This work provides in vivo evidence that a Cys-rich peptide, STIG1, functions as a signaling molecule for the pollen receptor kinase LePRK2 and contributes to the rapid pollen tube growth in the pistil. The finding that STIG1 binds to external PI(3)P on pollen tubes points to a role for PI(3)P in regulating polar cell growth and in mediating cell–cell interactions.
Abstract
The speed of pollen tube growth is a major determinant of reproductive success in flowering plants. Tomato (Solanum lycopersicum) STIGMA-SPECIFIC PROTEIN1 (STIG1), a small Cys-rich protein from the pistil, was previously identified as a binding partner of the pollen receptor kinase LePRK2 and shown to promote pollen tube growth in vitro. However, the in vivo function of STIG1 and the underlying mechanism of its promotive effect were unknown. Here, we show that a 7-kD processed peptide of STIG1 is abundant in the stigmatic exudate and accumulates at the pollen tube surface, where it can bind LePRK2. Antisense LePRK2 pollen was less responsive than wild-type pollen to exogenous STIG1 in an in vitro pollen germination assay. Silencing of STIG1 reduced both the in vivo pollen tube elongation rate and seed production. Using partial deletion and point mutation analyses, two regions underlying the promotive activity of the STIG1 processed peptide were identified: amino acids 80 to 83, which interact with LePRK2; and amino acids 88 to 115, which bind specifically to phosphatidylinositol 3-phosphate [PI(3)P]. Furthermore, exogenous STIG1 elevated the overall redox potential of pollen tubes in both PI(3)P-dependent and LePRK2-dependent manners. Our results demonstrate that STIG1 conveys growth-promoting signals acting through the pollen receptor kinase LePRK2, a process that relies on the external phosphoinositide PI(3)P.
INTRODUCTION
The pollen tube is one of the fastest growing cells; its speed (up to 240 μm/min in Tradescantia and Hemerocallis species; reviewed in Michard et al., 2009) is generally believed to be the result of natural selection (Mulcahy, 1979; Howden et al., 1998). Fast-growing pollen tubes are more likely to achieve fertilization and give rise to more vigorous progeny (Delph et al., 1998). During evolution, the increase in tube growth speed preceded the establishment of other floral traits that contribute to the reproductive success of diverse angiosperms (Williams, 2008). Consequently, there has been great interest in understanding how pollen tubes can achieve such fast growth rates.
Although mature pollen from many species can germinate and grow rapidly in a simple medium supplemented with Suc, boric acid, and calcium, the elongation rate in vitro falls far short of that in the pistil (Heslop-Harrison, 1987). Pistil tissues likely provide a more favorable environment and additional factors to facilitate germination and growth. Indeed, several growth-promoting factors have been identified from various plant species, including flavonols (Mo et al., 1992), unsaturated lipids (Wolters-Arts et al., 1998), a transmitting tissue–specific glycoprotein (Cheung et al., 1995), and a small unidentified component from styles, STYLE INTERACTOR FOR LePRKs (STIL) (Wengier et al., 2010), from species with wet stigmas and azadecalin-like molecules (Qin et al., 2011) from species with dry stigmas. However, the mechanisms by which these factors promote pollen tube growth remain largely unknown.
Pollen-specific receptor kinases (PRKs) have been implicated as candidate regulators for perceiving growth-promoting factors. For example, studies in tomato (Solanum lycopersicum) (Zhang et al., 2008) and Arabidopsis thaliana (Zhang and McCormick, 2007; Chang et al., 2013) PRKs demonstrated that they are involved in polarized pollen tube growth and also play roles in mediating pollen–pistil interactions (Wengier et al., 2003; Zhang et al., 2008). The pollen receptor kinases interact with pollen-specific guanine nucleotide exchange factors at the apical plasma membrane to regulate the activity of small GTPases called RAC/ROPs, which are key regulators of polarized tip growth in pollen tubes (reviewed in Zou et al., 2011). In tomato, LePRK2 and another pollen receptor kinase, LePRK1, associate in a high molecular weight complex in mature pollen (Wengier et al., 2003). Once pollen lands on the stigma, STIL and/or other components in the 3- to 10-kD fraction of style extracts specifically dephosphorylate LePRK2 and dissociate the LePRK complex (Wengier et al., 2003, 2010). It was hypothesized (Wengier et al., 2003) that the dissociation of the LePRK complex would induce the release of their cytoplasmic partners and therefore transduce signals to the pollen tube cytoplasm. In line with this hypothesis, antisense LePRK2 pollen tubes exhibited a reduced growth rate both in vitro and in the pistil and were defective in responding to the growth-promoting signal STIL (Zhang et al., 2008).
Three secreted proteins, LATE ANTHER TOMATO52 (LAT52) (Tang et al., 2002) and SHY (Guyon et al., 2004) from pollen and STIG1 from the stigma (Tang et al., 2004), were identified as binding partners for the extracellular portion of LePRK2. The female partner STIG1 is of special interest because, in an in vitro competition assay, it outcompeted LAT52 for binding to the LePRK2 extracellular domain (referred to as ECD2) and also stimulated in vitro pollen tube growth (Tang et al., 2004). Tomato STIG1 encodes a secreted protein of 143 amino acids with a conserved C-terminal Cys-rich domain. Although the functions of STIG1 homologs have been investigated in two closely related solanaceous species, petunia (Petunia hybrida) and tobacco (Nicotiana tabacum) (Verhoeven et al., 2005), as well as in Arabidopsis (Wrzaczek et al., 2009), a species with dry stigmas, the biological function of STIG1 is not conclusive. Both a STIG1 mutant in petunia and transgenic tobacco plants in which STIG1 was silenced had excess stigmatic exudate (Verhoeven et al., 2005), whereas a presumed null mutant of Arabidopsis STIG1 (grim reaper [gri]) exhibited significantly reduced seed set (Wrzaczek et al., 2009). Our aim, therefore, is to investigate the role of STIG1 in tomato reproduction and to study the molecular mechanism underlying its growth-promoting activity.
Here, we present evidence that tomato STIG1 functions as a peptide signaling molecule for LePRK2 in promoting pollen tube growth. We show that STIG1 is secreted and processed into an ∼7-kD peptide in the stigmatic exudate. This processed peptide contains a LePRK2 binding site and a newly identified phosphatidylinositol 3-phosphate [PI(3)P] binding motif; both are required for its growth-promoting activity. We used a redox-sensitive green fluorescent protein (GFP) to show that STIG1 elevated the overall redox potential of pollen tubes in a PI(3)P-dependent and LePRK2-dependent manner, indicating that STIG1-LePRK2 signaling promotes pollen tube growth by affecting cellular reactive oxygen species (ROS) production.
RESULTS
Secretion and Processing of STIG1 in Mature Stigmas
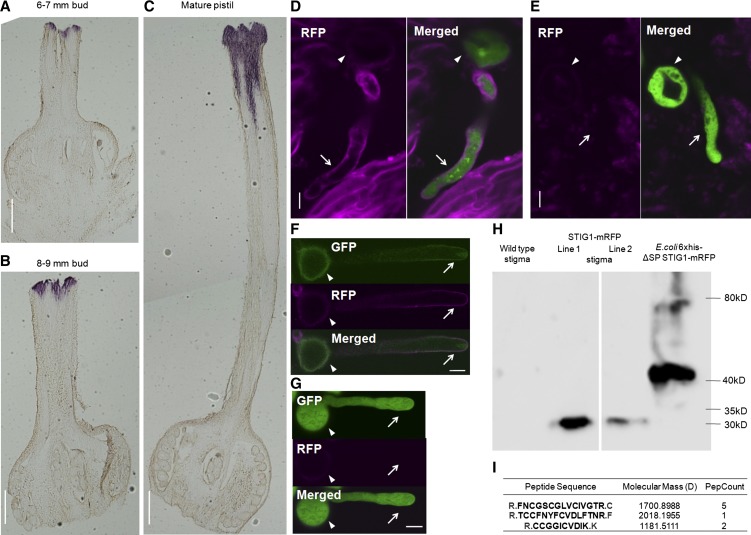
Tomato pollen tubes growing in vitro in optimized medium elongate at 2 to 6 μm/min, whereas it takes a pollen tube only 6 h to traverse a 1-cm style, at an average rate of 27 μm/min, 5-fold faster than the in vitro rate (Supplemental Figure 1). Previously, a role for recombinant tomato STIG1 in binding LePRK2 and promoting in vitro pollen tube growth was established (Tang et al., 2004). However, evidence for the action of STIG1 in vivo was lacking. Therefore, we decided to detect native STIG1 in vivo. First, the temporal and spatial expression patterns of STIG1 were investigated by in situ hybridization. In 6- to 7-mm young flower buds (developing flowers with green-yellowish petals, stages 13 to 15; Brukhin et al., 2003), the hybridization signal was confined to the superficial layers of the pistil (Figure 1A). In 8- to 9-mm flower buds (developing flowers with yellow, but still closed, petals, stages 16 to 18; Brukhin et al., 2003), the hybridization signal expanded within the stigmatic secretion zone (Figure 1B). In emasculated mature flowers, however, the signal extended from the secretion zone to the upper section of the style (Figure 1C). No signal was detected in the lower section of the style or the ovule. Thus, STIG1 expression was sustained throughout pistil maturation and was confined to the stigma and upper section of the style, consistent with a potential for STIG1 to affect pollen germination and early pollen tube growth.
Figure 1.
Tomato STIG1 Transcript Is Present in Mature Stigmas and STIG1 Is Processed in Stigmatic Exudate.
(A) to (C) In situ hybridization of STIG1 mRNA in a 6- to 7-mm flower bud (A), an 8- to 9-mm flower bud (B), and a mature pistil (C). Bars = 1 mm.
(D) and (E) Confocal images of eGFP-expressing pollen germinating in the stigma of a pistil expressing STIG1-mRFP (D) or a wild-type pistil (E).
(F) A representative ProLePRK2:LePRK2-eGFP pollen grain that had germinated on a transgenic tomato pistil expressing STIG1-mRFP for 1 h and then was washed from the stigma using germination medium.
(G) A representative eGFP-expressing pollen tube flushed from a wild-type stigma.
For (F) and (G), arrows and arrowheads indicate pollen tubes and pollen grains, respectively. Bars = 10 μm.
(H) Immunoblot analysis with anti-mRFP monoclonal antibody detects a 32-kD fusion protein in transgenic plants expressing STIG1-mRFP.
(I) STIG1 peptides in the 5- to 10-kD section of proteins from tomato stigma exudate, identified by ESI-MS. Trypsin digestion sites are shown in gray. PepCount indicates the number of matching peptides identified in ESI-MS.
To track the presence of STIG1 in mature stigmas, we generated transgenic plants expressing STIG1-mRFP (for monomeric red fluorescent protein) driven by the 35S promoter (Supplemental Figure 2B). Pistils from wild-type plants or from the transgenic plants were hand-pollinated with pollen expressing eGFP (for enhanced green fluorescent protein) under the control of the LAT52 promoter (Twell et al., 1990), then dissected and observed using confocal microscopy. The dim red autofluorescence around the pollen grains (Figures 1D and 1E, arrowheads) made it difficult to determine whether STIG1-mRFP was present there. However, a stronger fluorescent signal was detected evenly on the cell wall of pollen tubes growing in transgenic pistils (Figure 1D) but not in wild-type pistils (Figure 1E), from which we concluded that the red fluorescence was due to mRFP and not autofluorescence. To visualize the localization patterns of STIG1 and its binding partner LePRK2 at the same time, we also generated transgenic tomato plants overexpressing LePRK2-eGFP driven by its native promoter (Supplemental Figure 2C). Pollen grains from these plants were also hand-pollinated on pistils expressing STIG1-mRFP. Because LePRK2-eGFP pollen tubes were weakly fluorescent and to avoid background green autofluorescence from the stigma, short pollen tubes just germinating on the stigma were flushed from the pistils using small volumes of pollen germination medium, then imaged using confocal microscopy. STIG1-mRFP was distributed evenly at the cell wall and was not detected inside pollen tubes, while the LePRK2-eGFP signal showed membrane localization and also highlighted the apical clear zone, indicating that LePRK2-eGFP entered the secretory pathway (Figure 1F). An eGFP pollen tube that had been flushed from a wild-type stigma is also shown in Figure 1G, as a negative control. Note that there is dim red autofluorescence on the pollen grain but not on the pollen tube wall. The subcellular localizations of LeRPK2-GFP and STIG1-mRFP thus overlap at the pollen tube surface.
To determine the molecular mass of the STIG1-mRFP fusion protein, an anti-mRFP monoclonal antibody was used. This antibody recognized a 32-kD protein in stigmatic exudate (Figure 1H). This size was smaller than expected, given that the fusion protein should be 38.7 kD after cleavage of the signal peptide; mRFP alone is 25.8 kD. The 6xHis-ΔSp STIG1-mRFP expressed in Escherichia coli yielded a 41-kD band (the expected molecular mass), confirming antibody specificity (Figure 1H). This implies that a processed peptide of ∼7 kD is released from the Sl-STIG1 propeptide. Consistent with this, peptides derived from Nt-STIG1 (Supplemental Figure 3) or Ph-STIG1 (Verhoeven et al., 2005; see also Supplemental Figure 3) from stigmatic exudate of tobacco and petunia have been identified, and they all correspond to the conserved Cys-rich domains, while no peptide derived from the N terminus was found. To verify the existence of such a processed peptide in tomato, total stigmatic exudate was collected from mature unpollinated stigmas, and the proteins were subjected to Tricine-SDS-PAGE, a preferred electrophoretic system for the separation of proteins smaller than 20 kD. Sections of gels containing protein bands ranging between 3 and 5 kD, 5 and 10 kD, or 10 and 15 kD were excised and digested with trypsin, and the resulting peptides were sequenced by electrospray ionization–mass spectrometry (ESI-MS). In the 3- to 5-kD section, no STIG1 peptide was found; in the 5- to 10-kD section, three peptides were identified, all corresponding to the Cys-rich domain of STIG1 (Figure 1I; Supplemental Figure 3); in the 10- to 15-kD section, six peptides were identified, of which two corresponded to the N-terminal variable region and four to the Cys-rich domain (Supplemental Figure 3). These results suggest that a C-terminal peptide of around 7 kD is cleaved from the STIG1 propeptide in the stigmatic exudate and represents the major form of STIG1.
In Vivo Pollen Tube Growth and Seed Set Are Reduced in Transgenic STIG1 RNA Interference Plants
To directly address the function of STIG1 in pistils, transgenic tomato plants carrying an inverted repeat sequence against STIG1 were generated. STIG1 expression was significantly reduced in all nine T1 lines tested (Supplemental Figure 2A). Homozygotes obtained from four of these lines were used for further studies (Figure 2B). These transgenic plants grew normally, and no apparent changes of flower morphology were observed. As mature stigmas of tobacco plants silenced for STIG1 deposited more exudate (Verhoeven et al., 2005), we decided to look at the stigma morphology and exudate secretion in these RNA interference (RNAi) plants. Using conventional scanning electron microscopy, dense papilla cells of similar size protruding from the surface of mature stigmas in both wild-type and RNAi plants were observed (Supplemental Figure 4A), indicating that stigma maturation was not affected in these transgenic plants. Visualization of the stigmatic exudate was achieved using cryo-scanning electron microscopy; in mature stigmas of wild-type plants, although the intercellular spaces between papilla cells were filled with exudate, the tops of papilla cells were still visible; in STIG1 RNAi plants, however, patches of exudate would cover and mask the tops of papilla cells, suggesting that the stigmas accumulated more exudate (Supplemental Figure 4B).
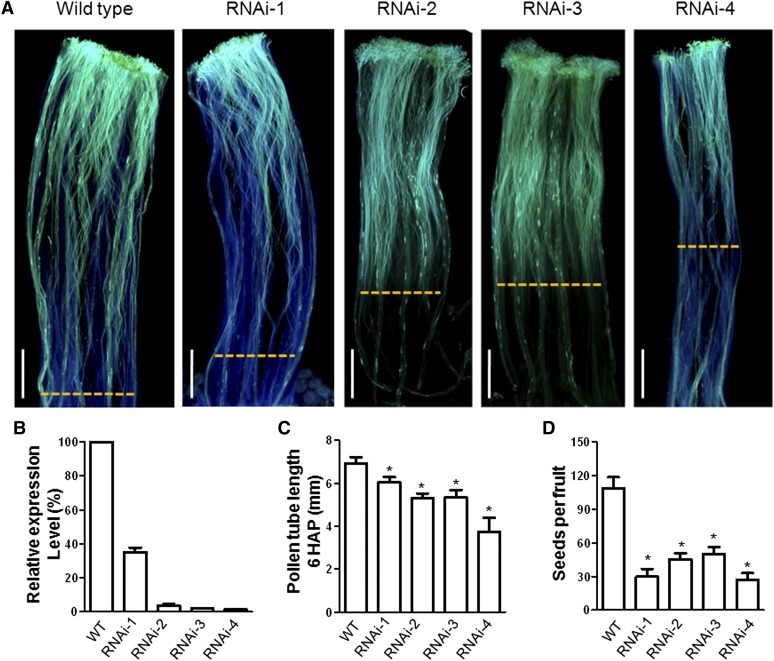
Figure 2.
Reduced Pollen Growth and Seed Content in STIG1 RNAi Plants.
(A) Wild-type pistils or transgenic pistils were hand-pollinated with wild-type pollen, dissected at 6 h, and stained with decolorized aniline blue to visualize pollen tubes. Yellow dashed lines indicate the growth front of pollen tubes. Bars = 1 mm.
(B) Quantitative RT-PCR of STIG1 mRNA levels, using total RNA of mature stigmas. n = 3 independent experiments.
(C) In vivo pollen tube lengths in (A). n = 3 independent experiments. At least six pistils were observed for each experiment.
(D) Seed content per fruit in self-pollinated STIG1 RNAi plants. n = 3 independent experiments. At least 10 fruits were harvested for each line in each experiment.
For (B) to (D), asterisks indicate significant differences from the wild type (P < 0.05, Student’s t test). Error bars indicate se.
We then examined pollen germination and pollen tube growth in these plants. When wild-type and transgenic STIG1 RNAi pistils were pollinated with wild-type pollen, the pollen germinated well on both stigmas. However, at 6 h after pollination, the average pollen tube length in transgenic pistils was shorter than in wild-type pistils (Figures 2A and 2C). Mature fruits from wild-type and STIG1 RNAi plants that were allowed to self-pollinate were harvested and their seeds were counted. STIG1 RNAi fruits had significantly fewer seeds than wild-type fruits (Figure 2D). Taken together, these findings indicate that the increased exudate in STIG1 RNAi plants did not affect in vivo pollen germination but that reduction of STIG1 did result in reduced pollen tube growth and somehow affected seed set.
Antisense LePRK2 Pollen Is Less Responsive to Exogenous STIG1 in Vitro
Because tomato STIG1 interacts with the extracellular domain of LePRK2 and stimulates pollen tube growth in vitro (Tang et al., 2004), we hypothesized that the stimulatory effect of STIG1 might depend on LePRK2 expression. To test this, liquid pollen germination assays were optimized to assess the promotive activity of recombinant GST-ΔSP STIG1. GST-ΔSP STIG1 within the range of 60 to 250 nM promoted tomato pollen tube growth in a dose-dependent manner (Figure 3A). At 500 nM, the promotive activity of recombinant STIG1 decreased, which might be because, at high concentrations, STIG1 tended to aggregate and precipitate in pollen germination medium. By contrast, tomato STIG1 showed no promotive activity toward tobacco pollen (Supplemental Figure 5). As the processed peptide identified in the stigmatic exudate would comprise the C-terminal Cys-rich domain of STIG1, we also tested the activity of this domain in vitro. Indeed, the Cys-rich domain, but not the N-terminal portion, promoted pollen tube growth and, at the same concentration, had more promotive activity than GST-ΔSP STIG1 (Figure 3C).
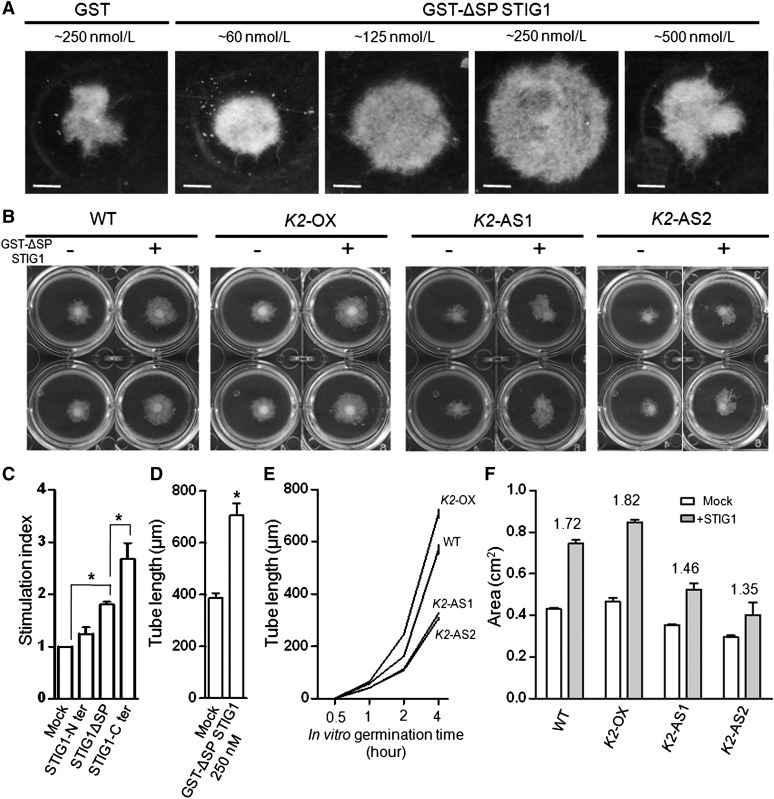
Figure 3.
Antisense LePRK2 Pollen Is Less Responsive Than Wild-Type Pollen to Exogenous STIG1 in Vitro.
(A) Purified recombinant GST-ΔSP STIG1 promotes pollen tube growth in a dose-dependent manner. Purified GST-ΔSP STIG1 of different concentrations was added to liquid germination medium at the onset of pollen germination. Images were acquired 18 h after germination. Bars = 0.5 cm.
(B) STIG1 pollen tube growth promotion assay with wild-type or transgenic LePRK2 pollen.
(C) Growth promotion effects of full-length or truncated STIG1 on tomato pollen tubes. An equal amount of recombinant protein (250 nM each) was used in this experiment. Stimulation index is defined as the fold change between the area of the pollen tube cluster with and without the corresponding protein. n = 3 independent experiments.
(D) Pollen tube lengths 8 h after germination in the presence of 250 nM GST-ΔSP STIG1 or 250 nM GST as a control.
(E) In vitro pollen tube lengths measured at different time points.
(F) Area of the pollen tube clusters in (B). Stimulation index is shown above each bar group.
For (C) to (F), asterisks indicate significant differences from the mock control (P < 0.05, Student’s t test). Error bars indicate se. K2-OX, ProLePRK2:LePRK2-eGFP; K2-AS1, AS-LePRK2 line1; K2-AS2, AS-LePRK2 line2.
If STIG1 promotes pollen tube growth by binding LeRPK2, the promotive effect should correlate with the LePRK2 expression level in pollen tubes. The tube growth of antisense LePRK2 pollen (previously developed by Zhang et al. [2008]), pollen overexpressing LePRK2-eGFP from its native promoter (Supplemental Figure 2C), and wild-type pollen was compared in the presence of 250 nM STIG1 (Figure 3B), a concentration at which the recombinant protein showed the most promotive effect (Figures 3A and 3D). In the absence of STIG1, the ProLePRK2:LePRK2-eGFP pollen tubes grown in vitro were significantly longer than wild-type pollen tubes, while the antisense LePRK2 pollen tubes were shorter (Figure 3E); consistent with such individual tube assessments, the ProLePRK2:LePRK2-eGFP pollen tubes formed the largest mat and the antisense LePRK2 mat was smallest (Figure 3B). The addition of STIG1 promoted the tube growth of all three pollen sources, but the stimulation of antisense LePRK2 pollen was weaker than for wild-type pollen or for ProLePRK2:LePRK2-eGFP pollen (Figure 3F). We concluded that the promotive effect of STIG1 at least partially relied on LePRK2.
Amino Acids F80N81Y82F83 in the Conserved Cys-Rich Domain of STIG1 Are Sufficient for Its Interaction with the Extracellular Domain of LePRK2
If the promotive effect of STIG1 depends on its direct interaction with LePRK2, we reasoned that the interaction site should be located in its C terminus (within the processed peptide). To identify which part of STIG1 was responsible for its interaction with LePRK2, we used yeast two-hybrid assays. A series of deletion or mutation fragments were fused to pGBKT7 and cotransformed with pGADT7-ECD2 (extracellular domain of LePRK2) in AH109 yeast cells. Interactions were determined by monitoring colony growth over 6 d on selective plates lacking Trp, Leu, His, and adenine (Figure 4B). When STIG1(16-143) (with the signal peptide removed) was used as the bait, colony growth was obvious, indicating a strong interaction. The bait vector (BD) alone, the N-terminal region STIG1(16-75) alone, or a short C-terminal region, STIG1(102-143), alone showed no interaction with ECD2. A longer C-terminal region, STIG1(76-143), interacted more strongly with ECD2 than did STIG1(16-143), as judged by growth and the number of transformants. The interacting domain was further delimited to amino acids F80N81Y82F83 in the C terminus, as STIG1(80-83) showed an interaction strength comparable to that of STIG1(16-143). Further single amino acid deletions within this region totally abolished the interaction, indicating that the tetrapeptide F80N81Y82F83 is the minimal peptide that is sufficient for interacting with ECD2. Several mutants of STIG1 were generated using site-directed mutagenesis. Consistent with the above findings, the point mutations F80A and N81A of full-length STIG1 significantly compromised their interaction with ECD2. In addition, two sextuple mutants, V85DL87EF88DR91EF92DI115D and Y82AF83AF88DR91EF92DI115D (these two mutants are discussed further below, in the phosphoinositide binding section), both showed slightly stronger interactions with ECD2 than did STIG1(16-143). In summary, in yeast, amino acids F80N81Y82F83 were sufficient for binding with ECD2, with Phe-80 and Asn-81 being the most important residues.
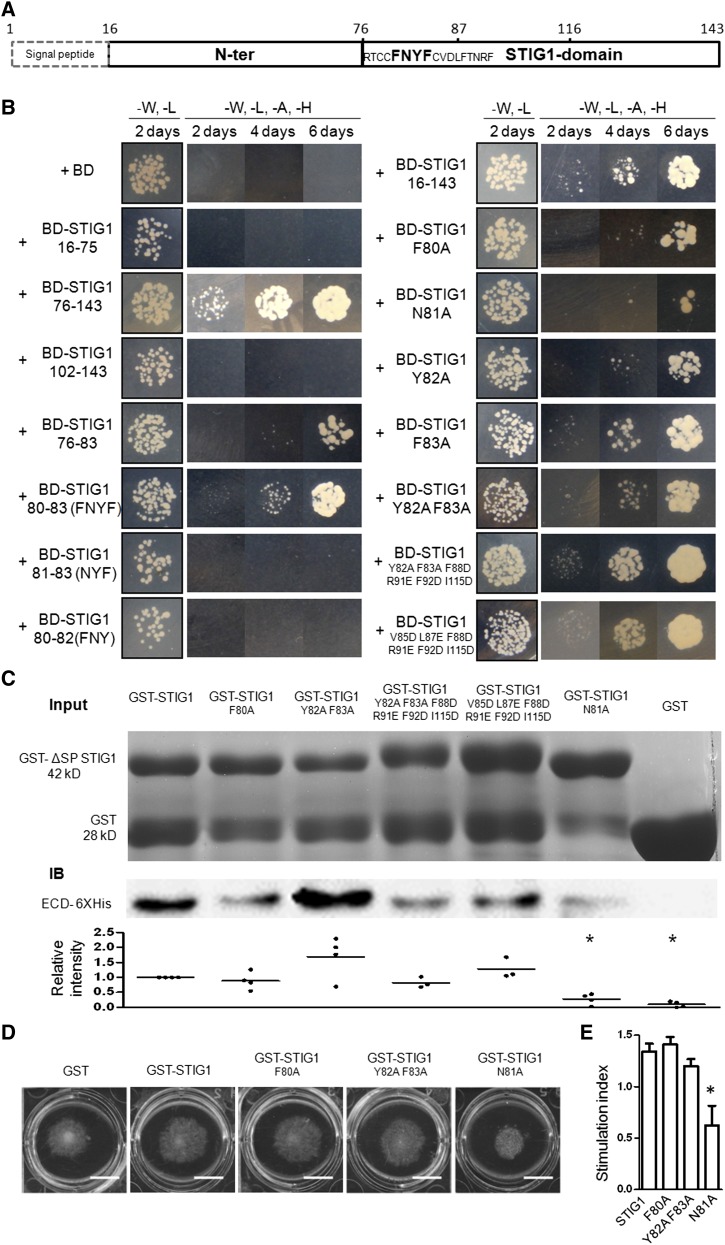
Figure 4.
Amino Acids F80N81Y82F83 in the Cys-Rich STIG1 Domain Are Necessary and Sufficient for Interaction with the Extracellular Domain of LePRK2.
(A) Schematic diagram of functional motifs/sites in STIG1. Numbers indicate amino acid positions. The FNYF motif is shown in boldface.
(B) Growth of yeast cells cotransformed with pGADT7-ECD2 and the listed constructs. Transformants were spotted on SD/-Leu-Trp or SD/-Leu-Trp-His-Ade medium.
(C) GST pull-down assay. Top panel, SDS-PAGE analysis of GST or GST fusion proteins. One-fifth of the corresponding proteins were loaded as an input control. Middle panel, proteins bound to Glutathione Sepharose 4B were separated by SDS-PAGE and detected with an anti-His monoclonal antibody. A representative gel is shown. Bottom panel, relative intensities in at least three experiments.
(D) Pollen tube growth promotion assay with wild-type or mutated GST-ΔSP STIG1. Bars = 1 cm.
(E) Pollen tube growth promotive effect of STIG1 and its mutants. Equal amounts of recombinant protein (250 nM each) were used. Error bars indicate se. n = 3 independent experiments. The asterisk indicates a significant difference from wild-type STIG1 (P < 0.05, Student’s t test).
To verify the binding affinities of the STIG1 mutants with ECD2, in vitro binding assays using GST (for glutathione S-transferase) fusion proteins and 6xHis-ECD2 were performed. GST (negative control) did not bind ECD2. One of the mutants, N81A, showed a significantly weaker interaction with ECD2 (Figure 4C). Other mutants either showed binding activity similar to that of STIG1 (F80A and Y82AF83AF88DR91EF92DI115D) or exhibited slightly stronger interaction (Y82AF83A and V85DL87EF88DR91EF92DI115D).
The above two sets of data together demonstrate that STIG1 bound to ECD2 through amino acids F80N81Y82F83 and that a specific mutation at Asn-81 (N81A) greatly compromised the interaction. To address the biological relevance of binding to LePRK2, the stimulatory effects of the N81A mutant and two other mutants were analyzed in pollen tube growth promotion assays (Figure 4D). The amino acid substitution at Asn-81 completely abolished growth-promoting activity, while the other two adjacent mutations (F80A and Y82AF83A) did not significantly affect the promotive effect of STIG1 (Figure 4E). Therefore, the pollen tube growth-promoting activity of STIG1 relies on direct interaction between STIG1 and LePRK2.
STIG1 Colocalized with a PI(3)P Biosensor on the Pollen Tube Surface
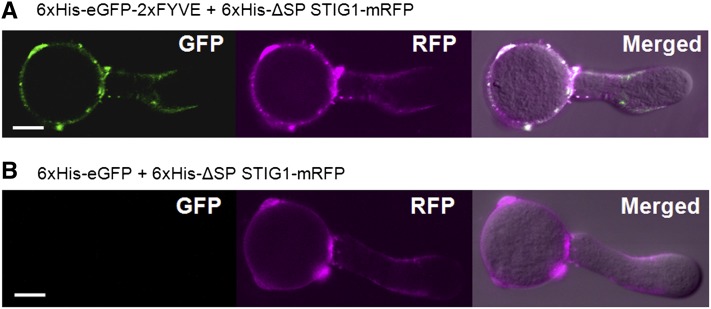
Transient expression of fluorescent reporter proteins in fast-growing pollen tubes by microprojectile bombardment (Twell et al., 1989) is a convenient and effective way to study protein localization (Cheung and Wu, 2007; Wang and Jiang, 2011). When transiently expressed in pollen tubes, STIG1-mRFP localized to numerous vesicular structures (Supplemental Figure 6), resembling the localization of PI(3)P in pollen tubes (Vermeer et al., 2006; Zhang et al., 2011). We wondered if similar localization patterns for STIG1 peptide and PI(3)P would occur in normal conditions (i.e., on the pollen tube surface). Previously, the presence of PI(3)P on the outer surface of root cells was detected using the highly specific biosensor 2xFYVE-GFP (Kale et al., 2010). We thus used a similar approach to test whether PI(3)P was also present on the pollen tube surface where STIG1 peptide accumulates (Figures 1D and 1F). When tomato pollen tubes were incubated with recombinant 2xFYVE-eGFP, the PI(3)P biosensor bound to the pollen tube surface unevenly: strong fluorescence was detected in the subapical region, moderate fluorescence was seen on the shank of pollen tubes, whereas little fluorescence was found in the tip region (Figure 5A). By contrast, recombinant eGFP alone did not bind the pollen tube surface (Figure 5B). Furthermore, recombinant ΔSP STIG1-mRFP showed a similar binding pattern and colocalized with 2xFYVE-eGFP on the pollen tube surface (Figures 5A and 5B). These results strongly indicate that PI(3)P is present on the outer surface of pollen tubes, where STIG1 can reach under normal conditions.
Figure 5.
STIG1 Colocalizes with the PI(3)P Biosensor 2xFYVE on the Outer Surface of Pollen Tubes When Provided Exogenously.
Tomato pollen tubes were incubated with recombinant 6xHis-ΔSP STIG1-mRFP and 6xHis-eGFP (A) or 6xHis-eGFP-2xFYVE (B). Bright-field images were overlaid with fluorescence images in the merged channel. Bars = 10 μm.
STIG1 Has Two Phospholipid Binding Motifs in the Conserved Cys-Rich Domain
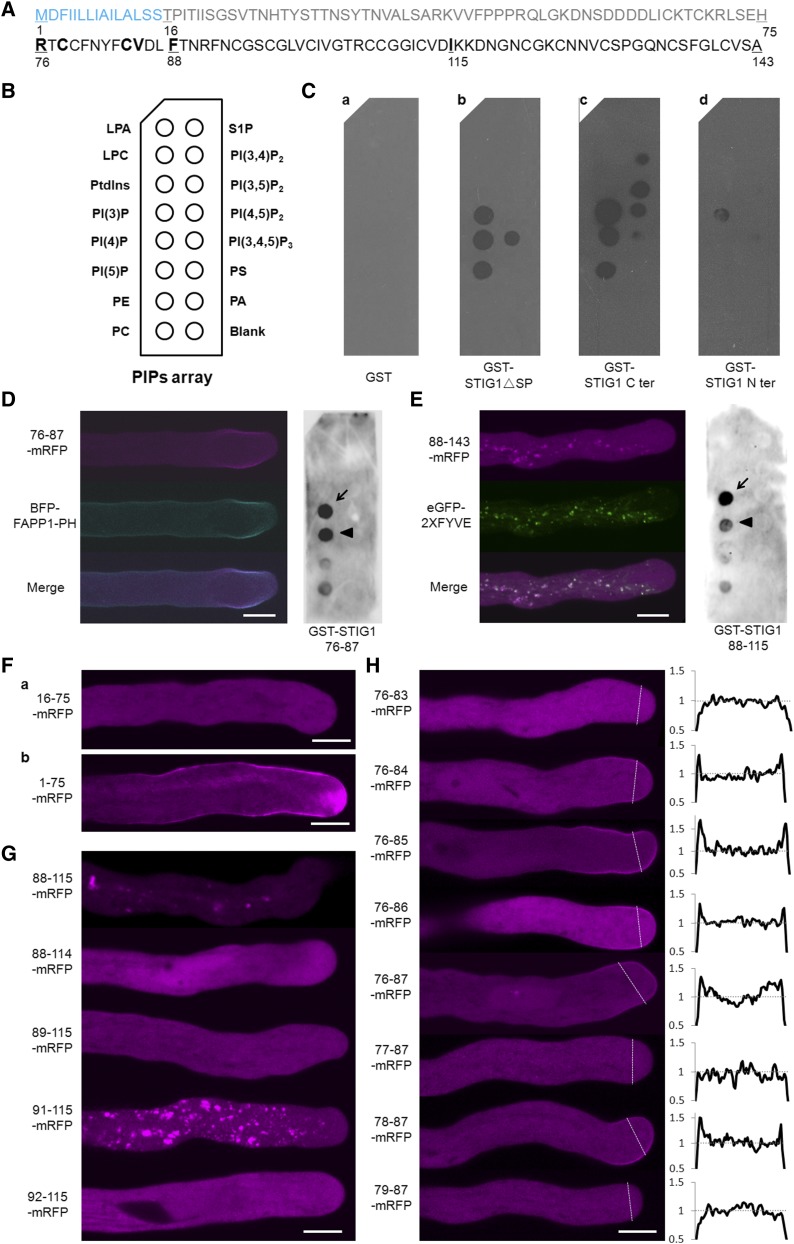
To test if STIG1 binds phospholipids directly, several GST fusion proteins were purified from E. coli and subjected to a protein–lipid overlay assay on which 14 phospholipids were spotted (Figure 6B). GST alone did not bind to any of the phospholipids (Figure 6C, a). GST-ΔSP STIG1 bound to three phosphatidylinositol monophosphates and to phosphatidylinositol 3,4,5-triphosphate (Figure 6C, b). The C-terminal Cys-rich domain also bound to the three phosphatidylinositol monophosphates and to two phosphatidylinositol biphosphates, phosphatidylinositol 3,5-biphosphate and phosphatidylinositol 4,5-biphosphate (Figure 6C, c). By contrast, the N-terminal region (amino acids 16 to 75; Figure 6A) showed only weak binding to PI(3)P (Figure 6C, d).
Figure 6.
Identification of Two Phospholipid Binding Motifs in the Conserved Cys-Rich Domain of STIG1.
(A) Amino acid sequence of STIG1. The signal peptide (blue), N terminus (gray), and C terminus (black) are indicated. Numbers indicate amino acid positions. Amino acids that play a positive role in phospholipid binding are shown in boldface.
(B) Schematic diagram of a PIP strip containing an array of immobilized phospholipids: lysophosphatidic acid (LPA), lysophosphocholine (LPC), phosphatidylinositol (PtdIns), PI(3)P, PI(4)P, phosphatidylinositol 5-phosphate [PI(5)P], phosphatidylethanolamine (PE), phosphatidylcholine (PC), sphingosine-1-phosphate (S1P), phosphatidylinositol 3,4-diphosphate [PI(3,4)P2], phosphatidylinositol 3,5-diphosphate [PI(3,5)P2], phosphatidylinositol 4,5-diphosphate [PI(4,5)P2], phosphatidylinositol 3,4,5-triphosphate [PI(3,4,5)P3], phosphatidylserine (PS), and phosphatidic acid (PA).
(C) Purified recombinant GST (a), GST-STIG1ΔSP (b), GST-STIG1 C-ter (c), and GST-STIG1 N-ter (d) were overlaid onto PIP strip membranes. Proteins bound to lipids were detected by immunoblotting with anti-GST monoclonal antibody. The images shown are representative of at least two dot-blot experiments.
(D) Representative pollen tube coexpressing amino acids 76 to 87 of STIG1 fused with mRFP and BFP-FAPP1-PH (left panel) and purified recombinant GST-STIG1 76 to 87 overlaid on a PIP strip (right panel).
(E) Representative pollen tube coexpressing amino acids 88 to 143 of STIG1 fused with mRFP and eGFP-2xFYVE (left panel) and purified recombinant GST-STIG1 88 to 115 overlaid on a PIP strip (right panel).
For (D) and (E), arrows and arrowheads indicate the PI(3)P spot and the PI(4)P spot, respectively. The images shown are representative of at least two dot-blot experiments.
(F) Representative pollen tubes expressing the N terminus of STIG1 (a) or the N terminus of STIG1 with STIG1 signal peptide (b).
(G) and (H) Representative pollen tubes transiently expressing different truncated versions of STIG1 within amino acids 88 to 143 (G) or amino acids 76 to 87 (H). The right panel in (H) show plots of relative pixel values along the lines drawn across the subapical regions of each pollen tube in the left panel.
For (H), the average pixel value of the cytosol in the subapical region was set to 1. All genes were driven by the LAT52 promoter and transiently expressed in tobacco pollen tubes. More than 15 pollen tubes were observed in each bombardment experiment. Bars = 10 μm.
Genetically encoded fluorescent phosphoinositide probes with high specificity are available to monitor the distribution and dynamics of various phosphoinositides in vivo (Vanhaesebroeck et al., 2001; Halet, 2005). To determine which part of STIG1 was responsible for lipid binding, we took advantage of the pollen tube bombardment assay and assessed the colocalization patterns between different STIG1 truncations and the PI(3)P marker eGFP-2xFYVE or the phosphatidylinositol 4-phosphate [PI(4)P] marker BFP-FAPP1-PH (He et al., 2011). We found that two adjacent regions in the conserved Cys-rich domain exhibited different lipid binding capacities. Amino acids 76 to 87 fused to mRFP preferentially localized at the subapical plasma membrane and colocalized with the PI(4)P marker BFP-FAPP1 (Figure 6D, left panel). In the lipid overlay assay, this motif showed equally strong binding with PI(3)P and PI(4)P (Figure 6D, right panel). Second, a truncation encompassing amino acids 88 to 143 localized to PI(3)P-positive vesicles (Figure 6E, left panel), as did a truncation encompassing amino acids 88 to 115 (Figure 6G, 88-115-mRFP). In the lipid overlay assay, the amino acid 88 to 115 truncation bound preferentially to PI(3)P (Figure 6E, right panel). Consistent with the lipid overlay assays, the N terminus alone (amino acids 16 to 75; Figure 6F, a) or with a signal peptide (amino acids 1 to 75; Figure 6F, b) showed localization patterns the same as those seen in the mRFP control tube (Supplemental Figure 6B), reinforcing the idea that the Cys-rich domain, rather than the N terminus, is responsible for lipid binding. Further deletion analysis showed that the positively charged residue Arg-91 and the hydrophobic residues Phe-88 and Ile-115 were important for the PI(3)P binding-mediated cytoplasmic punctate localization (Figure 6G). Similarly, we found that the positively charged amino acid Arg-76 and three hydrophobic amino acids in the PI(4)P binding region (Cys-78, Cys-84, and Val-85) promoted the subapical plasma membrane localization (Figure 6H).
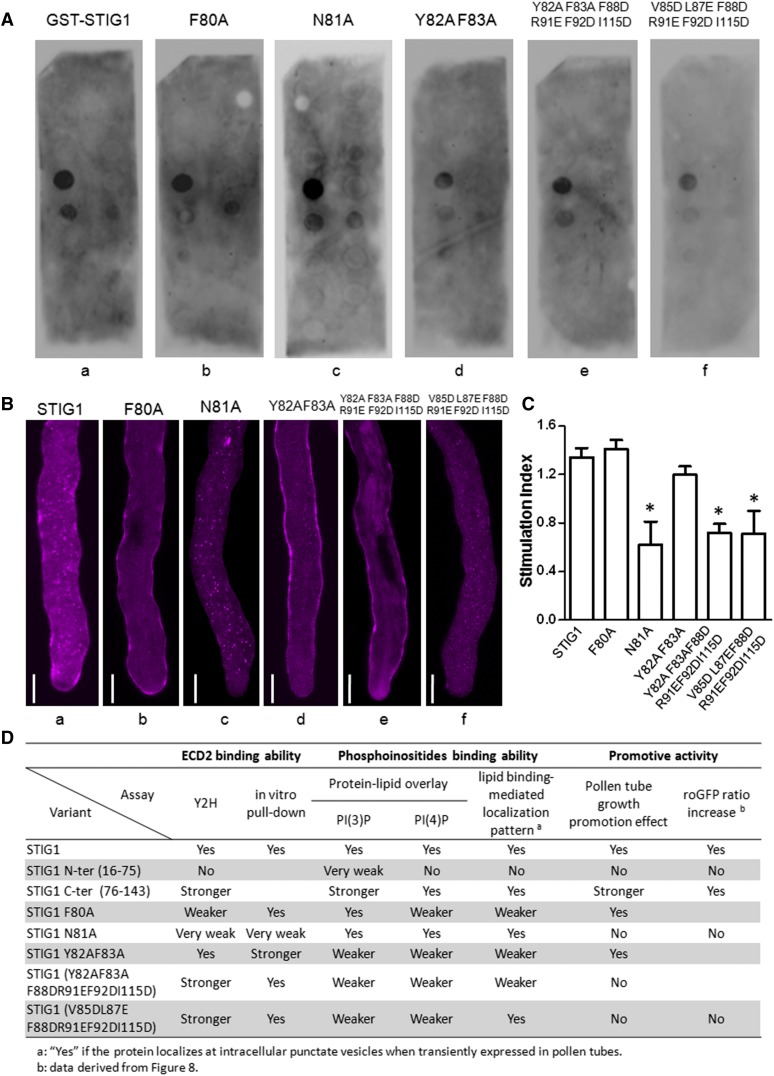
We further mutated the hydrophobic amino acids or positively charged amino acids in these two regions to Ala or to negatively charged residues and assessed how these mutations affected lipid binding. Mutant F80A showed weaker binding to PI(4)P, but its PI(3)P binding was not affected (Figure 7A, b), whereas mutant N81A exhibited binding affinities toward both lipids that were comparable to those of wild-type STIG1 (Figure 7A, a and c). The other three mutants (i.e., Y82AF83A, Y82AF83AF88DR91EF92DI115D, and V85DL87EF88DR91EF92DI115D) were compromised in PI(3)P binding and PI(4)P binding to different degrees (Figure 7A, d to f).
Figure 7.
The Pollen Tube Growth Promotive Effect of STIG1 Depends on Its Interaction with LePRK2 and on Its Phospholipid Binding Capacity.
(A) Phospholipid binding assay of recombinant GST-ΔSP STIG1 and its mutants. The data shown are representative of at least two independent experiments.
(B) Representative pollen tubes expressing STIG1-mRFP and its mutants. At least 10 pollen tubes were observed for each bombardment experiment. Bars = 10 μm.
(C) Pollen tube growth promotion effect of STIG1 and its mutants. Equal amounts of recombinant protein (250 nM each) were used. n = 3 independent experiments. Asterisks indicate significant differences from wild-type STIG1 (P < 0.05, Student’s t test). Error bars indicate se.
(D) Summary of the abilities of STIG1variants for LePRK2 interaction, phosphoinositide binding, and pollen tube growth promotive activities compared with wild-type STIG1. Yes, similar activity to LeSTIG1; No, no activity detected; blank, not tested; Y2H, yeast two-hybrid assay.
Secreted proteins with phospholipid binding motifs are translocated to the cytoplasm and localized on punctate vesicles when transiently expressed in pollen tubes (Supplemental Figure 6). Therefore, we speculated that the reduction of phospholipid binding capacity would result in the redistribution of STIG1 from the cytosol to the extracellular matrix. Indeed, when these mutants were transiently expressed in pollen tubes, two different localization patterns were observed. Mutants N81A and V85DL87EF88DR91EF92DI115D showed a localization pattern similar to that of STIG1; that is, the RFP signal was localized mostly at intracellular punctate vesicles and only a small portion of the fusion protein was secreted to the cell wall (Figure 7B, a, c, and f), suggesting that phospholipid binding was not affected. However, the RFP fusion protein of other mutants, including F80A, Y82AF83A, and Y82AF83AF88DR91EF92DI115D, aggregated significantly at the cell wall, and little signal was detected at punctate vesicles inside pollen tubes (Figure 7B, b, d, and e), indicating compromised phospholipid binding capacities for these mutants.
Taken together, we identified two regions in the C-terminal conserved Cys-rich domain of STIG1 that are sufficient for phosphoinositide binding: one is the PI(3)P-preferential binding site at amino acids 88 to 115 and the other is the PI(4)P-preferential binding site at amino acids 76 to 87.
The Promotive Effect of STIG1 Depends on the LePRK2 Binding Site and on Phosphoinositol Lipid Binding
Two functional sites were identified in the STIG1 peptide: the short PI(4)P binding site coincided with the ECD2 binding site, while the other site showed high binding specificity toward PI(3)P. We then asked if phosphoinositol lipid binding was relevant to the pollen tube growth promotive effect of STIG1. In addition, as the ECD2 binding site (amino acids 80 to 83) is included within the PI(4)P binding site (amino acids 76 to 87), we wondered if both ECD2 binding and phosphoinositol lipid binding contributed to the promotive effect of STIG1. To address these questions, we examined the pollen tube growth promotive activities of the substitution mutants mentioned above (Figure 7C; see also Figure 4D), which can distinguish phosphoinositol lipid binding from ECD2 binding. To summarize, we compared mutants with wild-type STIG1 in several aspects (Figure 7D). Mutant F80A, which showed weaker PI(4)P binding and lost the in vivo phospholipid binding–mediated cytoplasmic “punctate” localization pattern, was not compromised in the promotive activity. However, the N81A mutant, which showed diminished interaction between STIG1 and LePRK2 while maintaining phospholipid binding activities, could no longer promote the growth of tomato pollen tubes. These results showed that ECD2 binding, but not PI(4)P binding, is required for STIG1 to promote pollen tube growth. The remaining three mutants, namely Y82AF83A, Y82AF83AF88DR91EF92DI115D, and V85DL87EF88DR91EF92DI115D, also showed reduced or no pollen tube growth promotive activity (Figure 7C). Although these three mutants showed a general reduction in phospholipid binding, they did not affect the interaction between STIG1 and LePRK2. Considering that PI(4)P binding is not required for the STIG1 promotive effect, this result showed that PI(3)P binding, independent of the ECD2 interaction, contributed to the promotive effect of STIG1 on pollen tubes.
In summary, the interaction of STIG1 with LePRK2 and its phospholipid binding capacity, especially PI(3)P binding, are both required for STIG1 to promote pollen tube growth, and the contributions of these amino acid regions are independent of each other.
STIG1 Induces a PI(3)P-Dependent Increase in Intracellular Redox Potential
To understand the promotive effect of STIG1, it is necessary to study the downstream signaling events or cellular events that are triggered by STIG1. Activation of LePRK2 signaling likely involves dephosphorylation and results in its dissociation from LePRK1 (Wengier et al., 2003). Unlike STIL, STIG1 does not induce LePRK2 dephosphorylation (Wengier et al., 2010), but it was unknown if STIG1 affected LePRK1 and LePRK2 dissociation. We used a bimolecular fluorescence complementation (BiFC) assay to visualize the interaction between LePRK1 and LePRK2 on the plasma membrane (Supplemental Figure 7) and then coexpressed STIG1 with LePRK1 and LePRK2 BiFC plasmids in tobacco pollen tubes. Statistical analyses showed that STIG1 did not reduce the BiFC signal of the LePRK1 and LePRK2 interaction (Supplemental Figure 7). Therefore, STIG1 is not likely to induce the dissociation of the LePRK complex during pollen tube growth.
ROS are key components underlying the polarized growth machinery in both root hairs and pollen tubes (Foreman et al., 2003; Potocký et al., 2007). Previously, we showed that antisense LePRK2 pollen had an impaired response to Ca2+ for extracellular superoxide production (Zhang et al., 2008), suggesting that ROS production might be a downstream event of LePRK2 signaling. Therefore, we examined the effect of exogenous STIG1 on extracellular superoxide production using nitroblue tetrazolium (NBT), which is reduced by superoxide and forms a blue precipitate on the pollen tube surface (Supplemental Figures 8A and 8B). However, the application of full-length STIG1, its C terminus, or its N terminus did not significantly change the staining pattern of NBT (Supplemental Figure 8C), suggesting that the promotive effect of STIG1 might not affect extracellular superoxide production greatly.
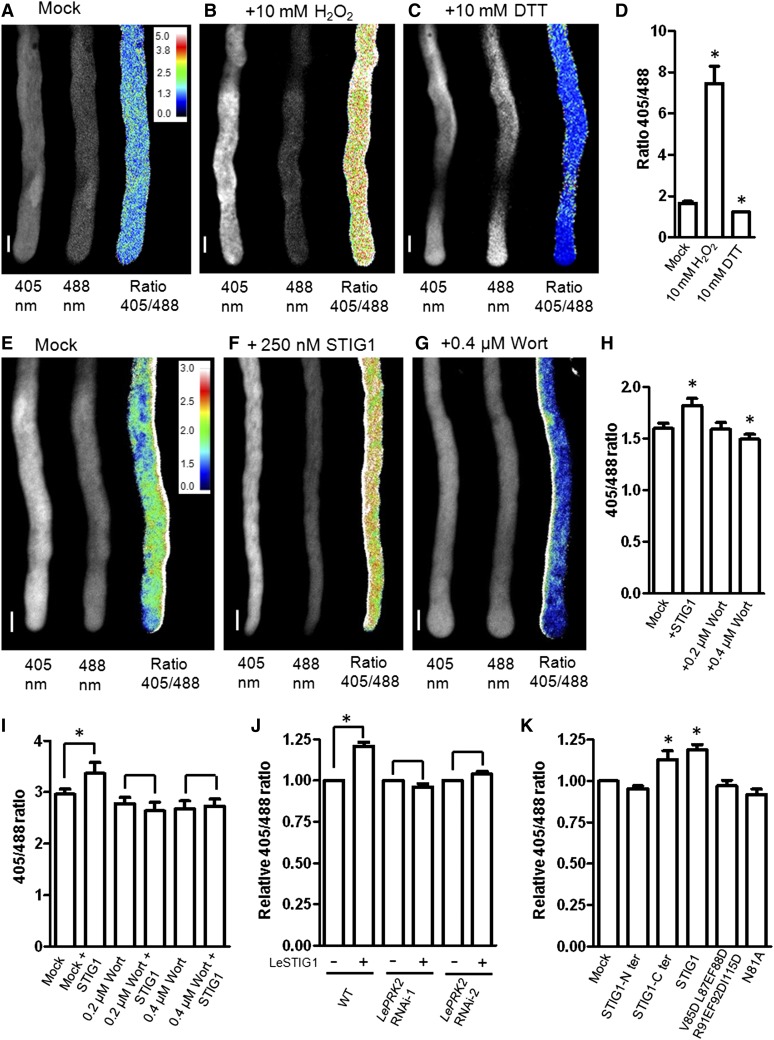
There is mounting evidence that PI(3)P plays a positive role in stimulating endocytosis and intracellular ROS production (Emans et al., 2002; Leshem et al., 2007; Lee et al., 2008). We wondered whether PI(3)P binding by STIG1 might affect intracellular ROS production. To test this, roGFP1, a ratiometric redox-sensitive GFP (Hanson et al., 2004), was expressed in pollen to enable dynamic measurements of the cellular redox status in vivo. Transgenic roGFP1 pollen responded quickly to redox changes induced by incubation with H2O2 or DTT, reflected by an immediate increase or decrease, respectively, of the 405:488 fluorescence ratio (Figures 8A to 8D). The addition of recombinant STIG1 to pollen germination medium induced a rapid intracellular ROS elevation within 3 min (Figure 8F).
Figure 8.
Exogenous STIG1 Elevates the Overall Redox Potential of in Vitro–Cultured Pollen Tubes in a PI(3)P-Dependent and LePRK2-Dependent Manner.
(A) to (C) roGFP transiently expressed in tobacco pollen tubes responds to redox changes induced by incubation with H2O2 (B) or DTT (C) relative to levels in mock-treated tubes (A).
(D) The 405:488 ratio of roGFP fluorescence in tobacco pollen tubes in (A) to (C). n > 6. Water was used as a mock control.
(E) to (G) The effects of DMSO (E), exogenous STIG1 (F), and the phosphoinositide 3-kinase inhibitor wortmannin (G) on the redox status of transgenic tomato pollen tubes expressing roGFP.
(H) The 405:488 ratio of roGFP fluorescence in tomato pollen tubes in (E) to (G). n > 6. DMSO was used as a mock control.
Three independent experiments were performed. Insets in (A) and (E) show the color scales for the ratio values. Bars = 10 μm.
(I) The 405:488 ratio of roGFP fluorescence in transgenic tomato pollen tubes treated with STIG1 alone or pretreated with wortmannin and then 250 nM STIG1. n > 6. Three independent experiments were performed. DMSO was used as a mock control.
(J) Intracellular ROS-promoting effects of exogenous STIG1 on roGFP-expressing pollen tubes in either the wild-type or the LePRK2 RNAi background. n > 6. Three independent experiments were performed.
(K) Effects of STIG1 deletion or substitution mutants on the redox status of transgenic tomato pollen tubes expressing roGFP. n > 6. Three independent experiments were performed.
For (J) and (K), equal amounts of recombinant protein (250 nM each) were used. The 405:488 ratio of mock-treated pollen tubes was set as 1.
Asterisks indicate significant differences from the mock control (P < 0.05, Student’s t test). Error bars indicate sd (D) or se ([H] to [K]).
Wortmannin is a specific inhibitor of phosphoinositide 3-kinases (Clague et al., 1995; Matsuoka et al., 1995), and in pollen tubes it disturbs PI(3)P production at concentrations below 30 μM (Zhang et al., 2010). Therefore, we tested the effect of wortmannin on intracellular ROS production in pollen tubes. As shown in Figure 8G, 0.4 μM wortmannin significantly reduced the redox potential of pollen tubes while 0.2 μM wortmannin did not significantly affect the redox potential (Figure 8H). Note that after 3 h of treatment with wortmannin, pollen tubes were shorter but the cytosol appeared normal (Supplemental Figure 9). Pretreatment with wortmannin, however, abolished the ROS increase induced by STIG1 (Figure 8I), suggesting that the intracellular ROS change in pollen tubes responding to STIG1 was a specific PI(3)P-dependent signaling event.
As antisense LePRK2 pollen tubes were less responsive to exogenous STIG1, we wanted to test the ROS stimulative effect of STIG1 on these pollen tubes. However, antisense LePRK2 pollen grains (Zhang et al., 2008) harbor a GFP-expressing cassette that is incompatible with roGFP imaging. Therefore, we generated two LePRK2 RNAi plants that contain an RFP reporter gene. Mature pollen of homozygotes from these lines had reduced LePRK2 expression, ∼1% (LePRK2 RNAi-1) and 15% (LePRK2 RNAi-2) of the levels in wild-type pollen (Supplemental Figure 2C). Moreover, LePRK2 RNAi pollen tubes grew slower in vitro, which recapitulated the phenotype (Zhang et al., 2008) of antisense LePRK2 pollen (Supplemental Figure 10). Homozygous LePRK2 RNAi pollen was then hand-pollinated on pistils of a heterozygous roGFP-expressing plant. F1 progeny with both the roGFP and roGFP/LePRK2 RNAi (RFP) constructs were analyzed. In pollen that did not carry the LePRK2 RNAi construct, exogenous STIG1 induced an increase in the 405:488 fluorescence ratio of roGFP. By contrast, no obvious redox change was triggered in pollen tubes with the LePRK2 RNAi construct (Figure 8J).
If the increased intracellular ROS production is indeed a downstream event triggered by STIG1 signaling, it should correlate with the growth stimulatory effect of STIG1. To test this, STIG1 deletion mutants or substitution mutants that can or cannot promote in vitro pollen tube growth were examined for their ability to stimulate intracellular ROS production. Consistent with our hypothesis, the STIG1 C-terminal Cys-rich domain faithfully induced an increase in intracellular redox potential, whereas the STIG1 N terminus did not (Figure 8K). Furthermore, two other mutants, with defects either in ECD2 binding (N81A) or PI(3)P binding (V85DL87EF88DR91EF92DI115D), were not able to stimulate intracellular ROS production (Figure 8K). Taken together, the binding of external PI(3)P and LePRK2 by STIG1 are both required for this downstream effect regarding intracellular ROS production and for the pollen tube growth promotive effect.
DISCUSSION
Here, we provide in vivo evidence that the pistil factor STIG1 functions as a signal that contributes to the rapid growth of tomato pollen tubes in the pistil. Intriguingly, in addition to a receptor binding site, a PI(3)P binding site exists in the processed STIG1 peptide. Several pieces of evidence support the notion that STIG1-LePRK2 signaling plays an important role in promoting pollen tube growth. First, STIG1 peptide, which is abundant in stigmatic exudate (Figure 1I), accumulates on the surface of pollen tubes, where it can bind to LePRK2 (Figures 1D and 1F). Second, reduced expression of either STIG1 or LePRK2 resulted in shorter pollen tubes in the pistil (Figure 2). Third, recombinant STIG1 promoted pollen tube growth in vitro, whereas antisense LePRK2 pollen was less responsive to exogenous STIG1 (Figure 3). Fourth, four amino acids in STIG1 determined the binding specificity to the extracellular domain of LePRK2 (Figure 4). Mutations in this region that affected the LePRK2–STIG1 interaction also impaired the growth promotive activity of STIG1 (Figures 4D and 7C).
The Cys-rich domain of STIG1 contains 14 conserved Cys residues (Supplemental Figure 11). Our results demonstrate that STIG1 undergoes proteolytic cleavage in the N-terminal variable region, and upon cleavage, a mature active peptide of around 7 kD comprises almost the entire Cys-rich domain. This is relevant, given that in the yeast two-hybrid assays, the Cys-rich domain alone had a much stronger binding affinity toward ECD2 than did full-length STIG1 (Figure 4B). More importantly, recombinant protein of this domain also showed a higher promotive activity in pollen tube growth assays (Figure 3C). Processing of precursor signaling peptides typically takes place at conserved dibasic motifs, which are recognition sites for subtilisin-like Ser proteases (Rholam and Fahy, 2009). Notably, there are two basic residues (K70R71) located at the end of the N-terminal variable region of STIG1 (Supplemental Figure 11) that might be involved in processing the STIG1 propeptide. The precise cleavage sites for two plant peptide hormones, At-RALF23 and At-PSK4, are at the C terminus of a Leu residue downstream of the dibasic motif (Srivastava et al., 2008, 2009). We suspect that STIG1 would be processed at Leu-72, resulting in a mature peptide of 71 amino acids (amino acids 73 to 143, 7.6 kD). Additional peptide analyses, in vitro peptide cleavage assays, or analyses with transgenic tomato expressing STIG1 with mutations in the dibasic site should help to determine the accurate cleavage site and to unravel the role of this dibasic motif in STIG1 processing. In the newly released tomato genome (Tomato Genome Consortium, 2012), there are 11 STIG1 domain–containing proteins (Supplemental Figure 12 and Supplemental Data Set 1). It is likely that the STIG1 family represents a class of signaling peptides, mediating different aspects of cell-to-cell communication.
Previous studies of STIG1 from different species showed different phenotypes (Verhoeven et al., 2005; Wrzaczek et al., 2009), leaving the role of STIG1 homologs an open question. In petunia, the loss of STIG1 did not affect in vivo pollen tube growth and seed set significantly (Verhoeven et al., 2005). In tomato, clear reductions in pollen tube growth and seed production were observed in STIG1 RNAi plants (Figure 2). The excess exudate found in all three solanaceous species with reduced STIG1 expression did not affect in vivo pollen germination (Figure 2A; Verhoeven et al., 2005). Unlike the lipid-rich, sticky stigmas in solanaceous species, Arabidopsis possesses dry stigmas. Nonetheless, the gri mutant also had reduced seed set (Wrzaczek et al., 2009), consistent with a role for STIG1 in pistils. It is worth noting that tomato STIG1 is different from its homologs in solanaceous species in several aspects. Despite the overall high sequence identities in their Cys-rich domains, Sl-STIG1 could not promote tobacco pollen tube growth in vitro (Supplemental Figure 5). More specifically, of the four amino acids (F80N81Y82F83) in Sl-STIG1 that are required for LePRK2 binding, there are one or two amino acid substitutions in the corresponding sites in the tobacco and petunia homologs (Y82A and F83S; Supplemental Figure 11). In addition, the expression of Sl-STIG1 was sustained throughout pistil maturation (Figure 1A), whereas in tobacco and petunia, STIG1 was highly expressed in very young and developing flowers and was not detected in mature flowers (Goldman et al., 1994; Verhoeven et al., 2005). Thus, our studies argue for a rapid evolution and functional diversification of the STIG1 homologs in pollen–pistil interactions.
The identification of phosphoinositide binding sites in Sl-STIG1 (Figures 5 and 6) raises the question of where the extracellular peptide might access PI(3)P or PI(4)P. It is generally considered that phosphoinositides are localized at the inner leaflet (cytoplasmic face) of cellular membranes (Roth, 2004). However, Kale et al. (2010) reported that PI(3)P is abundant on the outer surface of plant cell plasma membranes and further demonstrated that oomycete and fungal effectors harboring N-terminal RXLR motifs can be transferred into the cytoplasm of host plant cells via binding to external PI(3)P. Follow-up studies suggested that extracellular PI(3)P produced by Phytophthora pathogens might contribute to the PI(3)P pool during infection (Lu et al., 2013). In addition, the phosphatidylinositol monophosphate pool, especially PI(4)P, was detected in tomato apoplastic fluids and accumulated extracellularly in tomato cell suspensions upon xylanase treatment (Gonorazky et al., 2008, 2012). When incubated with pollen tubes, the PI(3)P biosensor eGFP-2xFYVE specifically bound to the pollen tube surface and colocalized with ΔSP STIG1-mRFP (Figure 5A and 5B). This observation supports the notion that STIG1 binds to PI(3)P exposed on the pollen tube outer membrane, where it also interacts with LePRK2. In transgenic tomato plants expressing STIG1-mRFP, the fusion protein accumulated evenly on the cell wall of pollen tubes growing in the pistils, while no fluorescence was detected inside pollen tubes (Figures 1D and 1F). This also supports the above hypothesis. However, we cannot exclude the possibility that STIG1 is endocytosed into pollen tubes, and it remains to be determined how PI(3)P is transported to the outer leaflet of the pollen tube plasma membrane.
We further provided two pieces of evidence suggesting that the PI(3)P binding of STIG1 peptide is functionally relevant. First, while mutations in the PI(4)P binding site did not or only slightly affected the promotive effect of STIG1 (Figure 7A, b and d), mutations in the PI(3)P binding motif resulted in a complete loss of its promotive activity (Figure 7A, e and f). Second, wortmannin treatment, which was shown to decrease external PI(3)P (Kale et al., 2010), diminished the intracellular ROS production induced by STIG1 (Figure 8I). PI(3)P is known to play a crucial role in determining the identities of endosomal compartments and in regulating almost every aspect of endosomal trafficking (Odorizzi et al., 2000; Di Paolo and De Camilli, 2006). There is support for PI(3)P acting in the regulation of endocytosis and ROS production in plants (Emans et al., 2002; Leshem et al., 2007; Lee et al., 2008). In roots, both increased endocytosis and ROS production triggered by salt stress are suppressed in Arabidopsis mutants that are defective in PI(3)P production (Leshem et al., 2007). Interestingly, the intracellular redox status of root cells in the elongation zone was more oxidized than that of cells in the root cap or root meristem (Jiang et al., 2006). Here, we showed that STIG1 elevated the overall cellular redox potential (Figure 8) and promoted pollen tube growth (Figure 3A), suggesting that higher elongation rates of pollen tubes are also accompanied by a more oxidized cellular redox status. Most importantly, mutant versions of STIG1, impaired either in PI(3)P binding or in LePRK2 binding, no longer promoted intracellular ROS production or in vitro pollen tube growth (summarized in Figure 7D). Thus, our study suggests a role for extracellular PI(3)P in mediating small peptide signal transduction and in regulating rapid cell elongation.
METHODS
Plant Material
Tomato (Solanum lycopersicum cv VF36) was grown under a light cycle of 12 h of light/12 h of dark. Temperature was maintained at 23 to 25°C during the day and 16 to 18°C during the night. Tobacco (Nicotiana tabacum cv Gexin No. 1) was grown at 28°C under a light cycle of 12 h of light/12 h of dark. Mature pollen was collected by vibrating anthers of open flowers with a biovortexer (BioSpec Products).
Pollen Bombardment, in Vitro Pollen Germination Assays, and Visualization of Pollen Tubes in Pistils
Pollen bombardment was performed as described (Twell et al., 1989). Briefly, ∼10 mg of tobacco pollen was bombarded with 5 μg of plasmids coated on 1-μm gold particles and then germinated in vitro in pollen germination medium [20 mM MES, pH 6.0, 3 mM Ca(NO3)2, 1 mM KCl, 0.8 mM MgSO4, 1.6 mM boric acid, 2.5% (w/v) Suc, and 24% (w/v) polyethylene glycol, molecular weight 4000]. The pollen-specific LAT52 promoter (Twell et al., 1990) was used in all bombardment assays. Both tobacco and tomato pollen were incubated at 25°C on six-well plates rotated horizontally at 150 and 60 rpm, respectively. BiFC was performed as described (Zhang and McCormick, 2007). Briefly, YC- or YN-containing plasmid (5 μg each) and control RFP plasmid (2 μg) were coated on gold particles. Pollen tubes were observed 3 to 8 h after bombardment, and images were captured using an Olympus BX51 microscope fitted with an Olympus DP71 digital camera or with a confocal microscope (Olympus Fluoview FV1000). In eGFP-2xFYVE and ΔSP STIG1-mRFP labeling experiments, tomato pollen tubes were cultured in a simplified medium [10% Suc, 1 mM Ca(NO3)2, 1 mM CaCl2, 1 mM MgSO4, and 1.6 mM boric acid] to avoid potential nonspecific binding caused by polyethylene glycol. Recombinant proteins (0.1 mg/mL) were added to the medium at the onset, and then pollen was allowed to germinate for 3 h before images were acquired. NBT staining of pollen tubes was performed as described (Zhang et al., 2008). Pollen tube lengths, pollen tube tip widths, and the intensity of formazan precipitation in pollen tube tips were measured using ImageJ (Rasband, 1997–2012). Decolorized aniline blue staining of pollen tubes in pistils was performed as described (Muschietti et al., 1994).
DNA Manipulation and the Generation of Transgenic Plants
The plasmids used for bombardment were derived from pLAT52:GFP or pLAT52:RFP, as described by Zhang et al. (2008). Fragments were amplified and inserted in frame at either the 5′ or 3′ end of the GFP or RFP coding sequence. For mutagenesis, the Mut Express II Fast Mutagenesis kit (Vazyme) was used. Intron-spliced hairpin RNA constructs were generated according to Wesley et al. (2001). The RNAi cassette for STIG1 included three parts: the 35S cauliflower mosaic virus promoter, an inverted repeat sequence against STIG1 cDNA spaced by the intron of LAT52, and the cauliflower mosaic virus 35S terminator. The RNAi cassette for LePRK2 included the LAT52 promoter and an inverted repeat sequence against the first 500-bp fragment of LePRK2 cDNA, spaced by the intron of LAT52, and the cauliflower mosaic virus 35S terminator. Fragments containing p35S:STIG1-mRFP, pLePRK2:LePRK2-eGFP, pLAT52:roGFP, the STIG1 RNAi cassette, or the LePRK2 RNAi cassette were inserted into the binary vector pCAMBIA2300 to generate the corresponding overexpression or RNAi construct. A separate mRFP gene driven by the LAT52 promoter was also included in the LePRK2 RNAi construct. Primers and cloning sites are provided in Supplemental Tables 1 and 2. Agrobacterium tumefaciens LBA4404 (Hoekema et al., 1983) carrying these plasmids was used to transform tomato as described (McCormick, 1991).
Scanning Electron Microscopy
For conventional scanning electron microscopy, mature pistils were fixed in FAA at 4°C for 2 h and dehydrated through a graded alcohol series of 50, 70, 80, 90, 95, and 100% ethanol, each for 10 min. The samples were then dried using liquid carbon dioxide as a transition fluid. Stigmas were dissected using glass needles and mounted on scanning electron microscopy stubs. Mounted specimens were sputter coated with palladium and examined with a scanning electron microscope (JEOL JSM-6360LV). For cryo-scanning electron microscopy, fresh pistils were glued onto scanning electron microscopy stubs and directly frozen in liquid nitrogen. The samples were sputter coated with 5-nm platinum in a cryo-preparation chamber and examined using the JEOL JSM-6360LV scanning electron microscope equipped with a cold stage (Quorum PP3010T).
Fusion Protein Purification and in Vitro Binding Assays
The coding sequences of the extracellular domain of LePRK2, eGFP, eGFP-2xFYVE, and ΔSP STIG1-mRFP were fused in frame with a 6-His tag in the pRSET-C vector (Invitrogen) and then expressed and purified under native conditions as described (Tang et al., 2002). STIG1 (with signal peptide removed) or its truncation/substitution mutants were fused with GST in the pGEX-4T3 vector (GE Healthcare). The resulting plasmids were transformed into Escherichia coli strain Rosetta (Novagen), and fusion protein production was induced with 0.1 mM isopropyl-β-d-thiogalactoside. The GST fusion proteins were then purified using Glutathione Sepharose 4B (GE Healthcare) following the manufacturer’s procedures. The concentrations of the fusion proteins were determined with UV light spectrophotometry. GST pull-down experiments were performed as described (Tang et al., 2004). GST (∼300 pmol), GST-ΔSP STIG1 or its mutants (∼100 pmol), and ∼100 pmol of 6xHis-ECD2 were used in these experiments. For lipid binding assays, PIP strips (P-6001; Echelon Biosciences) were blocked in a solution of 3% (w/v) fatty acid–free BSA or 4% (w/v) nonfat dry milk in Tris-buffered saline plus Tween 20 for 1 h and then incubated with 0.03 mg/mL GST fusion protein for 3 h with gentle agitation at room temperature. Bound GST fusion proteins were detected with an anti-GST monoclonal antibody (cw0082; CWbiotech) and visualized by secondary antibodies coupled to horseradish peroxidase (IH-0031; Dingguo Biotechnology) followed by enhanced chemiluminescence detection (Amersham Pharmacia Biotech). Experiments were performed at least two times with freshly purified proteins.
Yeast Two-Hybrid Assays
The MATCHMAKER GAL4 Two-Hybrid System (Clontech) was used. Yeast strain AH109 was cotransformed with pGADT7-ECD2 and pGBKT7 fused to appropriate deletion or mutation constructs of STIG1 using the rapid method of Gietz and Woods (2002). The transformants were spotted on SD medium lacking Trp/Leu (-W, -L) or SD medium lacking Trp/Leu/His/adenine (-W, -L, -H, -A) and examined for growth. Interaction strengths were scored visually based on the number of colonies and on growth rate.
Redox-Sensitive GFP Imaging and Ratiometric Analysis
Transgenic tomato plants expressing roGFP1 (Hanson et al., 2004) under the control of the pollen-specific promoter LAT52 were generated. In vitro–germinated transgenic pollen tubes were imaged using an Olympus confocal microscope (FV1000) equipped with lasers for 405- and 488-nm excitation. Images were acquired with a 20× lens (UPLSAPO; NA0.75) in multi-track mode with line switching and taking an average of four readings. In the first track, roGFP was excited at 405 nm. In the second track, roGFP was excited at 488 nm. For both excitation wavelengths, roGFP1 fluorescence was collected with a band-pass filter of 505 to 530 nm. Ratiometric analysis of fluorescence images was performed in Olympus Fluoview version 3.0a. The 405-nm image was divided by the 488-nm fluorescence intensity image to produce a ratio image on a pixel-by-pixel basis. Only ratios measured with identical settings were compared in absolute terms.
RNA Extraction, Quantitative RT-PCR, and in Situ Hybridization
Total RNA from stigmas was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. cDNA was synthesized using the SuperScript III system (Invitrogen). Quantitative real-time PCR of reverse-transcribed RNA was performed with SYBR Green I detection on an iCycler (Bio-Rad). The primers used to amplify a 150-bp fragment of STIG1 were 5′-ATCCTTCTCATCGCCATCCT-3′ and 5′-TAGCTGTCTGGGAGGAGGAA-3′. The primers used to amplify a 123-bp fragment of a tomato actin gene were 5′-GCGAGAAATTGTCAGGGACGT-3′and 5′-TGCCCATCTGGGAGCTCAT-3′. For in situ hybridization, the cDNA of STIG1 was subcloned into pBluescript SK+ vector for RNA probe synthesis. The antisense and sense RNA probes were synthesized by in vivo transcription using T7 and T3 RNA polymerase, respectively, using DIG RNA Labeling Mix (Roche). In situ hybridization experiments were performed as described (Cox and Goldberg, 1988; Langdale, 1993) using 10-μm sections of pistils.
Pharmacological Treatments
Wortmannin Ready Made Solution (Sigma-Aldrich) was supplied as a 10 mM solution in DMSO. Dilutions in DMSO were prepared and added to liquid pollen germination medium. Equivalent volumes of DMSO were added to the controls.
Protein Extraction and Peptide Analysis
Tomato stigmas were ground to powder in liquid nitrogen and then homogenized with extraction buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 10% [v/v] glycerol, 0.1% Nonidet P-40, 1 mM DTT, and 1× Protease Inhibitor Cocktail Complete [Boehringer Mannheim]). Proteins were separated by SDS-PAGE and used for immunoblots with anti-RFP monoclonal antibody (cw0298; CWbiotech). Stigma exudate was obtained as described (Verhoeven et al., 2005). Briefly, five stigmas from mature pistils were submerged in 20 μL of 50 mM NaAc, pH 4.5, in a 2-mL Eppendorf tube for 1 h with gentle agitation. A total of 200 stigmas were extracted, and the resulting exudate was pooled. The solution was then centrifuged for 20 min at 4°C to remove the aqueous phase and stored at −80°C. Protein samples from stigma exudate were separated using Tricine-SDS-PAGE. Sections of gels containing protein bands ranging between 3 and 5 kD, 5 and 10 kD, or 10 and 15 kD were excised and then digested with trypsin. All digested peptide mixtures were separated by reverse-phase HPLC followed by tandem mass spectrometry analysis on a surveyor liquid chromatography system (Thermo Finnigan). An in-house database was constructed with the FASTA protein sequences downloaded from the Sol Genomics Network (http://solgenomics.net/) containing predicted proteins of tomato species. Tandem mass spectrometry spectra were automatically searched against the database using BioworksBrowser rev. 3.1 (Thermo Electron).
Accession Numbers
GenBank accession numbers of the genes used in this article are as follows: AY376851 for Sl-STIG1, X77823 for Nt-STIG1, AF130352 for Ph-STIG1, NM_104192 for At-GRI (Arabidopsis Genome Initiative locus AT1G53130), U58473.1 for LePRK2, and U58474.1 for LePRK1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Tomato Pollen Tube Growth Rate Is Faster in Pistils Than in Optimized Germination Medium.
Supplemental Figure 2. Expression Levels of Target Genes in Transgenic Tomato Plants Generated in This Study.
Supplemental Figure 3. STIG1 Peptides Identified by Mass Spectrophotometry of Stigmatic Exudate of Tomato, Tobacco, and Petunia.
Supplemental Figure 4. Mature Stigmas of STIG1 RNAi Plants Have Normal Morphology but Accumulate More Exudate Than Wild-Type Stigmas.
Supplemental Figure 5. STIG1 Promotes Pollen Tube Growth of Tomato but Not Tobacco.
Supplemental Figure 6. STIG1 Localizes Both to the Pollen Tube Wall and to Intracellular Punctate Vesicles When Ectopically Expressed in Pollen Tubes.
Supplemental Figure 7. STIG1 Does Not Affect the BiFC Interaction between LePRK1 and LePRK2.
Supplemental Figure 8. Exogenous STIG1 Does Not Affect Extracellular Superoxide Production on the Pollen Tube Surface.
Supplemental Figure 9. Wortmannin Reduces Pollen Tube Length.
Supplemental Figure 10. LePRK2 RNAi Pollen Grew Shorter Tubes in Vitro.
Supplemental Figure 11. Amino Acid Alignment of STIG1 Homologs.
Supplemental Figure 12. STIG1 Domain–Containing Proteins in Tomato.
Supplemental Table 1. Detailed Information for Plasmid Construction.
Supplemental Table 2. Primer Sequences.
Supplemental Data Set 1. Text File of the STIG1 Alignment Used for Phylogenetic Analysis in Supplemental Figure 12.
Supplementary Material
Acknowledgments
We thank Yan Zhang for the kind gifts of the ProLAT52:2XFYVE-mRFP construct (Helling et al., 2006) and ProLAT52:ARA7-YFP (Zhang et al., 2010) and James Remington for the roGFP construct (Hanson et al., 2004). We thank Xiao-Shu Gao, Xiao-Yan Gao, Ji-Qin Li, and Xin Dong for help with roGFP imaging and cryo-scanning electron microscopy. We also thank Jorge Muschietti and Hong-Wei Xue for comments on the article. This work was supported by the Natural Science Foundation of China (Grant 30970266 to Dong Zhang and Grant 31170291 to W.-H.T.) and the Ministry of Science and Technology of China (Grant 2012AA10A302 to W.-H.T.).
AUTHOR CONTRIBUTIONS
W.-H.T. and W.-J.H. designed the research. W.-J.H. performed all experiments except for those in Supplemental Figure 7, which were performed by H.-K.L. H.-K.L. also provided yeast two-hybrid plasmids. W.-H.T., W.-J.H., and S.M. analyzed the data and wrote the article.
Glossary
- ROS
reactive oxygen species
- PI(3)P
phosphatidylinositol 3-phosphate
- ESI-MS
electrospray ionization–mass spectrometry
- RNAi
RNA interference
- PI(4)P
phosphatidylinositol 4-phosphate
- BiFC
bimolecular fluorescence complementation
- NBT
nitroblue tetrazolium
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Brukhin V., Hernould M., Gonzalez N., Chevalier C., Mouras A. (2003). Flower development schedule in tomato Lycopersicon esculentum cv. Sweet Cherry. Sex. Plant Reprod. 15: 311–320. [Google Scholar]
- Chang F., Gu Y., Ma H., Yang Z. (2013). AtPRK2 promotes ROP1 activation via RopGEFs in the control of polarized pollen tube growth. Mol. Plant 6: 1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. (2007). Structural and functional compartmentalization in pollen tubes. J. Exp. Bot. 58: 75–82. [DOI] [PubMed] [Google Scholar]
- Cheung A.Y., Wang H., Wu H.M. (1995). A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82: 383–393. [DOI] [PubMed] [Google Scholar]
- Clague M.J., Thorpe C., Jones A.T. (1995). Phosphatidylinositol 3-kinase regulation of fluid phase endocytosis. FEBS Lett. 367: 272–274. [DOI] [PubMed] [Google Scholar]
- Cox, K.H., and Goldberg, R.B. (1988). Analysis of gene expression. In Plant Molecular Biology: A Practical Approach, C.H. Shaw, ed (Oxford, UK: Oxford IRL Press), pp. 1–35. [Google Scholar]
- Delph L.F., Weinig C., Sullivan K. (1998). Why fast-growing pollen tubes give rise to vigorous progeny: The test of a new mechanism. Proc. R. Soc. B Biol. Sci. 265: 935–939. [Google Scholar]
- Di Paolo G., De Camilli P. (2006). Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657. [DOI] [PubMed] [Google Scholar]
- Emans N., Zimmermann S., Fischer R. (2002). Uptake of a fluorescent marker in plant cells is sensitive to brefeldin A and wortmannin. Plant Cell 14: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D., Davies J.M., Dolan L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Goldman M.H.S., Goldberg R.B., Mariani C. (1994). Female sterile tobacco plants are produced by stigma-specific cell ablation. EMBO J. 13: 2976–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonorazky G., Laxalt A.M., Dekker H.L., Rep M., Munnik T., Testerink C., de la Canal L. (2012). Phosphatidylinositol 4-phosphate is associated to extracellular lipoproteic fractions and is detected in tomato apoplastic fluids. Plant Biol. (Stuttg.) 14: 41–49. [DOI] [PubMed] [Google Scholar]
- Gonorazky G., Laxalt A.M., Testerink C., Munnik T., de la Canal L. (2008). Phosphatidylinositol 4-phosphate accumulates extracellularly upon xylanase treatment in tomato cell suspensions. Plant Cell Environ. 31: 1051–1062. [DOI] [PubMed] [Google Scholar]
- Guyon V., Tang W.H., Monti M.M., Raiola A., Lorenzo G.D., McCormick S., Taylor L.P. (2004). Antisense phenotypes reveal a role for SHY, a pollen-specific leucine-rich repeat protein, in pollen tube growth. Plant J. 39: 643–654. [DOI] [PubMed] [Google Scholar]
- Halet G. (2005). Imaging phosphoinositide dynamics using GFP-tagged protein domains. Biol. Cell 97: 501–518. [DOI] [PubMed] [Google Scholar]
- Hanson G.T., Aggeler R., Oglesbee D., Cannon M., Capaldi R.A., Tsien R.Y., Remington S.J. (2004). Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J. Biol. Chem. 279: 13044–13053. [DOI] [PubMed] [Google Scholar]
- He J., Scott J.L., Heroux A., Roy S., Lenoir M., Overduin M., Stahelin R.V., Kutateladze T.G. (2011). Molecular basis of phosphatidylinositol 4-phosphate and ARF1 GTPase recognition by the FAPP1 pleckstrin homology (PH) domain. J. Biol. Chem. 286: 18650–18657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling D., Possart A., Cottier S., Klahre U., Kost B. (2006). Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell 18: 3519–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison J. (1987). Pollen germination and pollen tube growth. Int. Rev. Cytol. 107: 1–78. [Google Scholar]
- Hoekema A., Hirsch P.R., Hooykaas P.J.J., Schilperoort R.A. (1983). A binary plant vector strategy based on separation of vir-region and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303: 179–180. [Google Scholar]
- Howden R., Park S.K., Moore J.M., Orme J., Grossniklaus U., Twell D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149: 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Schwarzer C., Lally E., Zhang S., Ruzin S., Machen T., Remington S.J., Feldman L. (2006). Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol. 141: 397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale S.D., et al. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142: 284–295. [DOI] [PubMed] [Google Scholar]
- Langdale, J.A. (1993). In situ hybridization. In The Maize Handbook, M. Freeling and V. Walbot, eds (New York: Springer-Verlag), pp. 165–180. [Google Scholar]
- Lee Y., Bak G., Choi Y., Chuang W.I., Cho H.T., Lee Y. (2008). Roles of phosphatidylinositol 3-kinase in root hair growth. Plant Physiol. 147: 624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem Y., Seri L., Levine A. (2007). Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant J. 51: 185–197. [DOI] [PubMed] [Google Scholar]
- Lu S., Chen L., Tao K., Sun N., Wu Y., Lu X., Wang Y., Dou D. (2013). Intracellular and extracellular phosphatidylinositol 3-phosphate produced by Phytophthora species is important for infection. Mol. Plant 6: 1592–1604. [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Bassham D.C., Raikhel N.V., Nakamura K. (1995). Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J. Cell Biol. 130: 1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, S. (1991). Transformation of tomato with Agrobacterium tumefaciens. In Plant Tissue Culture Manual, Fundamentals and Applications, Vol. B, K. Lindsey, ed (Amsterdam: Kluwer), pp. 1–9. [Google Scholar]
- Michard E., Alves F., Feijó J.A. (2009). The role of ion fluxes in polarized cell growth and morphogenesis: The pollen tube as an experimental paradigm. Int. J. Dev. Biol. 53: 1609–1622. [DOI] [PubMed] [Google Scholar]
- Mo Y., Nagel C., Taylor L.P. (1992). Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. USA 89: 7213–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy D.L. (1979). The rise of the angiosperms: A genecological factor. Science 206: 20–23. [DOI] [PubMed] [Google Scholar]
- Muschietti J., Dircks L., Vancanneyt G., McCormick S. (1994). LAT52 protein is essential for tomato pollen development: Pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 6: 321–338. [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S.D. (2000). Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 25: 229–235. [DOI] [PubMed] [Google Scholar]
- Potocký M., Jones M.A., Bezvoda R., Smirnoff N., Zárský V. (2007). Reactive oxygen species produced by NADPH oxidase are involved in pollen tube growth. New Phytol. 174: 742–751. [DOI] [PubMed] [Google Scholar]
- Qin Y., et al. (2011). Sulfinylated azadecalins act as functional mimics of a pollen germination stimulant in Arabidopsis pistils. Plant J. 68: 800–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband, W.S. (1997–2012). ImageJ. (Bethesda, MD: U.S. National Institutes of Health), http://imagej.nih.gov/ij/.
- Rholam M., Fahy C. (2009). Processing of peptide and hormone precursors at the dibasic cleavage sites. Cell. Mol. Life Sci. 66: 2075–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M.G. (2004). Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 84: 699–730. [DOI] [PubMed] [Google Scholar]
- Srivastava R., Liu J.X., Guo H., Yin Y., Howell S.H. (2009). Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 59: 930–939. [DOI] [PubMed] [Google Scholar]
- Srivastava R., Liu J.X., Howell S.H. (2008). Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 56: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Ezcurra I., Muschietti J., McCormick S. (2002). A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell 14: 2277–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kelley D., Ezcurra I., Cotter R., McCormick S. (2004). LeSTIG1, an extracellular binding partner for the pollen receptor kinases LePRK1 and LePRK2, promotes pollen tube growth in vitro. Plant J. 39: 343–353. [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D., Klein T.M., Fromm M.E., McCormick S. (1989). Transient expression of chimeric genes delivered into pollen by microprojectile bombardment. Plant Physiol. 91: 1270–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D., Yamaguchi J., McCormick S. (1990). Pollen-specific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109: 705–713. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Leevers S.J., Ahmadi K., Timms J., Katso R., Driscoll P.C., Woscholski R., Parker P.J., Waterfield M.D. (2001). Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70: 535–602. [DOI] [PubMed] [Google Scholar]
- Verhoeven T., Feron R., Wolters-Arts M., Edqvist J., Gerats T., Derksen J., Mariani C. (2005). STIG1 controls exudate secretion in the pistil of petunia and tobacco. Plant Physiol. 138: 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer J.E., van Leeuwen W., Tobeña-Santamaria R., Laxalt A.M., Jones D.R., Divecha N., Gadella T.W., Jr, Munnik T. (2006). Visualization of PtdIns3P dynamics in living plant cells. Plant J. 47: 687–700. [DOI] [PubMed] [Google Scholar]
- Wang H., Jiang L. (2011). Transient expression and analysis of fluorescent reporter proteins in plant pollen tubes. Nat. Protoc. 6: 419–426. [DOI] [PubMed] [Google Scholar]
- Wengier D., Valsecchi I., Cabanas M.L., Tang W.H., McCormick S., Muschietti J. (2003). The receptor kinases LePRK1 and LePRK2 associate in pollen and when expressed in yeast, but dissociate in the presence of style extract. Proc. Natl. Acad. Sci. USA 100: 6860–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengier D.L., Mazzella M.A., Salem T.M., McCormick S., Muschietti J.P. (2010). STIL, a peculiar molecule from styles, specifically dephosphorylates the pollen receptor kinase LePRK2 and stimulates pollen tube growth in vitro. BMC Plant Biol. 10: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590. [DOI] [PubMed] [Google Scholar]
- Williams J.H. (2008). Novelties of the flowering plant pollen tube underlie diversification of a key life history stage. Proc. Natl. Acad. Sci. USA 105: 11259–11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters-Arts M., Lush W.M., Mariani C. (1998). Lipids are required for directional pollen-tube growth. Nature 392: 818–821. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M., Brosché M., Kollist H., Kangasjärvi J. (2009). Arabidopsis GRI is involved in the regulation of cell death induced by extracellular ROS. Proc. Natl. Acad. Sci. USA 106: 5412–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Wengier D., Shuai B., Gui C.P., Muschietti J., McCormick S., Tang W.H. (2008). The pollen receptor kinase LePRK2 mediates growth-promoting signals and positively regulates pollen germination and tube growth. Plant Physiol. 148: 1368–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., McCormick S. (2007). A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 18830–18835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., He J., Lee D., McCormick S. (2010). Interdependence of endomembrane trafficking and actin dynamics during polarized growth of Arabidopsis pollen tubes. Plant Physiol. 152: 2200–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li S., Zhou L.Z., Fox E., Pao J., Sun W., Zhou C., McCormick S. (2011). Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J. 68: 1081–1092. [DOI] [PubMed] [Google Scholar]
- Zou Y., Aggarwal M., Zheng W.G., Wu H.M., Cheung A.Y. (2011). Receptor-like kinases as surface regulators for RAC/ROP-mediated pollen tube growth and interaction with the pistil. AoB Plants 2011: plr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.