT seedlings (Figure 5D), in which phyB is known to be localized to the cytoplasm (Nagy et al., 2000). These results indicate that PIL1 interacts with phyB in Arabidopsis under red light.
phyB Promotes the COP1–PIL1 Disassociation in Response to Red Light Exposure
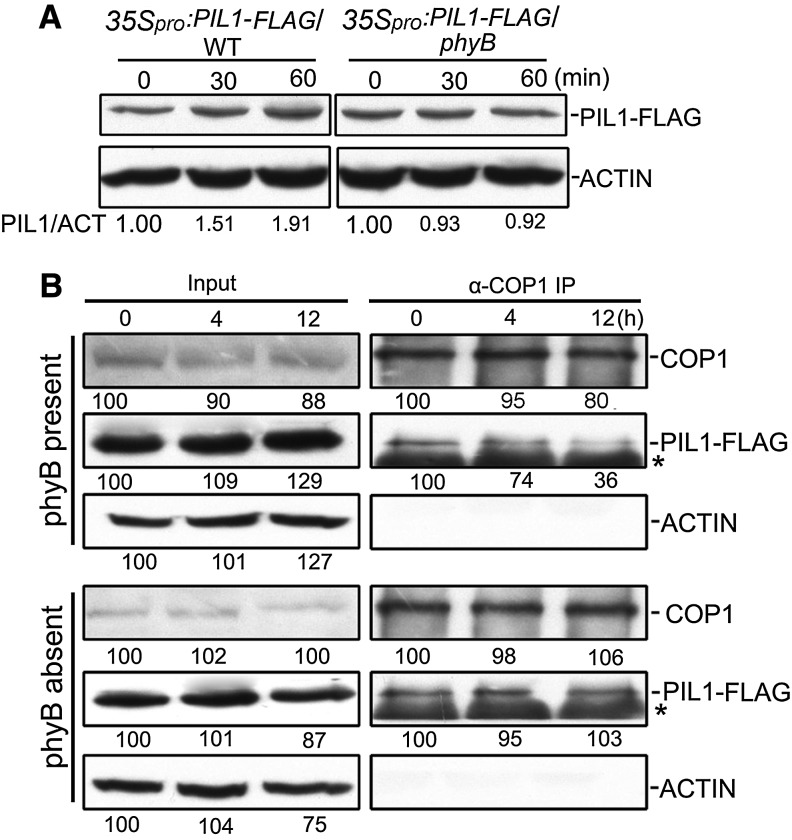
To examine whether phyB regulates PIL1 accumulation, we introgressed the 35Spro:PIL1-FLAG transgene from the wild type into the phyB mutant background by genetic crossing to generate the 35Spro:PIL1-FLAG#19/phyB transgenic plants. We analyzed PIL1 accumulation in 35Spro:PIL1-FLAG#19/WT and 35Spro:PIL1-FLAG#19/phyB seedlings grown in continuous darkness and red light and found that red light moderately induces PIL1-FLAG accumulation in the presence of phyB (35Spro:PIL1-FLAG#19/WT), but not in the absence of phyB (35Spro:PIL1-FLAG#19/phyB) (Supplemental Figure 9). Furthermore, we analyzed PIL1 accumulation dynamics in 35Spro:PIL1-FLAG#19/WT and 35Spro:PIL1-FLAG#19/phyB seedlings grown in the dark for 4 d and exposed to red light for 30 and 60 min, respectively. Red light exposure enhances PIL1 accumulation in the presence of phyB (35Spro:PIL1-FLAG#19/WT), but not in the absence of phyB (35Spro:PIL1-FLAG#19/phyB) (Figure 6A), suggesting that phyB is responsible for the red light-induced accumulation of PIL1.
Figure 6.
phyB Interferes with the COP1–PIL1 Interaction under Red Light in Plant Cells.
(A) PIL1 accumulated in a phyB-dependent manner. 35Spro:PIL1-FLAG#19/WT and 35Spro:PIL1-FLAG#19/phyB seedlings were grown in darkness for 4 d and then exposed to red light for the indicated time periods before total protein was extracted for immunoblot analysis with anti-FLAG and anti-ACTIN antibodies, respectively. PIL1/ACT indicates the band intensities of PIL1-FLAG normalized to ACTIN and is presented relative to that in darkness set at unity.
(B) Co-IP using anti-COP1 antibody in the extracts from 35Spro:PIL1-FLAG#19/WT (phyB present) and 35Spro:PIL1-FLAG#19/phyB seedlings (phyB absent) grown in continuous blue light for 4 d before being transferred to darkness for 4 h and then exposed to red light for the indicated time periods. The immunoprecipitates were probed with anti-COP1, anti-FLAG, and anti-ACTIN antibodies.
(C) Relative band intensities were normalized to the starting samples for each panel and shown below each lane. Asterisks show the heavy chain of IgG.
Since phyB promotes PIL1 stabilization and physically interacts with PIL1 (Figures 5 and 6A; Supplemental Figure 6), we then asked whether phyB might affect the COP1–PIL1 association. To test this possibility, we analyzed the association capacity of COP1 with PIL1 through Co-IP assays using dark-grown 35Spro:PIL1-FLAG#19/WT and 35Spro:PIL1-FLAG#19/phyB seedlings that were exposed to red light for different lengths of time, respectively. We confirmed equal loading by immunoblotting using an antibody against ACTIN (Figure 6B). The results demonstrate that, in the presence of phyB, the COP1–PIL1 interaction is reduced progressively with prolonged exposure to red light, with the amount of PIL1-FLAG coimmunoprecipitated with COP1 being reduced by ∼26 and 64% upon 4 and 12 h of red light irradiation compared with darkness, respectively (Figure 6B). In contrast, in the phyB mutant background, red light failed to induce the COP1–PIL1 dissociation (Figure 6B). These data indicate that the phyB-induced PIL1 accumulation is likely mediated through the phyB-promoted dissociation of COP1 from PIL1 under red light irradiation.
PIL1 Interacts with PIFs in Both Yeast and Plant Cells
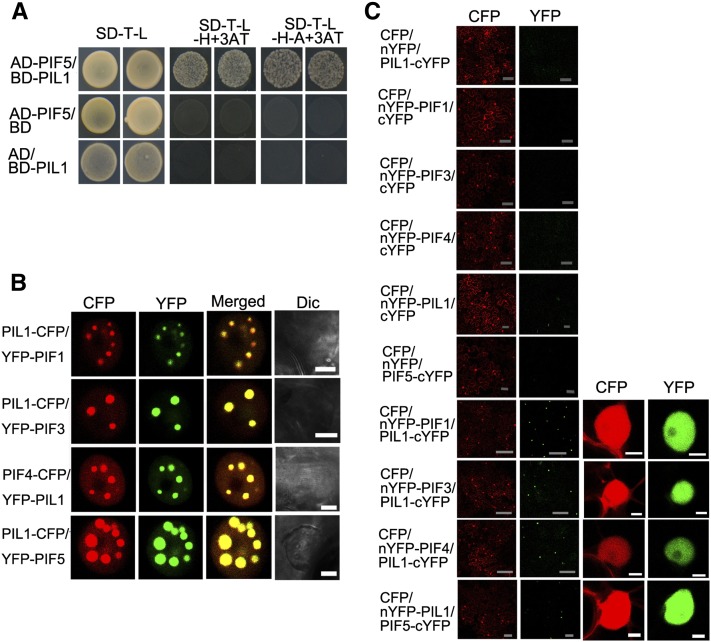
Since HFR1 interacts with PIFs and inhibits their activity (Fairchild et al., 2000; Hornitschek et al., 2009; Bu et al., 2011; Shi et al., 2013), we speculated that PIL1 might also directly interact with PIFs. To test this possibility, we performed a yeast two-hybrid assay with PIF5 fused to the GAL4 activation (AD) domain and PIL1 fused to the GAL4 binding (BD) domain. The results show that PIL1 interacts with PIF5 (Figure 7A).
Figure 7.
PIL1 Physically Interacts with PIFs.
(A) Yeast two-hybrid assay of the PIL1–PIF5 interaction. Yeast cells transformed with the indicated combinations of constructs were grown in nonselective (SD-T-L) or selective media with 2 mM 3AT (SD-T-L-H+3AT or SD-T-L-H-A+3AT).
(B) PIL1 and PIFs (PIF1, PIF3, PIF4, and PIF5) colocalize to NBs in tobacco cells. Dic, differential interference contrast. Bars = 5 μm.
(C) BiFC assay of the PIL1–PIFs (PIF1, PIF3, PIF4, and PIF5) interactions in tobacco leaf cells. CFP serves as an internal control. The left two panels show the wide field view to indicate the BiFC efficiency. The right two panels show a single nucleus at high magnification. Gray bars = 100 μm; white bars = 5 μm.
We then performed protein colocalization and BiFC assays to confirm the interactions between PIL1 and PIF5 and other PIFs. For the protein colocalization assay, PIL1-CFP was coexpressed with PIF1, PIF3, and PIF5 tagged with YFP (YFP-PIF1, YFP-PIF3, and YFP-PIF5). The results demonstrate that YFP-PIF1, YFP-PIF3, YFP-PIF5, and PIL1-CFP display NBs and that PIL1-CFP colocalizes with YFP-PIF1, YFP-PIF3, and YFP-PIF5 in the same NBs, respectively (Figure 7B; Supplemental Figure 10). We also transiently expressed PIF4 tagged with CFP (PIF4-CFP) and PIL1 tagged with YFP (YFP-PIL1), both of which form distinct NBs (Supplemental Figure 10). Coexpression of YFP-PIL1 and PIF4-CFP demonstrates that they colocalize in the same NBs (Figure 7B). For the BiFC assay, the N-terminal halves of YFP were fused to the N terminus of PIF1, PIF3, and PIF4, respectively. YFP fluorescence is detected in the nuclei of cells coexpressing PIL1-cYFP and nYFP-PIF1, PIL1-cYFP and nYFP-PIF3, or PIL1-cYFP and nYFP-PIF4 (Figure 7C). We also generated constructs expressing PIL1 fused to the C terminus of nYFP (nYFP-PIL1) and PIF5 fused to the N terminus of cYFP (PIF5-cYFP), respectively, and performed a BiFC assay. Strong YFP fluorescence was observed in cells coexpressing these two fusion proteins (Figure 7C). Taken together, these results demonstrate that PIL1 interacts with PIFs.
PIFs Are Genetically Epistatic to PIL1
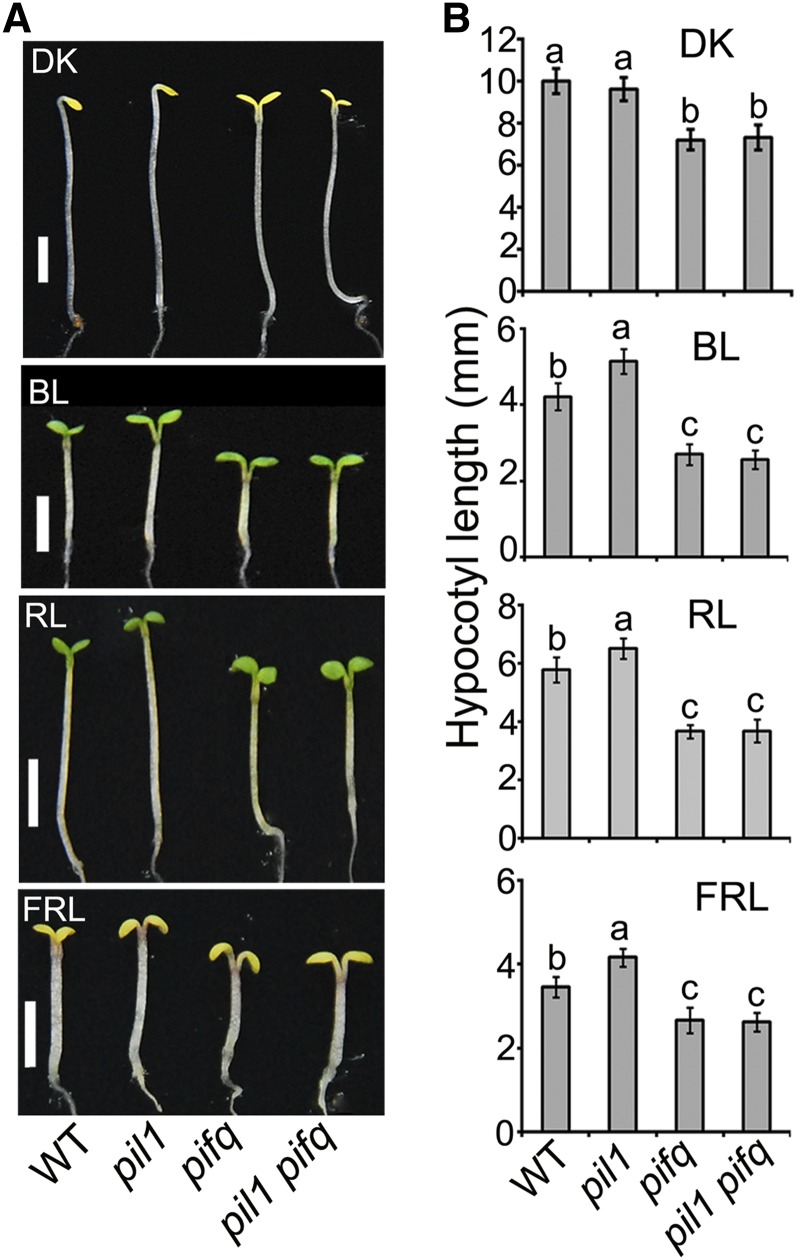
To explore the genetic interactions between PIL1 and PIFs, we generated a pil1 pif1 pif3 pif4 pif5 (pil1 pifq) quintuple mutant by crossing pil1-2 with pifq and analyzed its photomorphogenic phenotype in darkness, blue, red, and far-red light, respectively. Compared with the pil1 single mutant, the pil1 pifq quintuple mutant exhibits an expanded cotyledon phenotype in darkness and enhanced inhibition of hypocotyl elongation in blue, red, and far-red light, respectively, similar to pifq (Figure 8). These results indicate that pifq is epistatic over the pil1 mutant phenotype.
Figure 8.
PIFs Are Genetically Epistatic to PIL1.
(A) Photograph of representative seedlings of the indicated genotypes. Seedlings were grown under continuous light conditions for 4 d. DK, darkness; BL, blue light; RL, red light; FRL, far-red light. Bars = 2 mm.
(B) Quantification of hypocotyl lengths of the seedlings shown in (A). Error bars represent ±sd (n = 30). The letters “a” to “c” indicate statistically significant differences between means for hypocotyl lengths of the indicated genotypes, as determined by Tukey’s LSD test (P ≤ 0.01).
PIL1 and HFR1 Additively Inhibit the Transcription of PIF Direct-Target Genes
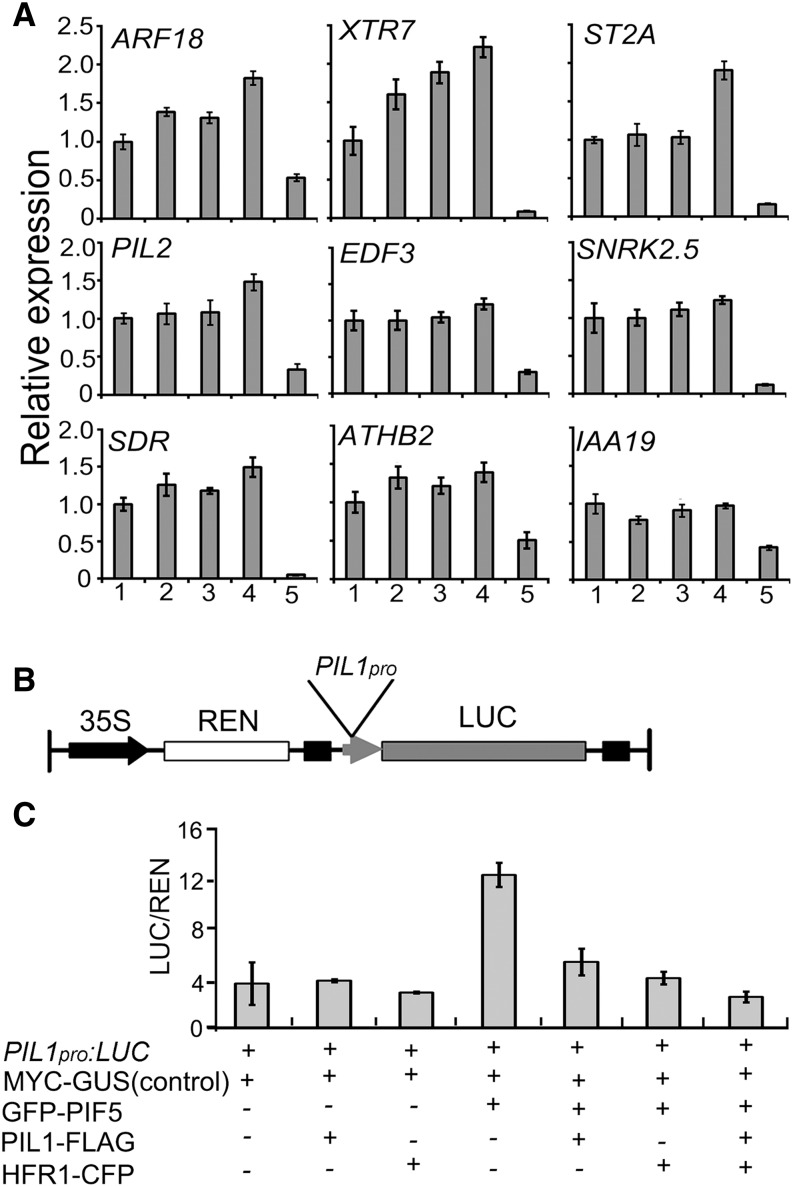
With the demonstrations that PIL1 interacts with HFR1, that both PIL1 and HFR1 interact with PIFs, and that HFR1 represses PIF1, PIF4, and PIF5 transcriptional activity (Fairchild et al., 2000; Hornitschek et al., 2009; Shi et al., 2013; Figure 4), we asked whether PIL1 might also regulate the expression of PIF direct-target genes. To test this possibility, we performed qRT-PCR to analyze up to nine direct targets of PIFs (Zhang et al., 2013), of which PIL2 and XTR7 are shown to regulate hypocotyl elongation (Penfield et al., 2010; Sasidharan et al., 2010), in wild-type, hfr1, pil1, pil1 hfr1, and pifq seedlings grown in darkness for 3 d and exposed to red light for 10, 20, and 30 min, respectively. All these genes are drastically downregulated in pifq (Figure 9A; Supplemental Figure 11). When exposed to red light for 20 min, more ARF18 and XTR7 transcripts were detected in pil1 and hfr1 than in the wild type, and even more transcripts were detected in the pil1 hfr1 double mutant than in pil1 and hfr1 single mutants (Figure 9A). The expression levels of EDF3, PIL2, ST2A, and SNRK2.5 in pil1 hfr1 were higher than those in the pil1 and hfr1 single mutants, though no differences were detected among pil1, hfr1, and the wild type (Figure 9A). We found that some PIF direct-target genes, including SDR, ATHB2, and IAA19, are not inhibited by PIL1 and HFR1 upon 20 min red light irradiation (Figure 9A). When 3-d-old dark-grown seedlings were exposed to red light for 10 min, the expression levels of ST2A, ARF18, and EDF3 were higher in pil1 hfr1 than in pil1 and hfr1 (Supplemental Figure 11A). However, the expression of these genes is not increased in hfr1, pil1, or hfr1 pil1 seedlings grown in continuous darkness or exposed to red light for 30 min, compared with the wild type (Supplemental Figures 11B and 11C). These results indicate that PIL1 and HFR1 might cooperatively inhibit the expression of some PIF direct-target genes within 20 min of red light irradiation.
Figure 9.
PIL1 and HFR1 Regulate the Transcription Level of PIF Direct-Target Genes in Vivo.
(A) qRT-PCR results showing the expression of PIF direct-target genes in multiple genotypes. Expression levels were normalized to an internal ACTIN2 control and are presented relative to the wild-type levels set at unity. Data are represented as the mean of biological triplicates ±sd (n = 3). All the seedlings were grown in darkness for 3 d and then transferred to red light for 20 min. 1, Wild type; 2, hfr1; 3, pil1; 4, pil1 hfr1; 5, pifq.
(B) Schematic presentation of the Dual-LUC assay reporter construct expressing LUC under the PIL1 promoter (PIL1pro).
(C) Tobacco leaves were infiltrated with strains harboring the PIL1pro:LUC reporter and effectors in the indicated combinations. The values were given by calculating the ratio of LUC activities to REN activities (LUC/REN). Error bars represent ±sd (n = 3).
We also performed qRT-PCR to analyze the expression of direct target genes of PIFs in seedlings of the wild type, 35Spro:PIL1-FLAG#19/WT, the hfr1 mutant overexpressing GFP-HFR1 (35Spro:GFP-HFR1/hfr1) (Yang et al., 2005), and pifq grown in darkness for 3 d and exposed to red light for 20 min, respectively. The results show that the expression levels of these genes except ARF18 in 35Spro:PIL1-FLAG#19/WT and 35Spro:GFP-HFR1/hfr1 are significantly lower than in the wild type (Supplemental Figure 12). Moreover, the expression of HFR1 and PIL1 is also reduced in 35Spro:PIL1-FLAG#19/WT and 35Spro:GFP-HFR1/hfr1, respectively (Supplemental Figure 12). Taken together, these data further suggest that both PIL1 and HFR1 are responsible for the inhibition of expression of some PIF direct-target genes upon 20 min of red light irradiation.
To explore the possible direct regulation of PIFs by PIL1 and HFR1, we utilized a transient transcription assay, the dual luciferase (Dual-LUC) assay, in tobacco. To do this, we isolated 2-kb promoter regions of PIL1 (PIL1pro), ST2A (ST2Apro), and IAA19 (IAA19pro), which are the well-known PIF direct-target genes (Hornitschek et al., 2009; Sun et al., 2013; Zhang et al., 2013), and made reporter constructs expressing LUC under the control of PIL1pro, ST2Apro, and IAA19pro, respectively (Figure 9B; Supplemental Figure 13A). The effector constructs expressing GFP-PIF5, HFR1-CFP, and PIL1-FLAG under the control of the 35S promoter were generated, respectively, and were transiently coexpressed in tobacco leaves with the reporter constructs in different combinations. The results show that PIF5 alone strongly stimulates the PIL1pro:LUC, ST2Apro:LUC, and IAA19pro:LUC reporter activity, respectively (Figure 9C; Supplemental Figures 13B and 13C), consistent with PIF5 being a positive regulator of these three genes (Hornitschek et al., 2009; Sun et al., 2013; Zhang et al., 2013). When PIL1 or HFR1 is coexpressed with PIF5, the activity of PIL1, ST2A, and IAA19 promoters is repressed (Figure 9C; Supplemental Figures 13B and 13C). When both PIL1 and HFR1 are coexpressed with PIF5, the activities of the PIL1 and ST2A promoters are further attenuated (Figure 9C; Supplemental Figure 13B). Taken together, these results indicate that PIL1 and HFR1 might act additively to repress the transcription of some of the PIF direct-target genes by directly interfering with the activities of PIF proteins.
DISCUSSION
COP1 Physically Interacts with PIL1 and Promotes Its Degradation in Darkness
PIL1 belongs to the bHLH transcription factor family and shares similarity to HFR1. The mutant alleles of pil1 (pil1-1, pil1-2, pil1-3, and pil1-4) have elongated hypocotyls, compared with the wild type, in red and far-red light (Khanna et al., 2006). In this study, we demonstrate that the pi11 mutant displays an elongated hypocotyl phenotype compared with the wild type, not only in red and far-red light, but in blue light as well (Figures 3 and 8; Supplemental Figures 1 and 2). Furthermore, we demonstrate that transgenic plants overexpressing PIL1 show a shortened hypocotyl phenotype under blue, red, and far-red light, respectively (Supplemental Figure 1). Moreover, pil1 hfr1 double mutant seedlings are taller than hfr1 single mutant in blue, red, and far-red light (Figures 3B and 3D), and loss function of PIL1 and HFR1 in the cop1 mutant background significantly attenuates the constitutively photomorphogenic phenotype of the cop1 mutant in the dark (Figures 3A and 3C). Based on the above evidence, we propose that, like HFR1, PIL1 is a positively acting transcription factor that promotes photomorphogenesis.
We analyzed PIL1 accumulation in the dark and light and found that PIL1 is degraded in the dark, and the 26S proteasome inhibitor can inhibit its degradation (Figures 2A, 2B, and 2E to 2G). Through combined approaches of yeast two-hybrid, in vivo protein colocalization, and Co-IP assays, we demonstrate that COP1 physically interacts with PIL1 (Figure 1). We further analyzed PIL1 protein levels in the wild type and cop1 mutant backgrounds in the dark and found that PIL1 accumulates at a much higher level in the cop1 mutant than in the wild type (Figures 2C and 2D). COP1 possesses intrinsic E3 activity and is shown to ubiquitinate several transcription factors, such as HY5, HFR1, and CO, and targets them for degradation (Osterlund et al., 2000; Saijo et al., 2003; Jang et al., 2005; Yang et al., 2005; L.J. Liu et al., 2008). We examined whether COP1 is able to ubiquitinate PIL1 by performing an in vitro ubiquitination analysis in the presence of MBP-COP1, E1, E2, and ubiquitin, but failed to detect the polyubiquitinated PIL1. Whether COP1 serves as an E3 ubiquitin ligase for PIL1 or functions with other components to ubiquitinate PIL1 in vivo will need further investigation. Taken together, our results suggest that PIL1 is a positive regulator of photomorphogenesis and COP1 directly interacts with PIL1 and promotes its degradation via the 26S proteasome-dependent pathway.
Photoactive phyB Physically Interacts with PIL1 and May Stabilize PIL1 through Inhibiting the COP1–PIL1 Association
Previous studies have shown that the APA and APB motifs are necessary for the interactions of PIFs with phyA and phyB, respectively (Khanna et al., 2004; Al-Sady et al., 2006; Shen et al., 2008). Though PIL1 also has a putative APB motif (Khanna et al., 2004; Leivar and Quail, 2011), a previous study did not demonstrate the interaction between PIL1 and Pfr phyB through an in vitro pull-down assay (Khanna et al., 2004). In this study, we analyzed the phyB–PIL1 interaction through combined approaches of yeast two-hybrid, in vivo protein colocalization, BiFC, and Co-IP (Figure 5; Supplemental Figure 6). The results suggest that PIL1 preferentially interacts with photoactive phyB and that the APB motif of PIL1 is required for the interaction with phyB. We analyzed PIL1 accumulation in the wild type and phyB mutant backgrounds and found that both initial red light irradiation (within 60 min) and continuous red light are able to enhance PIL1 accumulation in the wild type, but not in the phyB mutant (Figure 6A; Supplemental Figure 9). We sought to examine how phyB is able to stabilize PIL1 by analyzing the capacity of the COP1 and PIL1 interaction in the presence and absence of phyB upon various periods of red light exposure (Figure 6B). The results indicate that phyB is involved in promoting the COP1–PIL1 dissociation upon red light exposure. Since phyB interacts with both COP1 and PIL1 (Figure 5; Jang et al., 2010), whether the phyB–COP1 and/or phyB–PIL1 interaction is responsible for the negative regulation of the COP1–PIL1 interaction awaits further investigation.
It is interesting to note that, in contrast to PIFs, whose degradation is rapidly promoted by phyB in response to red light irradiation (Park et al., 2004; Shen et al., 2005, 2008; Al-Sady et al., 2006; Shen et al., 2007), PIL1 degradation is inhibited by phyB (Figure 6). In agreement with their contrasting protein degradation properties, the pifq mutant shows a constitutive photomorphogenic phenotype in the dark and enhanced responsiveness to light (Leivar et al., 2008, 2009), whereas the pil1 mutant displays reduced responsiveness to light (Figures 3 and 8; Supplemental Figures 1 and 2; Khanna et al., 2006). We carefully compared the amino acid sequences of PIL1 and PIFs and found that they share similarity only in the PIL and bHLH domains. Whether the amino acid sequence differences beyond these two domains in PIL1 and PIFs are responsible for the differences in the phyB-mediated regulation of protein stability remains to be determined.
It is worth noting that PIL1-FLAG in either 35Spro:PIL1-FLAG/WT or PIL1pro:PIL1-FLAG/pil1 transgenic plants increases upon 15 to 60 min red light irradiation, but declines after prolonged exposure (Figures 2F and 2G). Previous studies showed that nuclear phyB is unstable and decreases by ∼70% after 24 h red light treatment; specifically, a functional phyB N-terminal region containing a constitutively nuclear localized signal (NGG-NLS) has a half-life of <2 h (Jang et al., 2010). Upon red light exposure, the phyB–COP1 interaction may immediately exert inhibitory effects on COP1 and result in enhanced PIL1 accumulation, whereas upon prolonged red light illumination, the phyB–COP1 interaction may in turn trigger the degradation of phyB, and COP1 activity may increase, leading to the degradation of PIL1. Thus, dynamic changes in PIFs and PIL1 accumulation, PIL1 transcription, and phyB and COP1 activities in response to red light exposure regulate red light–mediated photomorphogenic development, and in these complex dynamics, PIL1, which accumulates to moderate levels upon initial red light irradiation, is likely responsible for mediating the photomorphogenic phenotype.
PIL1 and HFR1 Form Heterodimers and May Act Together to Suppress PIFs and Promote Photomorphogenic Development
In this study, we demonstrate that PIL1 and HFR1 act additively to promote photomorphogenesis, based on the fact that the pil1 hfr1 double mutant exhibits a greater reduction in the photomorphogenic phenotype than the pil1 or hfr1 single mutant under blue, red, and far-red light, respectively, and that the cop1 pil1 hfr1 triple mutant displays a more attenuated photomorphogenic phenotype than the cop1 pil1 or cop1 hfr1 double mutant in darkness, blue, red, and far-red light (Figure 3). Furthermore, through yeast two-hybrid, colocalization, and BiFC assays, we demonstrate that PIL1 physically interacts with HFR1 to form heterodimers (Figure 4). Therefore, we propose that PIL1 and HFR1 form heterodimers and act additively to promote photomorphogenesis.
It is shown that HFR1 forms heterodimers with PIFs and prevents PIFs from binding to DNA (Hornitschek et al., 2009; Shi et al., 2013). In this study, we also show that PIL1 physically interacts with PIFs (Figure 7) and that pifq is epistatic to the photomorphogenic phenotype of pil1 (Figure 8). Moreover, we demonstrate that PIL1, along with HFR1, acts to regulate the expression of some of the PIF direct-target genes likely through inhibiting the transcriptional activity of PIFs (Figure 9; Supplemental Figures 12 and 13). However, whether PIL1 and HFR1 act together to inhibit the binding capacity of PIFs to their target gene promoters requires further investigation. Specifically, our data show that PIL1 inhibits the transcription of PIF direct-target genes, such as ARF18 and XTR7, within 20 min of red light illumination, and the PIL1-FLAG protein in PIL1pro:PIL1-FLAG/pil1 plants accumulates to a slightly higher level upon 30 min red light irradiation and declines to the same level as in darkness after 60 min (Figures 2G and 9A). It is known that PIF degradation is induced rapidly upon 60 min red light exposure and that the half-life of PIFs is 5 to 20 min (Park et al., 2004; Shen et al., 2005, 2008; Al-Sady et al., 2006; Shen et al., 2007). Based on our results and these previous findings, we propose that the ratio of PIL1/PIFs increases dramatically upon 60 min red light exposure and that 20 min illumination might be sufficient to inhibit PIFs and the subsequent transcription of their direct-target genes, consistent with our notion that PIL1 mainly functions within 60 min of red light irradiation. Not all PIF direct-target genes are suppressed by PIL1 and/or HFR1 in the conditions tested in this study, indicating that the underlying regulatory mechanism is complex and may involve dynamic changes in phyB photoreceptor activity, PIF and PIL1 accumulation and PIL1 transcription, and may be affected by the possible complex feedback regulation resulting from these changes.
Action of PIL1 in Regulating Photomorphogenesis
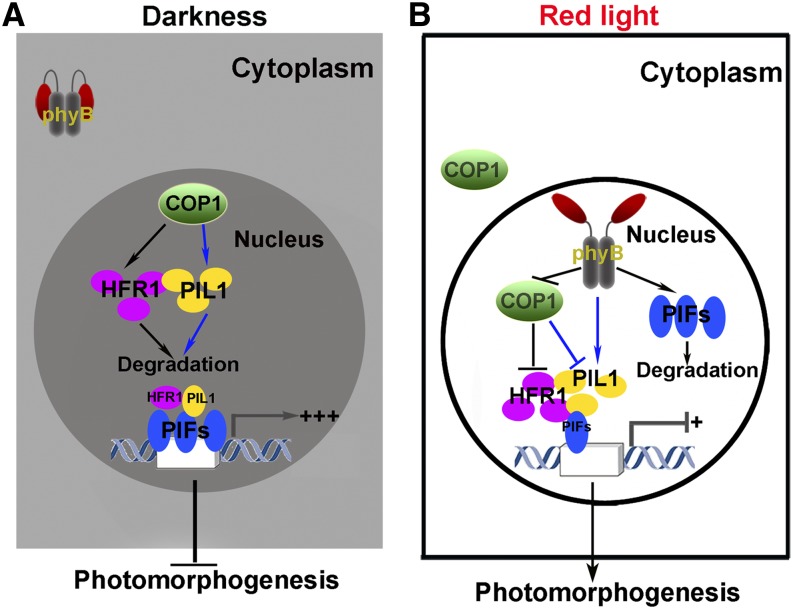
In this work, we demonstrate that PIL1 undergoes a dynamic accumulation during the transitions from dark to light and that the dynamic accumulation of PIL1 is involved in phyB- and COP1-mediated regulation of photomorphogenesis. Based on our findings and previous reports, we propose a mode of action of PIL1 in mediating photomorphogenesis upon red light irradiation. In darkness, phyB is localized in the cytoplasm and PIFs accumulate in the nucleus, whereas COP1 is localized to the nucleus, where it interacts with PIL1 and HFR1 and promotes their degradation via the 26S proteasome. Consequently, PIFs are not suppressed by PIL1 and HFR1 and are able to promote the expression of their direct-target genes and repress photomorphogenesis (Figure 10A). When exposed to red light, phyB is activated and imported into the nucleus, where it on the one hand interacts with PIFs and induces their degradation and on the other interacts with COP1 and PIL1, resulting in the accumulation of PIL1 presumably through different layers of regulation, including repressing COP1 activity, inhibiting the COP1–PIL1 association, and promoting the translocation of COP1 from the nucleus to the cytoplasm. The accumulated PIL1, together with HFR1, interacts with PIFs and inhibits the transcription of their direct-target genes, thus promoting photomorphogenesis (Figure 10B). Our model does not explain the mechanism by which PIL1 accumulation is dramatically enhanced under either continuous blue or far-red light or upon initial blue and far-red light exposure. It is possible that cryptochromes and phyA are involved in these processes, likely through repressing COP1, and this possibility needs to be investigated in future studies. In nature, where blue, red, and far red light are present, it is possible that PIL1 accumulation can be enhanced significantly by blue and far-red light. This possibility can be tested by investigating the accumulation of endogenous PIL1 under white light in the presence and absence of CRY1, CRY2, phyB, and phyA in future studies.
Figure 10.
Model of the Action of PIL1 in Regulating Photomorphogenesis.
(A) In darkness, phyB is inactive and localized to the cytoplasm and PIFs accumulate to a high level in the nucleus (shown by triple blue ellipsoids), while nuclear COP1 interacts with PIL1 and HFR1 and promotes their degradation. As a result, little HFR1 and PIL1 (shown by a single purple and yellow ellipsoid, respectively) interact with PIFs; thus, PIFs are highly active and strongly induce the transcription of their direct-target genes, repressing photomorphogenesis.
(B) When exposed to red light, phyB is activated and translocated into the nucleus, where it interacts with PIFs and promotes PIFs degradation. Meanwhile, phyB interacts with both COP1 and PIL1, resulting in the accumulation of HFR1 and PIL1 (shown by triple purple and yellow ellipsoids, respectively). The accumulated PIL1, along with HFR1, may interact with the remaining PIFs (shown by a single blue ellipsoid) to inhibit the transcription of PIF direct-target genes and promote photomorphogenesis. Prolonged red light exposure promotes the translocation of COP1 from the nucleus to the cytoplasm. Blue lines represent the direct interactions established in this study. Arrow and T-bar denote positive and negative regulation, respectively.
METHODS
Plant Materials
All plants used were of the Columbia ecotype. pil1-2, cop1-4, hfr1-201, pif1345 (pifq), and 35Spro:GFP-HFR1/hfr1 were described previously (Soh, 2000; Yamashino et al., 2003; Yang et al., 2005; Khanna et al., 2006; Kang et al., 2009; Jia et al., 2014).
Yeast Two-Hybrid Assay
For the LexA yeast two-hybrid system, yeast transformation, the calculation of relative β-galactosidase activities, and plate assays were performed as described previously (Yang et al., 2000; L.J. Liu et al., 2008). The GAL4 yeast two-hybrid assay was performed according to the manufacturer’s instructions (Matchmaker user’s manual; Clontech). At least three independent experiments were performed, and the result of one representative experiment is shown. The phyB–PIL1 interaction assay with PCB was performed as described (Shimizu-Sato et al., 2002), with minor modification. After heat treatment, the yeast cells were cultured for 3 h in darkness in liquid SD-T-L-H medium with 25 μM PCB (Scientific Frontier). The transformants were spread on SD-T-L-H plates with 25 μM PCB and incubated in continuous red light (3 μmol/s·m2) or darkness for 3 to 6 d.
Protein Extraction, Immunoblotting, and in Vivo Coimmunoprecipitation
Arabidopsis thaliana protein extracts were obtained with the following lysis buffer: 50 mM HEPES, pH 7.5, 150 mM KCl, 1 mM PMSF, 1× complete protease inhibitor cocktail (Roche), and 50 μM MG132 (Merck). PIL1-FLAG was detected with anti-FLAG monoclonal antibody (Sigma-Aldrich). Co-IP was performed by methods described previously (Shalitin et al., 2002; Zhu et al., 2008), with minor modifications. All the seedlings were grown in continuous blue light for 4 d and transferred to the indicated conditions and incubated in liquid Murashige and Skoog medium containing MG132. For Co-IP of PIL1 and COP1, 8 μL of anti-COP1 antiserum (Lian et al., 2011) was incubated with 20 μL of protein G-sepharose beads (bed volume; GE Healthcare) for 2 h at 4°C . Then, equal amounts of total protein (1 to 3 mg) in 1 mL of lysis buffer was added to the mixture and incubated for 40 min at 4°C. For Co-IP of phyB and PIL1, equal amounts of total protein (1 to 3 mg) in 1 mL were incubated with 25 μL FLAG-beads (Sigma-Aldrich) for 30 min at 4°C. All the immunoprecipitates were washed three times with lysis buffer before the concentrates were resuspended with 2× SDS sample buffer, boiled for 5 min, and then subjected to immunoblot analysis. All these experiments were independently repeated three times, and one representative result is shown. The immunoblots were quantified with ImageJ (http://rsb.info.nih.gov/ij).
Protein Colocalization and BiFC Assays
All constructs for colocalization and BiFC assays were obtained by PCR amplification of the related fragments and cloning into the corresponding vectors listed in Supplemental Table 1. The vectors of BiFC (pXY104 and pXY106) were described previously (Yu et al., 2008; Liu and Howell, 2010). These constructs were transformed into Agrobacterium tumefaciens strain GV3101 and infiltrated into tobacco (Nicotiana benthamiana) leaf epidermal cells at the indicated combinations (Figures 1E, 4C, 4D, 5B, 5C, 7B, and 7C; Supplemental Figures 7 and 10), and analyzed by confocal microscopy (Leica TCS SP5II) after 40 to 48 h. CFP served as the internal control in all BiFC analyses (Figures 4E, 5C, and 7C).
Dual-LUC Assay
The reporter constructs were obtained by PCR amplification of the related fragments (Supplemental Table 1) and cloned into the vector described previously (Hellens et al., 2005). The effector constructs were described above and listed in Supplemental Table 1. Infiltration and detection were performed according to the protocols described previously (H. Liu et al., 2008), with minor modifications. All overnight Agrobacterium cultures were collected, resuspended in Murashige and Skoog medium to OD600 = 0.6, and incubated at room temperature for 3 h. The reporter strain harboring PIL1pro:LUC or ST2Apro:LUC or IAA19pro:LUC was mixed with the effector strains harboring 35Spro:GFP-PIF5, 35Spro:PIL1-FLAG, and 35Spro:HFR1-CFP at the ratio of 1:5:2:2. The mixture that lacks any one of these three effector-comprising strains was supplemented with an equal amount of the control strain harboring 35Spro:MYC-GUS instead. The mixture of Agrobacterium suspensions was infiltrated into tobacco leaves. The leaf samples were collected after 2 d for the Dual-LUC assay using commercial Dual-Luciferase reaction reagents, according to the manufacturer’s instructions (Promega). Three biological repeats were measured for each sample.
Accession Numbers
Sequence data can be found in the Arabidopsis Genome Initiative Database under accession numbers At2g46970 (PIL1), At1G02340 (HFR1), At2g20180 (PIF1), At1g09530 (PIF3), At2g43010 (PIF4), At3g59060 (PIF5), At1g09570 (PHYA), At2g18790 (PHYB), At2g32950 (COP1), At3G15540 (IAA19), and At5g07010 (ST2A). Germplasm was used as: pil1-2 (SALK_025372).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Transgenic 35Spro:PIL1-FLAG Plants Display Inhibited Hypocotyl Elongation Phenotypes under Light Conditions.
Supplemental Figure 2. Expression of PIL1-FLAG under the Control of the PIL1 Native Promoter in the pil1 Mutant Rescues the Elongated Hypocotyl Phenotype.
Supplemental Figure 3. Immunoblot Analysis of the Protein Levels of COP1 and PIL1 Fragments in Yeast Cells.
Supplemental Figure 4. qRT-PCR Analysis Showing the Expression of PIL1 in PIL1pro:PIL1-FLAG/pil1 Seedlings.
Supplemental Figure 5. Immunoblot Analysis Showing PIL1-FLAG Accumulation in PIL1pro:PIL1-FLAG#15/pil1 Seedlings.
Supplemental Figure 6. PIL1 Preferentially Interacts with the Pfr Form of phyB.
Supplemental Figure 7. PIL1 and phyA Localize Together to NBs in Tobacco Cells.
Supplemental Figure 8. Immunoblot Analysis Showing Specificity of Anti-phyB Antibody.
Supplemental Figure 9. PIL1 Accumulation Is Promoted by phyB in Continuous Red Light Conditions.
Supplemental Figure 10. Negative Controls Showing No Colocalization of CFP with YFP-PIF1, YFP-PIF3, YFP-PIF5, or YFP-PIL1, and YFP with PIL1-CFP or PIF4-CFP in Vivo.
Supplemental Figure 11. qRT-PCR Analysis Showing the Expression of PIF Direct-Target Genes in Multiple Genotypes.
Supplemental Figure 12. Expression of PIF Direct-Target Genes Is Suppressed in Transgenic Plants Overexpressing PIL1 and HFR1.
Supplemental Figure 13. PIL1 and HFR1 Suppress ST2A and IAA19 Expression in the Dual-Luciferase Assay.
Supplemental Table 1. List of Vectors and Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank the ABRC for Arabidopsis mutant seeds, Yali He for assistance in statistic analysis, and Fangyuan Zhang for drawing the model. This work was supported by grants from the National Natural Science Foundation of China (90917014, 91217307, and 30830012 to H.-Q.Y. and 31170266 to H.-L.L.), China Innovative Research Team, Ministry of Education, and 111 Project (B14016).
AUTHOR CONTRIBUTIONS
Q.L. and H.-Q.Y. designed the research. Q.L. performed the experiments. H.-L.L. constructed some clones. Q.L., H.-L.L., and H.-Q.Y. analyzed data. H.-L.L., L.L., and K.-P.J. revised the article. Q.L., S.-B.H., and H.-Q.Y. wrote the article.
Glossary
- bHLH
basic helix-loop-helix
- NB
nuclear body
- Co-IP
coimmunoprecipitation
- qRT-PCR
quantitative RT-PCR
- BiFC
bimolecular fluorescence complementation
- PCB
phycocyanobilin
Footnotes
Online version contains Web-only data.
References
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222. [DOI] [PubMed] [Google Scholar]
- Briggs W.R., Olney M.A. (2001). Photoreceptors in plant photomorphogenesis to date. Five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol. 125: 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.A., Cloix C., Jiang G.H., Kaiserli E., Herzyk P., Kliebenstein D.J., Jenkins G.I. (2005). A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. USA 102: 18225–18230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q., Castillon A., Chen F., Zhu L., Huq E. (2011). Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis. Plant Mol. Biol. 77: 501–511. [DOI] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12: 514–521. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Daviere J.M., Rodriguez-Falcon M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blazquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Deng X.W., Caspar T., Quail P.H. (1991). cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5: 1172–1182. [DOI] [PubMed] [Google Scholar]
- Deng X.W., Matsui M., Wei N., Wagner D., Chu A.M., Feldmann K.A., Quail P.H. (1992). COP1, an arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a Gβ homologous domain. Cell 71: 791–801. [DOI] [PubMed] [Google Scholar]
- Deng X.W., Quail P.H. (1999). Signalling in light-controlled development. Semin. Cell Dev. Biol. 10: 121–129. [DOI] [PubMed] [Google Scholar]
- Duek P.D., Fankhauser C. (2003). HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J. 34: 827–836. [DOI] [PubMed] [Google Scholar]
- Elich T.D., Lagarias J.C. (1989). Formation of a photoreversible phycocyanobilin-apophytochrome adduct in vitro. J. Biol. Chem. 264: 12902–12908. [PubMed] [Google Scholar]
- Fairchild C.D., Schumaker M.A., Quail P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14: 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Chory J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13: 203–229. [DOI] [PubMed] [Google Scholar]
- Favory J.J., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28: 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M., Ulm R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17: 230–237. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U., Xu Y., Quail P.H. (1998). SPA1: a new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10: 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U., Tepperman J.M., Quail P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496–499. [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Lorrain S., Zoete V., Michielin O., Fankhauser C. (2009). Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- Jang I.C., Henriques R., Seo H.S., Nagatani A., Chua N.H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang J.Y., Seo H.S., Chua N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K.P., Luo Q., He S.B., Lu X.D., Yang H.Q. (2014). Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol. Plant 7: 528–540. [DOI] [PubMed] [Google Scholar]
- Kang C.Y., Lian H.L., Wang F.F., Huang J.R., Yang H.Q. (2009). Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.H.M. (1994). Photomorphogenesis in Plants. (Dordrecht, The Netherlands: Kluwer Academic Publishers). [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Shen Y., Toledo-Ortiz G., Kikis E.A., Johannesson H., Hwang Y.S., Quail P.H. (2006). Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 18: 2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubinger S., Fittinghoff K., Hoecker U. (2004). The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16: 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Tepperman J.M., Monte E., Calderon R.H., Liu T.L., Quail P.H. (2009). Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., Sun S.X., Li L., Yang H.Q. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25: 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. (2002). Blue light receptors and signal transduction. Plant Cell 14: S207–S225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25: 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., Lin C. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539. [DOI] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.J., Zhang Y.C., Li Q.H., Sang Y., Mao J., Lian H.L., Wang L., Yang H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863. [DOI] [PubMed] [Google Scholar]
- McNellis T.W., Deng X.-W. (1995). Light control of seedling morphogenetic pattern. Plant Cell 7: 1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Misera S., Deng X.W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E., Tepperman J.M., Al-Sady B., Kaczorowski K.A., Alonso J.M., Ecker J.R., Li X., Zhang Y., Quail P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101: 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Zhu L., Shen H., Huq E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 9433–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. (2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7: 708–711. [DOI] [PubMed] [Google Scholar]
- Nagy F., Schäfer E. (2000). Control of nuclear import and phytochromes. Curr. Opin. Plant Biol. 3: 450–454. [DOI] [PubMed] [Google Scholar]
- Nagy F., Kircher S., Schafer E. (2000). Nucleo-cytoplasmic partitioning of the plant photoreceptors phytochromes. Semin. Cell Dev. Biol. 11: 505–510. [DOI] [PubMed] [Google Scholar]
- Neff M.M., Fankhauser C., Chory J. (2000). Light: an indicator of time and place. Genes Dev. 14: 257–271. [PubMed] [Google Scholar]
- Ni M., Tepperman J.M., Quail P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667. [DOI] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466. [DOI] [PubMed] [Google Scholar]
- Park E., Kim J., Lee Y., Shin J., Oh E., Chung W.-I., Liu J.R., Choi G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45: 968–975. [DOI] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Halliday K.J. (2010). A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol. Biol. 73: 89–95. [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Su Y.-S., Lagarias J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57: 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I., Bou J., Sorin C., Devlin P.F., Martinez-Garcia J.F. (2006). Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 141: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Sullivan J.A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W. (2003). The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17: 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter M.G., Franklin K.A., Whitelam G.C. (2003). Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683. [DOI] [PubMed] [Google Scholar]
- Sasidharan R., Chinnappa C.C., Staal M., Elzenga J.T., Yokoyama R., Nishitani K., Voesenek L.A., Pierik R. (2010). Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 154: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Watanabe E., Tokutomi S., Nagatani A., Chua N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18: 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999. [DOI] [PubMed] [Google Scholar]
- Serino G., Deng X.-W. (2003). The COP9 signalosome: regulating plant development through the control of proteolysis. Annu. Rev. Plant Biol. 54: 165–182. [DOI] [PubMed] [Google Scholar]
- Shalitin D., Yang H., Mockler T.C., Maymon M., Guo H., Whitelam G.C., Lin C. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767. [DOI] [PubMed] [Google Scholar]
- Shen H., Moon J., Huq E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 44: 1023–1035. [DOI] [PubMed] [Google Scholar]
- Shen H., Zhu L., Castillon A., Majee M., Downie B., Huq E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20: 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Khanna R., Carle C.M., Quail P.H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145: 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Zhong S., Mo X., Liu N., Nezames C.D., Deng X.W. (2013). HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25: 3770–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J.M., Quail P.H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20: 1041–1044. [DOI] [PubMed] [Google Scholar]
- Shin J., Park E., Choi G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49: 981–994. [DOI] [PubMed] [Google Scholar]
- Soh M.S. (2000). REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12: 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Zhai Q., Li C. (2013). PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis. Plant Cell 25: 2102–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki G., Yanagawa Y., Kwok S.F., Matsui M., Deng X.-W. (2002). Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16: 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahleithner J.A., Li L.M., Lagarias J.C. (1991). Expression and assembly of spectrally active recombinant holophytochrome. Proc. Natl. Acad. Sci. USA 88: 10387–10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ma L.G., Li J.M., Zhao H.Y., Deng X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158. [DOI] [PubMed] [Google Scholar]
- Wei N., Kwok S.F., von Arnim A.G., Lee A., McNellis T.W., Piekos B., Deng X.-W. (1994). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6: 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G.C., Johnson E., Peng J., Carol P., Anderson M.L., Cowl J.S., Harberd N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T., Matsushika A., Fujimori T., Sato S., Kato T., Tabata S., Mizuno T. (2003). A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44: 619–629. [DOI] [PubMed] [Google Scholar]
- Yang H.Q., Tang R.-H., Cashmore A.R. (2001). The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Q., Wu Y.-J., Tang R.-H., Liu D., Liu Y., Cashmore A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827. [DOI] [PubMed] [Google Scholar]
- Yang J., Lin R., Sullivan J., Hoecker U., Liu B., Xu L., Deng X.W., Wang H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Li L., Guo M., Chory J., Yin Y. (2008). Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 7618–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.N., Wu Y., Tobias J.W., Brunk B.P., Deitzer G.F., Liu D. (2008). HFR1 is crucial for transcriptome regulation in the cryptochrome 1-mediated early response to blue light in Arabidopsis thaliana. PLoS ONE 3: e3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Maier A., Lee J.H., Laubinger S., Saijo Y., Wang H., Qu L.J., Hoecker U., Deng X.W. (2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Tepperman J.M., Fairchild C.D., Quail P.H. (2000). Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix–loop–helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 97: 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.