High salinity inhibits plant growth and can cause plant death. This work identified a root cell–expressed receptor-like kinase that mediates salt sensitivity by activating MAPK and upregulating ethylene synthesis, leading to an increase in the level of reactive oxygen species.
Abstract
High salinity causes growth inhibition and shoot bleaching in plants that do not tolerate high salt (glycophytes), including most crops. The molecules affected directly by salt and linking the extracellular stimulus to intracellular responses remain largely unknown. Here, we demonstrate that rice (Oryza sativa) Salt Intolerance 1 (SIT1), a lectin receptor-like kinase expressed mainly in root epidermal cells, mediates salt sensitivity. NaCl rapidly activates SIT1, and in the presence of salt, as SIT1 kinase activity increased, plant survival decreased. Rice MPK3 and MPK6 function as the downstream effectors of SIT1. SIT1 phosphorylates MPK3 and 6, and their activation by salt requires SIT1. SIT1 mediates ethylene production and salt-induced ethylene signaling. SIT1 promotes accumulation of reactive oxygen species (ROS), leading to growth inhibition and plant death under salt stress, which occurred in an MPK3/6- and ethylene signaling-dependent manner in Arabidopsis thaliana. Our findings demonstrate the existence of a SIT1-MPK3/6 cascade that mediates salt sensitivity by affecting ROS and ethylene homeostasis and signaling. These results provide important information for engineering salt-tolerant crops.
INTRODUCTION
High salinity inhibits plant growth and can cause plant death. The complex effects of salinity on the physiology and metabolism of plants include alterations in enzyme activity, ion homeostasis, osmotic balance, redox balance, signal transduction, and gene expression. Some of these changes are consequences of the direct impact of ions on normal protein and membrane function and can cause salt intolerance, while other changes represent adaptive cellular responses that counteract these negative effects, leading to increased tolerance to salt stress (Tuteja, 2007). For example, salt-induced activation of the salt overly sensitive signaling system increases the activity of Na+ extrusion antiporters and plant salt tolerance (Qiu et al., 2002; Munns and Tester, 2008), whereas salt-induced reactive oxygen species (ROS) production appears to contribute to salt stress (Zhu, 2002; Munns and Tester, 2008). The primary cellular targets of salt leading to these stress or adaptive responses remain unclear.
The internal changes in plants in response to external stimuli depend on a sophisticated signaling system. Cell surface–localized receptor-like kinases (RLKs) are the ideal candidates to initiate signaling pathways by perceiving and transmitting environmental signals to cellular machinery (Osakabe et al., 2013). Among the 610 RLKs in Arabidopsis thaliana and 1100 RLKs in rice (Oryza sativa) (Shiu et al., 2004), a small number have been characterized as receptors for phytohormones, polypeptides, and pathogens. Each of these RLKs can rapidly initiate signaling through the formation of oligomers and cross-phosphorylation of the intracellular serine/threonine kinase domain upon ligand binding (Diévart and Clark, 2004) and together they play diverse roles in plant development and pathogen resistance (Antolín-Llovera et al., 2012; Osakabe et al., 2013); some RLKs are also reported to function in drought and salt responses and tolerance (de Lorenzo et al., 2009; Ouyang et al., 2010; Marshall et al., 2012; Vaid et al., 2013). Rice SIK1(Os06g03970), a leucine-rich repeat RLK that is expressed most strongly in stem and panicle but which is not expressed in root, was found to be salt-inducible and a positive regulator of salt tolerance (Ouyang et al., 2010).
Lectin RLKs (LecRLKs), which are characterized by an N-terminal lectin domain resembling carbohydrate binding lectin proteins, represent the second largest subfamily of RLKs, with 75 members in Arabidopsis and 173 members in rice (Vaid et al., 2012). They are believed to be involved in saccharide signaling, and they play roles in self-incompatibility and in plant defenses against pathogens and pests, based on the known functions of lectin proteins in cell recognition. In addition, they are involved in plant salt stress and abscisic acid (ABA) responses (Vaid et al., 2013). However, whether these RLKs are involved in stress perception or whether they mediate salt signal relay has not been clarified.
The downstream events of RLK signaling involve protein phosphorylation. Mitogen-activated protein kinase (MAPK) cascades are a key convergence point for diverse signal transduction pathways, including those activated by developmental and environmental stimuli. MAPK modules play pivotal roles in many biological processes through diverse combinations; among them, MPK3, MPK4, and MPK6 are the three main kinases that mediate downstream signaling in Arabidopsis. Modification of the activity of these MAPKs causes changes in programmed development and stress sensitivity (Rodriguez et al., 2010). Rice MPK3 and MPK6 (also known as MPK5 and MPK1, respectively; Singh et al., 2012) are closely related to Arabidopsis MPK3 and MPK6 and function in biotic and abiotic stress responses (Xiong and Yang, 2003; Kishi-Kaboshi et al., 2010; Shen et al., 2010). Like in animal cells, genetic studies in Arabidopsis have demonstrated the functions of MAPK modules downstream of RLKs, including YODA-MKK4/5-MPK3/6 downstream of ERECTA-RLK in inflorescence architecture (Meng et al., 2012), the MEKK1-MKK4/MKK5-MPK3/MPK6 cascade downstream of FLS2-BAK1 in a pathogen-related response triggered by flg22 (Asai et al., 2002), and MPK3/6 activation induced by flg22 depending on LecRK-VI.2-1 (Singh et al., 2012). However, the mechanisms linking extracellular stimuli to activation of these modules remain unclear in plants.
Ethylene not only controls many aspects of plant physiology and development, it also regulates stress-related processes. Salt stress stimulates the production of ethylene, which can act as a secondary signal to initiate another round of signal transduction and modulate the responsive genes in whole plants for stress adaptation or stress sensitivity. The roles of ethylene in the regulation of salt tolerance are complex. Ethylene signaling components are essential for plant tolerance to salinity (Wang et al., 2002a), but increased 1-aminocyclopropane-1-carboxylate (ACC; an ethylene biosynthetic precursor) synthesis appears to reduce salt tolerance (Xu et al., 2008; Dong et al., 2011), suggesting that the control of ethylene homeostasis is an essential factor in the response to a salt challenge. The signaling pathway that links extracellular salt stress to intracellular ethylene production remains a mystery.
In this study, we characterized a LecRLK gene, Salt Intolerance 1 (SIT1), which mediates salt stress signal relay from the cell surface to intracellular MAPK modules. SIT1 is mainly expressed in root epidermal cells and rapidly activated by NaCl, which in turn activates MPK3/6. SIT1 promotes ethylene production and mediates salt-induced ethylene signaling. SIT1 promotes ROS accumulation, leading to plant death in a MPK6-, ethylene-, and ethylene signaling–dependent manner. Our results establish a SIT1-MPK3/6 cascade that mediates salt sensitivity by modulating ethylene and ROS homeostasis in rice.
RESULTS
The RLKs SIT1 and SIT2 Negatively Regulate Salt Tolerance in Rice
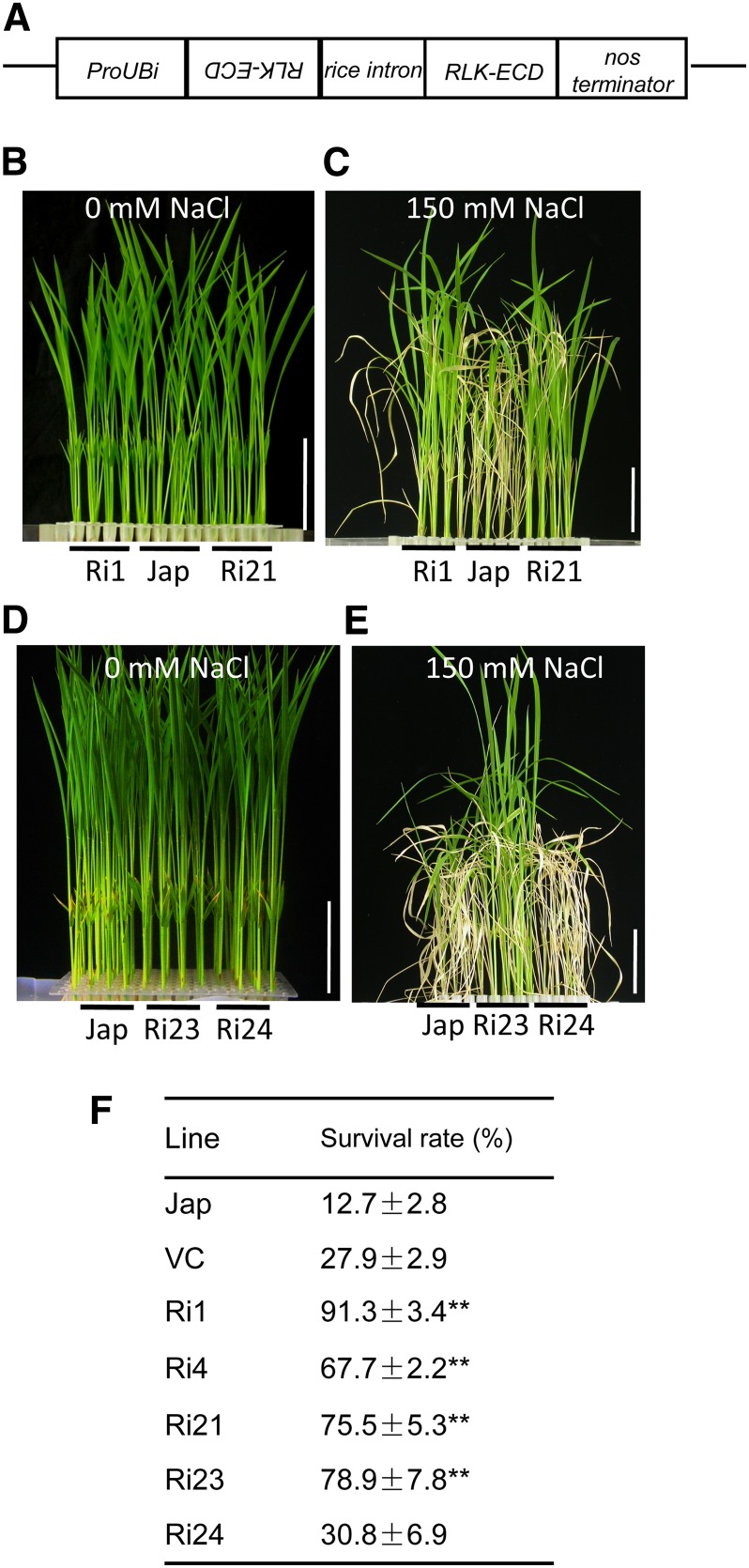
Transcriptional analyses of rice roots have revealed several salt-induced RLKs (see Methods). To study the function of these RLKs in the plant response to salt, we generated RNA interference (RNAi) transgenic rice plants for each RLK by introducing the vector pTCK303 (Wang et al., 2004), which harbored a fragment of the extracellular domain coding sequence (Figure 1A). T2 seedlings of the RNAi lines were assessed one-by-one under salt stress conditions. We found a LecRLK gene, SIT1 (Os02g42780), which contributes to salt sensitivity. Of five independent RNAi lines identified by DNA gel blotting (Supplemental Figure 1), four displayed increased salt tolerance (Figures 1B to 1F). Without NaCl treatment, the RNAi plants were indistinguishable from wild-type japonica (Jap) (Figures 1B and 1D). After treatment with 150 mM NaCl for 4 d and subsequent growth by regular hydroponic culture for 8 d, the RNAi plants showed significantly higher survival rates compared with Jap (Figures 1C and 1E) and control plants transformed with vector (Figure 1F). Ri24 was an exception. Under salt stress conditions, its performance was similar to that of Jap (Figure 1E) and its survival rate was similar to that of the control plants (Figure 1F). We also found that transformation with pTCK303 to produce RNAi lines had little positive effect on the salt tolerance of rice seedlings, as shown by the increased survival rates of the control plants compared with that of Jap (Figure 1F).
Figure 1.
Screening the Salt-Insensitive RNAi Plants.
(A) Diagrammatic representation of the RNAi construct used to knock down RLK gene expression. The RLK extracellular domain coding sequence (ECD) was used.
(B) to (E) Comparison of the phenotypes of representative SIT1-RNAi lines at T2 generation before ([B] and [D]) and after ([C] and [E]) salt treatment. Ten-day-old seedlings were used as described in Methods. Bars = 2 cm.
(F) Survival rate of wild-type (Jap), empty vector control (VC), and individual RNAi lines following salt exposure. Numbers represent the means ± se (Student’s t test, n ≥ 50 in [B] to [E]; **P < 0.01)
[See online article for color version of this figure.]
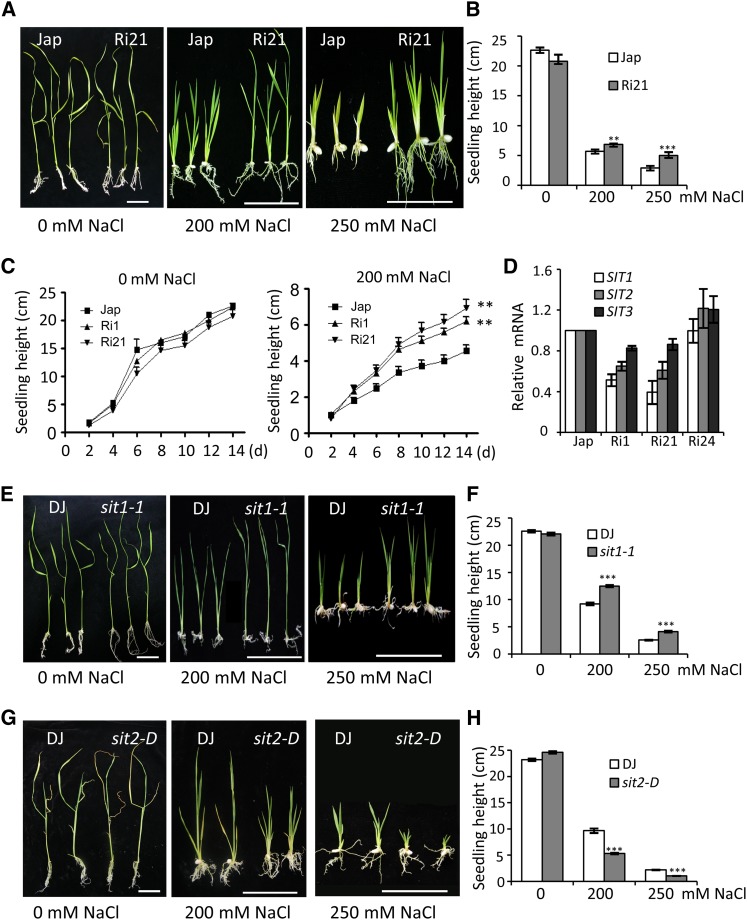
To further confirm the salt tolerance of the SIT1-RNAi seedlings and its correlation with reduced levels of SIT1, T2 progeny of Ri1 and Ri21, each of which harbored a single copy of the insert (Supplemental Figure 1), were screened for homozygous hygromycin resistance by genotyping and used in the following experiments. After 2 d of germination on regular medium, the seeds were grown on medium containing 200 or 250 mM NaCl. The shoots and roots of Jap plants were inhibited by increasing concentrations of NaCl, whereas the RNAi seedlings showed less inhibition (Figures 2A and 2B) and greater biomass accumulation under 250 mM NaCl stress condition (Supplemental Figure 2). A time-course analysis of the growth of the RNAi seedlings on medium containing 200 mM NaCl showed a higher growth rate compared with Jap plants. The Ri21 plants grew even more vigorously than the Ri1 plants (Figure 2C). Quantitative RT-PCR revealed dramatically reduced expression of SIT1 and its homolog SIT2 (Os04g44900), and slightly reduced expression of SIT3 (Os07g38810), in Ri1 and Ri21 plants but nearly normal levels of SIT1, SIT2, and SIT3 in Ri24 plants. Compared with Ri1, the Ri21 plants exhibited a lower level of SIT1, an equal level of SIT2, and a slightly higher level of SIT3 (Figure 2D). These results suggest that the improved salt tolerance of RNAi lines mainly relates to the downregulation of SIT1, SIT2, and, to a lesser extent, SIT3.
Figure 2.
SIT1 and SIT2 Contribute to Salt Sensitivity in Rice.
(A) The performance of RNAi seedlings compared with wild-type Jap on medium containing the indicated concentrations of NaCl.
(B) The height of the seedlings in (A).
(C) Time-course analysis of seedling height in the two SIT1-RNAi lines compared with wild-type Jap grown on medium with or without NaCl.
(D) Real-time PCR analysis of the SIT1, SIT2, and SIT3 mRNA levels in the RNAi seedlings.
(E) to (H) The performance of sit1-1 (E) and sit2-D (G) seedlings compared with wild-type DJ on medium containing the indicated concentrations of NaCl. (F) and (H) show the heights of the seedlings in (E) and (G), respectively. Error bars show the means ± se (Student’s t test, n ≥ 15 in [B], [F], and [H]; n = 12 in [C]); **P < 0.01 and ***P < 0.001. Bars = 2 cm.
To evaluate each gene’s contribution, we screened for SIT1 and SIT2 T-DNA insertion mutants and identified a null allele, sit1-1 (Supplemental Figures 3A to 3C), which also showed reduced salt sensitivity as evidenced by less growth inhibition in shoot than wild-type Dongjin (DJ) (Figures 2E and 2F), similar to Ri21.We also identified an activation-tagged allele for SIT2 (sit2-D) that expressed 5-fold more SIT2 mRNA than wild-type DJ (Supplemental Figures 3D to 3F). In contrast to the Ri21 and sit1-1 plants, sit2-D displayed increased salt sensitivity with greater inhibition in shoot growth (Figures 2G and 2H). We noted that sit1-1 mutant did not have stronger root system than that of DJ when grown on NaCl-containing medium (Figure 2E), unlike Ri21 roots in which SIT1 and SIT2 were reduced by around 50%. This could be due to the impairment in root development in the sit1-1 null mutant, which possessed a long but weak root system (Figure 2E, 0 mM NaCl). In addition, the discrepancy in root phenotype may result from difference in rice varieties. Together, these observations suggest that SIT1 and SIT2 each contribute to salt sensitivity and that their functions are not overlapping.
SIT1 and SIT2 Are Mainly Expressed in Root Epidermal Cells and Induced by Salt
SIT1 and SIT2 RNA expression was analyzed by quantitative RT-PCR in various tissues; the highest level of SIT1 and a relatively high level of SIT2 were detected in roots (Supplemental Figure 4A). SIT1 and SIT2 transcripts were mainly concentrated in the maturation zone, the region that produces lateral roots and root hairs. In RNAi plants, the transcript level of SIT1 was reduced more dramatically than that of SIT2 (Supplemental Figure 4B). These results suggest that SIT1 is a major contributor to root function and is responsible for the RNAi phenotype. We therefore focused on the function of SIT1. Rice plants transformed with a ProSIT1:GFP (green fluorescent protein) reporter construct exhibited a specific fluorescent signal in epidermal cells, which was enhanced by 200 mM NaCl treatment (Supplemental Figure 4C). Similar to the activation of the SIT1 promoter by salt, the mRNA level of SIT1 was highly increased by NaCl treatment, while ABA, ethylene (treatment with ACC), H2O2, and drought had a slight or little effect on SIT1 expression. SIT2 exhibited similarly induced expression (Supplemental Figure 4D). A SIT1-GFP fusion protein (diagramed in Supplemental Figure 4E) localized to the cell membrane in tobacco (Nicotiana tabacum) cells (Supplemental Figure 4F) and a SIT1-Myc fusion protein expressed in transgenic Arabidopsis cofractionated with cellular membranes (Supplemental Figure 4G). The localization of SIT1 at the surface of root cells suggests a direct function in the sensing of environmental signals.
Kinase Activity of SIT1 Is Required for Salt Sensitivity and Triggered by Sodium
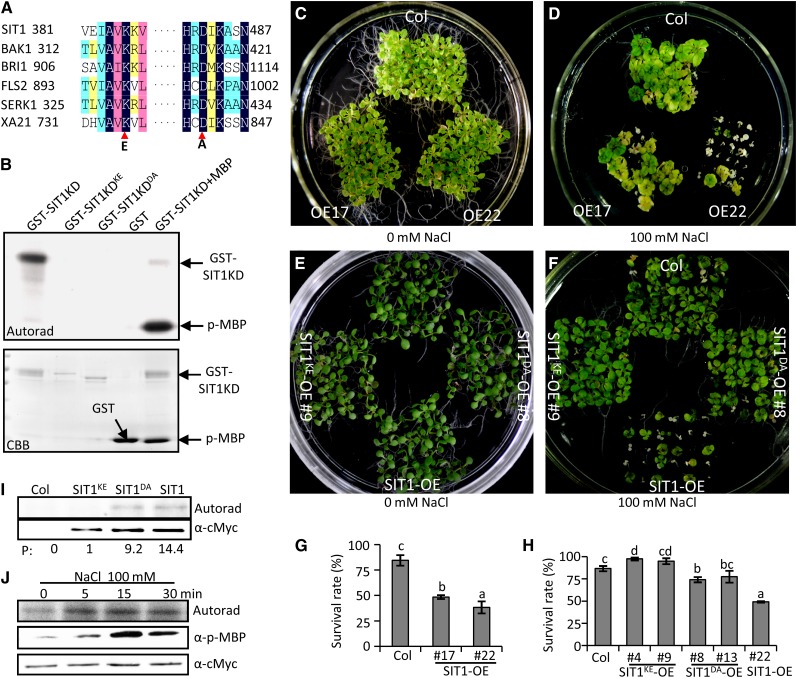
To determine whether SIT1 possesses kinase activity, the conserved lysine (Lys-386) in the ATP binding region and aspartic acid (Asp-482) in the activation loop of kinase domain (KD) were mutated to generate SIT1KDKE and SIT1KDDA, respectively (Figure 3A). The wild-type and mutated SIT1 kinase domains were expressed in Escherichia coli as glutathione S-transferase (GST) fusions. An in vitro kinase assay showed that the SIT1KD was able to catalyze the autophosphorylation and phosphorylation of myelin basic protein (MBP), but the mutated forms had no autophosphorylation activity (Figure 3B), indicating that SIT1 is an active RLK and that Lys-386 and Asp-482 are both necessary for its activity.
Figure 3.
SIT1 Kinase Activity Confers Salt Sensitivity.
(A) Alignment of a conserved motif from the KD of SIT1 with that from other known RLKs. Triangles indicate conserved residues in the active kinase that were mutated to the indicated amino acid in the following experiment.
(B) The GST-SIT1-KD fusion, but not mutated forms, showed autophosphorylation and substrate (MBP) phosphorylation. The upper panel shows autoradiography and the bottom panel Coomassie blue (CBB) staining of the gel.
(C) and (D) Arabidopsis seedlings overexpressing SIT1 were grown on 0.5× MS medium (C) and medium containing 100 mM NaCl for 15 d (D). OE17 and OE22 represent two transgenic lines.
(E) and (F) Arabidopsis seedlings overexpressing wild-type and mutant SIT1 were grown on 0.5× MS medium (E) and medium containing 100 mM NaCl for 15 d (F).
(G) and (H) Survival rates of the seedlings in (D) and (F), respectively, as determined by Student’s t test. a to d indicate significant differences. The bars represent the mean ± se of three biological repeats (n = 30/genotype).
(I) SIT1-Myc fusion proteins expressed in the Arabidopsis seedlings in (C) were immunoprecipitated and subjected to an in vitro autophosphorylation assay. The numbers at the bottom indicate the relative autoradiographic intensity (determined by normalization against the Myc fusion protein) and represent the kinase activity level.
(J) NaCl triggers SIT1 kinase activity. SIT1-Myc was immunoprecipitated from Arabidopsis after treatment with NaCl for the indicated times, then subjected to autophosphorylation and a substrate phosphorylation assay using MBP. Anti-p-MBP antibodies were used to probe for MBP phosphorylation. The bottom panels in (I) and (J) show the equal loading of SIT1-Myc.
[See online article for color version of this figure.]
To further assess the contribution of the kinase activity of SIT1 to salt sensitivity, we generated transgenic Arabidopsis expressing SIT1, SIT1KE, and SIT1DA as Myc fusions under the control of the 35S promoter (Supplemental Figure 5) and analyzed their survival rate on NaCl-containing medium. All of the Arabidopsis overexpression lines used in the study were homozygous and contained a single-copy insertion. On regular medium, little difference was noted among the seedlings (Figures 3C and 3E). On 0.5× Murashige and Skoog (MS) medium containing 100 mM NaCl, the transgenic plants showed normal germination, but the SIT1-overexpressing (OE) seedlings exhibited bleached cotyledons (Figure 3D) and a reduced survival rate (39 to 49%, compared with 87% for wild-type Columbia [Col]; Figure 3G). The SIT1DA-OE seedlings were also damaged, with a survival rate of ∼75%. In contrast, the SIT1KE-OE seedlings showed less damage, with around a 95% survival rate, and they grew slightly better than wild-type Col (Figures 3F and 3H). Similar results were obtained with seedlings grown on 150 mM NaCl-containing medium after germination (Supplemental Figure 6). To correlate these phenotypes with SIT1 kinase activity in vivo, SIT1 was immunoprecipitated from 10-d-old Arabidopsis seedlings grown under normal conditions and analyzed for kinase activity. Our results revealed SIT1 and SIT1DA phosphorylation and undetectable SIT1KE phosphorylation (Figure 3I). Since no SIT1DA autophosphorylation was detected in an in vitro assay (Figure 3B), the phosphorylation of SIT1DA in planta may be catalyzed by another kinase in the immunoprecipitated complex, and this activated SIT1DA contributed to the reduced survival rate of the transgenic Arabidopsis seedlings on salt medium, as compared with Col-0. Together, these results suggest that SIT1 kinase activity affects salt sensitivity.
To test whether SIT1 kinase activity is affected by NaCl, a SIT1-Myc fusion protein, expressed in Arabidopsis SIT1-OE#22 (Supplemental Figure 5), was immunoprecipitated from seedlings after a short period of NaCl treatment and analyzed for kinase activity. Our results show that SIT1 autophosphorylation and MBP phosphorylation by SIT1 both increased after 5 min of NaCl treatment and that the activity continued to increase after 15 min then decreased slightly after 30 min of treatment (Figure 3J). These results indicate that SIT1 is rapidly activated by NaCl.
SIT1 Mediates Salt Sensitivity by Activating MPK3/MPK6
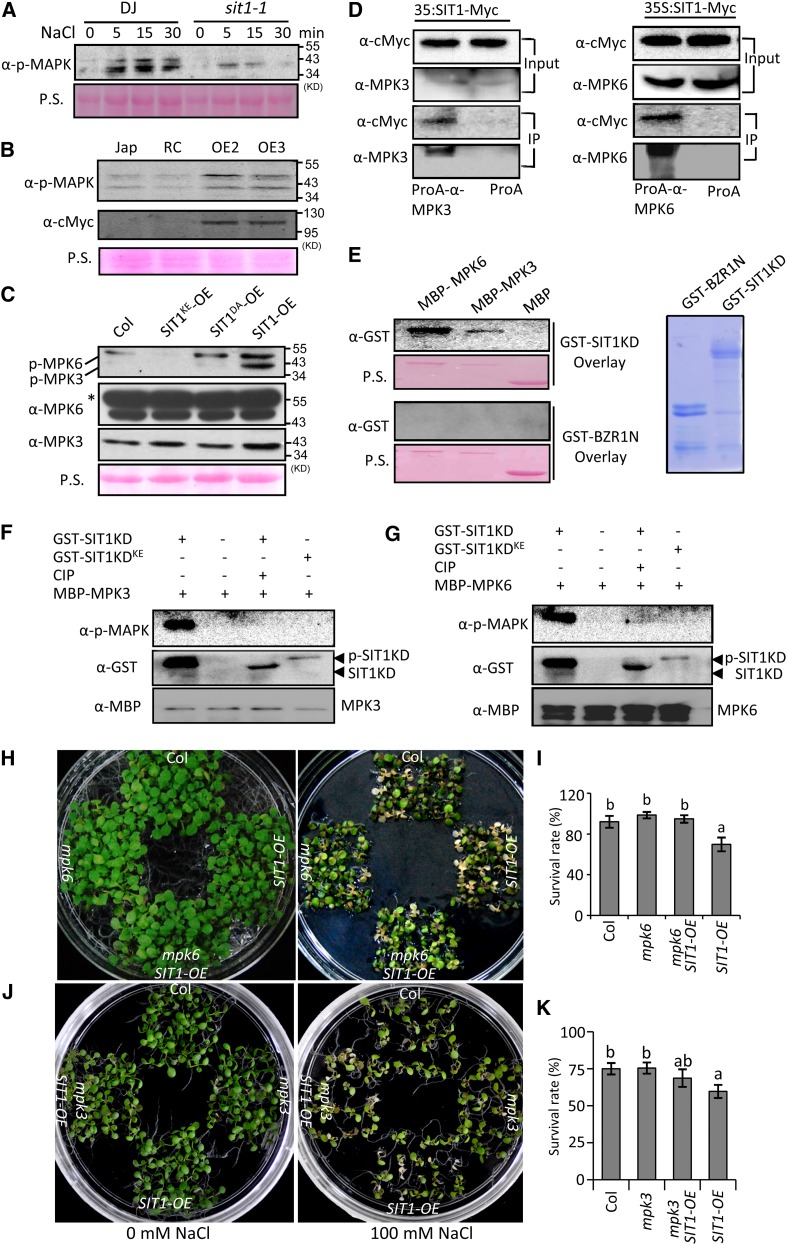
It has been reported that salt stress triggers a MAPK phosphorylation cascade (Kiegerl et al., 2000; Teige et al., 2004). To determine whether SIT1 is involved in NaCl-induced MAPK activation, we tested the phosphorylation status of MAPKs in rice plants and found that the level of phosphorylated MAPKs in DJ seedlings significantly increased after 5 and 15 min of treatment with 100 mM NaCl (Figure 4A). The kinetics of MAPK phosphorylation were similar to those of the autophosphorylation of SIT1 and substrate phosphorylation in SIT1-OE Arabidopsis seedlings after NaCl treatment (Figure 3J). In the sit1-1 mutant, by contrast, NaCl had a much weaker effect on MAPK phosphorylation (Figure 4A). These results indicate that the activation of MAPKs by NaCl requires SIT1.
Figure 4.
SIT1 Mediates Salt Sensitivity by Activating Rice MPK3/6.
(A) to (C) Immunoblot assay for MAPK phosphorylation using anti-p-MAPK antibodies. The sit1-1 mutant exposed to NaCl (A), SIT1-OE rice (B), and Arabidopsis seedlings expressing wild-type and mutated SIT1 (C). Proteins were extracted from 10-d-old rice roots ([A] and [B]) and Arabidopsis seedlings (C). RC in (B) indicates the relative control plants. Anti-MPK3 and -MPK6 antibodies were used in (C) to probe for MPK3 and MPK6. The asterisk indicates a nonspecific protein. Ponceau S (P.S.) staining indicates equal loading.
(D) Coimmunoprecipitation of SIT1 with Os-MPK3 or Os-MPK6. Protein was extracted from 30-d-old Jap rice leaves expressing SIT1-Myc and immunoprecipitated using anti-Os-MPK3 or -Os-MPK6 antibody-conjugated protein A or protein A only. The immunoblot was probed with anti-cMyc, -Os-MPK3, or -Os-MPK6 antibodies.
(E) The GST-SIT1-KD fusion but not GST-BZR1-N (Coomassie blue staining in the right panel) bound to Os-MPK6 or Os-MPK3 in a gel blot overlay. Ponceau S staining indicates the loading of MBP, MBP-Os-MPK6, and MBP-Os-MPK3. Bound proteins were probed with anti-GST-HRP antibodies.
(F) and (G) SIT1-KD, but not SIT1-KDKE, phosphorylates Os-MPK3 (F) and Os-MPK6 (G). Phospho-Os-MPK3 or -Os-MPK6, MBP-Os-MPK3 and MBP-Os-MPK6, GST-SIT1-KD, or GST-SIT1-KDKE, or CIP-treated SIT1-KD were detected using anti-p-MAPK, -MBP-HRP, and -GST-HRP antibodies, respectively.
(H) to (K) mpk6 ([H] and [I]) strongly suppressed while mpk3 ([J] and [K]) slightly suppressed the salt survival rate in SIT1-OE Arabidopsis plants. The indicated seedlings were grown on 0.5× MS medium supplemented with or without 100 mM NaCl for 15 (H) and 10 (J) d, and the salt survival rate was calculated ([I] for [H] and [K] for [J]). The data were analyzed using Student’s t test. Bars indicate the mean ± se of three biological repeats (n = 30/genotype). a and b indicate significant differences.
[See online article for color version of this figure.]
In contrast to the decreased activation of MAPKs in sit1-1, rice plants overexpressing a SIT1-Myc fusion showed higher levels of MAPK phosphorylation than wild-type Jap plants (Figure 4B). Consistent with this, SIT1 overexpression in Arabidopsis also caused the phosphorylation of MPK3 and MPK6. The overexpression of SIT1DA, which showed weak kinase activity in vivo (Figure 3I), caused a smaller increase in MAPK phosphorylation, whereas the MAPK phosphorylation signal was even lower in Arabidopsis plants overexpressing kinase-dead SIT1KE than in Col. The protein levels of MPK6 and MPK3 in these plants were nearly the same, as shown by immunoblotting (Figure 4C), indicating that the phosphorylation but not the protein expression of MAPKs affects SIT1 kinase activity.
To ascertain whether rice MPK3 (Os03g17700) and MPK6 (Os06g06090) are substrates of SIT1, we first performed a coimmunoprecipitation assay. As shown in Figure 4D, a SIT1-Myc fusion expressed in rice was immunoprecipitated by anti-MPK3 and -MPK6 antibodies, indicating that they are present in one protein complex. A subsequent gel blot assay further showed that GST-SIT1KD, but not the GST-N-terminal of brassinazole-resistant 1 (BZR1-N) (Wang et al., 2002b), which harbors several clusters of basic residues similar to the D domain of MAPKK (Wrzaczek and Hirt, 2001) and which was used as a control, could bind to MPK3 and MPK6 (Figure 4E). An in vitro phosphorylation assay using anti-pMAPK antibodies further revealed that SIT1KD, but not SIT1KDKE, could phosphorylate rice MPK3 and MPK6 (Figures 4F and 4G), regardless of the amount used (Supplemental Figures 7A and 7B). These results demonstrate that SIT1 can bind and activate rice MPK3 and MPK6 directly.
To evaluate whether MPK3 and MPK6 function downstream of SIT1 in Arabidopsis, we crossed a null allele, mpk6-3, which displays smaller rosette leaves and shorter siliques than the wild type (Bush and Krysan, 2007), with SIT1-OE Arabidopsis, which exhibits long, narrow leaves and longer siliques than Col. The mpk6 SIT1-OE double homozygotes showed a similar phenotype to mpk6 (Supplemental Figure 8), demonstrating that SIT1 functions through MPK6 in plant growth and development. Similarly, under salt stress, SIT1-induced salt sensitivity was MPK6 dependent. The survival rate was decreased in SIT1-OE Arabidopsis seedlings, but no significant difference in mpk6 SIT1-OE double homozygotes from that in mpk6 and Col when grown on medium containing 100 mM NaCl (Figures 4H and 4I). We also crossed SIT1-OE Arabidopsis with a null allele, mpk3-1, which shows subtle phenotypes with slightly reduced rosette leaves and a similar salt survival rate to that of Col. The mpk3 SIT1-OE double homozygotes showed an intermediate developmental phenotype (Supplemental Figure 8F) and a slightly higher salt survival rate than SIT1-OE plants (Figures 4J and 4K); however, the difference was not significant, suggesting that SIT1-induced salt sensitivity is, to a lesser extent, dependent on MPK3.
SIT1 Promotes ROS Production through MPK6 and MPK3
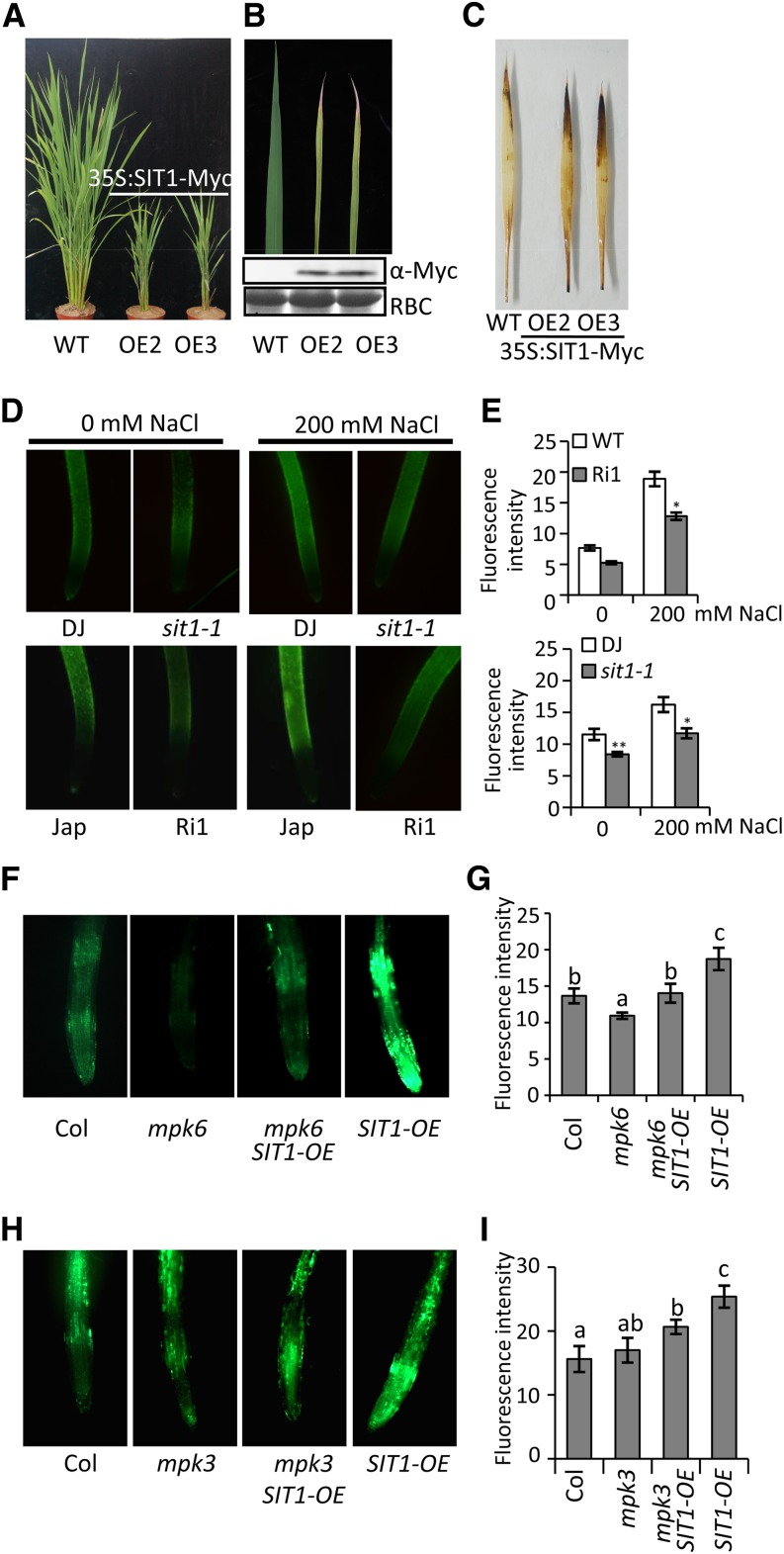
Salt causes an increase in ROS levels, resulting in oxidative stress (Munns and Tester, 2008; Miller et al., 2010), and MAPKs have been reported to mediate ROS production (Pitzschke et al., 2009). We found that the ROS level was higher in SIT1-OE rice leaves (Figures 5A to 5C) and lower in sit1-1 and RNAi roots with or without NaCl treatment (Figures 5D and 5E). The decreased ROS levels in rice occurred with increased peroxidase (POD) and glutathione reductase (GR) activity, but not superoxide dismutase (Supplemental Figures 9A and 9B), suggesting that the salt-induced overaccumulation of ROS is in part dependent on SIT1, which may suppress POD and GR activity through downstream effectors. We indeed found lower POD and GR activity in SIT1-OE Arabidopsis seedlings (Supplemental Figure 9C), and we observed increased ROS levels in their roots with or without NaCl treatment (Supplemental Figures 9D and 9E). However, the increased ROS level in the SIT1-OE Arabidopsis seedlings was abolished in mpk6 SIT1-OE double homozygotes (Figures 5F and 5G) and slightly decreased in mpk3 SIT1-OE double homozygotes (Figures 5H and 5I). Our results indicate that SIT1 promotes ROS accumulation mainly through MPK6 and to a lesser extent through MPK3.
Figure 5.
SIT1 Enhances ROS Accumulation in Rice and in Arabidopsis in an MPK6- and MPK3-Dependent Manner.
(A) to (C) Two representative rice plants overexpressing a SIT1-Myc fusion protein showed a dwarf stature (A), chlorotic withering, and early leaf senescence (B) at the tillering stage. Ponceau S staining of Rubisco (RBC) indicates equal loading.
(C) H2O2 accumulation in the second leaf of 10-d-old wild-type (Jap) and SIT1-OE rice plants was detected by 3,3′-diaminobenzidine staining.
(D) and (E) H2O2 accumulation in sit1-1 and SIT1-RNAi roots with or without 12 h of NaCl treatment. Representative images of CM-H2DCFDA staining (D) showing the ROS level. Quantification of the relative fluorescence intensity in the mature zone of the roots is shown in (E).
(F) to (I) mpk6 ([F] and [G]) largely and mpk3 ([H] and [I]) slightly suppressed the accumulation of ROS in SIT1-OE Arabidopsis plants. Representative images of CM-H2DCFDA staining show the ROS level in the indicated roots of 7-d-old Arabidopsis plants grown on 0.5× MS medium ([F] and [H]). Quantification of the relative fluorescence intensity is shown in (G) and (I), respectively. All data were analyzed by Student’s t test. Error bars represent the mean ± se of three biological repeats (n ≥ 8 in [E]; n ≥ 20 in [G] and [I]); *P < 0.05 and ** P < 0.01; a to c indicate significant differences.
Phylogenetic analysis revealed four closely related genes in Arabidopsis to SIT1 (Supplemental Figure 10A). At-SIT1, -2, -3, and -4 share ∼50% protein sequence identity with rice SIT1 (Supplemental Figure 10B). We obtained a T-DNA insertion mutant (sit3-D) that overexpressed Arabidopsis SIT3 (At4g02410) by 2-fold (Supplemental Figures 11A to 11C). sit3-D also displayed increased MPK3/MPK6 phosphorylation, a higher ROS level, and a lower salt survival rate than wild-type Col (Supplemental Figures 11D to 11G), suggesting that Arabidopsis SIT3 mediates salt sensitivity in a similar way to that of rice SIT1 and that the function of SIT1 is conserved between monocots and dicots.
SIT1 Promotes Ethylene Production
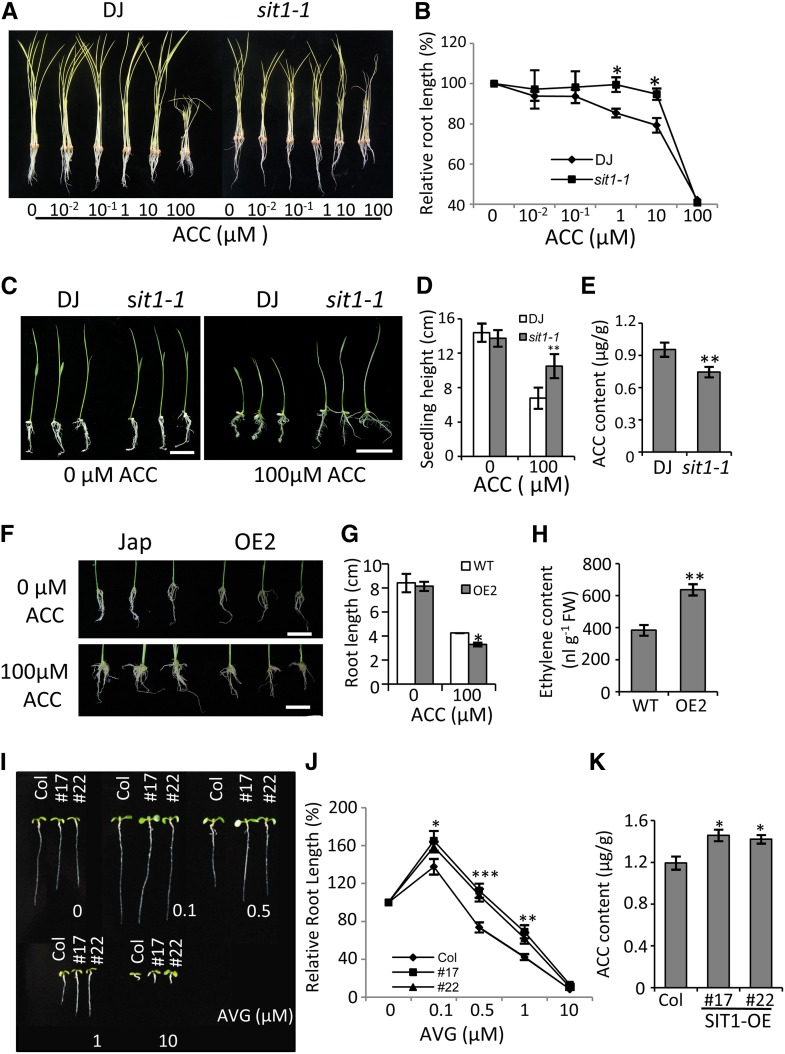
Abiotic stresses, including drought, salt, and cold, can trigger the generation of stress-related hormones and initiate endogenous hormone signaling, resulting in plant growth inhibition (Achard et al., 2006). We observed growth inhibition and early leaf senescence in SIT1-OE rice (Figures 5A and 5B) and delayed maturation in sit1-1 mutant rice (Supplemental Figure 12A). SIT1-OE Arabidopsis also displayed early leaf senescence (Supplemental Figure 12B). These pleiotropic phenotypes in rice and Arabidopsis are typical ethylene-related phenotypes, indicating that ethylene homeostasis was disturbed in these plants. We therefore investigated the sensitivity of sit1-1 and RNAi rice seedlings to ACC. Our results indicate reduced sensitivity to ACC in the sit1-1 and RNAi seedlings. Compared with the DJ wild type, the sit1-1 seedlings displayed longer roots and less root growth inhibition in response to increasing concentrations of ACC up to 100 μM under dark conditions (Figures 6A and 6B). A similar effect was observed in the shoots in response to 100 μM ACC under light conditions (Figures 6C and 6D). The RNAi seedlings showed a similar phenotype (Supplemental Figures 13A and 13B). These data suggest an ACC deficiency in sit1-1; this was confirmed by quantification of the ACC content, which was lower in sit1-1 than in DJ (Figure 6E). In contrast, root growth in the SIT1-OE rice plants was hypersensitive to inhibition by 100 μM ACC (Figures 6F and 6G), and the plants had a higher level of ethylene as compared with Jap (Figure 6H), which coincided with the early leaf senescence phenotype (Figure 5B).
Figure 6.
SIT1 Positively Regulates Ethylene Synthesis.
(A) and (B) Sit1-1 seedlings show reduced sensitivity to ACC inhibition. Three-day-old dark-grown seedlings were transferred to a hydroponic solution containing the indicated concentrations of ACC and allowed to grow for another 7 d in the dark (A), then the root length was measured (B).
(C) and (D) Seven-day-old light-grown sit1-1 seedlings displayed longer shoots than wild-type plants when grown on 0.5× MS agar medium containing 100 μM ACC, as measured in (D).
(E) The ACC contents in 10-d-old rice roots were measured using HPLC–electrospray ionization–tandem mass spectrometry.
(F) and (G) SIT1-OE rice roots showed increased sensitivity to ACC inhibition. Seven-day-old seedlings were transferred to 0.5× MS agar medium supplemented with or without 100 μM ACC and allowed to grow for another 10 d (F), then the root length was measured (G).
(H) The ethylene content of 7-d-old rice seedlings as measured using gas chromatography.
(I) and (J) Arabidopsis roots overexpressing SIT1 were more sensitive to growth promotion by a low concentration of AVG, and less sensitive to growth inhibition by a high concentration of AVG, as measured in (J). Seven-day-old seedlings grown on medium with the indicated concentrations of AVG were photographed.
(K) The ACC contents in 10-d-old Arabidopsis seedlings were measured by mass spectrometry. All data were analyzed by Student’s t test. Error bars indicate the means ± se of three biological repeats. n = 30/genotype in (B), (D), (G), and (J). *P < 0.05 and **P < 0.01.
Consistent with this, the SIT1-OE Arabidopsis plants possessed a higher ACC content (Figure 6K) and shorter roots than Col when grown on 0.5× MS medium. This root growth inhibition was suppressed by exposure to 0.1 μM aminoethoxyvinylglycine (AVG; an ACC synthase inhibitor), and the growth was stimulated due to reduced endogenous ethylene. Increasing the AVG concentration to further reduce the ethylene content resulted in root growth inhibition, but to a lesser extent in SIT1-OE seedlings due to their higher level of ACC compared with Col (Figures 6I and 6J). These data indicate that SIT1 positively regulates ethylene production.
SIT1-Induced ROS Accumulation Requires Ethylene and Ethylene Signaling
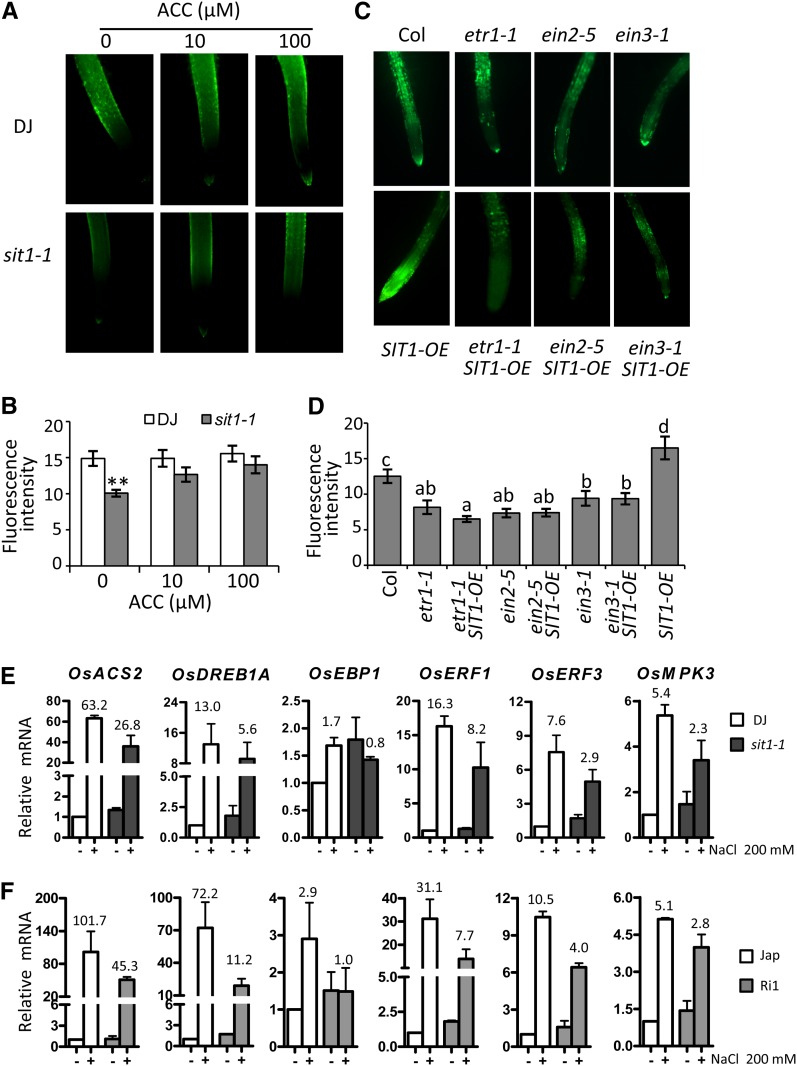
It has been reported that ethylene mediates plant stress responses by regulating ROS production (Mergemann and Sauter, 2000; Jung et al., 2009; Steffens and Sauter, 2009). To determine whether the reduced ROS level in sit1-1 rice is due to an ethylene deficiency, we added increasing amounts of ACC to the medium and found that the low level of ROS in sit1-1 roots gradually recovered, to a level near that seen in DJ exposed to 100 μM ACC (Figures 7A and 7B), suggesting that SIT1 promotes ROS production in an ethylene-dependent fashion. To assess whether ethylene signaling is essential for this process, we crossed SIT1-OE Arabidopsis with ethylene-insensitive mutants (etr1-1, ein2-5, and ein3-1). We found that the increased ROS level in SIT1-OE roots was abolished in etr1-1SIT1-OE, ein2-5SIT1-OE, and ein3-1SIT1-OE double homozygotes, in which the ROS level was similar to that in the respective single mutants (Figures 7C and 7D). These results demonstrate that SIT1-promoted ROS accumulation requires ethylene and ethylene signaling. We next detected salt-induced ethylene-responsive genes in rice and found that their expression was dramatically induced by NaCl in DJ and Jap, but to a lesser extent in sit1-1 and RNAi roots (Figures 7E and 7F), supporting our hypothesis that SIT1 is involved in salt-induced ethylene signaling.
Figure 7.
SIT1 Enhances ROS Production Depending on Ethylene and Ethylene Signaling.
(A) and (B) The reduced ROS in sit1-1 roots was partially recovered by exogenous ACC, as shown in (B).
(C) and (D) The increased ROS level in SIT1-OE Arabidopsis roots was abolished in crosses with ethylene signaling mutants, as shown in (D). The fluorescence intensity of CM-H2DCFDA staining represents the ROS level. Error bars indicate the mean ± se of three biological repeats (n ≥ 15). a to d indicate significant differences.
(E) and (F) The expression of salt-induced ethylene-responsive genes was decreased in sit1-1 (E) and RNAi (F) roots. Error bars indicate the mean ± se of three biological repeats. **P < 0.01. The numbers at the top indicate the fold change of mRNA level after salt challenge.
DISCUSSION
SIT1 Mediates NaCl Sensitivity through Kinase Activation
Many crops are sensitive to soil salinity. The occurrence of ion toxicity, osmotic stress, and oxidative damage in plants caused by exposure to salt involves several rounds of signaling events. The first round of signaling starts with the triggering of membrane sensors by environmental stimuli, which relay the signal through second messengers or a protein phosphorylation cascade, and ends with the transcriptional activation of primary responsive genes. Stress-induced hormones such as ABA and ethylene, in turn, initiate a second round of signaling events that propagate to other tissues (Xiong et al., 2002). Consequent alterations in enzyme activity and metabolites result in a disturbed redox balance, osmotic balance, and ion homeostasis, leading to growth inhibition. Based on our results, we propose that the RLK SIT1 is a primary target affected by high salinity or that it mediates salt stress signaling. SIT1 is preferentially expressed in root epidermal cells and is a membrane-localized RLK (Supplemental Figures 4C and 4F). The L-lectin domain in its extracellular region is predicted to bind monosaccharides and polypeptides, and it is regarded as a potential linker of the plasma membrane to the cell wall (André et al., 2005; Gouget et al., 2006). The rapid activation of SIT1 by NaCl treatment (Figure 3J) suggests that Na+ causes SIT1 activation directly or that its extracellular domain senses a signal released by Na+ from the cell wall. These features render SIT1 able to directly sense high salinity in soil. This is supported by the decreased salt sensitivity observed when SIT1 was knocked out or knocked down (Figures 1 and 2; Supplemental Figure 2) and the increased salt sensitivity observed when SIT1 was overexpressed (Figure 3D). Furthermore, SIT1 likely transmits the salt stress signal to downstream effectors since its kinase activity is required for salt sensitivity (Figure 3).
SIT1 Relays Salt Stress Signals by Activating MPK3/6
MAPK modules are conserved in eukaryotes and typically consist of MAP3K, MAP2K, and MAPK, which sequentially phosphorylate each other upon activation by external stimuli. In mammals, a cell membrane receptor tyrosine kinase mediates extracellular signaling, such as epidermal growth factor (Oda et al., 2005), and usually adaptors are required to link a membrane sensor to MAPK module activation upon ligand challenge (Morrison and Davis, 2003). Here, we present genetic and biochemical evidence showing that the RLK SIT1 relays salt stress signals by activating MPK3/6 in rice. First, SIT1 is an upstream mediator of salt stress signals. Indeed, a loss of SIT1 impaired salt-induced MPK3/6 activation in rice (Figure 4A), and the overexpression of SIT1 enhanced MPK3/6 phosphorylation (Figure 4B). Arabidopsis SIT3 activation seedlings (sit3-D) also displayed increased MPK3/MPK6 phosphorylation (Supplemental Figure 11D). Second, SIT1 kinase activity is positively correlated with MPK3/6 phosphorylation (Figure 4C), implying that rice MPK3/6 are direct targets of SIT1. This is supported by evidence showing that SIT1 can interact with rice MPK3/6 in vivo and in vitro and phosphorylate MPK3/6 in vitro (Figures 4D to 4G). Finally, SIT1-mediated salt sensitivity is dependent mainly on MPK6 and to a lesser extent on MPK3 (Figures 4H to 4K).
MPK3/6 are activated in response to multiple external stimuli, such as flg22 challenge (Asai et al., 2002), cold (Teige et al., 2004), heat (Evrard et al., 2013) and salt (Yu et al., 2010) stress in Arabidopsis, and salt, drought and cold stress in rice (Xiong and Yang, 2003). In addition to phosphorylation by upstream MKKs, MPKs are also regulated by crosstalk with other kinases (Moon et al., 2003; Ludwig et al., 2005; Takahashi et al., 2011). Our results suggest that once rice SIT1 is activated by high salinity, it phosphorylates MPK3/6, resulting in salt sensitivity. This is consistent with a report showing that increasing MPK3/6 activities by MKK9, an upstream activator, enhanced salt sensitivity (Xu et al., 2008). However, the findings of Teige et al. (2004) seem to contradict to our results. They reported that active MKK2, another upstream activator of MPK6, slightly reduced salt sensitivity when overexpressed and enhanced salt sensitivity when knocked out. This discrepancy could be due to the different downstream MAPKs that are targeted. MKK2 mainly acts on MPK4 and to lesser extent on MPK6, while SIT1 and MKK9 target MPK3/6 and function mainly through MPK6 (Figures 4H to 4K) (Xu et al., 2008). Therefore, different outcomes could be expected.
The substrates of MAPKs include transcription factors, transporters, and enzymes, which modulate multiple aspects of cellular physiology upon activation by MAPKs (MAPK Group, 2002; Liu and Zhang, 2004; Asai et al., 2008; Yoo et al., 2008), and the signaling specificity of MAPKs is maintained by spatiotemporal constraints and dynamic protein–protein interactions (Rodriguez et al., 2010). The notion that salt-induced SIT1 phosphorylation and MPK3/6 phosphorylation have very similar kinetics (Figures 3J and 4A) suggests the direct activation of MPK3/6 by SIT1. This may represent a rapid response of plant cells to salt stress that triggers a transient signaling or juxtamembranal event. The findings that constitutive expression of cotton (Gossypium hirsutum) MPK2 in tobacco (Zhang et al., 2011) and rice MPK5 (Xiong and Yang, 2003) in rice enhanced the drought and salt tolerance of transgenic plants may reflect spatial and temporal alterations in MAPK functions.
SIT1 Mediates Salt Sensitivity by Affecting Ethylene and ROS Homeostasis
Ethylene homeostasis is tightly controlled to maintain its dual functions in growth inhibition and growth stimulation. Under normal conditions, the ethylene concentration is low and only increased dramatically at defined developmental stages such as fruit ripening or during plant senescence (Lin et al., 2009). A variety of environmental stimuli, including pathogen exposure and salt stress, can induce ethylene production (Achard et al., 2006), in which the ACC synthase (ACS)–induced conversion of S-adenosyl methionine to ACC is a key step (Lin et al., 2009). However, the mechanism by which salt stress triggers this process remains elusive. We found that salt-induced ethylene production and subsequent ethylene signaling were partially SIT1 dependent. First, SIT1 positively regulates ACC synthesis. The ACC deficiency of sit1-1 rice (Figure 6E) led to delayed maturation at the harvesting stage (Supplemental Figure 12A), whereas overproduced ethylene or ACC in SIT1-OE rice or Arabidopsis (Figures 6H and 6K) led to early leaf senescence and growth inhibition (Figures 5A, 5B, and 6I; Supplemental Figure 12B; 0 μM AVG). Second, salt stress triggers a SIT1-MPK3/6 cascade (Figures 3J and 4A), and activated MPK3/MPK6 were reported to interact with and phosphorylate ACS, leading to stabilization and activation of the enzyme (Liu and Zhang, 2004). Third, a loss of SIT1 impaired salt-induced ethylene signaling, as evidenced by the significant decrease in fold changes of salt-induced ethylene-responsive genes in sit1-1 and RNAi rice plants, compared with wild-type DJ and Jap, respectively (Figures 7E and 7F). One such gene is rice ACS2, indicating that SIT1 also upregulates ACS gene expression in the presence of salt. In agreement with our results, active MKK9 also enhances ethylene production and upregulates ACS2 and ACS6 mainly through MPK6 and to a lesser extent through MAPK3 (Xu et al., 2008). MKK9-MPK6 activation causes premature senescence in whole Arabidopsis plants (Zhou et al., 2009). Since there are so many similarities between the rice SIT1-MPK3/6 cascade and Arabidopsis MKK9-MPK3/6 cascade, it would be interesting to examine whether or not SIT1 is involved in the rice MKK9-MPK3/6 phosphorylation cascade.
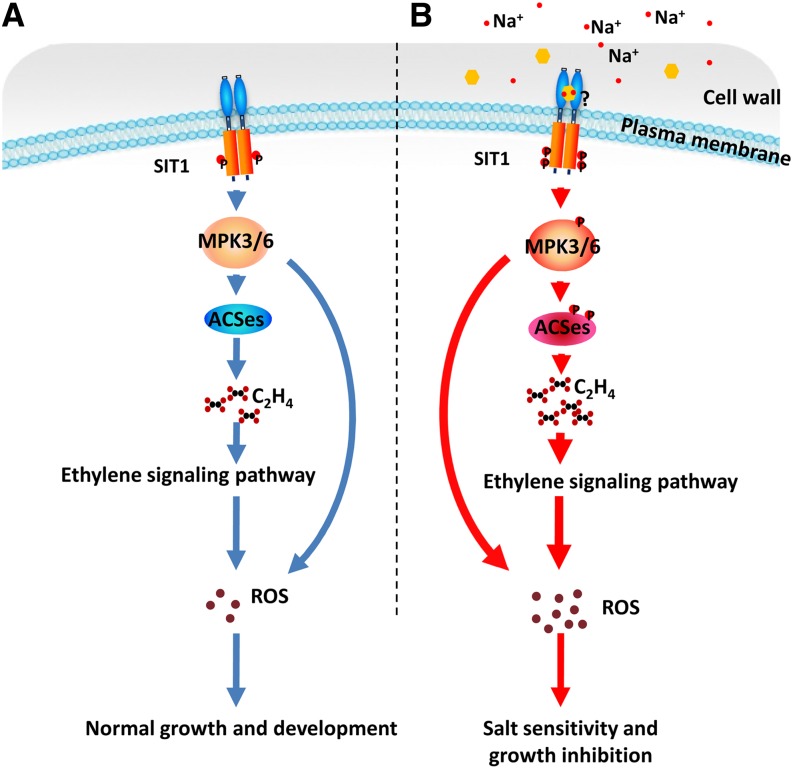
Salt stress induces the production of ROS, which either act as secondary messengers or cause oxidative damage to cellular organelles at high levels (Møller and Sweetlove, 2010). MAPKs, by activating NADPH (Asai et al., 2008), and ethylene, which is induced by stress and serves as a secondary signal, both promote ROS production (Mergemann and Sauter, 2000; Jung et al., 2009; Steffens and Sauter, 2009). These data are in accordance with our findings showing the reduced ROS level in mpk6 (Figure 5F) and the increased ROS level in ACC-treated sit1-1 (Figure 7A) and in Arabidopsis sit3-D mutant plants, which possess increased levels of MPK3/6 activity (Supplemental Figures 11D and 11E). Our genetic data further demonstrate that SIT1-promoted ROS accumulation is MPK3/6, ethylene, and ethylene signaling dependent (Figures 5F to 5H and 7A to 7D). Taken together, our results indicate that the SIT1-MPK3/6 phosphorylation cascade mediates salt sensitivity, at least in part, by affecting ethylene and ROS homeostasis, as summarized in Figure 8. Under normal conditions, SIT1 is less active and it maintains ethylene and ROS at moderate levels. This is essential for plant growth and development; exposure to high salinity activates SIT1 and induces SIT1 expression. MPK3/6 activation by SIT1 in turn promotes ROS or ethylene production by enhancing ACS function. Once initiated, ethylene signaling causes ROS overaccumulation, leading to growth inhibition and potentially plant death through oxidative damage.
Figure 8.
The SIT1-MPK3/6 Phosphorylation Cascade Mediates Salt Sensitivity in Rice.
(A) Under normal conditions, active SIT1, which is essential for rice growth and development, is very limited. Consequently, low levels of MAPK phosphorylation allow plants to maintain moderate levels of ethylene and ROS.
(B) During salt exposure, SIT1 is greatly activated, probably by a signaling molecule released by Na+ from the cell wall. Consequently, the MAPKs that have been activated by SIT1 promote ethylene and ROS overproduction, thereby inhibiting growth and even causing plant death due to oxidative stress.
Soil salinity is a serious threat to crop growth and agricultural productivity. Our findings demonstrate that salt sensitivity in rice is partly due to the salt-induced activation of SIT1. We noted that a low-level SIT1 is required for growth and development and that transient activation of SIT1 by salt enhanced the expression of stress resistance genes such as DREB1A (Dubouzet et al., 2003) (Figures 7E and 7F) but that constitutive expression or the knockout of SIT1 had negative effects on either salt tolerance or plant productivity. Therefore, the activity of SIT1 must be tightly controlled. These findings provide important new information for engineering salt-tolerant crops.
METHODS
Plant Materials and Growth Conditions
Two-week-old rice seedlings (Oryza sativa cv Nipponbare) were treated with 200 mM NaCl for various time periods and then subjected to a real-time PCR analysis (TaKaRa DRR041A) to examine the gene expression of RLKs selected based on chip data. Seven RLKs, including Os03g08550, Os09g38850, Os09g02250, Os04g51040, Os02g02120, and SIT1, were found to be induced by at least 3-fold (primers shown in Supplemental Table 1). RNAi constructs for these genes were made and RNAi plants were produced (primers shown in Supplemental Table 1). T2 seedlings were used to investigate the survival rate in the presence of NaCl. At least 20 individual RNAi lines were used. All seeds were germinated in water for 2 d, grown on a hydroponic culture solution (Ren et al., 2005) for 10 d, and treated with 150 mM NaCl-containing culture solution for another 4 d. After 8 d of recovery, the number of dead seedlings was determined. Homozygous sit1-1 (PFG_3A-09021.R) and sit2-D (PFG_3A-01613.R) T-DNA insertion mutants, produced in the Dongjin (DJ) background, were used. Seedlings were grown on 0.5× MS medium with or without NaCl at 25 and 28°C, respectively, in a light chamber under a 14-h/10-h light-dark cycle. Arabidopsis thaliana seeds were sown on 0.5× MS medium with or without 100 mM NaCl, kept for 2 d in the dark at 4°C, grown under a 16-h/8-h photoperiod at 23°C for 15 d, and then observed to determine the phenotype. All the experiments were repeated at least three times.
ROS Determination
For 3,3′-diaminobenzidine (Sigma-Aldrich) staining, 7-d-old rice leaves were used as described previously (Asano et al., 2012). For staining with 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA; Life Technologies), 5-d-old rice or Arabidopsis roots were used after 12 h incubation of seedlings in a media with or without 100 mM NaCl, as described previously (Borsani et al., 2005). The fluorescent intensity was determined with an Axio Imager (M2; Carl Zeiss); all pictures were analyzed using ImageJ 1.32j. For the quantitative measurement of H2O2, extracts from 5-d-old salt-treated or control seedlings using 20 mM sodium phosphate buffer (pH 6.5) (Shin and Schachtman, 2004) were analyzed with Amplex Red Hydrogen Peroxide/Peroxidase Assay Kit (Molecular Probes/Invitrogen) following the manufacturer’s instructions.
ACC and Ethylene Measurement
Roots of rice plants or whole Arabidopsis seedlings were harvested and ground in liquid nitrogen, then subjected to ACC measurement by HPLC–electrospray ionization–tandem mass spectrometry as described (Petritis et al., 2000). For ethylene measurement, 7-d-old wild-type (japonica) and SIT1-OE rice seedlings, three in each group, were placed in a 15-mL vial containing 3 mL of 0.5× MS medium and closed with a rubber cap on the top; the tubes were then allowed to sit for 24 h (16 h light/8 h dark) at 28°C. A total of 1 mL of the air from the headspace of each vial was taken to inject into the gas chromatograph.
Kinase Assay
The kinase domains of SIT1, SIT1KE, and SIT1DA expressed in Escherichia coli as GST fusions using pDEST15 (Invitrogen) were purified with glutathione agarose beads (GE Healthcare) and subjected to an in vitro kinase assay as described previously (Pu et al., 2012).
For in vitro phosphorylation assay of rice MPK3 and MPK6 by recombinant SIT1-KD, 1 μg of MBP-MPK3 and MBP-MPK6 fusion proteins expressed in E. coli were purified using amylose agarose beads (New England Biolabs) and incubated with 1 or 0.5 μg of GST-SIT1KD (Figures 4F and 4G) or with 2 μg of GST-SIT1KDKE (Supplemental Figure 7) in kinase reaction buffer (50 mM HEPES, 10 mM MgCl2, 5 mM MnCl2, and 1 mM ATP) at 30°C for 2 h. One of the reactions was supplemented with calf intestinal alkaline phosphatase (CIP) (M0290S; New England Biolabs) and incubated for 1 h at 37°C. The mixture was separated by 10% SDS-PAGE, and the blot was probed with anti-phospho-p44/42 MAPK antibodies (9101; Cell Signaling Technology) to detect the phosphorylated MAPKs and anti-MBP-HRP (E8038S; New England Biolabs) and anti-GST-HRP antibodies (HuaAn Biotechnology) to detect MBP-OsMPK3/6 and GST-SIT1-KD/GST-SIT1-KDKE, respectively. For the in vivo kinase assay, Arabidopsis seedlings expressing SIT1-Myc were treated with NaCl for the indicated time period. Next, the microsomes were collected, resuspended in NEB buffer (20 mM HEPES, 40 mM KCl, 250 mM sucrose, 1 mM PMSF, 1 mM DTT, and 1 mM sodium vanadate, pH 7.5), and ultrasonicated. The protein was solubilized with 0.1% Triton X-100, incubated with anti-cMyc agarose (Sigma-Aldrich) for 30 min, and then washed with 10 volumes of NEB buffer containing 0.1% Triton X-100. SIT1-Myc protein bound to the beads was subjected to a kinase assay using MBP (New England Biolabs) as the substrate. Each reaction contained 1 μg of fusion protein or 10 μL of beads in 50 mM HEPES-KOH (pH 7.5), 10 mM MgCl2, 5 mM MnCl2, 1 mM DTT, 0.1 mM ATP, 10 μCi of [γ-32P]ATP, and 3 μg of MBP. The mixture was allowed to react at 30°C for 30 min. Radioactive signals were detected using a phosphor imager. An MBP phosphorylation assay was conducted using anti-pMBP antibodies (05-429; Millipore).
Coimmunoprecipitation Assays
Coimmunoprecipitation were performed as described previously (Kim et al., 2011). Total protein was extracted from the leaves of 30-d-old 35S-SIT1-7MYC6HIS transgenic rice plants (Japonica) using NEB buffer (20 mM HEPES, pH 7.5, 40 mM KCl, and 1 mM EDTA) by centrifugation at 20,000g, 4°C for 20 min. A total of 10 μL of each supernatant was incubated with protein A beads (17-0780-01; GE Healthcare) bound to rice MPK3 or MPK6 antibodies (AbP80140-A-SE and AbP80147-A-SE; Beijing Protein Innovation) or with regular protein A beads. After 2 h, the beads were washed six times in wash buffer (20 mM HEPES, pH 7.5, 40 mM KCl, and 0.1% Triton X-100). An appropriate amount of 2× SDS sample buffer was then added to the beads, which were boiled for 10 min at 100°C. The proteins were then separated by 10% SDS-PAGE and detected by immunoblotting using anti-cMYC (SAB4700447; Sigma-Aldrich) and anti-Os-MPK3/6 antibodies (AbP80140-A-SE and AbP80147-A-SE; Beijing Protein Innovation).
Gel Blot Overlay Assay
The gel blot (polyvinylidene fluoride filter) containing MBP-Os-MPK3, MBP-Os-MPK6, and MBP was incubated with 2 μg of GST-SIT1-KD or GST-BZR1-N (BZR1 N-terminal 1 to 99 amino acids) as a control, followed by anti-GST-HRP (HuaAn Biotechnology).
Accession Numbers
Sequence data from this article can be found in the Rice and Arabidopsis Database under the following accession numbers: SIT1 (Os02g42780), SIT2 (Os04g44900), SIT3 (Os07g38810), At-SIT3 (At4g02410), Os-MAPK5 (Os03g17700), Os-MAPK1 (Os06g06090), Os-ACS2 (Os04g48850), Os-DREB1A (Os03g50885), Os-EBP1 (Os02g54160), Os-ERF1 (Os04g46220), Os-ERF3 (Os01g58420), Os-MAPK5 (Os03g17700), At-MAPK6 (At2g43790), At-MAPK3 (At3g45640), Os-18s (Os09g00998), Os-Actin (Os03g50890), mpk6-3 (salk-127507), mpk3-1 (salk-151594), Arabidopsis sit3D (salk-079614), sit1-1 (PFG_3A-09021.R), and sit2-1D (PFG_3A-01613.R).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. DNA Gel Blot Assay of SIT1-RNAi Rice Plants.
Supplemental Figure 2. Salt Tolerance Test of SIT1-RNAi Seedlings.
Supplemental Figure 3. Genotyping of the sit1-1 and sit2-D Mutants.
Supplemental Figure 4. Rice Expression Patterns of SIT1 and SIT2.
Supplemental Figure 5. Transgenic Arabidopsis Overexpressing Kinase-Active, but Not Kinase-Dead, SIT1 Exhibited an Obvious Phenotype.
Supplemental Figure 6. Investigation of the Survival Rates of SIT1-OE, SIT1KE-OE and SIT1DA-OE Seedlings on 150 mM NaCl-Containing Medium.
Supplemental Figure 7. SIT1 Phosphorylates Rice MPK3 and MPK6 in Vitro.
Supplemental Figure 8. Phenotypic Comparison of Arabidopsis Mutant mpk6 and mpk3 with the Double Homozygotes mpk6 SIT1-OE and mpk3 SIT1-OE.
Supplemental Figure 9. Anti-Oxidase Activity and ROS Level in the Mutants and Transgenic Plants.
Supplemental Figure 10. Plant Homologs of SIT1.
Supplemental Figure 11. Arabidopsis SIT3-Activation Seedlings with Increased MPK3 and MPK6 Phosphorylation and Higher Level of ROS Are Sensitive to NaCl.
Supplemental Figure 12. Phenotypic Comparisons among Wild-Type, sit1-1 Mutant, and SIT1-OE Plants.
Supplemental Figure 13. SIT1-RNAi Rice Seedlings Showed Reduced Sensitivity to Exogenous ACC.
Supplemental Table 1. Primers for RT-PCR and RNAi Constructs.
Supplemental Table 2. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Zhi-Yong Wang (Carnegie Institution for Science) for providing guidance and editing the article, Chong Kang (Institute of Botany, Chinese Academy of Sciences) for the RNAi construct PTCK303, Gynheung An (Postech Biotech Center, Republic of Korea) for sit1-1 and sit2-D T-DNA insertion seeds, Wen-Hua Zhang (Nanjing Agricultural University) for the mapk6, Jose M. Alonso (North Carolina State University) for the ein2-5, and the ABRC for the etr1-1 and ein3-1 seeds. This study was supported by the National Basic Research Program of China (2012CB114201), the National Science Foundation of China (31000126), and the National Science and Technology Major Project (2009ZX08009-017B and 2012ZX08001003-014).
AUTHOR CONTRIBUTIONS
C.-H.L. and G.W. performed most of the experiments. J.-L.Z. contributed data to Figures 4A, 4J, 4K, 5H, 5I, and Supplemental Figure 8. C.-H.L. and Y.-F.H. generated the RNAi transgenic rice and performed the large-scale screen. L.-F.A. performed the ACC quantification. L.-Q.Z. performed the protein purification. S.-W.Z. and Y.S. conceived of the project, designed the experiments, analyzed the data, and wrote the article together with C.-H.L. D.-Y.S. supervised and complemented the writing.
Glossary
- ROS
reactive oxygen species
- RLK
receptor-like kinase
- LecRLK
lectin RLK
- ABA
abscisic acid
- MAPK
mitogen-activated protein kinase
- ACC
1-aminocyclopropane-1-carboxylate
- RNAi
RNA interference
- MS
Murashige and Skoog
- Col
Columbia
- GR
glutathione reductase
- POD
peroxidase
- AVG
aminoethoxyvinylglycine
- ACS
ACC synthase
- CM-H2DCFDA
5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94. [DOI] [PubMed] [Google Scholar]
- André S., Siebert H.C., Nishiguchi M., Tazaki K., Gabius H.J. (2005). Evidence for lectin activity of a plant receptor-like protein kinase by application of neoglycoproteins and bioinformatic algorithms. Biochim. Biophys. Acta 1725: 222–232. [DOI] [PubMed] [Google Scholar]
- Antolín-Llovera M., Ried M.K., Binder A., Parniske M. (2012). Receptor kinase signaling pathways in plant-microbe interactions. Annu. Rev. Phytopathol. 50: 451–473. [DOI] [PubMed] [Google Scholar]
- Asai S., Ohta K., Yoshioka H. (2008). MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20: 1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T., Tena G., Plotnikova J., Willmann M.R., Chiu W.L., Gomez-Gomez L., Boller T., Ausubel F.M., Sheen J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Asano T., Hayashi N., Kobayashi M., Aoki N., Miyao A., Mitsuhara I., Ichikawa H., Komatsu S., Hirochika H., Kikuchi S., Ohsugi R. (2012). A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 69: 26–36. [DOI] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush S.M., Krysan P.J. (2007). Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. J. Exp. Bot. 58: 2181–2191. [DOI] [PubMed] [Google Scholar]
- de Lorenzo L., Merchan F., Laporte P., Thompson R., Clarke J., Sousa C., Crespi M. (2009). A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. Plant Cell 21: 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diévart A., Clark S.E. (2004). LRR-containing receptors regulating plant development and defense. Development 131: 251–261. [DOI] [PubMed] [Google Scholar]
- Dong H., Zhen Z., Peng J., Chang L., Gong Q., Wang N.N. (2011). Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J. Exp. Bot. 62: 4875–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet J.G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E.G., Miura S., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33: 751–763. [DOI] [PubMed] [Google Scholar]
- Evrard A., Kumar M., Lecourieux D., Lucks J., von Koskull-Döring P., Hirt H. (2013). Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. PeerJ 1: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouget A., Senchou V., Govers F., Sanson A., Barre A., Rougé P., Pont-Lezica R., Canut H. (2006). Lectin receptor kinases participate in protein-protein interactions to mediate plasma membrane-cell wall adhesions in Arabidopsis. Plant Physiol. 140: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.Y., Shin R., Schachtman D.P. (2009). Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiegerl S., Cardinale F., Siligan C., Gross A., Baudouin E., Liwosz A., Eklöf S., Till S., Bögre L., Hirt H., Meskiene I. (2000). SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12: 2247–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Burlingame A.L., Wang Z.Y. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 43: 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Kaboshi M., Okada K., Kurimoto L., Murakami S., Umezawa T., Shibuya N., Yamane H., Miyao A., Takatsuji H., Takahashi A., Hirochika H. (2010). A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 63: 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Zhong S., Grierson D. (2009). Recent advances in ethylene research. J. Exp. Bot. 60: 3311–3336. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig A.A., Saitoh H., Felix G., Freymark G., Miersch O., Wasternack C., Boller T., Jones J.D., Romeis T. (2005). Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proc. Natl. Acad. Sci. USA 102: 10736–10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAPK Group (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7: 301–308. [DOI] [PubMed] [Google Scholar]
- Marshall A., et al. (2012). Tackling drought stress: receptor-like kinases present new approaches. Plant Cell 24: 2262–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Wang H., He Y., Liu Y., Walker J.C., Torii K.U., Zhang S. (2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24: 4948–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergemann H., Sauter M. (2000). Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol. 124: 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Suzuki N., Ciftci-Yilmaz S., Mittler R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33: 453–467. [DOI] [PubMed] [Google Scholar]
- Møller I.M., Sweetlove L.J. (2010). ROS signalling—specificity is required. Trends Plant Sci. 15: 370–374. [DOI] [PubMed] [Google Scholar]
- Moon H., et al. (2003). NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc. Natl. Acad. Sci. USA 100: 358–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D.K., Davis R.J. (2003). Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu. Rev. Cell Dev. Biol. 19: 91–118. [DOI] [PubMed] [Google Scholar]
- Munns R., Tester M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59: 651–681. [DOI] [PubMed] [Google Scholar]
- Oda K., Matsuoka Y., Funahashi A., Kitano H. (2005). A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 1: 0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. (2013). Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J. Exp. Bot. 64: 445–458. [DOI] [PubMed] [Google Scholar]
- Ouyang S.Q., Liu Y.F., Liu P., Lei G., He S.J., Ma B., Zhang W.K., Zhang J.S., Chen S.Y. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62: 316–329. [DOI] [PubMed] [Google Scholar]
- Petritis K., Dourtoglou V., Elfakir C., Dreux M. (2000). Determination of 1-aminocyclopropane-1-carboxylic acid and its structural analogue by liquid chromatography and ion spray tandem mass spectrometry. J. Chromatogr. A 896: 335–341. [DOI] [PubMed] [Google Scholar]
- Pitzschke A., Djamei A., Bitton F., Hirt H. (2009). A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant 2: 120–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu C.X., Ma Y., Wang J., Zhang Y.C., Jiao X.W., Hu Y.H., Wang L.L., Zhu Z.G., Sun D., Sun Y. (2012). Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma, and fertility in rice, by promoting epidermal cell differentiation. Plant J. 70: 940–953. [DOI] [PubMed] [Google Scholar]
- Qiu Q.S., Guo Y., Dietrich M.A., Schumaker K.S., Zhu J.K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99: 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z.H., Gao J.P., Li L.G., Cai X.L., Huang W., Chao D.Y., Zhu M.Z., Wang Z.Y., Luan S., Lin H.X. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37: 1141–1146. [DOI] [PubMed] [Google Scholar]
- Rodriguez M.C., Petersen M., Mundy J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61: 621–649. [DOI] [PubMed] [Google Scholar]
- Shen X., Yuan B., Liu H., Li X., Xu C., Wang S. (2010). Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J. 64: 86–99. [DOI] [PubMed] [Google Scholar]
- Shin R., Schachtman D.P. (2004). Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc. Natl. Acad. Sci. USA 101: 8827–8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.H., Mayer K.F., Li W.H. (2004). Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., et al. (2012). Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiol. 160: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens B., Sauter M. (2009). Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell 21: 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F., Mizoguchi T., Yoshida R., Ichimura K., Shinozaki K. (2011). Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol. Cell 41: 649–660. [DOI] [PubMed] [Google Scholar]
- Teige M., Scheikl E., Eulgem T., Dóczi R., Ichimura K., Shinozaki K., Dangl J.L., Hirt H. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 15: 141–152. [DOI] [PubMed] [Google Scholar]
- Tuteja N. (2007). Mechanisms of high salinity tolerance in plants. Methods Enzymol. 428: 419–438. [DOI] [PubMed] [Google Scholar]
- Vaid N., Macovei A., Tuteja N. (2013). Knights in action: lectin receptor-like kinases in plant development and stress responses. Mol. Plant 6: 1405–1418. [DOI] [PubMed] [Google Scholar]
- Vaid N., Pandey P.K., Tuteja N. (2012). Genome-wide analysis of lectin receptor-like kinase family from Arabidopsis and rice. Plant Mol. Biol. 80: 365–388. [DOI] [PubMed] [Google Scholar]
- Wang K.L., Li H., Ecker J.R. (2002a). Ethylene biosynthesis and signaling networks. Plant Cell 14 (suppl.): S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Chen C.B., Xu Y.Y., Jiang R.X., Han Y., Xu Z.H., Chong K. (2004). A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 22: 409–417. [Google Scholar]
- Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002b). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2: 505–513. [DOI] [PubMed] [Google Scholar]
- Wrzaczek M., Hirt H. (2001). Plant MAP kinase pathways: how many and what for? Biol. Cell 93: 81–87. [DOI] [PubMed] [Google Scholar]
- Xiong L., Yang Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15: 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Schumaker K.S., Zhu J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (suppl.): S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Li Y., Wang Y., Liu H., Lei L., Yang H., Liu G., Ren D. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283: 26996–27006. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451: 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Nie J., Cao C., Jin Y., Yan M., Wang F., Liu J., Xiao Y., Liang Y., Zhang W. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188: 762–773. [DOI] [PubMed] [Google Scholar]
- Zhang L., Xi D., Li S., Gao Z., Zhao S., Shi J., Wu C., Guo X. (2011). A cotton group C MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant Mol. Biol. 77: 17–31. [DOI] [PubMed] [Google Scholar]
- Zhou C., Cai Z., Guo Y., Gan S. (2009). An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol. 150: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.