Active DNA demethylation, which represses gene silencing, is initiated by DNA demethylation enzymes such as REPRESSOR OF SILENCING1 (ROS1), but the regulation of these enzymes is poorly understood. This work shows that ROS5 regulates DNA demethylation by interacting with the histone acetyltransferase IDM1, creating a chromatin environment that facilitates ROS1 binding to erase DNA methylation.
Abstract
In Arabidopsis thaliana, active DNA demethylation is initiated by the DNA glycosylase REPRESSOR OF SILENCING1 (ROS1) and its paralogs DEMETER, DEMETER-LIKE2 (DML2), and DML3. How these demethylation enzymes are regulated, however, is poorly understood. Here, using a transgenic Arabidopsis line harboring the stress-inducible RESPONSIVE TO DEHYDRATION29A (RD29A) promoter–LUCIFERASE (LUC) reporter gene and the cauliflower mosaic virus 35S promoter (35S)–NEOMYCIN PHOSPHOTRANSFERASE II (NPTII) antibiotic resistance marker gene, we characterize a ROS locus, ROS5, that encodes a protein in the small heat shock protein family. ROS5 mutations lead to the silencing of the 35S-NPTII transgene due to DNA hypermethylation but do not affect the expression of the RD29A-LUC transgene. ROS5 physically interacts with the histone acetyltransferase ROS4/INCREASED DNA METHYLATION1 (IDM1) and is required to prevent the DNA hypermethylation of some genes that are also regulated by ROS1 and IDM1. We propose that ROS5 regulates DNA demethylation by interacting with IDM1, thereby creating a chromatin environment that facilitates the binding of ROS1 to erase DNA methylation.
INTRODUCTION
Transcriptional gene silencing is regulated by both DNA cytosine methylation and histone modifications in plants and mammals. In mammals, DNA methylation is mainly found at CG sites. By contrast, DNA methylation in plants occurs in any cytosine contexts, including CG, CNG, and CNN (where N is A, T, or C) (Law and Jacobsen, 2010). DNA METHYLTRANSFERASE1 is responsible for maintaining CG methylation, and CHROMOMETHLASE3 (CMT3) and CMT2 are responsible for maintaining CNG and CNN methylation, respectively (Lindroth et al., 2001; Jackson et al., 2002; Ebbs and Bender, 2006; Zemach et al., 2013). The RNA-directed DNA methylation (RdDM) pathway, which involves RNA polymerases IV and V, scaffold RNAs, 24-nucleotide small interfering RNAs, the methyltransferase DOMAINS REARRANGED METHYLASE2, and other proteins, is crucial for establishing and maintaining DNA methylation in short transposons and DNA repeat regions and along the edges of long transposons (Law and Jacobsen, 2010; Zemach et al., 2013). The chromatin remodeler DECREASE IN DNA METHYLATION1 (DDM1) is also vital to the maintenance of CG and non-CG methylation in the middle of long transposons and other DNA repeat regions (Vongs et al., 1993; Jeddeloh et al., 1999; Zemach et al., 2013).
DNA methylation can be actively erased through a base excision repair pathway initiated by the DNA glycosylase REPRESSOR OF SILENCING1 (ROS1) and its paralogs DEMETER (DME), DEMETER-LIKE2 (DML2), and DML3 (Zhu, 2009). ROS1 was first identified during a forward genetic screen for mutants with enhanced silencing of the stress-inducible RESPONSIVE TO DEHYDRATION29A (RD29A) promoter–LUCIFERASE (LUC) reporter gene and the cauliflower mosaic virus 35S promoter (35S)–NEOMYCIN PHOSPHOTRANSFERASE II (NPTII) antibiotic resistance marker gene (Gong et al., 2002). ROS1 mutations cause DNA hypermethylation and enhance the transcriptional gene silencing of many loci (Gong et al., 2002; Zhu et al., 2007). By screening for second site suppressor mutations of the ros1 mutant for the silenced RD29A-LUC, researchers have identified many components in the RdDM pathway (He et al., 2009a, 2009b, 2009c). ROS3, an RNA binding protein, also contributes to active DNA methylation and the prevention of transcriptional gene silencing at the RD29A-LUC and 35S-NPTII transgenes (Zheng et al., 2008). Recently, the DNA phosphatase ZDP was found to act downstream of ROS1 in one branch of the active DNA demethylation pathway (Martínez-Macías et al., 2012). ROS4/INCREASED DNA METHYLATION1 (IDM1) is also involved in the regulation of DNA demethylation (Li et al., 2012a; Qian et al., 2012). IDM1 encodes a histone acetyltransferase that binds methylated DNA at chromatin sites lacking histone H3K4me2 or H3K4me3 and that is able to acetylate H3 at Lys-18 and Lys-23, likely creating a chromatin environment that facilitates ROS1 function (Li et al., 2012a; Qian et al., 2012).
Although the RD29A-LUC transgenic line shows kanamycin resistance, the ddm1 mutation greatly increases 35S-NPTII expression (Li et al., 2012a), suggesting that both silencing and antisilencing factors contribute to the regulation of 35S-NPTII expression. To find additional players that have antisilencing roles, we previously performed a forward genetic screen to identify kanamycin-sensitive mutants in Arabidopsis thaliana. This screen led to the identification of ROS4/IDM1, several alleles of ROS1, and multiple components of the RdDM pathway (Li et al., 2012a). The RdDM pathway prevents the silencing of 35S-NPTII by positively regulating ROS1 expression (Li et al., 2012a). In this study, we characterized another ROS gene in Arabidopsis, ROS5, which encodes a member of the small heat shock–like protein (sHSP) family. However, ROS5 expression is not induced by heat shock and ROS5 does not function in response to heat stress. Our results indicate that ROS5 prevents DNA hypermethylation and thereby plays crucial antisilencing roles in regulating some genes.
RESULTS
ROS5 Encodes a sHSP Localized in the Nucleus
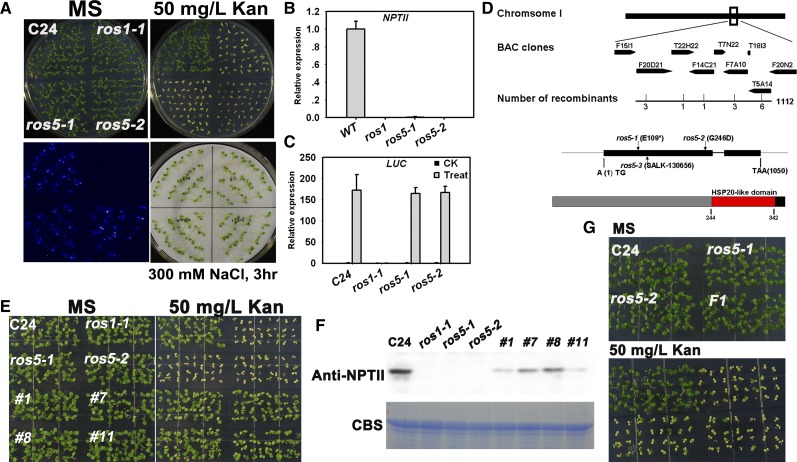
In this study, we isolated two kanamycin-sensitive mutant alleles, ros5-1 and ros5-2, from an ethyl methanesulfonate–mutagenized population of the C24 transgenic Arabidopsis line that carries the RD29A-LUC and 35S-NPTII transgenes (Li et al., 2012a). Both mutants were kanamycin sensitive and had a substantially reduced expression of NPTII, but the expression of the RD29A-LUC transgene or the endogenous RD29A gene was similar to that in the wild type. As a control, the ros1 mutation reduced the expression of both RD29A-LUC and 35S-NPTII (Figures 1A to 1C). Both the ros5-1 mutant and the ros5-1 ros1-1 double mutant did not exhibit any clear defects in growth and development in a growth room (Supplemental Figure 1). These results indicate that the ROS5 mutations lead to the silencing of 35S-NPTII but not RD29A-LUC.
Figure 1.
Identification and Characterization of the ros5 Mutants.
(A) The ros5 mutations lead to the silencing of 35S-NPTII but not to the silencing of the RD29A-LUC transgene. Seedlings grown on MS plates were subjected to luminescence imaging after treatment with 300 mM NaCl for 3 h. The plants were also grown on MS medium containing 50 mg/L kanamycin (Kan) and photographed after 7 d.
(B) Real-time PCR analysis of transcripts of NPTII in C24, ros1-1, ros5-1, and ros5-2. Three independent experiments were done with similar results, each with three technical replicates. One representative experiment is shown. Error bars represent se (n = 3).
(C) Real-time PCR analysis of transcripts of LUC in C24, ros1-1, ros5-1, and ros5-2 after treatment with 300 mM NaCl for 3 h. Three independent experiments were done with similar results, each with three technical replicates. One representative experiment is shown. Error bars represent se (n = 3).
(D) Diagram of the region identified by map-based cloning for ros5 mutations. In the identified region, a C-to-T mutation changes Glu to a stop codon in ros5-1 and a G-to-A mutation changes Gly to Asp in ros5-2. A T-DNA is inserted at position 20,453,546 of chromosome 1 in line SALK_130656 (ros5-3). The protein was analyzed at http://prosite.expasy.org. A conserved HSP20-like domain was found in the C-terminal region of ROS5.
(E) The kanamycin-sensitive phenotype is complemented by the ROS5 transgene in the ros5 mutants.
(F) Analysis of NPTII expression in complemented ros5 plants by protein gel blot analysis. A Coomassie blue–stained (CBS) gel was used as a loading control.
(G) Genetic analysis of the two ros5 alleles. The F1 seedlings of ros5-1 crossed with ros5-2 are sensitive on MS medium supplemented with 50 mg/L kanamycin.
We subjected the ROS5 gene to map-based cloning. We mapped the ros5-2 mutation to a region on the bottom of chromosome 1 and then narrowed the mutation to a small region between the BAC clones T22H22 and F14C21 (Figure 1D). We sequenced candidate genes in this region and found a G-to-A mutation, which changes the original amino acid Gly-246 to Asp in the first exon of AT1G54840 (Figure 1D). Another mutant allele, ros5-1, contains a C-to-T mutation, which changes the original amino acid Glu-109 to a stop codon in AT1G54840 (Figure 1D). To further confirm that AT1G54840 is the ROS5 gene, we cloned the wild-type genomic sequence of AT1G54840 and introduced it into the ros5-1 and ros5-2 mutants for a complementation assay. Four randomly selected transgenic lines were resistant to kanamycin and highly expressed the NPTII protein (Figures 1E and 1F). Additionally, we crossed ros5-1 with ros5-2 and found that the seedlings of the F1 generation were kanamycin sensitive (Figure 1G). These results confirmed that AT1G54840 is the ROS5 gene.
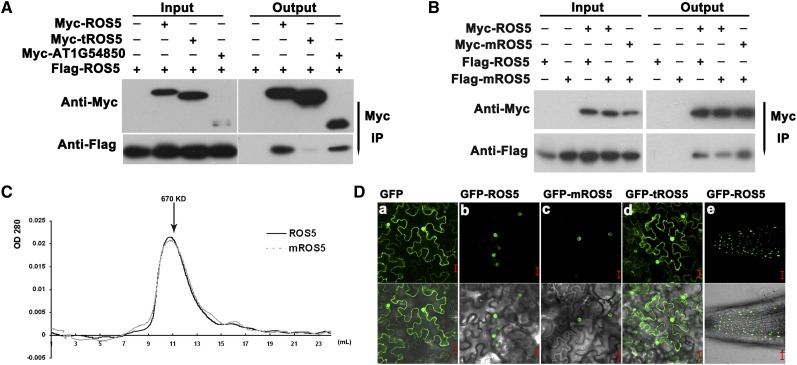
ROS5 encodes a putative protein of 349 amino acids with a conserved α-crystallin domain (ACD), which is found in most sHSP families (Supplemental Figure 2) (Scharf et al., 2001). The ACD contributes to the formation of homodimers and oligomers (Baranova et al., 2011). A coimmunoprecipitation assay using transiently expressed Myc-ROS5 and Flag-ROS5 in Arabidopsis protoplasts indicated that Myc epitope–tagged ROS5 coimmunoprecipitated Flag epitope–tagged ROS5 (Figure 2A). However, Myc epitope–tagged ROS5 without the ACD (Myc-tROS5) failed to coimmunoprecipitate Flag epitope–tagged ROS5 (Figure 2A), suggesting that the ACD plays a vital role in the formation of the ROS5 dimer/oligomer. ros5-2 carries a point mutation, which is conserved at the edge of the ACD (Figure 1D; Supplemental Figure 2). We wanted to know whether this mutation impairs the formation of the ROS5 dimer/oligomer. The result shows that the Myc epitope–tagged mutated ROS5 (G246D) (mROS5) could still coimmunoprecipitate Flag epitope–tagged mROS5 (Figure 2B). A gel filtration assay showed that both ROS5 and mROS5 formed a complex of ∼670 kD (Figure 2C), indicating that ROS5 may form an ∼16-mer complex and that the G246D mutant does not impair oligomer formation. The ACDs in wheat (Triticum aestivum) HSP16.9, a member of the sHSP family, can be formed as trimers of dimers and can be organized as a dodecamer double ring (van Montfort et al., 2001). Based on all of these results, we inferred that the mutation (G246D) may affect the interaction between ROS5 and other proteins or may affect the chaperone activity but not the oligomer formation of ROS5.
Figure 2.
Characterization of ROS5.
(A) Effect of the ACD on the dimerization of ROS5 as determined by coimmunoprecipitation in a protoplast transient assay. AT1G54850, a close homolog of ROS5, was used as a control. Immunoprecipitation (IP) was conducted with Anti-c-Myc-Agarose Affinity Gel (Sigma-Aldrich).
(B) Effect of the point mutation G246D (mROS5) on ROS5 dimerization as determined by coimmunoprecipitation in a protoplast transient assay. A Myc or Flag tag was fused to the N terminus of ROS5. Samples transformed with only Flag-ROS5 or Flag-mROS5 were used as controls. Immunoprecipitation was conducted with Anti-c-Myc-Agarose Affinity Gel.
(C) Effect of the point mutation G246D on the oligomer formation of ROS5 as determined by gel filtration. Wild-type ROS5 and the point mutation variant mROS5 expressed in Escherichia coli were both assayed at a concentration of 2.0 mg/mL. Protein elution was monitored by A280. Fraction numbers and the relevant molecular mass are shown.
(D) The subcellular localization of GFP-ROS5 (b), GFP-mROS5 (ros5-2) (c), and GFP-tROS5 (ROS5 without ACD) (d) in transient assays in tobacco epidermal cells. Subcellular localization of GFP-ROS5 was also investigated in stable transgenic ros5-1 plants carrying proROS5-GFP-ROS5 (e). GFP itself (a) was used as a control. Bars = 20 μm.
For determination of its subcellular localization, ROS5 was fused to the C terminus of green fluorescent protein (GFP) and stably expressed in Arabidopsis or transiently expressed in tobacco (Nicotiana benthamiana) epidermal cells under the control of a 35S promoter. Confocal microscopy of transgenic roots and of transiently expressed cells indicated that GFP-ROS5 was localized in the nucleus (Figure 2Db). As a control, GFP itself was expressed in the cytoplasm and nucleus (Figure 2Da). Additionally, the mROS5 and truncated ROS5 (tROS5), which lacks the ACD, were also fused to the C terminus of GFP and transiently expressed in tobacco. These results showed that mROS5 is also localized in the nucleus but that the truncated ROS5 is located in both the cytoplasm and nucleus (Figures 2Dc and 2Dd), suggesting that the point mutation (G246D) does not impair the subcellular localization of ROS5 and that the nuclear localization signal may be in the ACD. β‑Glucuronidase (GUS) staining of transgenic seedlings carrying ROS5 promoter–GUS indicated that the expression of ROS5 and ROS4/IDM1 overlap in cotyledons and hypocotyls in young seedlings (Supplemental Figure 3) (Li et al., 2012a).
ROS5 Is Required for Preventing DNA Hypermethylation at the 3′ Region of 35S-NPTII
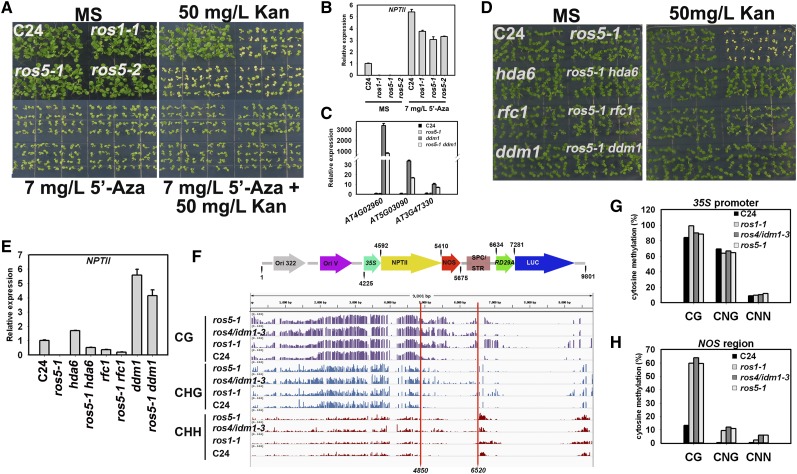
To determine whether 35S-NPTII silencing is related to DNA methylation, we tested the effect of 5′-aza-2′-deoxycytidine (5′-Aza), a cytosine methylation inhibitor, on the silenced 35S-NPTII in ros5. Addition of 5′-Aza to the medium caused ros5 mutants to become resistant to kanamycin (Figure 3A). Real-time PCR showed that the transcripts of NPTII in ros5 mutants were substantially increased after 5′-Aza treatment (Figure 3B). However, the transcript level of NPTII was lower in ros5 mutants than in the wild type and was even lower than in ros1-1 after 5′-Aza treatment, which is similar to the effect of ros4/idm1-3 mutation on 35S-NPTII expression under 5′-Aza treatment (Li et al., 2012a). A previous study of ros4/idm1-3 indicated that IDM1 is required for the expression of endogenous methylated genes when DNA methylation is inhibited by DDM1 mutation (Li et al., 2012a). We further tested whether the endogenous genes targeted by IDM1 can also be influenced by ROS5. We found that the transcript levels of the three IDM1-regulated genes were lower in ros5 ddm1 than in ddm1 (Figure 3C). As later we show that ROS5 physically interacts with IDM1, these results suggest that ROS5 works together with IDM1 to create a friendly chromatin environment that is required for DNA demethylation and is also possibly necessary for the expression of endogenous genes that are not targeted by DNA methylation or after DNA demethylation.
Figure 3.
ROS5 Is Required to Prevent DNA Hypermethylation.
(A) Addition of 7 mg/L 5′-Aza rescues the kanamycin-sensitive phenotype of ros5-1, ros5-2, and ros1-1 mutants on MS medium containing 50 mg/L kanamycin (Kan).
(B) Real-time PCR analysis of NPTII under treatment with 7 mg/L 5′-Aza in ros5-1, ros5-2, and ros1-1 mutants. Two independent experiments (each with three technical replicates) were conducted with similar results, and one representative experiment is shown. Error bars represent se (n = 3).
(C) Real-time PCR analysis of the transcripts of endogenous genes in wild-type C24, ros5-1, ddm1, and ros5-1 ddm1. Three independent experiments were done with similar results, each with three technical replicates. One representative experiment is shown. Error bars represent se (n = 3).
(D) Mutations of HDA6, RFC1, and DDM1 can release the silencing of 35S-NPTII in the ros5-1 mutant. Double mutants of ros5-1 hda6, ros5-1 rfc1, and ros5-1 ddm1 were photographed after growing for 7 d on MS medium containing 50 mg/L kanamycin.
(E) Real-time PCR analysis of NPTII expression in ros5-1 hda6, ros5-1 rfc1, and ros5-1 ddm1. Two independent experiments (each with three technical replicates) were conducted with similar results, and one representative experiment is shown. Error bars represent se (n = 3).
(F) DNA methylation status of the T-DNA region in the wild type, ros1-1, ros4/idm1-3, and ros5-1 as indicated by IGV snapshot. Whole-genome bisulfite sequencing data were mapped to the T-DNA sequence to obtain the methylation information. The region with significant hypermethylation is marked with red lines.
(G) Confirmation of T-DNA DNA methylation in the 35S promoter region by individual-locus bisulfite sequencing.
(H) Confirmation of T-DNA DNA methylation in the NOS terminator region by individual-locus bisulfite sequencing.
The silencing of 35S-NPTII in the ros1-1 mutant can be released by mutations in DDM1, HISTONE DEACETYLASE6 (HDA6), and some core DNA replication proteins (Liu et al., 2010; Li et al., 2012a). We wanted to know whether these ros1-1 suppressors could also suppress the release of silencing of 35S-NPTII by ros5. We generated double mutants of ros5-1 with ddm1, hda6, and dna replication factor c1 (rfc1) and found that ddm1, hda6, and rfc1 released the silencing of 35S-NPTII in ros5-1 (Figure 3D). Real-time PCR indicated that NPTII was strongly expressed in ddm1, moderately expressed in hda6, and weakly expressed in rfc1, but the expression of NPTII was less in the ros5 ddm1, ros5 hda6, and ros5 rfc1 double mutants than in the ddm1, hda6, and rfc1 single mutants (Figure 3E), suggesting that ROS5 has a role in promoting NPTII expression in the ddm1, hda6, and rfc1 mutant backgrounds.
According to whole-genome bisulfite sequencing of the RD29A-LUC transgenic C24 plant, DNA methylation was quite high in the 35S promoter and the 5′ region of NPTII but was low in the 3′ region of NPTII, the NOS terminator, and the RD29A-LUC region (Figure 3F). Mutations in ROS1, IDM1, and ROS5 all led to DNA hypermethylation in the 3′ region of the NPTII coding sequence and the NOS terminator (Figure 3F). However, only ros1-1 caused DNA hypermethylation in the RD29A promoter, which is consistent with previous results (Gong et al., 2002). Because DNA methylation in the 35S promoter is already high, these mutations only moderately increased the 35S DNA methylation level (Figure 3F). Individual-locus bisulfite sequencing in these mutants confirmed a slight increase in DNA methylation in the 35S promoter (Figure 3G) but a big increase in DNA methylation in the NOS terminator (Figure 3H). It seems that DNA methylation in the NOS terminator region is spread from the adjacent heavily methylated 35S promoter and part of the NPTII coding region and that ROS1, IDM1, and ROS5 are required to prevent this spreading. Studies in Arabidopsis and rice (Oryza sativa) suggest that DNA methylation in the 3′ region of the genes is more effective for repressing gene expression than that in the promoter region (Li et al., 2012b).
ROS5 Interacts with IDM1
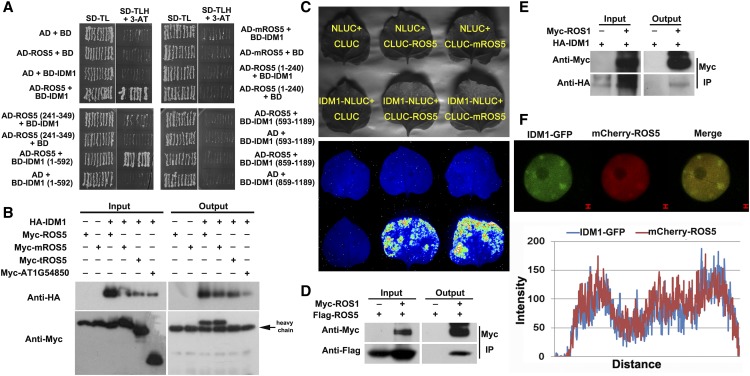
Because IDM1 and ROS5 were isolated from the same genetic screen, we wanted to know whether the ROS5 protein could interact with the IDM1 protein. A yeast two-hybrid assay showed that ROS5 interacted with IDM1 (Figure 4A). ROS5 lacking the N-terminal domain (1 to 240 amino acids) or the ACD (241 to 349 amino acids) could not interact with IDM1 (Figure 4A), suggesting that the complete form of ROS5 is essential for its interaction with IDM1. To investigate which region of IDM1 interacts with ROS5, we generated three truncated forms of IDM1 and performed the yeast two-hybrid assay. ROS5 interacted with IDM1 (1 to 592 amino acids) carrying the MBD domain but did not interact with the two truncated forms that lacked the MBD domain (Figure 4A), suggesting that the MBD domain of IDM1 is critical for its interaction with ROS5. Interestingly, the mutant mROS5 did not interact with IDM1 in yeast (Figure 4A), indicating that the Gly-246 residue of ROS5 is important for its interaction with IDM1.
Figure 4.
Analysis of the Interactions among ROS5, IDM1, and ROS1.
(A) Interaction of ROS5 with IDM1 as determined by yeast two-hybrid assay. The full-length or various truncated versions of ROS5 or IDM1 were analyzed by yeast two-hybrid assay for interaction. Note that mROS5 (ros5-2) did not interact with IDM1 in the assay.
(B) Interaction of ROS5 or mROS5 with IDM1 as indicated by coimmunoprecipitation in a protoplast transient assay. Three independent experiments were done with similar results.
(C) Interaction of ROS5 or mROS5 with IDM1 by firefly luciferase complementation imaging assay in tobacco leaves. Three independent experiments were done with similar results.
(D) Flag-ROS5 immunoprecipitated ROS1-Myc in the protoplast transient assay.
(E) HA-IDM1 immunoprecipitated ROS1-Myc the protoplast transient assay.
(F) Colocalization of mCherry-ROS5 with IDM1-GFP as indicated by transient assay in tobacco epidermal cells (top) and plot of a line showing a strong correlation (bottom). GFP was fused to the C terminus of ROS4/IDM1, and mCherry was fused to the N terminus of ROS5. Both IDM1-GFP and mCherry-ROS5 were cotransformed into tobacco epidermis cells. Bars = 1 μm.
To further validate the interaction between IDM1 and ROS5, we performed a coimmunoprecipitation assay, with AT1G54850, a homolog of ROS5, as a control. The result indicated that ROS5 but not AT1G54850 coimmuoprecipitated with IDM1 (Figure 4B), further confirming that IDM1 and ROS5 may interact and function in the same complex. As was the case in the yeast two-hybrid assay, ROS5 lacking the ACD (tROS5) could not coimmunoprecipitate IDM1, suggesting that the ACD of ROS5 is essential for the interaction of ROS5 with IDM1 in plant cells. However, mROS5 could still coimmunoprecipitate with IDM1. The in vivo interaction of ROS5 and IDM1 was validated by a firefly luciferase complementation imaging assay (Chen et al., 2008) in tobacco leaves. We found that both ROS5 and mROS5 interacted with IDM1 (Figure 4C). From these results, we suspected that the G246D mutation seemingly does not disrupt the interaction with IDM1 in plants. Although the yeast two-hybrid assay did not reveal a direct interaction between ROS1 and ROS5 or between ROS1 and IDM1, the coimmunoprecipitation assay indicated that Myc-ROS1 could coimmunoprecipitate with Flag-ROS5 or HA-IDM1 (Figures 4D and 4E), suggesting that these proteins might be in the same protein complex.
We also tested whether there might be a subnuclear colocalization of IDM1 and ROS5. ROS5 was fused to the C terminus of mCherry, and IDM1 was fused to the N terminus of GFP. The fused proteins were transiently coexpressed in tobacco epidermal cells under the control of the 35S promoter. As indicated by the strong yellow signals, IDM1-GFP and mCherry-ROS5 colocalized discrete subnuclear foci (Figure 4F).
Genome-Wide Effects of ROS5 on DNA Methylation
To assess the effects of ROS5 on DNA methylation, we performed whole-genome bisulfite sequencing in the wild type, ros5-1, ros1-1, and ros4/idm1-3 in the C24 background (7-d-old seedlings). The average depth of the sequenced methylomes is 20 to 30 times, with an error rate of 0.5 to 0.8%, indicating a good quality of the methylome data. The overall DNA methylation levels in the wild type, ros5-1, ros1-1, and ros4/idm1-3 are similar (Supplemental Figure 4). Furthermore, our methylome data (7-d seedlings) well matched most of the previously published data in both wild type and ros1-1 mutant 14-d seedlings (Supplemental Figure 5), and the C24 methylome largely matches with the Columbia methylome (Supplemental Figure 5).
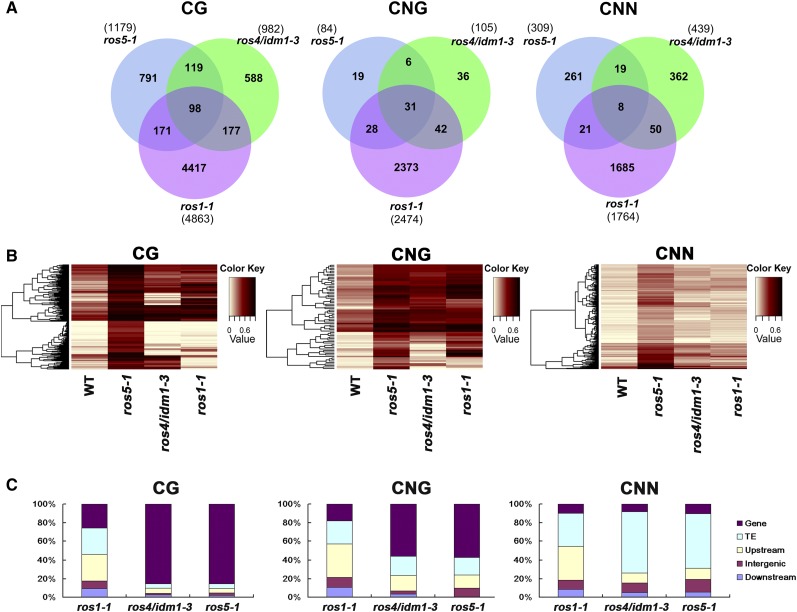
We identified differentially methylated regions (DMRs) caused by the ROS1, IDM1, and ROS5 mutations (Figure 5A; Supplemental Figure 6A and Supplemental Data Sets 1 to 7) using a previously described method (Stroud et al., 2013). More DMRs were identified in the CG context than in the CNG and CNN contexts, probably because of the cutoff that we used (CG, 0.4; CNG, 0.2; and CNN, 0.1). Many fewer loci were affected by ros4/idm1-3 and ros5 than by ros1-1 (Figures 5A and 5B; Supplemental Figures 6A to 6D and 7A and 7B). We identified overlapping hypermethylated DMRs between the ros5 and ros4/idm1-3 mutants (217 of 1179, 18.4% in the CG context; 37 of 84, 44% in the CNG context; and 27 of 309, 8.7% in the CNN context), between the ros5 and ros1 mutants (269 of 1179, 22.8% in the CG context; 59 of 84, 70% in the CNG context; and 29 of 309, 9.4% in the CNN context), and between the ros4/idm1-3 and ros1-1 mutants (275 of 982, 28% in the CG context; 73 of 105, 69.5% in the CNG context; and 58 of 439, 13.2% in the CNN context). Only a relatively few DMRs (98 in the CG context, 31 in the CNG context, and 8 in the CNN context) overlap in all three mutants (Figure 5A). To validate these results, we also conducted a heat map analysis of these three mutants. The heat-map analysis (Figure 5B; Supplemental Figures 7A and 7B) confirmed the overlapping of hypermethylated DMRs illustrated in Figure 5A. We also found that the loci affected by the ros1-1 mutation are distributed evenly on the chromosome; by contrast, CG and CNG methylation loci affected by the ros4/idm1-3 and ros5 mutations are mainly distributed in genic regions, while CNN methylation loci affected by ros4/idm1-3 and ros5 are mainly distributed in transposon regions (Figure 5C).
Figure 5.
Effect of ROS5 Mutation on Genome-Wide DNA Methylation.
(A) Numbers of overlapping hypermethylated DMRs among ros1-1, ros4/idm1-3, and ros5-1 mutants in the C24 accession in three contexts of the whole genome.
(B) Heat map showing the methylation levels of ros1-1 and ros4/idm1-3 in those regions that are hypermethylated DMRs in ros5-1 in three contexts. The color key is presented at right (light yellow indicates low methylation and black indicates high methylation).
(C) Compositions of the genomic locations of hypermethylated DMRs in ros1-1, ros4/idm1-3, and ros5-1 in three contexts. Detailed information is listed in Supplemental Data Set 7. TE, transposon elements.
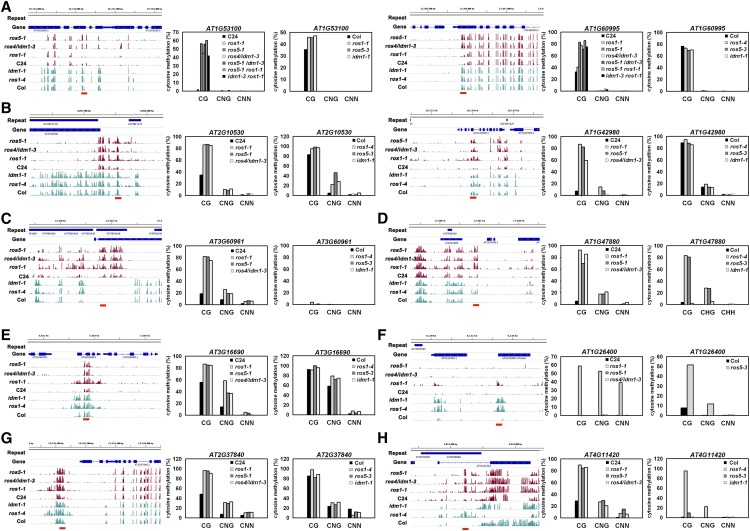
We selected 10 typical loci and determined their DNA methylation patterns by individual-locus bisulfite sequencing (7-d seedlings). The results confirmed the genome-wide bisulfite sequencing (Figures 6A to 6H). Among the loci, two (AT1G60995 and AT2G53100) (Figure 6A) are hypermethylated in ros5-1 and ros4/idm1-3 but not in ros1-1, suggesting that they are not regulated by ROS1. The DNA hypermethylation level was similar for the ros5-1 ros1-1 double mutant and the ros5-1 single mutant and was similar for the idm1-3 ros1-1 double mutant and the ros4/idm1-3 single mutant. Furthermore, the ros5-1 idm1-3 double mutant did not show an additive effect on DNA methylation level relative to the ros5-1 and ros4/idm1-3 single mutants, suggesting that ROS5 and IDM1 may function in the same pathway in the two regions. Both regions are hypermethylated in the CG context but not in non-CG contexts. In Columbia, however, ROS1, IDM1, and ROS5 do not significantly affect the DNA methylation at these two loci. For AT2G10530 and AT1G42980 (Figure 6B), CG methylation was higher in ros1-1, ros4/idm1-3, and ros5-1 than in the C24 wild type, but CG methylation did not differ among ros1-4, idm1-1, ros5-3, and the Columbia wild type. In AT3G60691 (Figure 6C), mutations in ROS1, IDM1, and ROS5 increased the CG, CNG, or CNN methylation level relative to the C24 wild type, but no methylation was found in this locus in Columbia. In AT1G47880 (Figure 6D), CG and CNG methylation was higher in ros1-1, ros4/idm1-3, and ros5-1 than in the C24 wild type, while CG and CNG methylation was higher in ros1-4 and ros5-3 than in Columbia but was similar in idm1-1 and in Columbia. In AT3G16690 (Figure 6E), ROS1, IDM1, and ROS5 mutations increased DNA methylation in C24, but the ROS1 mutation affected DNA methylation in a wider region than IDM1 and ROS5. In the Columbia accession, the increased DNA methylation in ros1-4 and idm1-1 was shifted to the left side of the same region in C24. AT1G26400 has been used as a DNA methylation marker for identifying the idm1-1 mutant in the Columbia accession (Qian et al., 2012). In the ros1-1 mutant (C24), AT1G26400 is also hypermethylated (Figure 6F). However, no clear methylation in AT1G26400 was found in ros5-1 or ros4/idm1-3 in the C24 accession. In AT2G37840 (Figure 6G), the DMR was regulated by ROS1, IDM1, and ROS5 in C24 but was not affected by the same mutations in Columbia. In AT4G11420 (Figure 6H), the DNA methylation in CG and non-CG contexts was higher in ros1-1, ros4/idm1-3, and ros5-1 than in the C24 wild type. In Columbia, however, the DNA methylation was increased by the ROS1 mutation but not by the IDM1 or ROS5 mutation. Given that ROS5 is expressed at a higher level in the early than in the later developmental stage, we compared the DNA methylation pattern in the NOS region (AT1G47880 and AT3G60961 between 7- and 14-d seedlings) and found that DNA methylation did not change, suggesting that DNA methylation is stably maintained in these loci (Supplemental Figure 8). These results suggest that ROS1, IDM1, and ROS5 are involved in preventing the hypermethylation in these overlapped loci (e.g., the 35S-NPTII transgene and DMRs overlapped by all three mutants). In the nonoverlapped DMRs, however, their close homologs might have redundant roles. Although most of the methylome patterns are similar between the C24 and Columbia accessions (Supplemental Figure 5), our results also indicate that some DMRs regulated by ROS1, IDM1, and ROS5 differ in the two accessions.
Figure 6.
DNA Methylation Patterns for Selected Genes.
Ten typical genes were selected according to the genome-wide DNA methylation information, and the pattern for each was confirmed by bisulfite sequencing in C24 and Columbia (Col).
(A) In AT1G53100 and AT1G60995, DNA methylation in CG was greatly increased by ROS1, IDM1, and ROS5 mutations in C24 but not in Columbia.
(B) In AT2G10530 and AT1G42980, DNA methylation in CG and CNG was increased by mutations in ROS1, IDM1, and ROS5 in C24 but not in Columbia.
(C) In AT3G60961, DNA methylation in CG, CNG, and CNN was increased by mutations in ROS1, IDM1, and ROS5 in C24 but was not clearly affected by the same mutations in Columbia.
(D) In AT1G47880, DNA methylation was increased by mutations in ROS1, IDM1, and ROS5 in C24, while mutations in ROS1 and ROS5 but not in IDM1 increased DNA methylation in Columbia.
(E) In AT3G16690, DNA methylation was increased by mutations in ROS1, IDM1, and ROS5 in C24, but ROS1 mutation had a greater effect than IDM1 or ROS5 mutation. In Columbia, the DNA methylation region affected by both ROS1 and ROS4/IDM1 mutations was shifted to the left side of the same region.
(F) In AT1G26400, DNA methylation was increased only by ROS1 mutation in C24 but was increased by all three mutations in Columbia.
(G) In AT2G37840, DNA methylation was increased by ROS1, IDM1, and ROS5 mutations in C24 but not in Columbia.
(H) In AT4G11420, DNA methylation was increased by ROS1, IDM1, and ROS5 mutations in C24 but was increased only by ROS1 mutation in Columbia.
[See online article for color version of this figure.]
Transcriptome Analysis of the ros5 Mutant
To investigate the role of ROS5 mutation in regulating endogenous genes, we conducted RNA sequencing (RNA-seq) using total RNAs isolated from 7-d-old seedlings with three biological replicates. The squared Pearson correlation coefficient between replicates for the wild type and the ros5-1 mutant ranges from 0.95 to 0.98, suggesting that biological replicates of RNA-seq data are reliable and valid. Using the RNA-seq data, we identified 81 differentially expressed genes (DEGs) from the ros5-1 mutant compared with wild-type C24 (Supplemental Data Set 8), among which the expression levels of 21 DEGs were more than 2-fold, but only 9 DEGs (including 2 kb of flanking) overlapped with DMRs (Supplemental Data Set 8). Among these nine DEGs, three genes showed decreased expression levels with higher methylation levels (Supplemental Figure 9), four genes showed increased expression with higher methylation levels (Supplemental Figure 10), one gene with increased expression levels showed lower methylation levels (Supplemental Figure 11A), and the remaining one gene contained both hypermethylated and hypomethylated DMRs (Supplemental Figure 11B). We also used real-time PCR to check the expression of six genes (AT1G60995, AT2G37840, AT4G11420, AT4G08930, AT4G09080, and AT5G49560) with DMRs in their promoter regions and found that the expression of four genes (AT1G60995, AT4G11420, AT4G09080, and AT5G49560) was slightly reduced in ros1-1, ros4, and ros5-1 relative to the wild type (Supplemental Figure 12), which could not be identified by RNA-seq in ros5-1 due to their small reduction in expression. Although DNA hypermethylation was mainly found in the 3′ region of 35S-NPTII in the ros5 mutant, ROS5 DMRs are not distributed in a biased manner in the 5′ and 3′ regions (Supplemental Figure 13). Based on these results, and given that the ros5-1 mutant has no observed developmental defects under normal growth conditions, we speculate that the ROS5 mutation might have a subtle effect on the regulation of endogenous genes under normal growth conditions. However, ROS5 might have important roles in regulating the expression of genes under specific conditions, such as transgene antisilencing or promoting gene expression when DNA methylation is erased in different cells/organs or during different developmental stages, regulating the genes not targeted by DNA methylation.
The Silencing of 35S-NPTII in ros5 May Involve Histone Modifications
IDM1 is a histone acetyltransferase that acetylates H3 at Lys-18 and Lys-23 (Li et al., 2012a; Qian et al., 2012). Because of the interaction between ROS5 and IDM1, we wanted to know whether the levels of H3K18ac and H3K23ac are affected in the ros5 mutant. A chromatin immunoprecipitation (ChIP) assay showed that the level of H3K18ac decreased significantly in the ros5 mutant, while the level of H3K23ac was only weakly reduced (Figure 7), suggesting that the ros5 mutation leads to the silencing of 35S-NPTII and to DNA hypermethylation partly through IDM1. We also used the publicly available H3K9me2 (heterochromatin marker) and H3K27me3 (euchromatin marker) data to check if any of these histone marks are enriched in ROS5 DMRs and found that H3K27me3 is enriched at a low ratio (H3K27me3:CG hypermethylation, 75:1137; H3K27me3:CNG hypermethylation, 15:70; H3K27me3:CNN hypermethylation, 10:300), but we found almost no H3K9me2 in ROS5 DMRs (Supplemental Figure 14), suggesting that most ROS5 DMRs are in euchromatin regions.
Figure 7.
Effect of the ROS5 Mutation on the Histone Modifications Mediated by IDM1.
ChIP analysis shows the histone acetylations at the 35S-NPTII transgene region and some DMRs. ChIP assay was conducted with anti-H3K18ac and anti-H3K23ac antibodies. ACTIN2, AT1G01260, and AT1G10950 were used as controls. Precipitations without antibodies were used as the negative control. Two independent experiments (each with three technical replicates) were conducted with similar results, and one representative experiment is shown. Error bars represent se (n = 3).
ROS5 Is Not Induced by Heat Stress and Does Not Compromise the Heat Tolerance of Plants
Many genes encoding sHSPs are rapidly induced by heat stress (Scharf et al., 2001; Waters, 2013). The presence of an HSP20-like domain in ROS5 indicates that the expression of ROS5 may be induced by heat stress. To determine whether the expression of ROS5 is heat inducible, we performed real-time PCR with RNA isolated from plants treated at 22 or 37°C. The expression of HSP101 was analyzed at the same time as a positive control, and the expression of AT1G54850 was used as a negative control. Heat stress induced the expression of HSP101 but did not induce the expression of ROS5 or AT1G54850 (Figure 8A).
Figure 8.
ROS5 Expression Is Not Induced by Heat Treatment, and ROS5 Mutations Do Not Affect Plant Thermotolerance.
(A) Effect of heat treatment on the expression of ROS5. HSP101 was used as a positive control, while AT1G54850 was used as a negative control. The seedlings were treated at 37°C for 1 h (heat) or 22°C for 1 h (control [Con]) before being grown for 2 d at 22°C. Three independent experiments (each with three technical replicates) were conducted with similar results, and one representative experiment is shown. Error bars represent se (n = 3).
(B) Analysis of the thermotolerance of ros5 mutants. Seeds were sown on MS medium for 2.5 d at 22°C in the dark and then were treated at 38°C for 1.5 h, at 45°C for 2 h, or sequentially at 38°C for 1 h, 22°C for 1 h, and 45°C for 1.5 h. After seedlings were grown at 22°C in the dark for an additional 2 d, hypocotyl lengths were measured. Two independent experiments (each with three technical replicates) were conducted with similar results, and one representative experiment is shown. Error bars represent se (n = 3).
The sHSPs help protect plants from heat and other stresses, and overexpression of sHSPs can enhance thermotolerance (Schöffl et al., 1998). Although ROS5 is not heat inducible, it is still possible that the thermotolerance of plants is compromised in the ros5 mutants. To investigate whether ROS5 is involved in thermotolerance in Arabidopsis, we tested the effect of heat stress on hypocotyl elongation. We hypothesized that if ROS5 is involved in thermotolerance, ros5-1 and ros5-2 should exhibit less tolerance of heat shock than the wild type. However, hypocotyl lengths did not differ significantly between wild-type and mutant plants in response to any temperature treatments (Figure 8B), indicating that ros5-1 and ros5-2 plants are not impaired in thermotolerance.
DISCUSSION
DNA methylation in repeats, transposons, and the promoter regions of some coding genes usually causes a condensed chromatin environment that results in their silencing (Furner and Matzke, 2011). Active DNA demethylation occurs during different developmental stages or under different environmental stimuli and causes some genes to be activated and specific transposons to move (Furner and Matzke, 2011; Yu et al., 2013). Active DNA demethylation is catalyzed by ROS1 and its paralogs DME, DML2, and DML3 (Gong et al., 2002). A previous study suggested that ROS1 randomly slides along DNA to find a specific target (Ponferrada-Marín et al., 2012). However, how these enzymes identify their chromatin targets to remove DNA methylation is poorly understood (Zheng et al., 2008). Based on genetic screens and biochemical analysis, several proteins, including ROS3 and ROS4/IDM1 (Li et al., 2012a; Qian et al., 2012), the DNA phosphatase ZDP (Martínez-Macías et al., 2012), the DNA repair protein XRCC1 (Martínez-Macías et al., 2013), and SSRP1, a component of the FACT histone chaperone (Ikeda et al., 2011), are involved in active DNA demethylation. Except for ZDP, which directly participates in base excision repair after ROS1, the other proteins are all indirectly involved in active DNA demethylation. These other proteins contribute to active DNA methylation, probably by reducing chromatin compaction and thereby facilitating ROS1 binding (ROS3 and IDM1) or by stimulating ROS1/DME activity (XRCC1 and SSRP1). Besides ROS1 and its paralogs for DNA demethylation, other proteins, such as the histone H3K9 demethylase INCREASE IN BONSAI METHYLATION1 (Saze et al., 2008; Rigal et al., 2012), the RNA binding protein ANTISILENCING1 (Wang et al., 2013), and the putative chromatin regulator ENHANCED DOWNY MILDEW2 (Lei et al., 2014), are found to prevent CNG methylation mainly in gene body regions. In this study, we identified a sHSP, ROS5, that interacts with IDM1 and has functions in antisilencing and in the regulation of active DNA demethylation.
The major families of conserved HSPs/chaperones that exist in a wide diversity of organisms are Hsp70 (DnaK), Hsp60, Hsp90, Hsp100, and the sHSPs (HSP20) (Waters, 2013). HSPs play crucial roles in protein refolding, assembly, trafficking, stability, and degradation and in preventing protein aggregation under stress and nonstress conditions (Scharf et al., 2001; Wang et al., 2004; Siddique et al., 2008; Bondino et al., 2012; Waters, 2013). The Arabidopsis genome contains 19 classical sHSPs, which are divided into 11 subfamilies according to their intracellular localization (Scharf et al., 2001; Siddique et al., 2008; Waters, 2013). The current working model is that the sHSPs work with other chaperones to prevent irreversible protein aggregation and to resolubilize aggregated proteins. An analysis of the Arabidopsis genome has identified 25 new members with one or more ACDs that are unrelated to the 11 subfamilies of the classical sHSPs; these are also referred to as ACD proteins, although their biological roles are largely unknown (Scharf et al., 2001). One of these ACD proteins, RESTRICTED TEV MOVEMENT2, is required for restricting the long-distance movement of Tobacco etch virus in Arabidopsis (Whitham et al., 2000).
ROS5 is an ACD sHSP that may assemble into a 16-oligomer complex. Although most HSPs exhibit heat-inducible expression, some are constitutively expressed (Scharf et al., 2001; Waters, 2013). ROS5 is not induced by heat or other abiotic stresses. We could not detect any general chaperone activity of ROS5 in preventing protein unfolding and aggregation in an in vitro assay by using the common chaperone substrates (Supplemental Figure 15). These results suggest that ROS5 itself is unlikely to possess typical chaperone roles. ROS5 physically interacts with the histone acetyltransferase IDM1, and its ACD is essential for the interaction. Our genetic and bisulfite sequencing analyses indicated that IDM1 and ROS5 work in the same pathway to regulate DNA demethylation. We speculate that the interaction between ROS5 and IDM1 might increase IDM1 protein folding or stability or that ROS5 might work with other chaperone proteins to achieve its function in assisting IDM1 activity. Both IDM1 and ROS5 are required for the expression of genes when DNA methylation is inhibited (in ddm1 or following addition of the DNA methylation inhibitor 5′-Aza), suggesting that both IDM1 and ROS5 also work downstream of DNA methylation to regulate gene expression. IDM1 is mainly responsible for two histone modifications, H3K18ac and H3K23ac. In the ros5 mutant, we found that H3K18ac is reduced and that H3K23ac is slightly reduced, suggesting that ROS5 partially affects IDM1 function. ROS5 might work together with IDM1 to create a less compact chromatin environment that helps ROS1 find its DNA methylation targets. Genetic analysis indicates that IDM1 and ROS5 targets partially overlap with ROS1 targets. Because ROS1 has several homologs, it is possible that loci not targeted by ROS1 are regulated by the ROS1 homologs. Our coimmunoprecipitation data also showed that ROS5 or IDM1 can coimmunoprecipitate with ROS1, although a yeast two-hybrid assay failed to detect a direct interaction between ROS1 and ROS5 or IDM1, suggesting that these proteins form a complex.
Our whole-genome bisulfite sequencing analysis indicates that the DMRs regulated by ROS1, IDM1, and ROS5 in C24 are largely different from those regulated by the same genes in the Columbia accession. In Columbia, a previous study with the idm1-1 mutant identified 1098 DNA hypermethylated loci and 75 DNA hypomethylated loci in the whole genome (Qian et al., 2012). Fewer DMRs are caused by ros4/idm1-3 in C24 than by idm1-1 in the Columbia accession in all three contexts (Supplemental Figures 16A and 16B). Furthermore, there is only a slight overlap of DMRs in ros4/idm1-3 (C24) and idm1-1 (Columbia) in each context. The DNA methylation patterns in ros1-1 (C24) and ros1-4 (Columbia) (Supplemental Figures 17A and 17B) are also largely different. These results suggest that different accessions substantially differ in DNA methylation and that the same proteins target different genomic regions in the two accessions, although they may have similar regulatory mechanisms.
METHODS
Plant Growth Conditions and Mutant Screening
Arabidopsis thaliana seeds sterilized with 0.5% NaClO were sown onto Murashige and Skoog (MS) medium containing 2% (w/v) Suc and 0.8% (w/v) agar. After 3 d at 4°C, the plates were transferred to a growth chamber at 22°C under long-day conditions (23 h of light/1 h of dark). Generally, 7-d-old seedlings grown on MS medium were transferred to soil and cultivated in a greenhouse at 20°C under long-day conditions (16 h of light/8 h of dark). For the transient protoplast assay, 3-week-old seedlings grown in the growth chamber were used to prepare protoplasts (Sheen, 2001).
Wild-type C24 and ros1-1 mutant plants carry the homozygous T-DNA loci 35S-NPTII and RD29A-LUC (Gong et al., 2002). The mutants ros4/idm1-3, ddm1, rfc1, and hda6 were obtained from previous screenings conducted in our laboratory (Li et al., 2012a). An ethyl methanesulfonate–mutagenized population in the wild-type C24 background was generated and screened for kanamycin-sensitive mutants as described previously (Li et al., 2012a). Briefly, wild-type seeds can grow well on MS medium containing 50 mg/L kanamycin. We screened mutants that showed sensitive phenotypes on MS medium plates containing 50 mg/L kanamycin and transferred the mutants into soil for recovery before they were dead. Then, the kanamycin-sensitive phenotypes of the mutants were confirmed with the next-generation seeds on MS medium containing 50 mg/L kanamycin. The T-DNA insertion lines SALK_037306 (idm1-1) and SALK_130656 (ros5-3) were obtained from the Arabidopsis stock center.
Map-Based Cloning and Mutant Complementation
We generated a mapping population by crossing a ros5-2 mutant (C24 accession) with a Columbia accession (gl1). In the segregating F2 population, ros5 mutants were first selected based on the kanamycin-sensitive phenotype and then checked by PCR (using primers 35S-NPTII-F and 35S-NPTII-R) to confirm that all selected mutants contained the 35S-NPTII transgene. About 600 plants carrying the 35S-NPTII transgene and showing the kanamycin-sensitive phenotype were selected and used for mapping. For complementation, we cloned the ROS5 genomic sequence (∼3.6 kb, from −2055 to +1513 bp containing the ∼2-kb promoter, the coding region, and the 400-bp 3′ region) into the SalI and KpnI sites of pCAMBIA1391 using two primers (complemental genomicseq-F and complemental genomicseq-R). The construct was introduced into ros5-2 mutants using Agrobacterium tumefaciens GV3101. Primers used for the construct are listed in Supplemental Data Set 9.
Bisulfite Sequencing
About 500 ng of DNA was treated with the EZ Methylation-Gold Kit (Zemo Research) following the manufacturer’s protocol. The treated DNA was eluted with 20 μL of H2O2. About 2 μL of bisulfite-treated DNA was used in the PCR with the specific primers listed in Supplemental Data Set 9. Amplified products were subcloned into the pMD18-T vector (TaKaRa), and ∼10 independent clones from each sample were sequenced for each region.
RNA Analysis
For real-time PCR assays, total RNA was isolated using Trizol reagent (Invitrogen), and contaminating DNA was digested with RNase-free DNaseI (TaKaRa). A 4-μg quantity of RNA was used for the first-strand cDNA synthesis with M-MLV reverse transcriptase (Promega) in a 20-μL reaction volume. Then, 2 μL of the cDNA reaction mixture (diluted 10 times) was used as a template in a 20-μL PCR with SYBR Green Master mix (TaKaRa); the PCR was conducted on an ABI 7300 machine (Life Technologies) with three technical replicates. Specific primers used for each gene are listed in Supplemental Data Set 9. UBIQUITIN-CONJUGATING ENZYME28 was used as an internal reference control.
Coimmunoprecipitation
ROS4/IDM1 was cloned with primers IDM1-SacI-F and IDM1-AvrII-R and fused into the SacI and SpeI sites of a modified pCAMBIA1307 vector containing the HA tag coding sequence. Wild-type and mutated ROS5 (carrying a mutation of Gly-246 to Asp) were cloned with primers ROS5-KpnI-R and ROS5-SalI-F, and both of them were fused into the SalI and KpnI sites of a modified pCAMBIA1300 containing either the Myc or Flag coding sequence. ROS5 lacking the sequence coding the HSP20-like domain was cloned with primers tROS5-KpnI-R and ROS5-SalI-F and AT1G54850 was cloned with primers AT1G54850-SalI-F and AT1G54850-KpnI-R, then both of them were fused into the SalI and KpnI sites of a modified pCAMBIA1300 vector containing the Myc tag coding sequence. The full-length cDNA of ROS1 was cloned with primers ROS1-SpeI-F and ROS1-SalI-R and then fused into the SpeI and SalI sites of a modified vector named 326-N-Myc.
Plasmids were isolated and then purified by CsCl gradient centrifugation. Purified plasmids were transformed into Arabidopsis mesophyll protoplasts. After they were incubated overnight in a light chamber, the transformed protoplasts were harvested and homogenized in 1 mL of immunoprecipitation buffer (50 mM Tris, pH 7.6, 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40, 0.5 mM DTT, 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitor cocktail [one tablet per 50 mL]; Roche). The homogenized solution was incubated on a rotary shaker for 15 min and then centrifuged at 16,000g for 10 min at 4°C. After the supernatant was transferred to a new 1.5-mL tube, 20 μL of Anti-HA-Agarose Affinity Gel or Anti-C-Myc-Agarose Affinity Gel (Sigma-Aldrich) was added. After the preparation was incubated on a rotary shaker at 4°C for 3 h, the agarose beads were washed five times with 1.5 mL of immunoprecipitation buffer. The immunoprecipitated products were then detected by protein gel blot. Primers for the above constructs are listed in Supplemental Data Set 9.
Analysis of Seedling Thermotolerance
A seedling thermotolerance assay was performed as described previously with some modifications (Whitham et al., 2000). Briefly, wild-type C24, ros5-1, and ros5-2 seeds were sterilized and plated on MS medium containing 2% (w/v) Suc and 1% (w/v) agar. The plates were wrapped with aluminum foil to exclude light. After 3 d at 4°C, the plates were transferred to a growth chamber at 22°C and kept vertical for 2.5 d. The plates were then subjected to one of three treatments. In the 45°C treatment, the plates were kept at 45°C for 2 h before they were returned to 22°C. In the 38°C treatment, the plates were kept at 38°C for 1.5 h before they were returned to 22°C. In the 38/22/45°C treatment, the plates were kept at 38°C for 1.5 h, returned to 22°C for 2 h for recovery, and then kept at 45°C for 2 h before they were returned to 22°C. Two days after the treatments were applied, hypocotyl lengths were measured.
Yeast Two-Hybrid Assays
To determine whether ROS4/IDM1 and ROS5 interact, we constructed AD-ROS5 and BD-IDM1. AD-ROS5 was generated by cloning the full-length cDNA of ROS5 into the NdeI and BamHI sites of pGADT7 vector using primers ROS5-NdeI-F and ROS5-BamHI-R, and BD-IDM1 was generated by cloning the full-length cDNA of IDM1 into the NcoI and XmaI sites of pGBKT7 vector using primers ROS4/IDM1-NcoI-F and ROS4/IDM1-XmaI-F. To identify which domain of IDM1 is required for the interaction with ROS5, we generated three truncated IDM1 constructs: BD-IDM1 (1 to 592), BD-IDM1 (593 to 1189), and BD-IDM1 (859 to 1189). BD-IDM1 (1 to 592) was constructed by cloning 1 to 1776 bp of IDM1 cDNA into NcoI and PstI sites of pGBKT7 vector with primers ROS4/IDM1-NcoI-F and ROS4/IDM1-PstI-F; BD-IDM1 (593 to 1189) was constructed by cloning 1777 to 3567 bp of IDM1 cDNA into SfiI and XmaI sites of pGBKT7 vector with primers ROS4/IDM1-SfiI-1F and ROS4/IDM1-XmaI-R; BD-IDM1 (859 to 1189) was constructed by cloning 2575 to 3567 bp of IDM1 cDNA into SfiI and XmaI sites of pGBKT7 vector with primers ROS4/IDM1-SfiI-2F and ROS4/IDM1-XmaI-R. To identify which domain of ROS5 is essential for the interaction with IDM1, we generated two truncated ROS5 constructs: AD-ROS5 (1 to 240) and AD-ROS5 (241 to 349). AD-ROS5 (1 to 240) was constructed by cloning 1 to 840 bp of ROS5 cDNA into NdeI and BamHI sites of pGADT7 vector with primers ROS5-NdeI-F and ROS5-BamHI-1R; AD-ROS5 (241 to 349) was constructed by cloning 841 to 1047 bp of ROS5 cDNA into NdeI and BamHI sites of pGADT7 vector with primers ROS5-NdeI-1F and ROS5-BamHI-R. Mutated ROS5 (carrying the G246D mutation) cDNA was also cloned into the NdeI and BamHI sites of pGADT7 vector with primers ROS5-NdeI-F and ROS5-BamHI-R. Primers used to generate the constructs are listed in Supplemental Data Set 9.
Corresponding constructs were then cotransformed into the yeast strain AH109. Positive strains carrying pGADT7 and pGBKT7 vectors were screened on synthetic dropout medium lacking Trp and Leu. Ten independent clones of the positive strains were then selected to test the interaction on synthetic dropout medium lacking Trp, Leu, and His and supplemented with 10 mM 3-aminotriazole, which increases the stringency of selection. If the two proteins interacted, the selected positive strains survived on synthetic dropout medium lacking Trp, Leu, and His.
Whole-Genome Bisulfite Sequencing and Data Analysis
Genomic DNA was isolated from 7-d seedlings to generate MethylC-seq libraries. After bisulfite treatment and library preparation, MethylC-seq was performed using HiSeq 2000 (Illumina).
For data analysis, low-quality bases (q < 13) of reads were filtered using SolexaQA software (Cox et al., 2010). Clean reads were mapped to the reference genome sequence of Arabidopsis (TAIR10) with Bismark (Krueger and Andrews, 2011). Only uniquely mapped reads were retained. PCR duplicate reads were removed. The ratio of C to CT was used to indicate methylation level. DMRs for each mutant were identified by comparing their methylation levels in the three contexts with those of the wild type. Specifically, the reference genome was divided into 100-bp bins. Bins with at least four cytosines that are each covered by at least four reads were retained. By comparing the number of called Cs and Ts in the mutant and the wild type for each bin, we selected bins with the absolute difference of methylation level of 0.4, 0.2, and 0.1 for CG, CNG, and CNN, respectively, and Benjamini-Hochberg–corrected false discovery rate < 0.01 (Fisher’s exact test). The selected bins within 200 bp of each other were merged into DMRs. Heat maps were drawn using heatmap.2 in the gplots package in R. Complete linkage hierarchical clustering with Euclidean distance as a distance measure was used to sort the rows.
To investigate DMR enrichment, genes (including flanking 2-kb regions) overlapped with DMRs were selected for this analysis. For upstream and downstream 2-kb regions, the regions were divided into 20 bins. Gene bodies were divided into 60 bins. Methylation levels were calculated for each bin and plotted.
ChIP Assay
The ChIP assay was performed as described previously (Li et al., 2012a). The antibodies used were anti-H3K18ac (Millipore 07-354) and anti-H3K23ac (Millipore 07-355). The ChIP product was finally dissolved in 500 μL of distilled, deionized water, and a 5-μL aliquot of the product was used in each real-time PCR. The primers used are listed in Supplemental Data Set 9.
Analysis of ROS5 Localization and Colocalization of ROS5 and ROS4/IDM1
For the analysis of ROS5 localization, a vector expressing the GFP-ROS5 fused protein was generated with a modified pCAMBIA1300 vector containing GFP coding sequence, and the full-length cDNA of ROS5 was cloned with primers (ROS5-SalI-F and ROS5-KpnI-R) and fused into the SalI and KpnI sites of the modified pCAMBIA1300 vector. For mROS5 localization, the vector that expressed the GFP-mROS5 fused protein was constructed. The cDNA of mROS5 was cloned from the ros5-2 mutant with primers (ROS5-SalI-F and ROS5-KpnI-R) and fused into the modified pCAMBIA1300 vector with the same method used for the GFP-ROS5 construct. To investigate the effect of the ACD on ROS5 localization, we used the same procedure with primers (ROS5-SalI and tROS5-KpnI-R) to generate a construct that expressed tROS5 lacking the ACD.
For analysis of the colocalization of ROS4/IDM1 and ROS5, a vector expressing the mCherry-ROS5 fused protein was generated using a modified pCAMBIA1300 containing the mCherry coding sequence. The full-length cDNA of ROS5 was cloned with primers (ROS5-SalI-F and ROS5-KpnI-R) and fused into the SalI and KpnI sites of the modified pCAMBIA1300 vector. A vector expressing IDM1-GFP was obtained from a previous study (Li et al., 2012a). Transient assay and image acquisition were performed as described previously (Li et al., 2012a). Primers for these constructs are listed in Supplemental Data Set 9.
Firefly Luciferase Complementation Imaging Assay
To investigate the interaction between ROS5 and IDM1, three constructs was generated: IDM1-NLUC, CLUC-ROS5, and CLUC-mROS5. IDM1-NLUC was constructed by cloning the IDM1 cDNA into the BamHI site of the pCAMBIA-NLUC vector with primers (IDM1-NLUC-F and IDM1-NLUC-R), and the wild type ROS5 and point mutation (mROS5) cDNAs were cloned with primers (ROS5-CLUC-KpnI-F and ROS5-CLUC-PstI) and fused into KpnI and PstI sites of the pCAMBIA-CLUC vector. All constructs were transformed into Agrobacterium strain GV3101, which was then used to transform Nicotiana benthamiana as follows: the single clone of each strain was incubated with 5 mL of YEB supplemented with 50 mg/L kanamycin and 50 mg/L rifampicin at 28°C for ∼24 h. Then, the bacteria were collected and resuspended with 3 mL of injection buffer (10 mM MES, pH 5.7, 10 mM MgCl2, and 250 μM acetosyringone) at a final OD600 (OD600 = 2.0 for IDM1 and OD600 = 0.3 for ROS5 and its variant mROS5), then the resuspended solutions were kept at 28°C for 3 h before being injected into N. benthamiana leaves. After 3 d, 1 mM luciferin was sprayed onto the lower epidermis and kept in the dark for 5 min, then a CCD camera (1300B; Roper) was used to capture the fluorescence signal at −110°C.
Protein Purification and Gel-Exclusion Chromatography
To prepare the purified wild-type and mutant ROS5 protein, the full-length cDNAs of the wild-type and mutant ROS5 were cloned with primers (ROS5-BamHI-F and ROS5-XhoI-R) and fused into the BamHI and XhoI sites of the pET30a vector. The constructs were transformed into Escherichia coli BL21 (DE3). Generally, an independent clone was selected, incubated overnight at 37°C in 5 mL of Luria-Bertani liquid medium, transferred to 200 mL of Luria-Bertani medium, and incubated to an A260 of 0.6 at 37°C. Expression was then induced by the addition of 0.2 mM isopropyl thio-β-d-galactoside. After 8 h, cells were harvested and homogenized by ultrasonification in 30 mL of lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, and 2 mM PMSF). Then, the homogenized solution was centrifuged at 5000g at 4°C for 20 min. The supernatant was transferred to a new 50-mL tube, and 100 μL of nickel–nitrilotriacetic acid agarose was added to the supernatant. After incubation on a rotary shaker at 4°C for 2 h, the nickel–nitrilotriacetic acid agarose was washed three times in 20 mL of wash buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 50 mM imidazole, and 2 mM PMSF). Then, the nickel–nitrilotriacetic acid agarose was transferred to a new 1.5-mL tube, and protein was eluted twice with 600 μL of elution buffer (20 mM Tris, pH 7.5, 300 mM NaCl, 5 mM MgCl2, and 250 mM imidazole). Finally, the two eluates were combined.
To investigate the oligomeric states, we separately applied purified wild-type and mutant ROS5 to Superdex-200 (GE Healthcare) in a buffer (20 mM Tris, pH 7.5, 300 mM NaCl, and 5 mM MgCl2). Sample aliquots of 1 mL with a protein concentration of 2 mg/mL were injected and separated on the chromatography system.
A genomic DNA fragment containing −2055 to −1 bp of ROS5 was cloned with primers (genomicseq-F and native pro-NcoI-R) and fused into the SalI and NcoI sites of the pCAMBIA1391 vector. Then, the construct was transformed into the C24 wild type using Agrobacterium strain GV3101. Transgenic T2 plants resistant to hygromycin were used for histochemical GUS staining as described previously (Li et al., 2012a).
Aggregation Protection Assay
The cDNA of Hsp18.5 was cloned with primers (Hsp18.5-KpnI-F and Hsp18.5-BamHI-F) and fused into the KpnI and BamHI sites of pET28a vector. The expression and purification of Hsp18.5 was conducted as described above. The aggregation protection assay was conducted as described previously (Basha et al., 2006) with few modifications. Briefly, 5 µg of porcine mitochondrial MDH (Roche) was incubated with gradually increased amounts of sHSP (ROS5 or Hsp18.5) for 2 h at 45°C in a 50-μL reaction buffer (25 mM HEPES, pH 7.5, 150 mM KCl, 5 mM MgCl2, and 2 mM DTT). Control samples (CK) containing only MDH were not treated with heating. After heat treatment, all samples were cooled on ice for 5 min and then centrifuged for 15 min at 16,000g at 4°C. After centrifugation, the supernatant was transferred to a new 1.5-mL tube and mixed with 10 μL of 6× SDS loading buffer. Then, the pellet was washed one time with 1 mL of reaction buffer and centrifuged for 15 min at 16,000g at 4°C. After the supernatant was removed, the pellet was dissolved in 50 μL of 1× SDS loading buffer. Finally, the pellet and supernatant were analyzed by 12% SDS-PAGE and stained with Coomassie Brilliant Blue.
Identification of DEGs
Three biological replicates were used for RNA-seq in both C24 and ros5-1. Forty thousand reads for each library were selected and aligned to the reference genome sequence of Arabidopsis (TAIR10) using Bowtie (Langmead et al., 2009). Properly mapped paired end reads were used to calculate the means and sd of inner sizes of the RNA-seq library. Reads from RNA-seq data were mapped to the reference genome sequence of Arabidopsis (TAIR10) using TopHat (Trapnell et al., 2012). Fragments per kilobase per million reads of genes for each biological replicate were determined using Cufflinks (Trapnell et al., 2012). DEGs were identified requiring q < 0.05 and log2 (fold change) > 2 or < −2 using Cuffdiff (Trapnell et al., 2012). Genes with fragments per kilobase per million reads > 1 in either the ros5-1 mutant or the wild type were defined as expressed.
Analysis of Histone Modification Enrichment of ROS5 DMRs Using Publicly Available ChIP Sequencing Data
ChIP sequencing data (H3K9me2, H3K27me3, and input) were downloaded from the National Center for Biotechnology Information Short Read Archive under accession number SRP002100. Reads were first aligned to the reference genome using Bowtie (Langmead et al., 2009). Histone modification peaks were identified using MACS (Zhang et al., 2008).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: AT3G14980 (ROS4/IDM1), AT1G54840 (ROS5), AT2G36490 (ROS1), AT5G66750 (DDM1), AT5G63110 (HDA6), AT5G22010 (RFC1), AT1G74310 (HSP101), and AT3G18780 (ACTIN2). The original data sets of bisulfite sequencing and RNA sequencing have been deposited in the Gene Expression Omnibus database with the accession number SRP042060.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Growth Phenotypes of the Wild Type (C24), ros5-1, ros1-1, and ros1-1 ros5-1 in Soil.
Supplemental Figure 2. Sequence Alignment of ROS5 among Members of the HSP20 Family.
Supplemental Figure 3. Analysis of the ROS5 Expression Pattern in a Transgenic Plant Carrying the ROS5-GUS Transgene.
Supplemental Figure 4. DNA Methylation in Three Contexts (CG, CNG, and CNN) across the Five Arabidopsis Chromosomes in C24, ros1-1, ros4/idm1-3, and ros5-1 Mutants.
Supplemental Figure 5. Comparison of Measured DNA Methylation Levels in This Study with Those of Previous Studies.
Supplemental Figure 6. Overlap of Hypomethylated DMRs in ros1-1, ros4/idm1-3, and ros5-1 in Three Contexts (CG, CNG, and CNN) as Illustrated by Heat Maps.
Supplemental Figure 7. Overlap of Hypermethylated DMRs among ros1-1, ros4/idm1-3, and ros5-1 in the C24 Accession as Indicated by Heat Maps.
Supplemental Figure 8. DNA Methylation Comparison of the NOS Region, AT1G47880, and AT3G60961 between 7-d and 14-d Seedlings.
Supplemental Figure 9. Integrative Genomics Viewer (IGV) Snapshot of Three Genes with Decreased Expression and Increased Methylation.
Supplemental Figure 10. IGV Snapshot of Four Genes with Increased Expression and Increased Methylation.
Supplemental Figure 11. IGV Snapshots of the Gene with Increased Expression Containing Both Decreased and Increased Methylation and of the Gene with Increased Expression and Increased Methylation.
Supplemental Figure 12. Analysis of the Expression of Genes with DMRs in Their Promoter Regions by Real-Time PCR.
Supplemental Figure 13. DNA Methylation Levels of ROS5 and the Wild Type along the Gene 5′ Region, the Gene Body, and the Gene 3′ Region.
Supplemental Figure 14. The Histone Modifications H3K27me3 and H3K9me2 at the DMRs of the ros5-1 Mutant Using Publicly Available Data.
Supplemental Figure 15. ROS5 Is Ineffective for Routine Chaperone Activity.
Supplemental Figure 16. Methylation Patterns of idm1-1 in Columbia and ros4/idm1-3 in C24 as Indicated by Heat Maps.
Supplemental Figure 17. Methylation Patterns of ros1-4 in Columbia and ros1-1 in C24 as Indicated by Heat Maps.
Supplemental Data Set 1. List of Hypermethylated DMRs in ros1-1 in Three Contexts.
Supplemental Data Set 2. List of Hypermethylated DMRs in ros5-1 in Three Contexts.
Supplemental Data Set 3. List of Hypermethylated DMRs in ros4/idm1-3 in Three Contexts.
Supplemental Data Set 4. List of Hypomethylated DMRs in ros1-1 in Three Contexts.
Supplemental Data Set 5. List of Hypomethylated DMRs in ros5-1 in Three Contexts.
Supplemental Data Set 6. List of Hypomethylated DMRs in ros4/idm1-3 in Three Contexts.
Supplemental Data Set 7. Detailed Information on the Genomic Locations of Hypermethylated DMRs in ros1-1, ros5-1, and ros4/idm1-3 in Three Contexts.
Supplemental Data Set 8. Differentially Expressed Genes in ros5-1.
Supplemental Data Set 9. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Jian-Kang Zhu (Shanghai Center for Plant Stress Biology, Purdue University) for providing idm1-1 and ros1-4 mutants, Mei Zhang and Jian Chen (China Agricultural University) for technical help in deep sequencing, and Zengqin Deng (China Agricultural University) for technical help in gel filtration. This work was supported by the Natural Science Foundation of China (Grants 31330041 and 31121002).
AUTHOR CONTRIBUTIONS
Y.Z. and Z.G. conceived this project and designed all experiments. Y.Z., S.X., X.L., and C.W. performed experiments, and Y.Z., S.X., Z.C., J.L., and Z.G. analyzed the data. Y.Z., S.X., and Z.G. wrote the article.
Glossary
- RdDM
RNA-directed DNA methylation
- ACD
α-crystallin domain
- 5′-Aza
5′-aza-2′-deoxycytidine
- DMR
differentially methylated region
- RNA-seq
RNA sequencing
- DEG
differentially expressed gene
- ChIP
chromatin immunoprecipitation
- sHSP
small heat shock–like protein
- MS
Murashige and Skoog
- PMSF
phenylmethylsulfonyl fluoride
- IGV
Integrative Genomics Viewer
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Baranova E.V., Weeks S.D., Beelen S., Bukach O.V., Gusev N.B., Strelkov S.V. (2011). Three-dimensional structure of α-crystallin domain dimers of human small heat shock proteins HSPB1 and HSPB6. J. Mol. Biol. 411: 110–122. [DOI] [PubMed] [Google Scholar]
- Basha E., Friedrich K.L., Vierling E. (2006). The N-terminal arm of small heat shock proteins is important for both chaperone activity and substrate specificity. J. Biol. Chem. 281: 39943–39952. [DOI] [PubMed] [Google Scholar]
- Bondino H.G., Valle E.M., Ten Have A. (2012). Evolution and functional diversification of the small heat shock protein/α-crystallin family in higher plants. Planta 235: 1299–1313. [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M.P., Peterson D.A., Biggs P.J. (2010). SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs M.L., Bender J. (2006). Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18: 1166–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furner I.J., Matzke M. (2011). Methylation and demethylation of the Arabidopsis genome. Curr. Opin. Plant Biol. 14: 137–141. [DOI] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T., David L., Zhu J.K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814. [DOI] [PubMed] [Google Scholar]
- He X.J., Hsu Y.F., Pontes O., Zhu J., Lu J., Bressan R.A., Pikaard C., Wang C.S., Zhu J.K. (2009a). NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 23: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.J., Hsu Y.F., Zhu S., Liu H.L., Pontes O., Zhu J., Cui X., Wang C.S., Zhu J.K. (2009b). A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 23: 2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.J., Hsu Y.F., Zhu S., Wierzbicki A.T., Pontes O., Pikaard C.S., Liu H.L., Wang C.S., Jin H., Zhu J.K. (2009c). An effector of RNA-directed DNA methylation in Arabidopsis is an ARGONAUTE 4- and RNA-binding protein. Cell 137: 498–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Kinoshita Y., Susaki D., Ikeda Y., Iwano M., Takayama S., Higashiyama T., Kakutani T., Kinoshita T. (2011). HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev. Cell 21: 589–596. [DOI] [PubMed] [Google Scholar]
- Jackson J.P., Lindroth A.M., Cao X., Jacobsen S.E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560. [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes T.L., Richards E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22: 94–97. [DOI] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. (2011). Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., et al. (2014). Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns. Proc. Natl. Acad. Sci. USA 111: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Qian W., Zhao Y., Wang C., Shen J., Zhu J.K., Gong Z. (2012a). Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 11425–11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., et al. (2012b). Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics 13: 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth A.M., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080. [DOI] [PubMed] [Google Scholar]
- Liu Q., Wang J., Miki D., Xia R., Yu W., He J., Zheng Z., Zhu J.K., Gong Z. (2010). DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 22: 2336–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Macías M.I., Córdoba-Cañero D., Ariza R.R., Roldán-Arjona T. (2013). The DNA repair protein XRCC1 functions in the plant DNA demethylation pathway by stimulating cytosine methylation (5-meC) excision, gap tailoring, and DNA ligation. J. Biol. Chem. 288: 5496–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Macías M.I., Qian W., Miki D., Pontes O., Liu Y., Tang K., Liu R., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T., Zhu J.K. (2012). A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol. Cell 45: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponferrada-Marín M.I., Roldán-Arjona T., Ariza R.R. (2012). Demethylation initiated by ROS1 glycosylase involves random sliding along DNA. Nucleic Acids Res. 40: 11554–11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W., et al. (2012). A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336: 1445–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigal M., Kevei Z., Pélissier T., Mathieu O. (2012). DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J. 31: 2981–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H., Shiraishi A., Miura A., Kakutani T. (2008). Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 319: 462–465. [DOI] [PubMed] [Google Scholar]
- Scharf K.D., Siddique M., Vierling E. (2001). The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperones 6: 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöffl F., Prändl R., Reindl A. (1998). Regulation of the heat-shock response. Plant Physiol. 117: 1135–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Siddique M., Gernhard S., von Koskull-Döring P., Vierling E., Scharf K.D. (2008). The plant sHSP superfamily: Five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfort R.L., Basha E., Friedrich K.L., Slingsby C., Vierling E. (2001). Crystal structure and assembly of a eukaryotic small heat shock protein. Nat. Struct. Biol. 8: 1025–1030. [DOI] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R.A., Richards E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928. [DOI] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Shoseyov O., Altman A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9: 244–252. [DOI] [PubMed] [Google Scholar]
- Wang X., Duan C.G., Tang K., Wang B., Zhang H., Lei M., Lu K., Mangrauthia S.K., Wang P., Zhu G., Zhao Y., Zhu J.K. (2013). RNA-binding protein regulates plant DNA methylation by controlling mRNA processing at the intronic heterochromatin-containing gene IBM1. Proc. Natl. Acad. Sci. USA 110: 15467–15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters E.R. (2013). The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64: 391–403. [DOI] [PubMed] [Google Scholar]
- Whitham S.A., Anderberg R.J., Chisholm S.T., Carrington J.C. (2000). Arabidopsis RTM2 gene is necessary for specific restriction of tobacco etch virus and encodes an unusual small heat shock-like protein. Plant Cell 12: 569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Lepère G., Jay F., Wang J., Bapaume L., Wang Y., Abraham A.L., Penterman J., Fischer R.L., Voinnet O., Navarro L. (2013). Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 110: 2389–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Pontes O., Zhu J., Miki D., Zhang F., Li W.X., Iida K., Kapoor A., Pikaard C.S., Zhu J.K. (2008). ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature 455: 1259–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Kapoor A., Sridhar V.V., Agius F., Zhu J.K. (2007). The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr. Biol. 17: 54–59. [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2009). Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43: 143–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.