A signaling process derived from carotenoids in ζ-carotene desaturase mutants regulates the expression of a variety of chloroplast- and nucleus-encoded genes and dramatically affects early leaf development in Arabidopsis. The signaling molecule involved in this process is an apocarotenoid whose synthesis requires the activity of the carotenoid cleavage dioxygenase CCD4.
Abstract
In addition to acting as photoprotective compounds, carotenoids also serve as precursors in the biosynthesis of several phytohormones and proposed regulatory signals. Here, we report a signaling process derived from carotenoids that regulates early chloroplast and leaf development. Biosynthesis of the signal depends on ζ-carotene desaturase activity encoded by the ζ-CAROTENE DESATURASE (ZDS)/CHLOROPLAST BIOGENESIS5 (CLB5) gene in Arabidopsis thaliana. Unlike other carotenoid-deficient plants, zds/clb5 mutant alleles display profound alterations in leaf morphology and cellular differentiation as well as altered expression of many plastid- and nucleus-encoded genes. The leaf developmental phenotypes and gene expression alterations of zds/clb5/spc1/pde181 plants are rescued by inhibitors or mutations of phytoene desaturase, demonstrating that phytofluene and/or ζ-carotene are substrates for an unidentified signaling molecule. Our work further demonstrates that this signal is an apocarotenoid whose synthesis requires the activity of the carotenoid cleavage dioxygenase CCD4.
INTRODUCTION
Chloroplasts are essential for plant survival, and they differentiate from a proplastid in response to environmental and developmental cues. Chloroplast differentiation requires the participation of many proteins with diverse structural, metabolic, and regulatory functions. Most of these proteins are nucleus encoded and are imported into the developing organelle (Barkan and Goldschmidt-Clermont, 2000). In this form, the nucleus regulates essential aspects of chloroplast development (anterograde regulation).
Multiple lines of evidence have demonstrated that the developing chloroplast also signals to the nucleus its metabolic and developmental status (retrograde regulation). Retrograde feedback mechanisms coordinate gene expression in both compartments to ensure that appropriate levels of protein complexes are present during chloroplast differentiation and function. Multiple signaling pathways are recognized as participating in retrograde communication. Some of these pathways participate during chloroplast development, while others are involved in the operational control of the organelle (Pogson et al., 2008). All of these pathways modulate the expression of a differential set of nuclear genes, many involved in organelle functionality. The crosstalk of these pathways is essential for dynamic acclimation of the plant to fluctuating developmental and environmental conditions (Pogson et al., 2008).
Genetic screens using inhibitors such as norflurazon or lincomycin that repress carotenoid biosynthesis or plastid translation, respectively, have been invaluable toward identifying mutants in signaling pathways impaired in retrograde regulation (Susek et al., 1993; Sullivan and Gray, 1999; Gray et al., 2003; Nott et al., 2006). The characterization of these mutants, and more recently the use of genomic approaches, have led to the identification of the genetic components of different retrograde signaling pathways (Kleine et al., 2007; Koussevitzky et al., 2007; Sun et al., 2011; Kindgren et al., 2012).
Although retrograde regulation has been a subject of study for over 30 years, clarity is still needed with respect to the identity of the signals that initiate these signaling pathways. The characterization of retrograde mutants provided evidence that the precursor of chlorophyll Mg-protoporphyrin IX acts as a signal for the tetrapyrrole pathway (Mochizuki et al., 2001; Strand et al., 2003), but such function is still being debated (Mochizuki et al., 2008). Other molecules, such as heme, 1O2, and H2O2, have also been suggested as possible signals for different retrograde pathways (Kim et al., 2009; Woodson et al., 2011), but conclusive demonstration of these functions is still lacking. Experimental evidence of true retrograde signals exists for 3′-phosphoadenosine 5′-phosphate, an abiotic stress–induced molecule that was shown to move from the plastid to the nucleus in response to abiotic stresses and for which a nuclear target was identified (Estavillo et al., 2011). Two plastid-derived isoprenoid derivatives, methylerythritol cyclodiphosphate and β-cyclocitral, have also been shown to induce retrograde signaling, but their receptor(s) or sites of action in the nucleus are unknown (Ramel et al., 2012; Xiao et al., 2012). Methylerythritol cyclodiphosphate is an intermediate of the methylerythritol phosphate (MEP) pathway that accumulates under abiotic stress (Xiao et al., 2012). β-Cyclocitral is a volatile apocarotenoid derived from β-carotene that accumulates in response to 1O2 and light stresses and regulates nuclear gene expression (Ramel et al., 2012, 2013). These findings indicate that the nature of retrograde signaling is largely undefined territory.
Many apocarotenoids are generated by the cleavage action of an ancient family of oxidative enzymes that belong to the carotenoid cleavage deoxygenases (CCD) (Walter et al., 2010). In most plant species, the CCD family comprises multiple members, nine in the case of Arabidopsis thaliana. Although some of these enzymes have the capacity to cleave different carotenoids, substrate specificity has been found for several of these enzymes (Auldridge et al., 2006b). The 9-cis-epoxy-carotenoid dioxygenase (NCED) subfamily has been shown to cleave 9-cis-isomers of epoxycarotenoids to yield the abscisic acid (ABA) precursor (Tan et al., 2003). By contrast, CCD7 and CCD8 are involved in the production of strigolactone (SL) (Ruyter-Spira et al., 2013). The substrate specificities of some other members, such as Arabidopsis CCD1 and CCD4, are less clear.
In this work, we show that loss of function of the ζ-carotene desaturase (ZDS; encoded by ZDS/CLB5/SPC1/PDE181) arrests chloroplast biogenesis at a very early stage of development. It is well known that various compounds derived from carotenoids have signaling functions that modulate developmental and stress responses in many organisms (Walter et al., 2010; Cazzonelli, 2011). Plants produce different carotenoids, all derived from a common 40-carbon phytoene precursor synthesized by the action of phytoene synthase (PSY). A series of desaturation and isomerization reactions catalyzed by two evolutionarily related desaturases (phytoene desaturase [PDS] and ZDS) and two isomerases (15-cis-ζ-carotene isomerase and carotenoid isomerase [CRTISO]) convert phytoene to all-trans-lycopene (Cazzonelli and Pogson, 2010).
ZDS catalyzes the second set of desaturation reactions subsequent to those catalyzed by PDS. While mutations in PSY, PDS, and ZDS result in bleached seedlings, here we show that loss-of-function alleles of ZDS also exhibit dramatic alterations in leaf morphology and in the expression of a variety of chloroplast proteins and genes required for leaf development. These phenotypes were not observed in other carotenoid-deficient mutants, including mutants deficient in the closely related desaturase, PDS. We present biochemical and genetic data to demonstrate that molecular and morphological changes are caused by the accumulation of specific linear carotenes in zds mutants and that the process responsible for these phenotypes requires CCD4 activity. We propose that a cis-carotene signal regulates the expression of a variety of chloroplast- and nucleus-encoded genes and dramatically affects early leaf development.
RESULTS
The CLB5 Mutation Disrupts ZDS Activity
To identify genes that mediate or regulate chloroplast development, several mutants that affect chloroplast biogenesis were selected (Gutiérrez-Nava et al., 2004). chloroplast biogenesis5 (clb5) was of particular interest, as it arrests chloroplast development at a very early stage of proplastid-to-chloroplast differentiation. Three independent clb5 alleles were isolated, and in all of them the albino phenotype segregated as a single recessive locus. clb5-1 was derived from an ethyl methanesulfonate collection, and clb5-2 was produced by fast neutron mutagenesis. clb5-3 was selected from a T-DNA mutant collection, but the mutation responsible for the albino phenotype did not segregate with the T-DNA. Cosegregation analysis of PCR-based molecular markers showed that the clb5 mutation was linked to the upper arm of chromosome 3 (Gutiérrez-Nava et al., 2004).
Fine mapping using a final population of 1610 chromosomes located the CLB5 gene between the simple sequence length polymorphism (SSLP) markers F7018(C) and T19J14(11), which border 13 annotated loci (Figure 1A). One of the loci represents the ZDS gene (also referred as PIGMENT DEFECTIVE EMBRYO181 [PDE181] and SPONTANEOUS CELL DEATH1 [SPC1]), involved in carotenoid biosynthesis (Figure 1B), and mutant alleles segregate for pigment-defective plants (McElver et al., 2001; Dong et al., 2007). Progeny from crosses between clb5-1 and spc1-2 (SALK_033039/spc1-2) heterozygous mutants produced ∼25% albino seedlings, demonstrating that these mutants were allelic.
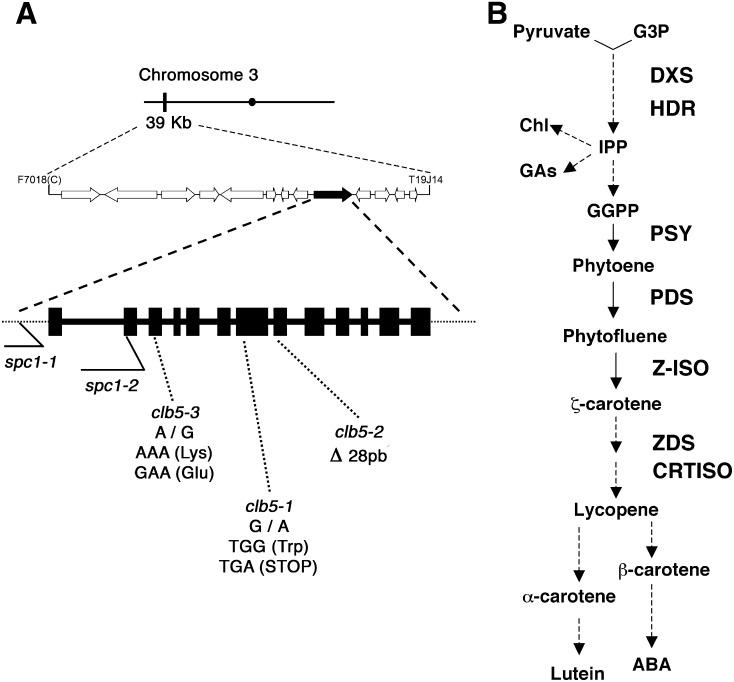
Figure 1.
Mapping of the clb5-1 Mutation.
(A) Representation of the chromosome (black bar) and ZDS gene location (black arrow). The structure of the ZDS gene is shown with 13 exons (black boxes) and introns (line). The locations of the mutations for each clb5 and the previously reported spc1 alleles are indicated.
(B) Simplified diagram of the carotenoid biosynthetic pathway. Chl, chlorophyll; GAs, gibberellins; GGPP, geranylgeranyl diphosphate; G3P, glyceraldehyde-3-phosphate; HDR, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase; IPP, isopentenyl pyrophosphate; Z-ISO, 15-cis-ζ-carotene isomerase.
Confirmation that all of the clb5 phenotypes are attributable to a defect in ZDS gene expression was obtained from molecular complementation. A full-length cDNA of the wild-type ZDS gene regulated by the cauliflower mosaic virus (CaMV) 35S promoter was introduced into clb5-1 heterozygous plants. The clb5-1 mutation results in the loss of a BamHI restriction site present in the wild-type allele and was used as a polymorphic marker to genotype the CaMV35S:ZDS T2 transgenic green progeny (Supplemental Figure 1). Plants homozygous for the clb5-1 mutation (Figure 2B) displayed a phenotype indistinguishable from wild-type plants (Figure 2A) in the presence of the transgene (Figure 2C), which demonstrates that a mutation in the ZDS locus was responsible for the arrested chloroplast phenotype.
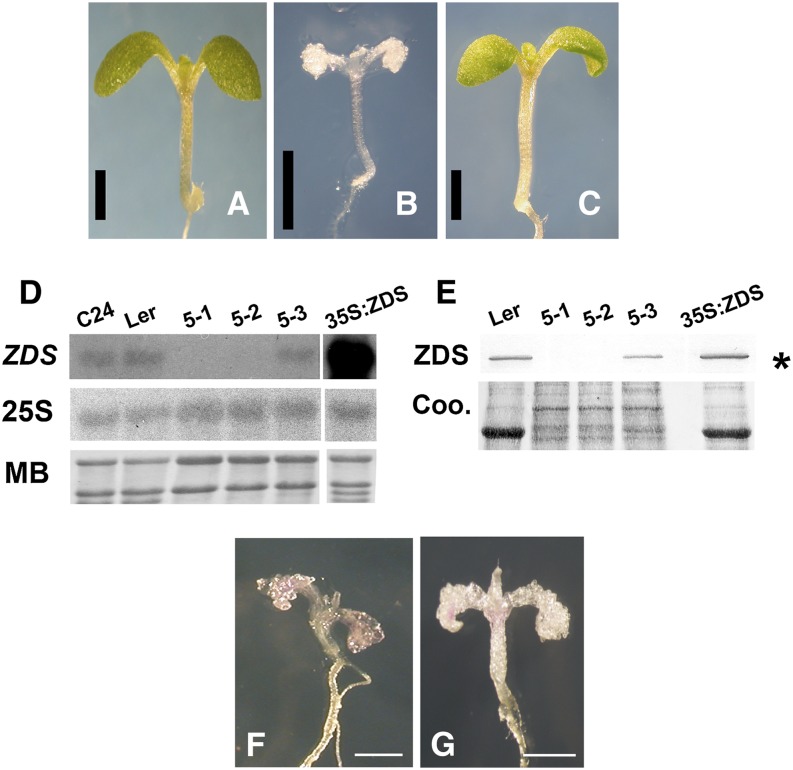
Figure 2.
Molecular Complementation of clb5 and Characteristics of the clb5 Alleles.
(A) to (C) Phenotypes of a representative 8-d-old wild-type Ler seedling (A), a 16-d-old clb5-1 mutant (B), and an 8-d-old T2 transgenic clb5-1 homozygous complemented line (C). Bars = 1 mm.
(D) RNA gel blot analysis of ZDS transcript from 8-d-old wild-type (C24 and Ler ecotypes) and complemented mutant (35S:ZDS) plants and from 15-d-old clb5 seedlings. Fifteen micrograms of total RNA was loaded in each lane. Hybridization of the same membrane with the 25S rRNA and the methylene blue (MB)–stained gels are shown as loading controls. The RNA gel blots are representative of two independent biological experiments.
(E) Expression of the ZDS protein in the clb5 alleles. ZDS protein was immunodetected in protein extracts from 8-d-old wild-type (Ler) and complemented transgenic (35S:ZDS) plants or from 15-d-old clb5 alleles. Ten micrograms of Ler, 5 μg of 35S:ZDS, and 15 μg of the clb5 mutant total protein extracts were run in each lane. A Coomassie blue (Coo)–stained gel run in parallel is shown as a loading control. The asterisk marks the ZDS protein. The protein blot is representative of three independent biological experiments.
(F) and (G) Phenotypes of 10-d-old clb5-1 (F) and clb5-2 (G) alleles. Bars = 0.5 mm.
[See online article for color version of this figure.]
Next, we sequenced the ZDS gene locus and identified the precise mutation for each of the clb5 alleles. clb5-1 has a G-to-A transition at nucleotide 693 that generates a stop codon and results in a truncated protein of 230 amino acids. In the clb5-2 allele, a 28-bp deletion at the beginning of the eighth exon removes the acceptor sequence of the intron/exon splicing boundary. Finally, in the clb5-3 mutant, we found an A-to-G transition that results in a Lys-to-Glu amino acid substitution (Figure 1A). This Lys residue is highly conserved in the ZDS proteins of monocots, dicots, and other photosynthetic organisms (Supplemental Figure 2). This residue lies within the proposed NADP binding region (Linden et al., 1994).
ZDS transcript was not detected in the clb5-1 and clb5-2 mutant alleles (Figure 2D), nor did a polyclonal antibody to Arabidopsis ZDS detect any protein (Figure 2E). From these data, we conclude that both clb5-1 and clb5-2 are null ZDS mutant alleles. By contrast, lower relative levels of RNA (Figure 2D) and protein (Figure 2E) were reproducibly detected in independent biological experiments in clb5-3 plants, consistent with its point mutation. The overexpression of ZDS in the clb5-1 mutant allele showed higher levels of ZDS mRNA and protein when compared with wild-type Landsberg erecta (Ler) (Figures 2D and 2E), which supports our complementation studies. The albino phenotype displayed by the two clb5 null alleles (Figures 2F and 2G; Supplemental Figure 3) was similar to that reported for the spc1-2 mutant (Dong et al., 2007), although an unreported and strikingly obvious needle-like leaf developmental phenotype was also apparent in the spc1-2 mutant. In contrast with the report of Dong et al. (2007), the zds/clb5 mutant alleles are able to continue growing when maintained in in vitro conditions, albeit at a much slower rate than wild-type plants (Supplemental Figure 4). The abnormalities in the leaf morphology of the zds/clb5 null alleles are not exclusive to the first leaves and are maintained in the subsequent leaves produced through the plant’s development (Supplemental Figure 4). Therefore, our data clearly confirm that lesions in the ZDS gene are responsible for the albino phenotype and for the developmental abnormalities in the leaves of the zds/clb5 alleles.
The chloroplast subcellular localization of ZDS was corroborated with a fusion containing the 120 amino acids of the N-terminal region of ZDS and the green fluorescent protein reporter (ZDS:GFP) and transiently expressed in Arabidopsis protoplasts (Supplemental Figure 4). The ZDS:GFP fusion is detected as a needle-like fluorescence within the chloroplasts (Supplemental Figures 5A and 5B). These fibrillar structures resemble those reported for plastoglobuli, which are lipoprotein organellar particles with a dynamic morphology that change in response to environmental conditions, and where the ZDS protein has been detected (Bréhélin et al., 2007; Shumskaya et al., 2012). However, future studies using the complete protein are required to confirm this possibility. Accordingly, protein quantification by antibody detection showed higher levels of the ZDS protein in photosynthetic tissues, which is consistent with the function of this protein (Supplemental Figure 6). Furthermore, the protein expression pattern observed is also consistent with the mRNA transcript expression profile reported previously (Dong et al., 2007).
Loss of ZDS Alters Leaf and Chloroplast Development
We were surprised by the severity of leaf developmental defects observed in the two zds/clb5 null alleles (Figures 2F and 2G; Supplemental Figure 3). To address this systematically, we conducted a more extensive analysis of the clb5 morphology in comparison with other albino mutants with defects in carotenoid biosynthesis (Figure 3A). Two of these mutants, cloroplastos alterados (cla1-1) and clb6-1, affect the MEP pathway, which provides the precursors for carotenoids (Figure 3A). The psy1 and pds3-1 mutants disrupt the expression of the PSY and PDS proteins required in the biosynthesis of phytoene and ζ-carotene, respectively, the latter being the direct substrate of ZDS (Figure 3A). clb5-1 mutant seedlings developed small, bumpy cotyledons that curve downward. The first leaves of clb5 are radial, needle-like projections and lack differentiation between petiole and blade (Figure 3B). A similar phenotype was observed in the clb5-2 null mutant allele (Supplemental Figure 3). By contrast, the cla1, clb6, psy1, and pds3 albino mutants all display more normal leaf morphology with a clearer distinction between the petiole and leaf blade. Consistent with our previous results (Gutiérrez-Nava et al., 2004), clb5-1 displays the most severe seedling morphology of the mutants evaluated (Figure 3B).
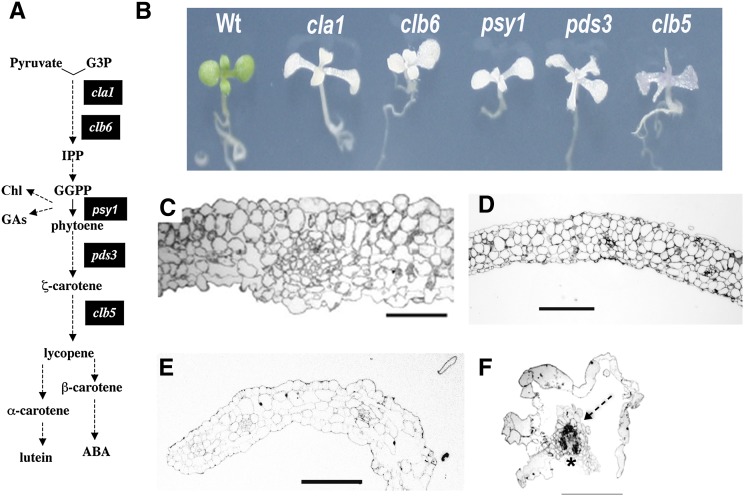
Figure 3.
Morphological Analyses of the clb5 Mutant.
(A) Simplified diagram of the carotenoid pathway as described in Figure 1. The black boxes indicate the steps of the pathway altered in mutants used in this study.
(B) Morphology of albino mutants that affect carotenoid biosynthesis. Plants were grown in a 16/8-h light/dark period for 8 d for the wild type and 15 d for the mutants.
(C) to (F) Light micrographs of transverse sections of wild-type (C), clb6 (D), pds3 (E), and clb5-1 (F) seedling leaves. In (F), vascular tissue is marked with an asterisk and mesophyll tissue is marked with a dashed arrow. Plants were grown on MS medium for 21 d, and the second leaf of a representative seedling for each phenotype was fixed for light microscopic analysis. Bars = 100 μm.
In general, the leaves of the zds/clb5 null alleles have a translucent appearance in comparison with other albino mutants (Figure 3B). This phenotype appears to be associated with an extensive vitrification of the epidermal cells, which look water-soaked and swollen, resulting in an irregular surface (Supplemental Figure 7A). Although preliminary characterization of this mutant led us to conclude that the leaves in clb5 lacked trichomes (Gutiérrez-Nava et al., 2004), further analysis of a higher number of plants demonstrated that occasional trichomes develop in the leaves of the clb5-1 mutant, albeit in a much lower number than other albino mutants or the wild-type plants (Supplemental Figure 7B). In spite of the uneven leaf surface, differentiated stomata can be found in these leaves, although in a lower number than in wild-type leaves (Supplemental Figure 7C). There are also profound alterations in the internal anatomy of clb5 leaves. Transverse sections prepared at the midpoint of the first leaf blade showed that the clb5-1 leaves (Figure 3F) consist of an inner cylinder of vascular tissue, surrounded in most places by a single layer of epidermal cells, compared with the more typical structure seen in the wild type, clb6, and pds3 (compare Figures 3C and 3E). clb5-1 lacks most of the mesophyll tissue and instead has empty spaces between the epidermis and the vascular tissue and completely lacks palisade mesophyll.
To analyze in more detail the defects of zds/clb5 associated with leaf development, next we analyzed the expression of several markers associated with defining leaf development. Each marker gene, fused to β-glucuronidase (GUS), was introgressed into the clb5-1 background, and reporter gene expression was analyzed in homozygous clb5-1 seedlings. Reporter gene fusions of PHABULOSA (gPHB:GUS) and FILAMENTOUS FLOWER (pFIL:GUS) were used to determine the polarity specification (Donnelly et al., 1999; Sabatini et al., 1999; Nelissen et al., 2003; Gillmor et al., 2010). Relative to the wild type (Figure 4C), the mutation in clb5-1 did not affect the expression pattern of the pFIL:GUS reporters in cotyledons (Figure 4D). However, the normal adaxial expression of gPHB:GUS in the first true leaves (Figure 4A) was not detected in the clb5-1 mutant, where expression is restricted to the vascular tissue (Figure 4B). In the case of the abaxially expressed pFIL:GUS marker (Figure 4D), no expression was observed in clb5-1 mutant leaves, in contrast with the pattern observed in the wild-type plants (Figure 4C). Consequently, although polarity in cotyledons appears normal, the clb5-1 primary leaves display disregulated PHB and FIL expression, indicating that leaf polarity is compromised.
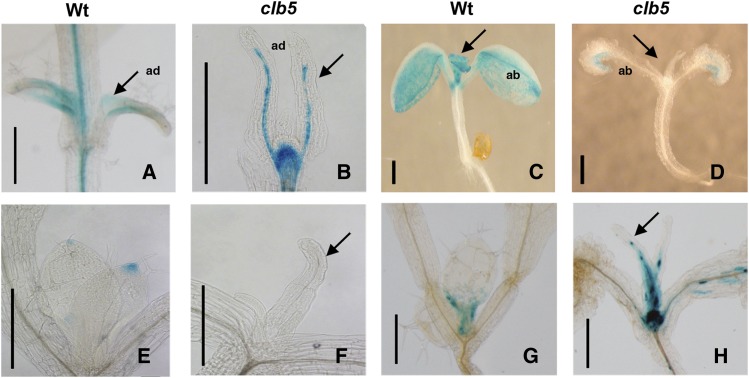
Figure 4.
Expression Pattern of Leaf Developmental Markers in the clb5-1 Mutant.
GUS expression pattern is shown for the gPHB-GUS adaxial marker in 7-d-old wild-type (A) and 10-d-old clb5-1 (B) plants, the pFIL:GUS ventral marker in 7-d-old wild-type (C) and 10-d-old clb5-1 (D) plants, the DR5:GUS synthetic auxin reporter in wild-type (E) and 14-d-old clb5-1 (F) plants, and pCYCB1:D-box-GUS in 6-d-old wild-type (G) and 14-d-old clb5-1 (H) plants. Arrows indicate primary leaves. ab, abaxial; ad, adaxial. Bars = 0.5 mm.
Auxin is also a central factor that contributes to final leaf shape. Auxin accumulation and/or responsiveness in the clb5-1 leaf were analyzed through the expression of the auxin-inducible synthetic DR5:GUS reporter, which normally shows maximal expression at the edges of a developing leaf (Scarpella et al., 2010). Relative to wild-type leaf primordia, where GUS activity was detected in the tip of the leaf (Figure 4E), the clb5-1 mutants displayed no detectable DR5:GUS expression in the leaf tip (Figure 4F).
Leaf morphology also depends on cell division and expansion. To estimate the transition between cell proliferation and cell expansion, we followed the expression of the pCYCB1:D-box-GUS construct (Donnelly et al., 1999). As reported previously, pCYCB1:D-box-GUS expression in 6-d-old wild-type leaves is mostly observed in the base of the leaf (Figure 4G). In the clb5-1 mutants, the expression of pCYCB1:D-box-GUS was detected throughout the entire developing leaf (Figure 4H). The misexpression of pCYCB1:D-box-GUS further confirms that the zds/clb5 mutation disrupts leaf blade morphogenesis.
Loss of ZDS Disrupts the Expression of Several Chloroplast- and Nucleus-Encoded Plastid Proteins
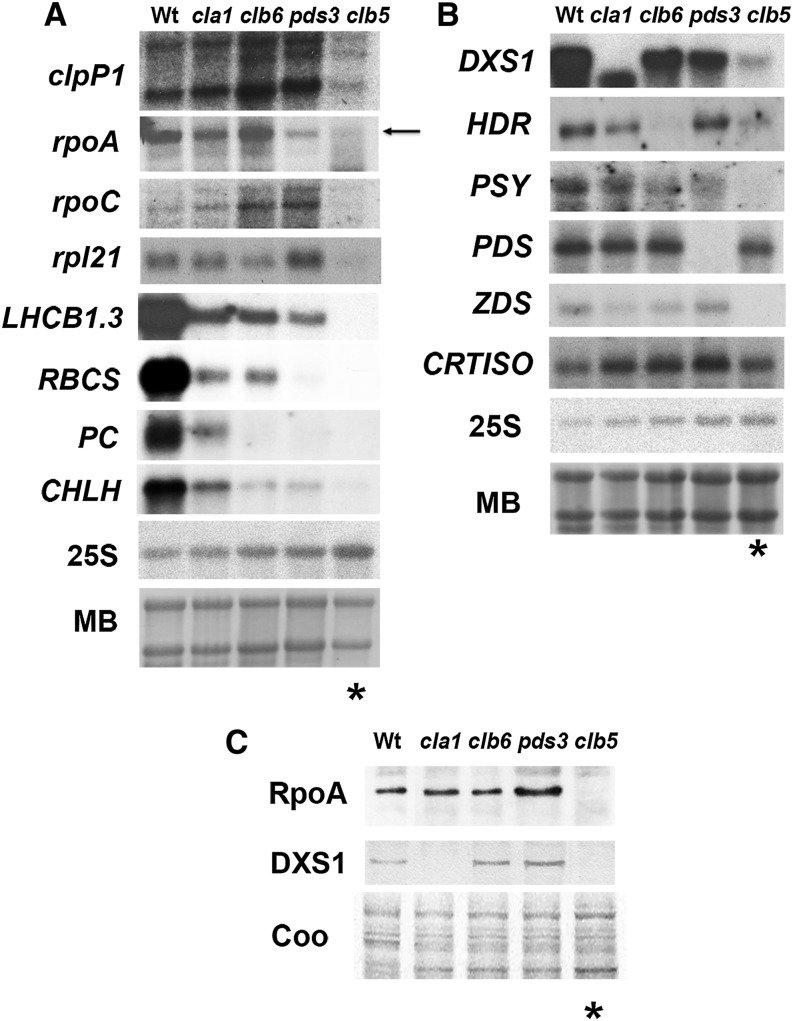
The accumulation of nucleus- and chloroplast-encoded transcripts synthesized at specific stages of chloroplast development was severely decreased in the clb5-1 mutant when compared with the wild type and other albino mutants (Figure 5). As shown in Figure 5A, the accumulation of transcripts expressed during early chloroplast development in the wild type, preceding the expression of photosynthetic genes, is barely detectable in the clb5-1 seedlings. This includes two subunits of the plastid-encoded RNA polymerase (rpoA and rpoC), the catalytic subunit of the ClpP protease (clpP1), and a nucleus-encoded ribosomal protein (rpl21) (Figure 5A). A similar scenario is observed with nucleus-encoded genes required for photosynthetic activity (Figure 5A), including light-harvesting LHCB1.3, the small subunit of Rubisco (RBCS), plastocyanin (PC), and subunit H of the Mg-chelatase complex (CHLH). These results are consistent with previous reports showing the downregulation of RBCS and LHC1 gene expression (Gutiérrez-Nava et al., 2004; Dong et al., 2007).
Figure 5.
Expression of Nucleus- and Chloroplast-Encoded Transcripts and Proteins.
(A) Expression of chloroplast marker genes: clpP1, rpoA, rpoC, rpl21, LHCB1.3, RBCS, PC, and CHLH.
(B) Nucleus-encoded genes involved in the carotenoid biosynthetic pathway: DXS1, HDR, PSY, PDS, ZDS, and CRTISO probes.
Each lane contains 15 μg of total RNA from 8-d-old wild-type and 15-d-old mutant seedlings. The hybridization of the 25S rRNA and methylene blue (MB)–stained gels are shown as loading controls. These data are representative of two independent biological experiments.
(C) Analysis of the protein levels of RpoA and nucleus-encoded DXS1. Total protein extracts were isolated from 8-d-old wild-type (Ler) and 15-d-old cla1-1, clb6, pds3, and clb5-1 mutant seedlings. Immunoblotting was performed with specific antibodies against the RpoA and DXS1 proteins. Each lane contains 15 μg of total protein extract. A Coomassie blue–stained gel run in parallel with the same samples is shown as a loading control (Coo).
Asterisks mark the clb5 mutant lanes. The protein blots shown are representative from three independent biological replicates.
We also analyzed the transcript accumulation of some nucleus-encoded genes directly involved in carotenoid biosynthesis. Transcript levels of genes required for the biosynthesis of isoprenoid precursors (DXS1 and HDR) and for the PSY enzyme, a limiting step in the biosynthetic carotenoid pathway (Figure 1B), are downregulated in the clb5-1 mutant (Figure 5B). The deregulation of gene expression in clb5-1 is not global, as the accumulation of PDS or CRTISO transcripts remains unaltered in clb5-1 in comparison with other albino mutants and wild-type plants.
Because of the substantial posttranscriptional regulation of proteins in chloroplasts, we also checked whether the RNA abundance defects observed in clb5-1 are also reflected at the protein level. We confirmed that the abundance of the α-subunit of the chloroplast-encoded RNA polymerase (RpoA) and the 1-deoxy-d-xylulose 5-phosphate synthase (DXS) enzymes is almost undetectable in clb5-1 compared with the wild type and other albino mutants (Figure 5C), which demonstrates that the changes observed at the transcription level are also reflected at the protein level.
The Accumulation of Specific Carotenes Is Responsible for the clb5 Mutant Phenotype and Affects Gene Expression
The hormones ABA and SL are two signals derived from carotenoids that affect plant development, including leaf morphology (Liu et al., 2012; Ramel et al., 2013). Thus, we next asked whether or not treatment with these hormones could restore the leaf phenotype of clb5. We found that the exogenous addition of these two hormones does not alleviate the finger-like leaf morphology typically observed in the clb5 mutant (Supplemental Figure 8). However, the presence of ABA (0.5 and 1 μM) severely retarded the development of wild-type and mutant seedlings compared with its absence, supporting the action of this hormone in this assay. Also, the presence of SL resulted in the production of higher numbers of leaves in the clb5-1 and pds3 albino mutants in a shorter time, supporting the effectiveness of the SL treatment.
Carotenoids also serve as essential photoprotectors, and carotenoid-deficient mutants are subject to extensive photooxidative stress (Triantaphylidès and Havaux, 2009). It has also been documented that reactive oxygen species (ROS) might act as a signal to modulate plastid-to-nucleus communications and development (Pogson et al., 2008; Bräutigam et al., 2009), and the clb5 phenotypes could be due to the accumulation of ROS. We tested this possibility by quantifying the levels of H2O2 and O2− in the clb5 mutant. We also analyzed the expression of the ROS-responsive gene ZAT12 (Davletova et al., 2005) and the photosystem II–associated protein EXECUTER1 (EX1 gene), which is an essential component of O2− signaling (Lee et al., 2007). We could not detect any major differences in the ROS levels or in the expression levels of the ZAT12 and EX1 genes in the clb5-1 mutant compared with other carotenoid-deficient mutants (Supplemental Figure 9). In summary, the clb5 finger-like leaf morphology does not appear to be a consequence of low ABA or SL levels, nor is it a result of the overaccumulation of ROS.
Other possibilities to explain the phenotypic alterations associated with clb5 are that ZDS directly regulates gene expression and development through an activity on a substrate other than carotenoids or that the absence of ZDS enzyme generates or inhibits the production of a specific carotenoid-derived signal. For example, in addition to hormones, an oxidative cleavage product of β-carotene acts as signaling molecule in response to oxidative stress, and cis-carotenes have been implicated as signals that mediate gene expression in Arabidopsis (Ramel et al., 2012). To distinguish between these possibilities, clb5-1 seedlings were grown in the presence or absence of fluridone. Fluridone inhibits PDS activity, which functions in the enzymatic step prior to ZDS (Figure 1B) (Bartels and Watson, 1978). We reasoned that if a signal is derived from phytofluene and/or ζ-carotenoids that accumulate in ZDS mutants, then the needle-like leaf morphology observed in clb5-1 mutants may be partially complemented in the presence of fluridone.
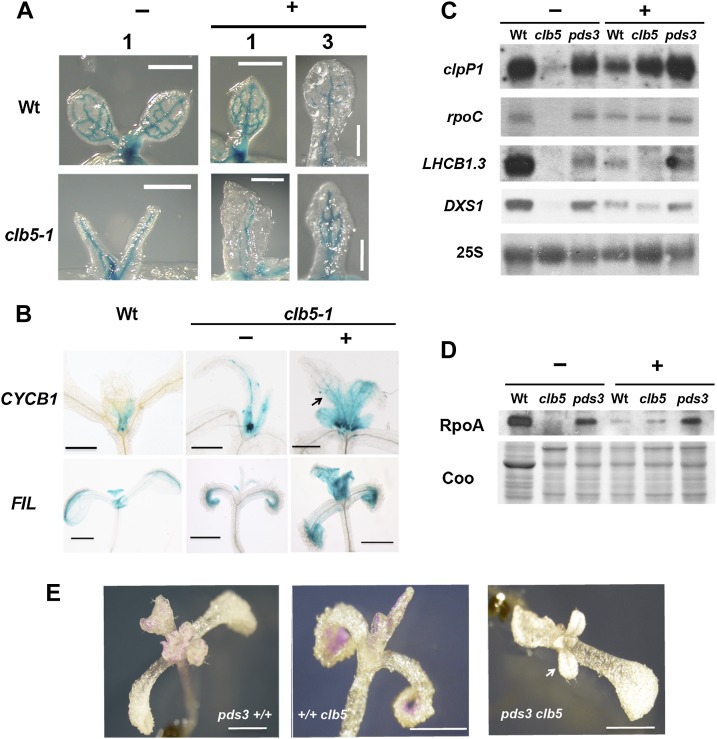
Homozygous clb5-1 and pds3 mutants and wild-type controls were transferred to medium with fluridone 4 d after germination and evaluated when fluridone impact was clearly detectable by bleaching in the wild-type plant leaves (15 d after germination; Supplemental Figure 10). The first true leaves of the clb5-1 mutant seedlings grown in fluridone displayed a more normal morphology, with a flatter lamina in comparison with the finger-type projections of those grown without the PDS inhibitor (Supplemental Figure 10). The pds3 mutant showed no noticeable impact upon leaf phenotype when grown under the same treatment (Supplemental Figure 10). To further evaluate the improvements in clb5 leaf morphology, we followed the expression of pREV:GUS (Emery et al., 2003), a vascular-specific marker introduced into the clb5-1 mutant background. Compared with the finger-type structures observed without fluridone, a more normal leaf shape and more complex vascular system were observed, especially in the third leaf after fluridone treatment (Figure 6A). In accordance with this improvement, the expression patterns of the leaf developmental markers CYCB1 and FIL (Donnelly et al., 1999; Nelissen et al., 2003) were also improved (Figure 6B). In the presence of fluridone, the pCYCB1,1:D-box-GUS marker in the treated clb5 plants was detected toward the base of the leaves and more closely resembled the pattern observed in wild-type plants (Figure 6B). Similarly, pFIL-GUS expression was detected in the first true leaves of wild-type and fluridone-treated clb5 plants, in contrast with the clb5 seedlings growing in the absence of fluridone (Figure 6B).
Figure 6.
Inhibition of PDS Improves Leaf Development and Gene Expression of the clb5-1 Mutant.
(A) Morphology of the first (1) or third (3) leaf in 8-d-old wild-type or 15-d-old clb5 seedlings treated with (+) or without (−) fluridone. The GUS expression pattern corresponds to the pREV:GUS vascular marker.
(B) GUS expression pattern of the pCYCB1:D-box-GUS and pFIL:GUS markers of 8-d-old wild-type and 15-d-old clb5-1 seedlings treated with (+) or without (−) fluridone.
(C) Transcript levels of clpP1, rpoC, LHCB1.3, and DXS1 genes. Each lane contains 5 μg of total RNA. The 25S rRNA hybridization is shown as a loading control.
(D) RpoA protein levels of Ler wild-type, clb5-1, and pds3 mutant seedlings treated with (+) or without (−) fluridone. Each lane contains 10 μg of total protein extract from the same developmental age as in (C). The blot was developed using RpoA antibody. A Coomassie blue–stained gel run in parallel with the same samples is shown as a loading control (Coo). The gels shown are representative from three independent biological replicates.
(E) Seedling morphology of 20-d-old single pds3 (pds3+/+) and clb5-1 (+/+clb5) mutants and double pds3 clb5 mutants.
Bars = 1 mm.
The partial suppression of the clb5 needle-like leaf phenotype by fluridone also correlates with a higher accumulation of the clpP1 and rpoC transcripts, both expressed during early chloroplast development. Transcriptional recovery was also observed for the nucleus-encoded DXS1, matching that of pds3, and to lesser extent for the LHCB1.3 and RBCS genes (Figure 6C), which are expressed during later stages of plastid development. The level of the RpoA protein was partially restored by fluridone treatment to levels similar to those detected in the fluridone-treated wild-type seedlings (Figure 6D). By contrast, no difference in the RpoA protein levels was observed in the pds3 seedlings in the absence or presence of fluridone (Figure 6D). Taken together, these data support the notion that it is not the ZDS protein itself but rather the accumulation of phytofluene and ζ-carotenoids or their derivatives that is responsible for the needle-like leaf morphology as well as the gene expression defects observed in the clb5 mutant.
To further support that the phenotypes observed in clb5 are caused by the accumulation of specific carotenoids and are not a secondary effect of the fluridone inhibitor used in the previous analyses, a double pds3 clb5 mutant was generated (Supplemental Figure 11). In agreement with our previous results, the leaf phenotype displayed by pds3 clb5 double mutants appeared similar to the pds3 allele, showing a flat lamina and the presence of trichomes, which was very different from the finger-type projections observed in clb5 (Figure 6E). These data conclusively demonstrate that the leaf defects observed in the clb5 mutant result from the accumulation of phytofluene and ζ-carotenoids as a consequence of the absence of ZDS.
Deficiency in ZDS activity results in the accumulation of higher levels of cis-carotenoids (Dong et al., 2007). We used HPLC to confirm that ζ-carotene was the predominant carotenoid in clb5-1. In wild-type plants grown under standard light conditions, the pigment profile consists of β-carotene plus the three oxygenated xanthophylls, lutein, violaxanthin, and neoxanthin (Supplemental Figure 12). In the pds3 mutant, no significant levels of chlorophyll or carotenoid pigments are detected, with the exception of phytoene (assayed spectrophotometrically at 286 nm) (Supplemental Figure 12), as reported previously (Qin et al., 2007). In contrast, the most abundant carotenoid in clb5-1 was ζ-carotene, with trace amounts of phytofluene and cis-isomers of ζ-carotene also detected (Supplemental Figure 12). This is similar to previously published spectral data for ZDS mutants (Matthews et al., 2003; Conti et al., 2004; Dong et al., 2007).
CCD4 Is Required to Promote clb5 Mutant Leaf and Expression Phenotypes
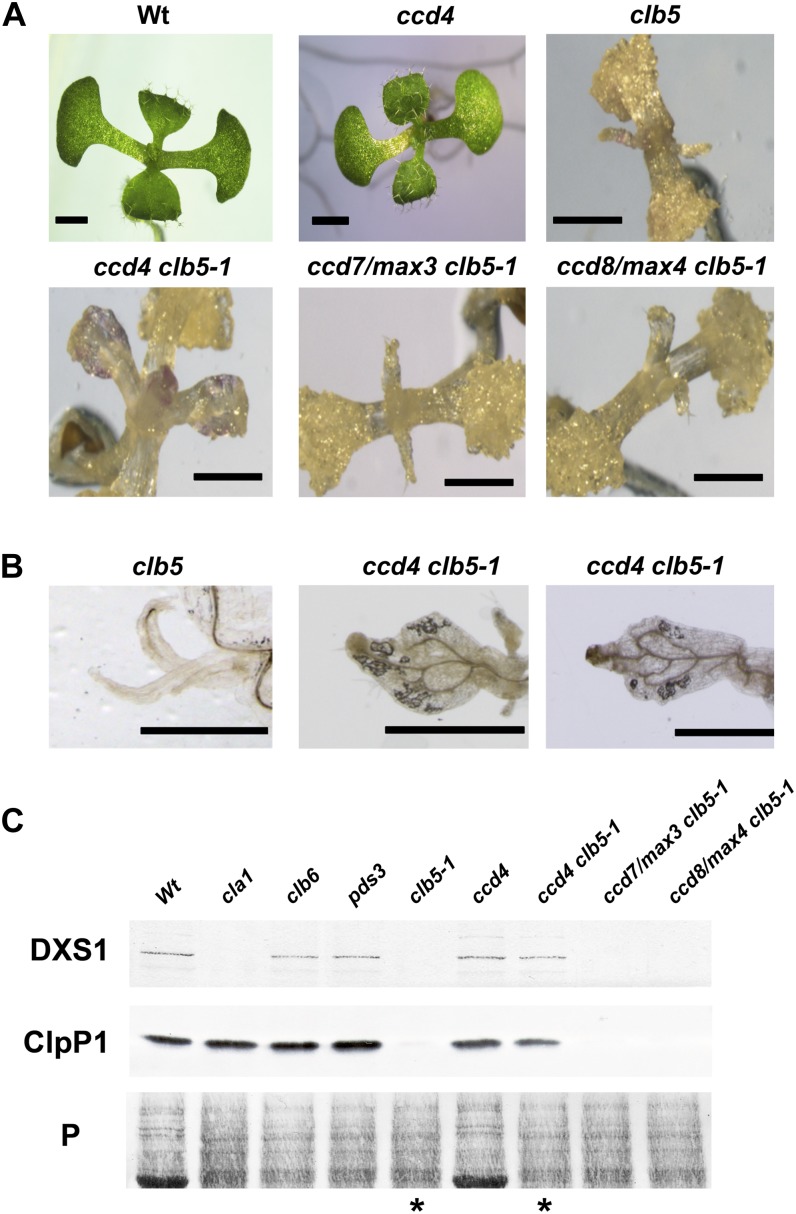
It is known that carotenoids are substrates of enzymatic cleavage, producing different apocarotenoids with diverse signaling functions, such as hormones and volatiles that influence plant development (Walter et al., 2010; Van Norman et al., 2014). Many apocarotenoids are produced by the action of the family of CCD enzymes that catabolize particular carotenoid substrates (Auldridge et al., 2006b; Ramel et al., 2012). However, undertaking reverse-phase HPLC will not detect modified cleavage products such as ABA or SL. Thus, to explore the involvement of CCDs in the phenotype of the clb5 mutant, we generated double mutants between clb5-1 and the three CCD genes (ccd4, ccd7/max3, and ccd8/max4) that encode plastid-localized enzymes. Interestingly, the leaf morphology of the segregating albino ccd4 clb5 double mutant appeared more normal in development when compared with the clb5-1 plant (Figure 7A). In contrast with the finger-type projections characteristic of the clb5-1 leaf, the ccd4 clb5-1 leaf has a closer appearance to the wild type, with a flatter lamina and better vascular system development (Figure 7B). The improvement in leaf morphology was not observed in either the ccd7/max3 clb5-1 or the ccd8/max4 clb5-1 plant, demonstrating that this effect is unique to CCD4.
Figure 7.
Role of CCDs in the Morphological and Expression Defects of the clb5-1 Mutant.
(A) Morphology of seedlings from 8-d-old wild-type and ccd4 mutant seedlings or 16-d-old clb5-1, ccd4 clb5-1, ccd7/max3 clb5-1, and ccd8/max4 clb5-1 mutant seedlings. Bars = 1 mm.
(B) Close-ups of the primary leaf of 16-d-old clb5 and two representative ccd4 clb5-1 leaves. Bars = 1 mm.
(C) Analysis of the protein levels of the nucleus-encoded DXS1 and the chloroplast-encoded ClpP1. Total protein extracts were isolated from 8-d-old wild-type (Ler) and ccd4 mutant seedlings or from 15-d-old cla1-1, clb6, pds3, clb5-1, ccd4 clb5-1, ccd7/max3 clb5-1, and ccd8/max4 clb5-1 mutant seedlings. Immunoblotting was performed with polyclonal antibodies against the DXS1 and ClpP proteins as described in Methods. Each lane contains 15 μg of total protein extract. The Ponceau-stained membrane is shown as a loading control (P). Asterisks mark the lanes corresponding to clb5-1 and ccd4 clb5-1 mutants. The protein gel shown is representative from three independent biological replicates.
[See online article for color version of this figure.]
To further investigate a role for ccd4 in complementing clb5, the expression levels of the DXS1 and ClpP1 proteins were analyzed in these double mutants. As observed previously (Figure 5B), the accumulation of DXS1 was barely detected in the clb5-1 mutant, in contrast with the wild type and other albino mutants (clb6 and pds3). By contrast, DXS1 is detectable in the protein extracts from the albino ccd4 clb5 double mutant (Figure 7C). Similarly, the expression of the ClpP1 subunit, which is barely detectable in the clb5 mutant, is now expressed in the ccd4 clb5 mutant, at levels similar to the other albino mutants and wild-type plants (Figure 7C). In agreement with the leaf phenotype, no detectable DXS1 or ClpP1 was found in either ccd7/max3 clb5-1 or ccd8/max4 clb5-1 seedlings (Figure 7C). Taken together, these results confirm a key role of the CCD4 enzyme in the generation of an apocarotenoid responsible for the morphological and expression defects observed in the clb5-1 mutant.
DISCUSSION
Carotenoids are accessory pigments and photoprotectors, limiting photooxidative damage during photosynthesis. These compounds are also precursors of ABA and SL and volatiles with signaling functions (Walter et al., 2010; Liu et al., 2012; Ramel et al., 2013). Impaired carotenoid biosynthesis can result in pleiotropic phenotypes due to photooxidative damage and hormone deficiencies (Matthews et al., 2003; Conti et al., 2004; Dong et al., 2007).
Several lines of evidence indicate that, in addition to these known defects caused by carotenoid deficiency and oxidative stress, deficiency in ZDS activity results in the accumulation of a molecule that impairs normal leaf development and the expression of a variety of nucleus- and chloroplast-encoded genes. First, although ZDS affects the third step in carotenoid biosynthesis, chloroplast development in clb5 is arrested at earlier stages when compared with mutants in the MEP pathway, which provides the carotenoid precursors, or in psy and pds mutants, which affect the initial steps of carotenogenesis. Second, carotenoid deficiency per se cannot account for the phenotypes. PDS is the first desaturase of carotenoid biosynthesis, and it carries out a very similar reaction to ZDS (Cazzonelli and Pogson, 2010). Pigment analysis of the pds3 mutant showed undetectable levels of all carotenoids except phytoene. However, pds3 mutants do not display the finger-like leaf morphology observed in clb5. Also, the expression of genes and proteins, including those characteristic of early chloroplast development (rpoA and rpoC), is mildly affected in pds3 compared with clb5. Third, neither ABA nor SL can alleviate the leaf defects of the clb5-1 mutant. Fourth, PDS inhibitors such as fluridone or the mutation of the PDS gene largely suppress clb5-specific leaf phenotypes and gene expression defects. The evidence is strong in linking a carotenoid or carotenoid derivative, and not a new activity for ZDS, in conferring clb5 phenotypes. These findings led us to conclude that a molecule generated from the accumulated phytofluene or ζ-carotene isomers is responsible for altering the expression of a number of chloroplast-related genes and for disrupting leaf development.
Pigment analysis showed that clb5 seedlings accumulate phytofluene and ζ-carotene isomers. However, given the low levels of the phytofluene and ζ-carotenoid species that accumulate in the clb5 mutant, the lack of appropriate standards, and the observation that most hormones and signals are produced at very low levels, it would be a substantial challenge to identify the signal responsible for the clb5 phenotypes by HPLC fractionation. Indeed, the structure of SL is very different from that of the apocarotenoid it is derived from (Alder et al., 2012), so genetic evidence is fundamental to identifying the nature of the signal.
It is well established that carotenoids are subject to extensive enzymatic and nonenzymatic modifications, including oxidation and cleavage. Many apocarotenoids produced through these remodeling processes have signaling activities, as they can move to different subcellular compartments in which they perform various biological functions, including modulating developmental processes such as lateral root pattering (Bouvier et al., 2005; Walter et al., 2010; Van Norman et al., 2014). We reasoned that it was possible that an apocarotenoid derived from the linear carotenoids that accumulate in clb5 could generate a signal responsible for the described clb5 phenotypes. From the nine CCD genes in Arabidopsis, five participate in the biosynthesis of ABA (NCED2, -3, -5, -6, and -9) and two in the synthesis of SL (CCD7 and CCD8). The functions of the cytosolic CCD1 and plastidic CCD4 enzymes are less clear, although they have been implicated in the production of some plant volatiles (Walter et al., 2010; Ramel et al., 2012). Apparently, both enzymes can cleave linear carotenoids at C9-C10, C9′, and C10′ (Vogel et al., 2008; Huang et al., 2009).
One of the most significant findings of this work is that the absence of CCD4 rescues the phenotypic and gene expression defects of the clb5-1 mutant. This is a specific function of CCD4, as it was not observed with CCD7 or CCD8. We demonstrate that CCD4 has the capacity to recognize and cleave one or more of the carotenes that accumulate in clb5 and that this cleavage product, or a derivative thereof, is responsible for the morphological and gene expression defects observed in the clb5 mutant. This result also supports that the clb5 phenotypes result from the accumulation of phytofluene and ζ-carotene isomers and not from the accumulation of a new toxic metabolite. However, this finding contrasts with previous data using in vitro Escherichia coli assays, where it was found that CCD4 does not accept ζ-carotene as the preferred substrate (Huang et al., 2009). One possible explanation is that ζ-carotene itself might not be the direct substrate of CCD4 but instead phytofluene or even a not-yet identified compound derived from these cis-linear carotenoids. An alternative explanation is that the substrate specificity of CCD4 differs between the in vitro and in vivo studies. Interestingly, a recent analysis in tomato (Solanum lycopersicum) speculates that two distantly related CRTISO genes have the capacity to use cis-ζ-carotenes and transform them to all-trans-ζ-carotene, initiating a new metabolic branch (Fantini et al., 2013). Putative orthologs of these genes (CRTISO-L) also exist in Arabidopsis, opening the possibility that all-trans-ζ-carotene could also be a substrate for the CCD4 enzyme. Interestingly, recent work with tomato mutants provided evidence of a regulatory signal produced from linear cis-carotenoids, such as neurosporene and prolycopene, that regulate PSY expression (Kachanovsky et al., 2012).
CCD4-related genes are found in diverse plant species, supporting an important role for this gene (Ahrazem et al., 2010). Recent evidence indicated that CCD4 is important in the turnover of β-carotenoids during seed maturation and leaf senescence in Arabidopsis (Gonzalez-Jorge et al., 2013). Accordingly, the expression analysis through quantitative RT-PCR and from transgenic plants showed that the CCD4 genes are preferentially expressed in late seed development and flowers, although low expression levels were also detected in mature leaves (Huang et al., 2009; Ahrazem et al., 2010). Moreover CCD4-related genes in other plants have undergone gene duplications that appear to be subjected to stringent regulation. However, so far, no leaf phenotype associated with the ccd4 mutant has been reported. Clearly, a more detailed analysis of the expression pattern of CCD4 during leaf development and of the phenotype of the ccd4 mutant could shed light on a possible signaling role of the apocarotenoid derived from this enzymatic activity.
When and where does the signal act? It is tempting to speculate that the molecule responsible for the defects described in this work involves a signal that regulates gene expression and leaf development, although we cannot exclude that (1) a clb5-generated apocarotenoid does not become synthesized during normal Arabidopsis development, (2) the observed defects result from the fortuitous interaction of this molecule with one or more plant targets, and (3) the apocarotenoid is produced during a particular developmental stage or in response to an environmental change. In fact, based on the pleiotropic phenotypes that result from this clb5 carotenoid-derived signal, one could speculate that the production of this signal is probably regulated in response to specific physiological conditions. In this sense, it is interesting that proteomic studies demonstrated that CCD4 colocalizes with ZDS in plastoglobules, which are the site of high carotenoid accumulation under stressful conditions (Ytterberg et al., 2006). Whether the CCD4 colocalization or cell-specific expression patterns are important for the generation of this signal is a matter for future research. In addition, recent data showed that the rate-limiting enzymes of the carotenoid pathway, such as PSY, display a particular expression pattern (Van Norman et al., 2014).
From a physiological point of view, plastid-derived regulation of gene expression and development might provide an important feedback mechanism that reflects the capacity of the organelle to establish a functional photosynthetic apparatus in response to environmental cues such as light. Indeed, the SAL1–3′-phosphoadenosine 5′-phosphate retrograde signaling pathway that operates during abiotic stress also alters developmental processes such as flowering time and petiole length (Estavillo et al., 2011). It is interesting that during evolution, the CrtI-type desaturases capable of catalyzing the entire desaturation steps to produce lycopene present in archea, bacteria, and fungi were substituted in cyanobacteria and maintained in all plants by four enzymes, including the PDS and ZDS desaturases (Sandmann, 2009). Thus, this substitution provides the capacity of these enzymes to link carotenogenesis with the photosynthetic electron chain via the plastoquinone pool (Foudree et al., 2010). Perhaps the reactions of these two desaturases are strategic in their capacity to perceive the metabolic status and functionality of the organelle. One could speculate that a signal derived from the intermediate linear cis-carotenoid molecules might be an important link to the metabolic status of the organelle by adjusting the development of complex body plans present in vascular plants (Norris et al., 1995).
The severe abnormality of leaf morphology and anatomy in clb5 is striking compared with other albino mutants. However, this is not exclusive to clb5, since it is well recognized how important the chloroplast status is in defining leaf architecture (Pyke et al., 2000). Several mutants affected in chloroplast development and functionality display defects in specific aspects of plant development. For example, altered differentiation and division of the palisade cells has been observed in several mutants that affect chloroplast biogenesis, such as dag and scabra3 (Chatterjee et al., 1996; Hricová et al., 2006), and in carotenoid biosynthesis mutants, such as ghost (Scolnik et al., 1987). Such defects have been attributed to photooxidative damage induced by altered carotenoid profiles that, in turn, regulate terminal stages of mesophyll cell differentiation (Woodson and Chory, 2008). The fact that several mutants affected in chloroplast biogenesis display defects in leaf anatomy and in chloroplast- and nucleus-encoded genes further supports the existence of a plastid-derived signal that regulates leaf development. Clearly, the plastid status can affect the differentiation of a particular cell type and the leaf shape. Early studies have demonstrated that the genotype of the plastids in evening primrose (Oenothera biennis) can underpin leaf shape (Tiller and Bock, 2014).
Leaf development is a process that involves the differentiation of specialized cells along different axes to achieve the correct size and shape. It is well known that this process requires the participation of a variety of regulators, including hormones, small RNAs, and transcription factors, that function in a multilayered regulatory network to influence development (Tsukaya, 2013). The morphological abnormalities observed in clb5 occur earlier in development and are more severe than the ones observed in mutants like cla1 and pds3. This finding, together with the altered expression profile observed in clb5, indicates that the disruption of ZDS generates a molecule in the chloroplast that affects the earlier stages of chloroplast and leaf development. Our analysis of the expression pattern of the adaxial PHB and abaxial FIL markers demonstrated that the finger-type leaves in the clb5 mutant display defects in the dorsoventrality differentiation of the leaf, resembling an abaxialized leaf (Liu et al., 2012). Interestingly, HD-ZIPIII–type transcription factors required for adaxial specification contain a START domain, related to the ABA binding domain from the PYR ABA receptor, that is predicted to bind a yet unidentified ligand (Liu et al., 2012). It is tempting to speculate that the apocarotenoid derived from cis-ζ-carotenes could interact with some of these transcription factors, a possibility that remains to be addressed in the future. It is worth noting that these defects are restricted to the primary leaves and are not observed in the cotyledons, exemplifying the differences in the developmental regulation between these two organs with respect to plastid communication.
A consideration of mutants that have leaf development phenotypes similar to clb5 brought us to enlarged fil expression domain2 (enf2), which is a pale seedling that harbors a mutation in a chloroplast protein (ENF2) necessary for correct plastid gene expression and exhibits narrow, serrated leaf defects (Tameshige et al., 2013). Similar to clb5, the enf2 mutant displays defects in the expression of the abaxial FIL gene and in the leaf lamina morphology. The authors demonstrated that the enf2 defects correlate with an abaxialization of the leaf that, in turn, affects expansion of the lamina. Our analyses demonstrated major defects in the expansion of clb5 lamina, which contain fewer mesophyll cells with big air spaces and bumpy epidermal cells, all phenotypes that are consistent with abaxialized leaves (Liu et al., 2012; Tameshige et al., 2013). An additional similarity of profound defects in chloroplast gene expression and in organelle protein synthesis was also observed between these mutants. According to this work, impairment in plastid transcription or translation results in a retrograde signal that modulates leaf development (Tameshige et al., 2013). In summary, the plastid plays a critical role in defining leaf morphology.
Another mutant with morphological similarities with clb5 is bypass1 (bps1). bps1 affects a protein of unknown function that generates a mobile signal generated by the root that arrests leaf development (Van Norman et al., 2011). Similar to clb5, the phenotype of bps1 is partially rescued by fluridone, suggesting the participation of carotenoids (Van Norman and Sieburth, 2007). However, unlike bps1, we never observed major root defects in clb5. At this point, any connection between bps1 and clb5 is speculative.
In conclusion, we show that, in addition to the known ABA and SL hormones, linear carotenoids can generate an apocarotenoid that plays important roles in the regulation of leaf development and in the expression of a variety of chloroplast- and nucleus-encoded genes. This molecule originates from phytofluene and ζ-carotenoids and requires the action of the CCD4 tailoring enzyme. This study exemplifies the importance of the carotenoid pathway in generating regulatory signals that, besides their well-established functions, can clearly affect major developmental processes such as leaf development and function as feedback signals responding to the status of organelle development.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana Columbia-0 and Ler ecotypes were used in this study. Seed growth under sterile conditions was done in 1× Murashige and Skoog (MS) medium with Gamborg vitamins (Phytotechnology Laboratories) supplemented with 1% (w/v) Suc, solidified with 0.8% (w/v) phytoagar, and for adult plants in Metro-Mix 200 (SunGro). Seedlings were grown under a 16/8-h light/dark cycle (120 μmol m−2 s−1) at 22°C. To break dormancy, seeds were vernalized at 4°C for 4 d. Seeds segregating for pds3 (ZHJ070204) were provided by Li-Jia Qu (Peking University), and the spc1-2 (SALK_033039) mutant was obtained from the ABRC. Seeds of pFIL:GUS and pREV:GUS lines were provided by John Bowman (Monash University), pCYCB1,1:D-box-GUS by Dirk Inzé (Ghent University), and gPHB:GUS by Stewart Gillmor (Gillmor et al., 2010). Homozygous lines for each marker were crossed with clb5 mutant heterozygous plants. The presence of the corresponding transgene and the clb5 mutation was corroborated through GUS histological staining and the albino phenotype in the F2 generation. For ABA and SL treatments, clb5 heterozygous seedlings were germinated in MS medium and transferred to MS normal medium or MS medium supplemented with 0.5 μM ABA, 1 μM ABA, or 0.5 μM GR24 SL 2 d after germination, where they were grown in a 16/8-h light/dark period until morphology was analyzed (Supplemental Figure 8).
Mutant Complementation and Double Mutant Generation
The full ZDS cDNA was obtained by RT-PCR using the oligonucleotides ZDS5 and ZDS3 (Supplemental Table 1). Independent homozygous lines for the transgene were selected, and the presence of the clb5 mutation was corroborated by the absence of a BamHI site in a fragment amplified with the ZDST5 and ZDST3 primers (Supplemental Figure 1). For generation of the pds3 clb5 double mutant, single heterozygous pds3 and clb5-1 mutants were crossed and the F2 progeny were genotyped by PCR using genomic DNA. To genotype the clb5-1 allele, the CLB5 gene was amplified using ZDS-3F and ZDS-4R oligonucleotides (Supplemental Table 1). The 586-bp product was digested with BamHI and analyzed on a 1% agarose gel (Supplemental Figures 1 and 11). PDS3 genotyping was done as reported (Qin et al., 2007). The ccd4 clb5-1 double mutant was generated by crossing the homozygous ccd4 mutant (SALK_097984C), donated by the van Wijk lab (Cornell University), with heterozygous clb5-1. ccd4 homozygous mutant plants were genotyped using the CD4FW and LB T-DNA oligonucleotides (Supplemental Table 1). Double mutants between ccd7/max3 and ccd8/max4 were obtained by crossing the homozygous ccd mutants with the clb5-1 heterozygous plants. Homozygous ccd7 and ccd8 plants were selected based on the presence of the branching phenotype (Auldridge et al., 2006a). The presence of the clb5 mutation in all these lines was verified through the segregation of albino embryos.
Histochemical GUS Staining
The GUS histochemical assay was performed as reported previously (Jefferson, 1987). Seedlings were mounted in 50% glycerol and analyzed using a light microscope (Nikon Eclipse E600).
Herbicide Treatment
Plants were germinated for 4 d in MS medium, and wild-type, clb5-1, and pds3 albino seedlings were transferred to medium containing 5 μM fluridone (Phytotechnology Laboratories). Plants were collected after 8 d for the wild-type control and after 15 d for the wild-type, clb5-1, and pds3 fluridone-treated seedlings.
Genetic Mapping and Sequence Analysis
A mapping population was generated by crossing CLB5+/− plants with the Columbia-0 ecotype. Fine-map positions were assigned by examining recombination frequencies between the gene of interest and linked flanking SSLP markers in a population of 805 F2 clb5 albino and heterozygous plants (CLB5+/−) (Lukowitz et al., 2000). SSLP markers were designed based on the insertion/deletion database produced by the Cereon Arabidopsis polymorphism database (https://www.arabidopsis.org/browse/Cereon/index.html). Alignment of At-ZDS with the ZDS from photosynthetic organisms (Supplemental Figure 2) was performed using InterPro functional analysis (http://www.ebi.ac.uk/interpro/).
Gene Expression Analyses
Total RNA was isolated using TRIZOL (Invitrogen). For RNA gel blot analysis, the RNA was fractionated by electrophoresis and transferred onto Hybond N+ nylon membranes (GE Biosciences). Hybridizations and washes were performed in high-stringency conditions according to standard procedures using 32P-radiolabeled probes produced with the Megaprime DNA labeling system (Amersham). The fragments used as probes were obtained by PCR using specific oligonucleotides (Supplemental Table 1). Specific fragments were used for the Arabidopsis PC, RCBS, and LHCB1.3 genes. The RPL21 probe was obtained from the 146E8 clone from the ABRC. As a loading control, final hybridization was performed using the 25S rRNA with a short time exposure. For quantitative real-time RT-PCR (Supplemental Figure 9), total RNA was treated with RNase-free DNase I (Promega) and repurified with the RNA cleanup protocol in the RNeasy kit (Qiagen). First-strand cDNA was synthesized with MMLV reverse transcriptase (Invitrogen) using 3 μg of total RNA and oligo(dT) primer, according to the manufacturer. Two microliters from a 1:10 dilution of the first-strand solution was used for the subsequent quantitative PCR with FastStart DNA MasterPLUS SYBR Green I in the LightCycler carousel-based system (Roche). The primers used for EX1 and ZAT12 genes are described in Supplemental Table 1. All relative transcript levels are referred to EF1. Normalized values come from three technical replicates from two independent experiments. Tukey statistical analysis was used to evaluate the confidence of the data (http://www.r-project.org/).
Antibody Preparation
Polyclonal antibodies were generated against the protein product of the ZDS cDNA cloned in the pDEST17 vector (Invitrogen). The recombinant proteins were purified under denaturing conditions from Escherichia coli crude extracts according to the QIAexpressionist protocol (Qiagen). For antibody generation, 50 μg of the gel-purified ZDS protein was used to immunize New Zealand rabbits. The ZDS antibodies were purified by immune adsorption to nitrocellulose according to a published protocol (Harlow and Lane, 1988). The specificity of the antibodies against the ZDS protein was confirmed by analysis of the clb5 mutants.
Protein Gel Blot Analysis
Total protein samples were obtained from frozen tissues in SDS sample buffer (0.125 M Tris-HCl, pH 6.8, 20% [v/v] glycerol, 4% [w/v] SDS, and 2% [v/v] 2-mercaptoethanol). The protein concentration was determined with Bradford reagent (Bio-Rad). Proteins were separated by SDS-PAGE. To verify equal protein loading, a parallel gel was run and stained with Coomassie Brilliant Blue R 250. The proteins were transferred onto nitrocellulose (Hybond C; GE Biosciences). Immunodetection was performed using the following antibody dilutions: 1:500 of ZDS, 1:1000 of DXS, 1:5000 of ClpP (Uniplastomic), and 1:2000 of RpoA (donated by Yuzuru Tozawa, Ehime University). An anti-rabbit immunoglobulin horseradish peroxidase conjugate or an anti-mouse alkaline phosphatase was used as the secondary antibody (Invitrogen). The detected proteins were visualized using an enhanced chemiluminescence detection kit (Thermo Scientific) or a BCIP/NBT substrate kit (Invitrogen). A minimum of two independent biological experiments were done for each of the protein analyses included in this article.
Light Microscopy
Tissues were fixed with 6% (w/v) glutaraldehyde and 2% paraformaldehyde in PBS (pH 7.2) for 18 h. After dehydration in a graded series of ethanol, samples were embedded in epoxy resin, and 0.5- to 0.7-μm semithin sections were obtained. The sections were stained with 1% (w/v) toluidine blue and observed in a bright field with a light microscope (Nikon Eclipse E600).
Pigment Analysis
Carotenoid pigments were quantified in 15-d-old mutants and 8-d-old wild-type plants grown in MS medium under standard conditions (150 μmol m−2 s−1) or in the presence of 10 μM norflurazon (Supplemental Figure 12). Pigments were extracted from frozen tissues (50 to 100 mg); samples were prepared in 500 μL of acetone:ethyl acetate (3:2, v/v). HPLC scans of carotenoids were extracted from pds3 and clb5-1 mutants as well as the wild type. Pigments were monitored at λmax of 400 nm (major carotenoids and chlorophyll) and 286 nm (phytoene). There was a minimum of three replicates per mutant and treatment.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: ZDS (At3g04870), PDS (At4g14210), DXS1 (At4g15560), HDR (At4g34350), PSY (At5g17230), CCD4 (At4g19170), CCD7 (At2g44990), CCD8 (At4g32810), PC (At1g76100), RCBS (At1g67090), LHCB1.3 (At1g29930), and RPL21 (At1g33680).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Genotyping of the clb5-1 Complemented Plants.
Supplemental Figure 2. Amino Acid Alignment of the Conserved Region of ZDS.
Supplemental Figure 3. Phenotypes of the Different clb5 Alleles.
Supplemental Figure 4. Phenotype of an 8-Week-Old clb5-1 Mutant Plant.
Supplemental Figure 5. Subcellular Localization of ZDS.
Supplemental Figure 6. ZDS Protein Abundance in Arabidopsis Tissues.
Supplemental Figure 7. Morphological Analysis of the clb5-1 Leaves.
Supplemental Figure 8. Effect of the ABA and Strigolactone Hormones over the clb5 Leaf Morphology.
Supplemental Figure 9. Analysis of the ROS in the clb5 Mutant.
Supplemental Figure 10. Fluridone Treatment.
Supplemental Figure 11. Genotyping of the Double pds3 clb5 Mutant.
Supplemental Figure 12. HPLC Chromatograms of Carotenoids.
Supplemental Table 1. Oligonucleotides Used in This Work.
Supplementary Material
Acknowledgments
We dedicate this work to the memory of Carolina San Román. We thank Virginia Walbot and Kenneth Luehrsen for helpful comments and Guadalupe Zavala and Andrés Saralegui for assistance with light and confocal microscopy. We thank the ABRC for providing seeds and TAIR for organizing Arabidopsis resources. This work was supported by the Consejo Nacional de Ciencia y Tecnología (Grant CONACYT127546 and fellowships to A.-O.A.-V., E.L., and S.D.l.T.), the Dirección General de Asuntos para el Personal Académico-UNAM (Grants IN208211-3 and IN207214), the Howard Hughes Medical Institute, and the Australian Research Council Centre of Excellence in Plant Energy Biology (Grants CE14100008 and DP130102593).
AUTHOR CONTRIBUTIONS
A.-O.A.-V. mapped and identified the mutant and performed expression analysis, leaf marker analysis, mutant phenotypic characterization, electron microscopy analysis, and pigment analysis. E.C. performed expression analysis, double mutant generation, and analysis. E.L. performed double mutant generation and analysis, leaf marker generation, and analysis. N.N. performed pigment analysis and double mutant generation. C.S.R. performed antibody generation and protein analysis. S.D.l.T performed transcriptional analysis and inhibitor analysis. M.R.-V. performed protein analysis. M.d.l.L.G.-N. generated the mapping population. C.I.C. discussed and edited the article. B.J.P. supervised, discussed, and edited the article. P.L. designed the project, supervised, discussed the results, and wrote the article.
Glossary
- MEP
methylerythritol phosphate
- ABA
abscisic acid
- SL
strigolactone
- SSLP
simple sequence length polymorphism
- Ler
Landsberg erecta
- ROS
reactive oxygen species
- MS
Murashige and Skoog
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Ahrazem O., Trapero A., Gómez M.D., Rubio-Moraga A., Gómez-Gómez L. (2010). Genomic analysis and gene structure of the plant carotenoid dioxygenase 4 family: A deeper study in Crocus sativus and its allies. Genomics 96: 239–250. [DOI] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., Ghisla S., Bouwmeester H., Beyer P., Al-Babili S. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351. [DOI] [PubMed] [Google Scholar]
- Auldridge M.E., Block A., Vogel J.T., Dabney-Smith C., Mila I., Bouzayen M., Magallanes-Lundback M., DellaPenna D., McCarty D.R., Klee H.J. (2006a). Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 45: 982–993. [DOI] [PubMed] [Google Scholar]
- Auldridge M.E., McCarty D.R., Klee H.J. (2006b). Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 9: 315–321. [DOI] [PubMed] [Google Scholar]
- Barkan A., Goldschmidt-Clermont M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572. [DOI] [PubMed] [Google Scholar]
- Bartels P.G., Watson C.W. (1978). Inhibition of carotenoid synthesis by fluridone and norflurazon. Weed Sci. 26: 198–203. [Google Scholar]
- Bouvier F., Isner J.C., Dogbo O., Camara B. (2005). Oxidative tailoring of carotenoids: A prospect towards novel functions in plants. Trends Plant Sci. 10: 187–194. [DOI] [PubMed] [Google Scholar]
- Bräutigam K., et al. (2009). Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell 21: 2715–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréhélin C., Kessler F., van Wijk K.J. (2007). Plastoglobules: Versatile lipoprotein particles in plastids. Trends Plant Sci. 12: 260–266. [DOI] [PubMed] [Google Scholar]
- Cazzonelli C. (2011). Carotenoids in nature: Insights from plants and beyond. Funct. Plant Biol. 38: 833–847. [DOI] [PubMed] [Google Scholar]
- Cazzonelli C.I., Pogson B.J. (2010). Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 15: 266–274. [DOI] [PubMed] [Google Scholar]
- Chatterjee M., Sparvoli S., Edmunds C., Garosi P., Findlay K., Martin C. (1996). DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 15: 4194–4207. [PMC free article] [PubMed] [Google Scholar]
- Conti A., Pancaldi S., Fambrini M., Michelotti V., Bonora A., Salvini M., Pugliesi C. (2004). A deficiency at the gene coding for zeta-carotene desaturase characterizes the sunflower non dormant-1 mutant. Plant Cell Physiol. 45: 445–455. [DOI] [PubMed] [Google Scholar]
- Davletova S., Schlauch K., Coutu J., Mittler R. (2005). The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 139: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Deng Y., Mu J., Lu Q., Wang Y., Xu Y., Chu C., Chong K., Lu C., Zuo J. (2007). The Arabidopsis Spontaneous Cell Death1 gene, encoding a zeta-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signalling. Cell Res. 17: 458–470. [DOI] [PubMed] [Google Scholar]
- Donnelly P.M., Bonetta D., Tsukaya H., Dengler R.E., Dengler N.G. (1999). Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev. Biol. 215: 407–419. [DOI] [PubMed] [Google Scholar]
- Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13: 1768–1774. [DOI] [PubMed] [Google Scholar]
- Estavillo G.M., et al. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini E., Falcone G., Frusciante S., Giliberto L., Giuliano G. (2013). Dissection of tomato lycopene biosynthesis through virus-induced gene silencing. Plant Physiol. 163: 986–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudree A., Aluru M., Rodermel S. (2010). PDS activity acts as a rheostat of retrograde signaling during early chloroplast biogenesis. Plant Signal. Behav. 5: 1629–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor C.S., Park M.Y., Smith M.R., Pepitone R., Kerstetter R.A., Poethig R.S. (2010). The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Jorge S., et al. (2013). Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25: 4812–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J.C., Sullivan J.A., Wang J.H., Jerome C.A., MacLean D. (2003). Coordination of plastid and nuclear gene expression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Nava M.d.l.L., Gillmor C.S., Jiménez L.F., Guevara-García A., León P. (2004). CHLOROPLAST BIOGENESIS genes act cell and noncell autonomously in early chloroplast development. Plant Physiol. 135: 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Immunizations. (Plainview, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Hricová A., Quesada V., Micol J.L. (2006). The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol. 141: 942–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F.C., Molnár P., Schwab W. (2009). Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 60: 3011–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5: 387–405. [Google Scholar]
- Kachanovsky D.E., Filler S., Isaacson T., Hirschberg J. (2012). Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis-carotenoids. Proc. Natl. Acad. Sci. USA 109: 19021–19026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Lee K.P., Baruah A., Nater M., Göbel C., Feussner I., Apel K. (2009). 1O2-mediated retrograde signaling during late embryogenesis predetermines plastid differentiation in seedlings by recruiting abscisic acid. Proc. Natl. Acad. Sci. USA 106: 9920–9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P., Kremnev D., Blanco N.E., de Dios Barajas López J., Fernández A.P., Tellgren-Roth C., Kleine T., Small I., Strand A. (2012). The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 70: 279–291. [DOI] [PubMed] [Google Scholar]
- Kleine T., Kindgren P., Benedict C., Hendrickson L., Strand A. (2007). Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719. [PubMed] [Google Scholar]
- Lee K.P., Kim C., Landgraf F., Apel K. (2007). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 10270–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden H., Misawa N., Saito T., Sandmann G. (1994). A novel carotenoid biosynthesis gene coding for zeta-carotene desaturase: Functional expression, sequence and phylogenetic origin. Plant Mol. Biol. 24: 369–379. [DOI] [PubMed] [Google Scholar]
- Liu T., Reinhart B.J., Magnani E., Huang T., Kerstetter R., Barton M.K. (2012). Of blades and branches: Understanding and expanding the Arabidopsis ad/abaxial regulatory network through target gene identification. Cold Spring Harb. Symp. Quant. Biol. 77: 31–45. [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C.S., Scheible W.R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123: 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews P.D., Luo R., Wurtzel E.T. (2003). Maize phytoene desaturase and zeta-carotene desaturase catalyse a poly-Z desaturation pathway: Implications for genetic engineering of carotenoid content among cereal crops. J. Exp. Bot. 54: 2215–2230. [DOI] [PubMed] [Google Scholar]
- McElver J., et al. (2001). Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Brusslan J.A., Larkin R., Nagatani A., Chory J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98: 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Tanaka A., Masuda T., Nagatani A. (2008). The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 105: 15184–15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H., Clarke J.H., De Block M., De Block S., Vanderhaeghen R., Zielinski R.E., Dyer T., Lust S., Inzé D., Van Lijsebettens M. (2003). DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 15: 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris S.R., Barrette T.R., DellaPenna D. (1995). Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7: 2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Jung H.S., Koussevitzky S., Chory J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57: 739–759. [DOI] [PubMed] [Google Scholar]
- Pogson B.J., Woo N.S., Förster B., Small I.D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13: 602–609. [DOI] [PubMed] [Google Scholar]
- Pyke K., Zubko M.K., Day A. (2000). Marking cell layers with spectinomycin provides a new tool for monitoring cell fate during leaf development. J. Exp. Bot. 51: 1713–1720. [DOI] [PubMed] [Google Scholar]
- Qin G., Gu H., Ma L., Peng Y., Deng X.W., Chen Z., Qu L.J. (2007). Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17: 471–482. [DOI] [PubMed] [Google Scholar]
- Ramel F., Birtic S., Cuiné S., Triantaphylidès C., Ravanat J.L., Havaux M. (2012). Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiol. 158: 1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F., Mialoundama A.S., Havaux M. (2013). Nonenzymic carotenoid oxidation and photooxidative stress signalling in plants. J. Exp. Bot. 64: 799–805. [DOI] [PubMed] [Google Scholar]
- Ruyter-Spira C., Al-Babili S., van der Krol S., Bouwmeester H. (2013). The biology of strigolactones. Trends Plant Sci. 18: 72–83. [DOI] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472. [DOI] [PubMed] [Google Scholar]
- Sandmann G. (2009). Evolution of carotene desaturation: The complication of a simple pathway. Arch. Biochem. Biophys. 483: 169–174. [DOI] [PubMed] [Google Scholar]
- Scarpella E., Barkoulas M., Tsiantis M. (2010). Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. 2: a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnik P.A., Hinton P., Greenblatt I.M., Giuliano G., Delanoy M.R., Spector D.L., Pollock D. (1987). Somatic instability of carotenoid biosynthesis in the tomato ghost mutant and its effect on plastid development. Planta 171: 11–18. [DOI] [PubMed] [Google Scholar]
- Shumskaya M., Bradbury L.M., Monaco R.R., Wurtzel E.T. (2012). Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation, and activity. Plant Cell 24: 3725–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A., Asami T., Alonso J., Ecker J.R., Chory J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421: 79–83. [DOI] [PubMed] [Google Scholar]
- Sullivan J.A., Gray J.C. (1999). Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11: 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Feng P., Xu X., Guo H., Ma J., Chi W., Lin R., Lu C., Zhang L. (2011). A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2: 477. [DOI] [PubMed] [Google Scholar]
- Susek R.E., Ausubel F.M., Chory J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799. [DOI] [PubMed] [Google Scholar]
- Tameshige T., Fujita H., Watanabe K., Toyokura K., Kondo M., Tatematsu K., Matsumoto N., Tsugeki R., Kawaguchi M., Nishimura M., Okada K. (2013). Pattern dynamics in adaxial-abaxial specific gene expression are modulated by a plastid retrograde signal during Arabidopsis thaliana leaf development. PLoS Genet. 9: e1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B.C., Joseph L.M., Deng W.T., Liu L., Li Q.B., Cline K., McCarty D.R. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35: 44–56. [DOI] [PubMed] [Google Scholar]
- Tiller N., Bock R. (2014). The translational apparatus of plastids and its role in plant development. Mol. Plant in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantaphylidès C., Havaux M. (2009). Singlet oxygen in plants: Production, detoxification and signaling. Trends Plant Sci. 14: 219–228. [DOI] [PubMed] [Google Scholar]
- Tsukaya H. (2013). Leaf development. The Arabidopsis Book 11: e0163, /10.1199/tab.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman J.M., Sieburth L.E. (2007). Dissecting the biosynthetic pathway for the bypass1 root-derived signal. Plant J. 49: 619–628. [DOI] [PubMed] [Google Scholar]
- Van Norman J.M., Murphy C., Sieburth L.E. (2011). BYPASS1: Synthesis of the mobile root-derived signal requires active root growth and arrests early leaf development. BMC Plant Biol. 11: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman J.M., Zhang J., Cazzonelli C.I., Pogson B.J., Harrison P.J., Bugg T.D., Chan K.X., Thompson A.J., Benfey P.N. (2014). Periodic root branching in Arabidopsis requires synthesis of an uncharacterized carotenoid derivative. Proc. Natl. Acad. Sci. USA 111: E1300–E1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J.T., Tan B.C., McCarty D.R., Klee H.J. (2008). The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J. Biol. Chem. 283: 11364–11373. [DOI] [PubMed] [Google Scholar]
- Walter M.H., Floss D.S., Strack D. (2010). Apocarotenoids: Hormones, mycorrhizal metabolites and aroma volatiles. Planta 232: 1–17. [DOI] [PubMed] [Google Scholar]
- Woodson J.D., Chory J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9: 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.D., Perez-Ruiz J.M., Chory J. (2011). Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Savchenko T., Baidoo E.E., Chehab W.E., Hayden D.M., Tolstikov V., Corwin J.A., Kliebenstein D.J., Keasling J.D., Dehesh K. (2012). Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535. [DOI] [PubMed] [Google Scholar]
- Ytterberg A.J., Peltier J.B., van Wijk K.J. (2006). Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140: 984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.