Abstract
Each year millions of pulmonary nodules are discovered by computed tomography and subsequently biopsied. As the majority of these nodules are benign, many patients undergo unnecessary and costly invasive procedures. We present a 13-protein blood-based classifier that differentiates malignant and benign nodules with high confidence, thereby providing a diagnostic tool to avoid invasive biopsy on benign nodules. Using a systems biology strategy, 371 protein candidates were identified and a multiple reaction monitoring (MRM) assay was developed for each. The MRM assays were applied in a three-site discovery study (n = 143) on plasma samples from patients with benign and Stage IA cancer matched on nodule size, age, gender and clinical site, producing a 13-protein classifier. The classifier was validated on an independent set of plasma samples (n = 104), exhibiting a high negative predictive value (NPV) of 90%. Validation performance on samples from a non-discovery clinical site showed NPV of 94%, indicating the general effectiveness of the classifier. A pathway analysis demonstrated that the classifier proteins are likely modulated by a few transcription regulators (NF2L2, AHR, MYC, FOS) that are associated with lung cancer, lung inflammation and oxidative stress networks. The classifier score was independent of patient nodule size, smoking history and age, which are risk factors used for clinical management of pulmonary nodules. Thus this molecular test can provide a powerful complementary tool for physicians in lung cancer diagnosis.
Introduction
Computed tomography (CT) identifies millions of pulmonary nodules annually with many being undiagnosed as either malignant or benign (1–3). In many cases, histopathological diagnosis by biopsy techniques such as fine needle aspiration is impossible (due to nodule location) or inconclusive (due to small nodule size). The vast majority of these nodules are benign, but nevertheless many patients with benign nodules undergo unnecessary procedures. It is estimated that only 20% of patients with lung nodules undergoing biopsy or surgery actually have a malignant lung nodule (4). Consequently, there is a high unmet need for a non-invasive clinical test that can discriminate between benign and malignant nodules (5, 6).
The performance and development requirements for a diagnostic test to mitigate the use of invasive and costly medical procedures for lung nodule evaluations are as follows:
First, physicians require a negative test result (i.e. “benign”) to be correct with high probability (over 90%) to ensure malignant nodules are not accidentally eliminated, that is, high negative predictive value (NPV), which is the percentage of correct negative test results. A NPV of 90% reduces the post-test probability of cancer to 10% or lower – a two-fold reduction in cancer risk from the 20% pre-test probability of cancer among patients selected for invasive procedures.
Second, the diagnostic test must frequently provide actionable results for clinical utility and economic benefit. This corresponds to the specificity of the test that is the percentage of benign nodules correctly called benign (i.e. negative) by the test. Specificity indicates the fraction of patients with benign tumors that can be identified confidently by the test. High impact tests such as Oncotype DX for treatment stratification of breast cancers has reported actionable results in approximately 34% of cases (7).
Third, the diagnostic test must be developed and validated on intended use samples from multiple independent sites without demographic bias on key clinical parameters such as age, nodule size, gender, etc. Intended use samples are defined to be radiologically discovered and pathologically confirmed malignant or benign nodules with a diameter of less than 30 mm (Stage IA cancers). The intended use population has a high occurrence of current and former smokers as this is a significant risk factor for lung cancer.
Fourth, development and validation studies should conform to rigorous guidelines for test development such as those recently provided by the Institute of Medicine (IOM) (8).
Previous biomarker studies on lung cancer (9–15) have not achieved optimal development and performance requirements, in particular, the requirement of achieving a NPV of 90% on a multisite validation study with only Stage IA samples.
We present here a 13-protein plasma test, or classifier, meeting the performance and development requirements stated above. We utilized multiple reaction monitoring (MRM, also known as selected reaction monitoring in literature) mass spectrometry (MS) to measure the concentrations of candidate proteins in plasma (16, 17). The benefits of MRM assays include high protein specificity, large multiplexing capacity, high sensitivity (mid atomole), significant dynamic range (106), and both rapid and reliable assay development and deployment. MRM has been used for clinical testing of small molecule analytes for many years, and recently in the development of biologically relevant assays (18–23).
Results
Selection of biomarker candidates for assay development
To identify lung cancer biomarkers in blood that are shed or secreted from lung tumor cells, proteins over-expressed on the cell surface or over-secreted from lung cancer tumor cells relative to normal lung cells were identified from freshly resected lung tumors using organelle isolation techniques combined with mass spectrometry (24, 25)(Table S1). In addition, an extensive literature search for lung cancer biomarkers was performed using public and private resources (Table S2). Both the tissue-sourced biomarkers and literature-sourced biomarkers were required to have evidence of presence in blood (Table S3 and Fig. S1). More details are provided in Supplementary Materials.
Table 1 presents the steps taken in the refinement of these initial 388 protein candidates down to the set of 13 classifier proteins used for validation and performance assessment. The results are presented in the same sequence.
Table 1.
Steps in refining the 388 candidates down to the 13-protein classifier.
| Number of Proteins | Refinement | Data |
|---|---|---|
| 388 | Lung cancer associated protein candidates sourced from tissue and literature. | Table S3 |
| 371 | Number of the 388 protein candidates successfully developed into a MRM assay. | Table S5 |
| 190 | Number of the 371 MRM protein assays detected in plasma. | Table S5 |
| 125 | Number of the 190 MRM protein assays detected in at least 50% of cancer or 50% of benign discovery samples. | - |
| 36 | Number of the 125 detected proteins that were cooperative. | Table S7 |
| 21 | Number of the 36 cooperative proteins with robust MRM assays (i.e. no interfering signals, good signal-to-noise, etc.) | Table S7 |
| 13 | Number of the 21 robust and cooperative proteins with stable logistic regression coefficients. | Table 3 |
Development of MRM assays
Standard synthetic peptide techniques were used to develop a 371-protein multiplexed MRM assay from the 388 protein candidates. On average over 4 MRM transitions, i.e. predetermined and highly specific mass products (17), per protein were developed (Table S4–S5). Statistical correlation techniques were used to establish a transition-to-protein error rate below 5% ensuring high quality MRM assay development (see Supplementary Materials and Fig. S2). Synthetic peptides could not be developed or confidently identified for 17 candidates. The 371-protein MRM assay was applied to plasma samples from patients with pathologically confirmed benign or malignant nodules to determine how many of the 371 proteins could be detected in plasma. A total of 190 MRM assays were able to detect their target proteins in plasma (51% success rate; see also Table S4 and Fig S2). This success rate compared favorably to similar efforts (16%) to develop large scale MRM assays for the detection of diverse cancer markers in blood (26).
Classifier discovery
A total of 143 samples were obtained from three clinical sites (Table 2). All samples were selected to be consistent with intended use, specifically, having nodule size between 4–30 mm. Cancer and benign samples were pathologically confirmed. After clinical data monitoring, six samples were later found to have nodule size outside the 4–30mm range. Benign and cancer samples were pairwise matched on age, gender, nodule size and clinical site to avoid bias during MRM analysis. One benign sample was lost due to experimental deviations. See Table S6 for the experimental design.
Table 2.
Clinical characteristics of subjects and nodules in the discovery and validation studies.
| Characteristics | Cancer | Benign | p-value | Cancer | Benign | p-value |
|---|---|---|---|---|---|---|
|
| ||||||
| Discovery Study | Validation Study | |||||
|
|
||||||
|
Subjects
|
72 | 71 | 52 | 52 | ||
| Age (year)* | 65 (59–72) | 64 (52–71) | 0.46† | 63 (60–73) | 62 (56–67) | 0.03† |
| Gender | 1.00‡ | 0.85‡ | ||||
| Male | 29 | 28 | 25 | 27 | ||
| Female | 43 | 43 | 27 | 25 | ||
| Smoking history Status | 0.006‡ | 0.006‡ | ||||
| Never§ | 5 | 19 | 3 | 15 | ||
| Former | 60 | 44 | 38 | 29 | ||
| Current | 6 | 6 | 11 | 7 | ||
| No data | 1 | 2 | 0 | 1 | ||
| Pack-year*¶ | 37 (20–52) | 20 (0–40) | 0.001† | 40 (19–50) | 27 (0–50) | 0.09† |
|
Nodules
|
||||||
| Size (mm)* | 13 (10–16) | 13 (10–18) | 0.69† | 16 (13–20) | 15 (12–22) | 0.68† |
| Source | 1.00‡ | 0.89‡ | ||||
| IUCPQ|| | 14 | 14 | 13 | 12 | ||
| New York | 29 | 28 | 6 | 9 | ||
| Pennsylvania | 29 | 29 | 14 | 13 | ||
| Vanderbilt | 0 | 0 | 19 | 18 | ||
| Histopathology | ||||||
| Benign diagnosis | ||||||
| Granuloma | - | 48 | - | 26 | ||
| Hamartoma | - | 9 | - | 6 | ||
| Scar | - | 2 | - | 2 | ||
| Other | - | 12 | - | 18 | ||
| Cancer diagnosis | ||||||
| Adenocarcinoma | 41 | - | 25 | - | ||
| Squamous cell | 3 | - | 15 | - | ||
| Large cell | 0 | - | 2 | - | ||
| Bronchioloalveolar (BAC) | 3 | - | 0 | - | ||
| Adenocarcinoma/BAC | 21 | - | 5 | - | ||
| Other | 4 | - | 5 | - | ||
Data shown are median values with quartile ranges indicated in parentheses.
Mann-Whitney test.
Fisher’s exact test.
A never smoker is defined as an individual who has a lifetime history of smoking less than 100 cigarettes.
A pack-year is defined as the product of the total number of years of smoking and the average number of packs of cigarettes smoked daily. Pack-year data were not available for 4 cancer and 6 benign subjects in the discovery set and 2 cancer and 3 benign subjects in the validation set.
IUCPQ is the Institute Universitaire de Cardiologie et de Pneumologie de Quebec.
The 371-protein MRM assay was applied to the 143 discovery samples and the resulting transition data were analyzed to derive a 13-protein classifier fit to a logistic regression model (Table 3). The steps in the refinement from the 371 proteins to the 13 proteins participating in the classifier are summarized in Table 1. The key step in this refinement was the identification of 36 cooperative proteins. A protein was deemed cooperative if found more frequently on best performing panels than expected by chance alone. This strategy was motivated by the intent to capture the integrated behavior of proteins within lung cancer-perturbed networks. This was a defining step in the discovery of the classifier as the most cooperative proteins were often not the proteins with best individual performance. Full details of the estimation procedure and discovery process appear in Materials and Methods; sample classifier scores are listed in Table S6; the 36 cooperative proteins are provided in Table S7. The applicability of logistic regression modeling to the discovery dataset is provided in Table S8.
Table 3.
The 13-protein logistic regression classifier.
| Protein (Human) | Transition | Coefficient* | 95% CI* | p-value† | Tissue biomarker‡ | Biomarker in literature |
|---|---|---|---|---|---|---|
| LRP1 | TVLWPNGLSLDIPAGR_855.00_400.20 | −1.59 | (−2.39, −1.20) | 9.64E-05 | EPI | (38) |
| BGH3 | LTLLAPLNSVFK_658.40_804.50 | 1.73 | (1.18, 2.53) | 4.37E-03 | (39) | |
| COIA1 | AVGLAGTFR_446.26_721.40 | −1.56 | (−2.48, −1.16) | 3.90E-04 | (40) | |
| TETN | LDTLAQEVALLK_657.39_330.20 | −1.79 | (−2.95, −1.21) | 6.71E-03 | (41) | |
| TSP1 | GFLLLASLR_495.31_559.40 | 0.53 | (0.35, 0.93) | 1.13E-02 | (42) | |
| ALDOA | ALQASALK_401.25_617.40 | −0.80 | (−1.21, −0.51) | 1.03E-02 | Secreted, EPI | (43) |
| GRP78 | TWNDPSVQQDIK_715.85_260.20 | 1.41 | (0.91, 2.24) | 6.24E-03 | Secreted, EPI, ENDO | (44) |
| ISLR | ALPGTPVASSQPR_640.85_841.50 | 1.40 | (0.85, 2.21) | 1.09E-02 | ||
| FRIL | LGGPEAGLGEYLFER_804.40_913.40 | 0.39 | (0.24, 0.66) | 1.63E-02 | Secreted, EPI, ENDO | (45) |
| LG3BP | VEIFYR_413.73_598.30 | −0.58 | (−0.97, −0.28) | 6.24E-02 | Secreted | (46) |
| PRDX1 | QITVNDLPVGR_606.30_428.30 | −0.34 | (−0.64, −0.10) | 1.54E-01 | EPI | (47) |
| FIBA | NSLFEYQK_514.76_714.30 | 0.31 | (0.16, 0.53) | 4.10E-02 | (48) | |
| GSLG1 | IIIQESALDYR_660.86_338.20 | −0.70 | (−1.51, −0.19) | 1.60E-01 | EPI, ENDO | |
| Constant | 36.16 | (19.72, 59.01) |
Based on 20,000 Monte Carlo cross validation permutations with a 20% hold-out rate: “Coefficient” is the corresponding median values from the permutations, “95% CI” is the corresponding 95% BCa confidence intervals (49).
Based on likelihood ratio test on the whole discovery dataset, comparing the models with or without the corresponding proteins.
See Supplementary Materials and Table S3 for details.
Classifier performance in discovery study
We assessed the performance of the classifier in the discovery study in terms of negative predictive value (NPV) and specificity (SPC) (Fig. 1). When the classifier predicts that a patient from the intended use population has a benign tumor, the NPV is the probability that this prediction is true. NPV can be calculated from the classifier’s sensitivity, specificity and the estimated cancer prevalence in the intended use population (27). Specificity is the fraction of the benign nodules that the classifier is able to detect with high confidence. Thus, the higher the specificity at a given high NPV, the more patients with benign nodules can be rescued from unnecessary invasive procedures.
Fig. 1.
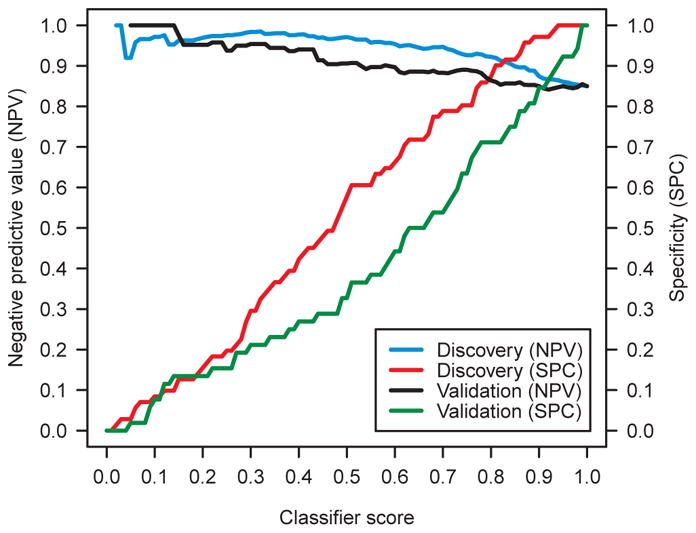
Performance of the classifier on the discovery samples (n=143) and validation samples (n=104). Negative predictive value (NPV) and specificity (SPC) are presented in terms of classifier score. A cancer prevalence of 15% was assumed.
We assumed a cancer prevalence of 15% based on estimates from the literature (4). Note that this prevalence is larger than the prevalence for the much smaller nodules studied in the National Lung Screening Trial (NLST) study (28). The classifier generated a cancer score, ranging from 0 to 1. Any reference value in this range can be defined so that a sample was predicted to be benign if its classifier score was not above the reference value, or malignant otherwise. We plotted in Fig. 1 the NPV and the specificity of the classifier on the discovery samples as a function of the reference value.
Table 4 reports the classifier’s performance on the discovery samples for multiple lung cancer prevalences with selected reference values. For each prevalence, the reference value was selected that corresponded to a discovery NPV of 95%. For a cancer prevalence of 15%, the reference value of 0.60 was selected and the classifier had a NPV of 95% +/−2% and a specificity of 66% +/−11% on the discovery samples, where 95% confidence intervals (CIs) were reported (see Supplementary Materials). The receiver operating characteristic (ROC) curve and the corresponding area under the curve (AUC) of the classifier on the discovery samples were reported in Fig. S3.
Table 4.
Performance of the classifier in discovery and validation at three cancer prevalences.
| Dataset | Prevalence (%) | Reference Value | Sensitivity (%) | Specificity (%) | NPV* (%) | PPV (%) |
|---|---|---|---|---|---|---|
| Discovery (n=143) | 15 | 0.60 | 82 | 66 | 95 | 30 |
| 20 | 0.46 | 90 | 49 | 95 | 31 | |
| 25 | 0.42 | 93 | 45 | 95 | 36 | |
|
| ||||||
| Validation (n=104) | 15 | 0.60 | 71 | 44 | 90 | 18 |
| 20 | 0.46 | 83 | 29 | 87 | 23 | |
| 25 | 0.42 | 90 | 27 | 89 | 29 | |
|
| ||||||
| Vanderbilt (n=37) | 15 | 0.60 | 79 | 56 | 94 | 24 |
| 20 | 0.46 | 89 | 33 | 93 | 25 | |
| 25 | 0.42 | 100 | 28 | 100 | 32 | |
The area under the receiver operating characteristic (ROC) curve (AUC) was 0.82, 0.60 and 0.74 in discovery, in validation and for Vanderbilt samples, respectively. Note that partial AUC is the preferred metric for tests maximizing NPV (27).
NPV is negative predictive value.
PPV is positive predictive value.
Classifier validation
The 13-protein classifier was fully defined before validation was performed, including the identity of the proteins, the logistic regression model, and the reference value used to classify a nodule as benign or malignant. We followed precisely the bright line demarcation of a locked down omics test as defined in the Institute of Medicine (IOM) omics report (8).
A total of 52 cancer and 52 benign samples (Table 2) were used to validate the performance of the 13-protein classifier. All validation samples were from different patients than the discovery samples. In addition, 36% of the validation samples were sourced from a new fourth clinical site, Vanderbilt University. The remaining validation samples were selected randomly from the discovery sites. Samples were consistent with intended use and matched as in the discovery study. The classifier was applied to the validation samples and analyzed (see Materials and Methods). See Table S9 for the experimental design and classifier scores of individual samples.
The performance of the classifier on the validation samples was summarized in Fig. 1, Table 4 and Fig. S3, along with the corresponding performance on the discovery samples. All reference values in Table 4 were selected in discovery and applied directly in validation. For cancer prevalence 15%and reference value 0.60, the classifier had NPV 90% +/−5 % and specificity 44% +/−13 % on the validation samples. The NPV and specificity on the Vanderbilt samples were 94% and 56%, respectively, providing a strong sign that the classifier was not overfit to the discovery sites.
Fig. 2 presents the application of the classifier to all 247 discovery and validation samples. We compared the clinical risk factors of smoking (measured in pack years) and nodule size (proportional to the diameter of each circle) to the classifier score assigned to each sample. Nodule size did not appear to increase with the classifier score. Indeed, both large and small nodules were spread across the classifier score spectrum. To quantify this observation, the Pearson correlation (R) between the classifier score and nodule size, smoking history pack-year and age were calculated (Table 5). The largest R2 was 0.05, indicating that all correlations were either non-existing or very weak. The implication of this observation is remarkable. The classifier provides information on the disease status of pulmonary nodules that is independent of the three currently used risk factors for malignancy (age, smoking history and nodule size)(29, 30), and thus provides incremental molecular information of added clinical value. For a similar plot of nodule size vs. classifier score, see Fig. S4. See Table S10 for more details illustrating impact of clinical characteristics on classifier score.
Fig. 2.
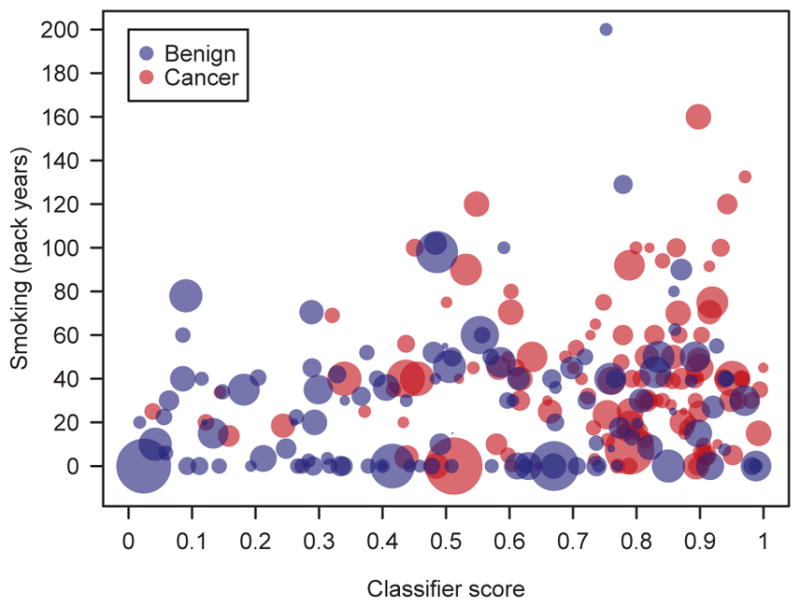
Multivariate analysis of clinical (smoking, nodule size) and molecular (classifier score) factors as they relate to cancer and benign samples (n=247) in the discovery and validation studies. Smoking is measured by pack-years on the vertical. Nodule size is represented by circle diameter.
Table 5.
Pearson correlation between classifier score and clinical risk factors.
| Risk factor | All samples | Cancer samples | Benign samples |
|---|---|---|---|
| Age (year) | 0.190 | 0.015 | 0.227 |
| Smoking history (pack year) | 0.185 | 0.089 | 0.139 |
| Nodule size (mm) | −0.071 | −0.081 | −0.035 |
The molecular foundations of the classifier
By design, the 13 classifier proteins were selected from a candidate list of 388 proteins, each with either empirical evidence or literature support of differential expression in lung cancer tissue, or both (Table 3). The only exception was the protein ISLR whose function is currently not well-characterized in the literature despite the fact that it was identified as a lung cancer biomarker in the automatic literature search.
To better understand the specific role each of these 13 proteins have in lung cancer, and their relationship to each other, they were submitted for pathway analysis using Ingenuity Systems (IPA). First, the transcription regulators most likely to cause a modulation of these 13 proteins were identified. With standard IPA analysis parameters, the four most significant nuclear transcription regulators were FOS (proto-oncogene c-Fos), NF2L2 (nuclear factor erythroid 2-related factor 2), AHR (aryl hydrocarbon receptor) and MYC (myc proto-oncogene protein): See Materials and Methods for details. These proteins regulate 12 of the 13 classifier proteins, with ISLR being the exception (see below).
FOS is common to many forms of cancer. NF2L2 and AHR are associated with lung cancer, oxidative stress response and lung inflammation. MYC is associated with lung cancer and oxidative stress response. These four transcription regulators and the 13 classifier proteins, collectively, are also highly associated (p-value 1.0×10−7) with the same three biological networks, namely, lung cancer, lung inflammation and oxidative stress response. This is summarized in Fig. 3 where the classifier proteins (green), transcription regulators (blue) and the three merged networks (orange) are depicted. Only ISLR is not connected through these three networks to other classifier proteins, although it is connected through cancer networks not specific to lung. In summary, the modulation of the 13 classifier proteins can be linked back to a few transcription regulators highly associated with lung cancer, lung inflammation and oxidative stress response networks; three biological processes reflecting aspects of lung cancer.
Fig. 3.
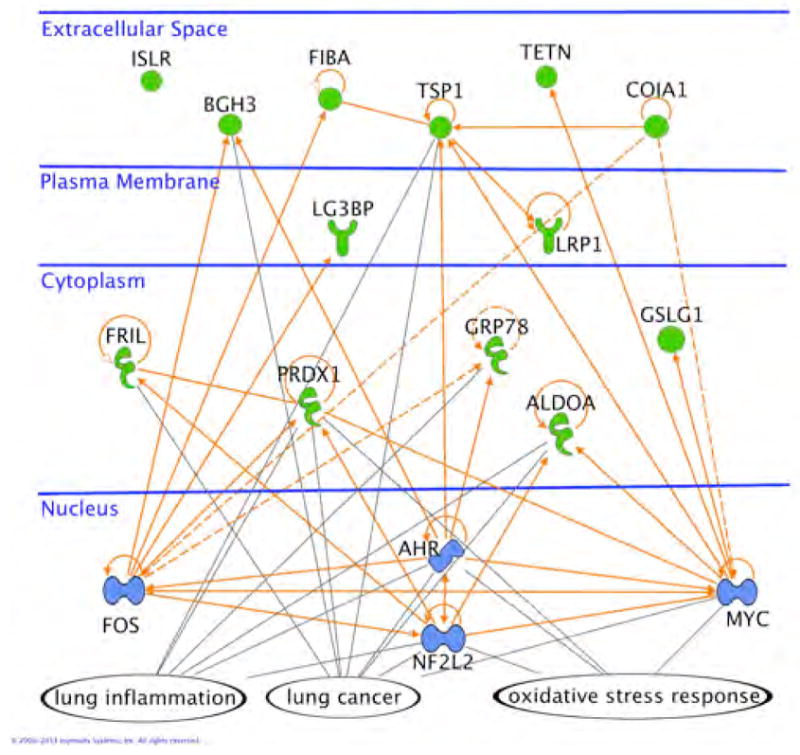
The 13 classifier proteins (green), 4 transcription regulators (blue) and the three networks (orange lines) of lung cancer, oxidative stress response and lung inflammation. All references are human UniProt identifiers.
Derivation of the classifier
MRM technology enabled the simultaneous exploration of a large number of lung cancer relevant proteins. The definitive step in the derivation of the classifier was the identification of the most cooperative protein biomarkers. Typically, proteins are shortlisted in the discovery process by filtering on individual diagnostic performance. To contrast the difference between filtering proteins based on strong individual performance as opposed to frequency on high performance panels, we calculated a p-value for each of the 21 MRM-robust cooperative proteins using the Mann-Whitney non-parametric test (31). Only 1 of these 21 proteins had a p-value below 0.10 (Table S7). In addition, the cooperative protein score for each of these 21 proteins was calculated. The cooperative protein score measured the frequency at which each protein appeared on high performance panels (see Materials and Methods). Importantly, the p-values and cooperative scores of the 21 MRM-robust cooperative proteins were not correlated (Pearson correlation −0.24, p-value 0.30).
Most informative proteins
Which proteins in the classifier were most informative? To answer this question all possible classifiers were constructed from the set of 21 MRM-robust cooperative proteins (Table S7) and their performance measured. The frequency of each protein among the 100 best performing panels was determined (see “Frequency” column of Table S7). Four proteins (LRP1, COIA1, ALDOA, LG3BP) were highly enriched with 95% of the 100 best classifiers having at least three of these four proteins (p-value < 1.2 ×10−41). The conclusion was that high performance panels of cooperative proteins for pulmonary nodule characterization were similar in composition to one another with a preference for a set of particularly informative (cooperative) proteins. It is expected that optimizations of the classifier on different technological platforms (e.g. sample processing pipelines, mass spectrometry instrumentation, etc.) may result in changes in optimal classifier parameters such as the logistic regression protein coefficients.
Discussion
Classifier score independent of clinical risk factors
Recent work has demonstrated that there is frequently not a correlation between an individual protein’s blood concentration and tumor size (32), as is the case for our signature. In fact, even if individual protein concentrations increased or decreased with tumor size, our classifier score is combination of these 13 protein concentrations, and so, would not necessarily correlate to tumor size. Of clinical importance is that the classifier provides a new metric, independent of current diagnostic risk factors (29, 30), for assessing the molecular status of a lung nodule.
Limitations of work
The main limitation of the work presented is that both discovery and validation studies were conducted using retrospective samples. A prospective study on intended use samples is required to further validate the utility of the classifier for clinical use.
A second limitation is that the molecular classifier presented here is not integrated with clinical risk factors such as nodule size, age and smoking history for the purpose of a single classifier for lung nodule diagnosis. Although an integrated classifier would be ideal, in practice pulmonologists vary broadly in the use of clinical risk factors, and so, it is actually preferable to have a molecular diagnostic test that produces a score independent of clinical risk factors.
Materials and Methods
Discovery study design
A retrospective, multi-center, case-control study was performed using K2-EDTA plasma aliquots previously obtained from subjects who provided informed consent and contributed biospecimens in studies approved by the Ethics Review Board (ERB) or the Institutional Review Boards (IRB) at the IUCPQ or New York University and the University of Pennsylvania, respectively. In addition, plasma samples were provided by study investigators after review and approval of the sponsor’s study protocol by the respective institution’s ERB or IRB, as required. Sample eligibility for the proteomic analysis was based on the satisfaction of the study inclusion and exclusion criteria (Supplementary Materials). Each cancer-benign sample pair was matched in best efforts by gender, nodule size (±10 mm), age (±10 years), smoking history pack-years (±20 pack-years), and by center. The study was powered with a probability of 92% to detect 1.5 fold differences in protein abundance between malignant and benign lung nodules. See Table S6 for more details.
Analysis of plasma samples using MRM-MS
The protocol for MRM-MS analysis of plasma aliquots included immunodepletion on IgY14-Supermix resin columns (Sigma), denaturation, trypsin digestion, and de-salting, followed by reversed-phase liquid chromatography and MRM-MS analysis of the obtained peptide samples (see details in Supplementary Materials).
Development of MRM assays
MRM assays for candidate proteins were developed based on synthetic peptides (17, 26, 33). More details are provided in Supplementary Materials.
Identification of endogenous normalizing proteins
The following criteria were used to identify a transition of a normalization protein: (A) highest median intensity of all transitions from the same protein; (B) detected in all samples; (C) ranking high, as a normalizer, in reducing median technical coefficient of variation (CV); (D) ranking high in reducing median column drift (defined in Supplementary Materials); and (E) possession of low median technical CV and low median biological CV, i.e. median CV of transition intensities that were measured on clinical samples. Six endogenous normalizing proteins were identified (Table S11 and Fig. S5).
Normalization of raw MRM-MS data
Six normalization transitions (Table S11) were used to normalize raw MRM-MS data to reduce sample-to-sample intensity variations within same study. A scaling factor was calculated for each sample so that the intensities of the six normalization transitions of the sample were aligned with the corresponding median intensities of all human plasma standard (HPS) samples. Assuming that Ni,s was the intensity of a normalization transition i in sample s and N̂i the corresponding median intensity of all HPS samples, then the scaling factor for sample s was given by Ŝ/Ss, where
| (1) |
was the median of the intensity ratios and Ŝ was the median of Ss over all samples in the study. For each transition of each sample, its normalized intensity was calculated as
| (2) |
where Ii,s was the raw intensity.
Logistic Regression Model
The logistic regression classification method (34, 35) was used to combine a panel of transitions into a classifier and to calculate a classification score between 0 and 1 for each sample. The score (Ps) of a sample was determined as
| (3) |
where Ǐi,s was the logarithmically transformed (base 2), normalized intensity of transition i in sample s, βi was the corresponding logistic regression coefficient, α was a classifier-specific constant, and N was the total number of transitions in the classifier. A sample was classified as benign if Ps was less than or equal to a reference value or cancer otherwise.
Lung nodule classifier development
The classifier development included the following steps:
Normalization of raw MRM-MS data was performed to reduce sample-to-sample intensity variations using a panel of six endogenous proteins as described above. After normalization, MRM-MS data were filtered down to transitions having the highest intensities of the corresponding proteins and satisfying the criterion for detection in a minimum of 50% of the cancer or 50% of the benign samples. A total of 125 proteins satisfied these criteria of reproducible detection. Missing values were replaced by half the minimum detected values of the corresponding transitions in all samples.
Remaining transitions were then used to identify proteins, defined as cooperative proteins, that occurred with high frequency on top-performing protein panels. The cooperative proteins were derived using the following estimation procedure as it was not computational feasible to evaluate the performance of all possible protein panels.
Monte Carlo cross validation (MCCV) (36) was performed on 1×106 panels, each panel comprised of 10 randomly selected proteins and fitted to a logistic regression model, as described above, using a 20% holdout rate and 102 sample permutations. The receiver operating characteristic (ROC) curve of each panel was generated and the corresponding partial area under the ROC curve (AUC) but above the boundary of sensitivity being 90%, defined as the partial AUC (27, 37), was used to assess the performance of the panel. By focusing on the performance of individual panels at high sensitivity region, the partial AUC allowed for the identification of panels with high and reliable performance on NPV. The candidate proteins that occurred in the top 100 performing panels with a frequency greater than that expected by chance were identified empirically as cooperative proteins. For each protein the cooperative score was defined as its frequency on the 100 high performance panels divided by the expected frequency. Highly cooperative proteins had a score of 1.75 or higher (one-sided p value < 0.05), cooperative proteins had a score higher than 1 while non-cooperative proteins had a score of 1 or less. Note that one million panels were sampled to ensure that the 100 top performing panels were exceptional (empirical p value ≤ 10−4). In addition, panels of size 10 were used in this procedure based on empirical evidence that larger panels did not change the resulting list of cooperative proteins. This also avoided overfitting the logistic regression model. In total, 36 cooperative proteins were identified, including 15 highly cooperative proteins.
Raw chromatograms of all transitions of cooperative proteins were manually reviewed. Proteins with low signal-to-noise ratios and/or showing evidence of any interference were removed from further consideration. In total, 21 cooperative and robust proteins were identified.
Remaining candidate proteins were then evaluated in an iterative, stepwise procedure to derive the final classifier. In each step, MCCV was performed using a holdout rate of 20% and 104 sample permutations to train the remaining candidate proteins to a logistic regression model and to assess the variability, i.e. stability, of the coefficient derived for each protein by the model. The performance of the model was assessed by partial AUC (27). The protein having the least stable coefficient was identified and removed. This procedure was repeated until no protein was left. During this procedure a total of 21 logistic regression classifiers were developed and their respective performances were obtained. Among the 21 classifiers the 13-protein classifier had the optimal performance and was selected as the classifier for validation. Seven of the 13 proteins in the final classifier were highly cooperative.
Proteins in the final classifier were further trained to a logistic regression model by MCCV with a holdout rate of 20% and 2×104 sample permutations. Results for the lung nodule classifier development are summarized in Table S7.
Lung nodule classifier validation
The design of the validation study was identical to that of the discovery study, but involved plasma samples associated with independent subjects not evaluated in the discovery study. Additional specimens were obtained from Vanderbilt University with similar requirements for patient consent, IRB approval, and satisfaction of HIPAA requirements. Of the 104 total cancer and benign samples in the validation study, half were analyzed immediately after the discovery study, while the other half were analyzed later. The study was powered to observe the expected 95% CI of the NPV associated with reference value 0.60. See Supplementary Materials and Table S9 for more details.
The raw MRM-MS dataset in the validation study was normalized in the same way as the discovery dataset. Variability between the discovery and the validation studies was mitigated by utilizing human plasma standard (HPS) samples in both studies as external calibrator: See Supplementary Materials for details. Missing data in the validation study were then replaced by half the minimum detected values of the corresponding transitions in the discovery study. Normalized, calibrated transition intensities were applied to the logistic regression model of the final classifier learned previously in the training phase, from which classifier scores were assigned to individual samples. The performance of the lung nodule classifier on the validation samples was then assessed based on the classifier scores.
IPA pathway analysis
Standard parameters were used. Specifically, in the search for nuclear transcription regulators, requirements were p-value < 0.01 with a minimum of 3 proteins modulated. Significance was determined using a right-tailed Fisher’s exact test using the IPA Knowledge Database as background.
Statistical analysis
All statistical analyses were performed with STATA, MATLAB and/or R. Specific tests and analysis details are indicated at appropriate sections.
Supplementary Material
Acknowledgments
We thank the reviewers for many insightful and constructive comments. We also thank the subjects who contributed biospecimens during translational research studies and to the research staff at each of the participating institutions, and also to the research team at Caprion Proteomics. We also thank Michel Laviolette and François Maltais at the IUCPQ for their helpful discussions, and Adam Callahan, Sherri Rogalski, Ed Gonterman and Mi-Youn Brusniak at Integrated Diagnostics (Indi) for their many contributions to this work.
Funding: Supported by Integrated Diagnostics.
Footnotes
Author contributions:
Individual co-author contributions, in addition to manuscript drafting, review and approval, are as follows.
Concept and Design: XJL, MD, SWH, ScL, NDP, SL, PPM, HP, WNR, AV, KCF, LH, PK
Data Acquisition: ScL, HB, MS, OG, JL, RA, DC
Data Analysis: XJL, CH, PYF, MD, SWH, LWL, MM, HB, PK
Data Interpretation: XJL, MD, SWH, NDP, KCF, LH, PK
Competing interests:
XJL, CH, PYF, SWH, LWL, MM, ScL, KCF and PK are current and/or past employees of and have equity interest in Integrated Diagnostics (Indi); MD, HB, MS, OG, JL, RA and DC are consultants and/or performed contracted work for Indi; LH is a board member with equity of Indi.
XJL, CH, MD, KCF and PK filed patent applications directed toward the compositions, methods, kits and processes described in the US and in foreign jurisdictions.
Data and materials availability:
Raw data in mzML format can be downloaded from SRMAtlas (http://www.peptideatlas.org/PASS/PASS00261).
References and Notes
- 1.Schauer DA, Linton OW. National Council on Radiation Protection and Measurements report shows substantial medical exposure increase. Radiology. 2009 Nov;253:293. doi: 10.1148/radiol.2532090494. [DOI] [PubMed] [Google Scholar]
- 2.Stern SH, Kaczmarek R, Spelic VDC, Suleiman OH. Nationwide Evaluation of X-Ray Trends (NEXT) 2000–01 Survey of Patient Radiation Exposure from Computed Tomographic (CT) Examinations in the United States. 2001. [Google Scholar]
- 3.Wiener RS, Schwartz LM, Woloshin S, Welch HG. Population-based risk for complications after transthoracic needle lung biopsy of a pulmonary nodule: an analysis of discharge records. Annals of internal medicine. 2011 Aug 2;155:137. doi: 10.1059/0003-4819-155-3-201108020-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacMahon H, Austin JH, Gamsu G, Herold CJ, Jett JR, Naidich DP, Patz EF, Jr, Swensen SJ, Fleischner S. Guidelines for management of small pulmonary nodules detected on CT scans: a statement from the Fleischner Society. Radiology. 2005 Nov;237:395. doi: 10.1148/radiol.2372041887. [DOI] [PubMed] [Google Scholar]
- 5.Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers--blood-based strategies to detect and monitor cancer. Nature reviews clinical oncology. 2011 Mar;8:142. doi: 10.1038/nrclinonc.2010.220. [DOI] [PubMed] [Google Scholar]
- 6.Miller YE. Minimizing unintended consequences of detecting lung nodules by computed tomography. American journal of respiratory and critical care medicine. 2008 Nov 1;178:891. doi: 10.1164/rccm.200808-1257ED. [DOI] [PubMed] [Google Scholar]
- 7.Kelly CM, Krishnamurthy S, Bianchini G, Litton JK, Gonzalez-Angulo AM, Hortobagyi GN, Pusztai L. Utility of oncotype DX risk estimates in clinically intermediate risk hormone receptor-positive, HER2-normal, grade II, lymph node-negative breast cancers. Cancer. 2010 Nov 15;116:5161. doi: 10.1002/cncr.25269. [DOI] [PubMed] [Google Scholar]
- 8.Micheel C, Nass SJ, Omenn GS Institute of Medicine (U.S.) Evolution of translational omics: lessons learned and the path forward. National Academies Press; Washington, D.C: 2012. Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials; p. xv.p. 338. [PubMed] [Google Scholar]
- 9.Pecot CV, Li M, Zhang XJ, Rajanbabu R, Calitri C, Bungum A, Jett JR, Putnam JB, Callaway-Lane C, Deppen S, Grogan EL, Carbone DP, Worrell JA, Moons KG, Shyr Y, Massion PP. Added value of a serum proteomic signature in the diagnostic evaluation of lung nodules. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012 May;21:786. doi: 10.1158/1055-9965.EPI-11-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostroff RM, Bigbee WL, Franklin W, Gold L, Mehan M, Miller YE, Pass HI, Rom WN, Siegfried JM, Stewart A, Walker JJ, Weissfeld JL, Williams S, Zichi D, Brody EN. Unlocking biomarker discovery: large scale application of aptamer proteomic technology for early detection of lung cancer. PloS one. 2010;5:e15003. doi: 10.1371/journal.pone.0015003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bigbee WL, Gopalakrishnan V, Weissfeld JL, Wilson DO, Dacic S, Lokshin AE, Siegfried JM. A multiplexed serum biomarker immunoassay panel discriminates clinical lung cancer patients from high-risk individuals found to be cancer-free by CT screening. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2012 Apr;7:698. doi: 10.1097/JTO.0b013e31824ab6b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rom WN, Goldberg JD, Addrizzo-Harris D, Watson HN, Khilkin M, Greenberg AK, Naidich DP, Crawford B, Eylers E, Liu D, Tan EM. Identification of an autoantibody panel to separate lung cancer from smokers and nonsmokers. BMC cancer. 2010;10:234. doi: 10.1186/1471-2407-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam S, Boyle P, Healey GF, Maddison P, Peek L, Murray A, Chapman CJ, Allen J, Wood WC, Sewell HF, Robertson JF. EarlyCDT-Lung: an immunobiomarker test as an aid to early detection of lung cancer. Cancer Prev Res (Phila) 2011 Jul;4:1126. doi: 10.1158/1940-6207.CAPR-10-0328. [DOI] [PubMed] [Google Scholar]
- 14.Chapman CJ, Healey GF, Murray A, Boyle P, Robertson C, Peek LJ, Allen J, Thorpe AJ, Hamilton-Fairley G, Parsy-Kowalska CB, MacDonald IK, Jewell W, Maddison P, Robertson JF. EarlyCDT(R)-Lung test: improved clinical utility through additional autoantibody assays. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2012 Oct;33:1319. doi: 10.1007/s13277-012-0379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yildiz PB, Shyr Y, Rahman JS, Wardwell NR, Zimmerman LJ, Shakhtour B, Gray WH, Chen S, Li M, Roder H, Liebler DC, Bigbee WL, Siegfried JM, Weissfeld JL, Gonzalez AL, Ninan M, Johnson DH, Carbone DP, Caprioli RM, Massion PP. Diagnostic accuracy of MALDI mass spectrometric analysis of unfractionated serum in lung cancer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2007 Oct;2:893. doi: 10.1097/JTO.0b013e31814b8be7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nature biotechnology. 2009 Jul;27:633. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Molecular systems biology. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cima I, Schiess R, Wild P, Kaelin M, Schuffler P, Lange V, Picotti P, Ossola R, Templeton A, Schubert O, Fuchs T, Leippold T, Wyler S, Zehetner J, Jochum W, Buhmann J, Cerny T, Moch H, Gillessen S, Aebersold R, Krek W. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2011 Feb 22;108:3342. doi: 10.1073/pnas.1013699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin S, Zhou Y, Lok AS, Tsodikov A, Yan X, Gray L, Yuan M, Moritz RL, Galas D, Omenn GS, Hood L. SRM targeted proteomics in search for biomarkers of HCV-induced progression of fibrosis to cirrhosis in HALT-C patients. Proteomics. 2012 Apr;12:1244. doi: 10.1002/pmic.201100601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng X, Hood BL, Sun M, Conrads TP, Day RS, Weissfeld JL, Siegfried JM, Bigbee WL. Lung cancer serum biomarker discovery using glycoprotein capture and liquid chromatography mass spectrometry. Journal of proteome research. 2010 Dec 3;9:6440. doi: 10.1021/pr100696n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Addona TA, Shi X, Keshishian H, Mani DR, Burgess M, Gillette MA, Clauser KR, Shen D, Lewis GD, Farrell LA, Fifer MA, Sabatine MS, Gerszten RE, Carr SA. A pipeline that integrates the discovery and verification of plasma protein biomarkers reveals candidate markers for cardiovascular disease. Nature biotechnology. 2011 Jul;29:635. doi: 10.1038/nbt.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whiteaker JR, Lin C, Kennedy J, Hou L, Trute M, Sokal I, Yan P, Schoenherr RM, Zhao L, Voytovich UJ, Kelly-Spratt KS, Krasnoselsky A, Gafken PR, Hogan JM, Jones LA, Wang P, Amon L, Chodosh LA, Nelson PS, McIntosh MW, Kemp CJ, Paulovich AG. A targeted proteomics-based pipeline for verification of biomarkers in plasma. Nature biotechnology. 2011 Jul;29:625. doi: 10.1038/nbt.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hood LE, Omenn GS, Moritz RL, Aebersold R, Yamamoto KR, Amos M, Hunter-Cevera J, Locascio L. New and improved proteomics technologies for understanding complex biological systems: addressing a grand challenge in the life sciences. Proteomics. 2012 Sep;12:2773. doi: 10.1002/pmic.201270086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunet S, Thibault P, Gagnon E, Kearney P, Bergeron JJ, Desjardins M. Organelle proteomics: looking at less to see more. Trends in cell biology. 2003 Dec;13:629. doi: 10.1016/j.tcb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 25.Yates JR, 3rd, Gilchrist A, Howell KE, Bergeron JJ. Proteomics of organelles and large cellular structures. Nature reviews Molecular cell biology. 2005 Sep;6:702. doi: 10.1038/nrm1711. [DOI] [PubMed] [Google Scholar]
- 26.Huttenhain R, Soste M, Selevsek N, Rost H, Sethi A, Carapito C, Farrah T, Deutsch EW, Kusebauch U, Moritz RL, Nimeus-Malmstrom E, Rinner O, Aebersold R. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Science translational medicine. 2012 Jul 11;4:142ra94. doi: 10.1126/scitranslmed.3003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford University Press; New York: 2003. [Google Scholar]
- 28.The National Lung Screening Trial Research. Church TR, Black WC, Aberle DR, Berg CD, Clingan KL, Duan F, Fagerstrom RM, Gareen IF, Gierada DS, Jones GC, Mahon I, Marcus PM, Sicks JD, Jain A, Baum S. Results of initial low-dose computed tomographic screening for lung cancer. The New England journal of medicine. 2013 May 23;368:1980. doi: 10.1056/NEJMoa1209120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swensen SJ, Silverstein MD, Ilstrup DM, Schleck CD, Edell ES. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Archives of internal medicine. 1997 Apr 28;157:849. [PubMed] [Google Scholar]
- 30.Gould MK, Ananth L, Barnett PG SCSG Veterans Affairs. A clinical model to estimate the pretest probability of lung cancer in patients with solitary pulmonary nodules. Chest. 2007 Feb;131:383. doi: 10.1378/chest.06-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann HB, Whitney DR. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Annals of Mathematical Statistics. 1947;18:50. [Google Scholar]
- 32.Beer LA, Wang H, Tang HY, Cao Z, Chang-Wong T, Tanyi JL, Zhang R, Liu Q, Speicher DW. Identification of multiple novel protein biomarkers shed by human serous ovarian tumors into the blood of immunocompromised mice and verified in patient sera. PloS one. 2013;8:e60129. doi: 10.1371/journal.pone.0060129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nature methods. 2010 Jan;7:43. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 34.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer; 2009. [Google Scholar]
- 35.Hosmer DW, Lemeshow S, Sturdivant RX. Wiley series in probability and statistics. 3. Applied logistic regression. pp. pages cm. [Google Scholar]
- 36.Xu QS, Liang YZ. Monte Carlo cross validation. Chemometrics and Intelligent Laboratory Systems. 2001;56:1. [Google Scholar]
- 37.McClish DK. Analyzing a portion of the ROC curve. Medical decision making: an international journal of the Society for Medical Decision Making. 1989 Jul-Sep;9:190. doi: 10.1177/0272989X8900900307. [DOI] [PubMed] [Google Scholar]
- 38.Meng H, Chen G, Zhang X, Wang Z, Thomas DG, Giordano TJ, Beer DG, Wang MM. Stromal LRP1 in lung adenocarcinoma predicts clinical outcome. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011 Apr 15;17:2426. doi: 10.1158/1078-0432.CCR-10-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao L, Ji W, Zhang L, Ou G, Feng Q, Zhou Z, Lei M, Yang W, Wang L. Changes of circulating transforming growth factor-beta1 level during radiation therapy are correlated with the prognosis of locally advanced non-small cell lung cancer. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2010 Apr;5:521. doi: 10.1097/JTO.0b013e3181cbf761. [DOI] [PubMed] [Google Scholar]
- 40.Iizasa T, Chang H, Suzuki M, Otsuji M, Yokoi S, Chiyo M, Motohashi S, Yasufuku K, Sekine Y, Iyoda A, Shibuya K, Hiroshima K, Fujisawa T. Overexpression of collagen XVIII is associated with poor outcome and elevated levels of circulating serum endostatin in non-small cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004 Aug 15;10:5361. doi: 10.1158/1078-0432.CCR-04-0443. [DOI] [PubMed] [Google Scholar]
- 41.Okano T, Kondo T, Kakisaka T, Fujii K, Yamada M, Kato H, Nishimura T, Gemma A, Kudoh S, Hirohashi S. Plasma proteomics of lung cancer by a linkage of multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis. Proteomics. 2006 Jul;6:3938. doi: 10.1002/pmic.200500883. [DOI] [PubMed] [Google Scholar]
- 42.Papadaki C, Mavroudis D, Trypaki M, Koutsopoulos A, Stathopoulos E, Hatzidaki D, Tsakalaki E, Georgoulias V, Souglakos J. Tumoral expression of TXR1 and TSP1 predicts overall survival of patients with lung adenocarcinoma treated with first-line docetaxel-gemcitabine regimen. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009 Jun 1;15:3827. doi: 10.1158/1078-0432.CCR-08-3027. [DOI] [PubMed] [Google Scholar]
- 43.Lin CC, Chen LC, Tseng VS, Yan JJ, Lai WW, Su WP, Lin CH, Huang CY, Su WC. Malignant pleural effusion cells show aberrant glucose metabolism gene expression. The European respiratory journal. 2011 Jun;37:1453. doi: 10.1183/09031936.00015710. [DOI] [PubMed] [Google Scholar]
- 44.Urban P, Bilecova-Rabajdova M, Marekova M, Vesela J. Progression of apoptic signaling from mesenteric ischemia-reperfusion injury to lungs: correlation in the level of ER chaperones expression. Molecular and cellular biochemistry. 2012 Mar;362:133. doi: 10.1007/s11010-011-1135-4. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Asmitananda T, Gao L, Gai D, Song Z, Zhang Y, Ren H, Yang T, Chen T, Chen M. Biomarkers in the lung cancer diagnosis: a clinical perspective. Neoplasma. 2012;59:500. doi: 10.4149/neo_2012_064. [DOI] [PubMed] [Google Scholar]
- 46.Marchetti A, Tinari N, Buttitta F, Chella A, Angeletti CA, Sacco R, Mucilli F, Ullrich A, Iacobelli S. Expression of 90K (Mac-2 BP) correlates with distant metastasis and predicts survival in stage I non-small cell lung cancer patients. Cancer research. 2002 May 1;62:2535. [PubMed] [Google Scholar]
- 47.Li S, Wang R, Zhang M, Wang L, Cheng S. Proteomic analysis of non-small cell lung cancer tissue interstitial fluids. World journal of surgical oncology. 2013 Aug 5;11:173. doi: 10.1186/1477-7819-11-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones JM, McGonigle NC, McAnespie M, Cran GW, Graham AN. Plasma fibrinogen and serum C-reactive protein are associated with non-small cell lung cancer. Lung Cancer. 2006 Jul;53:97. doi: 10.1016/j.lungcan.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 49.Efron B, Tibshirani R. Bootstrap Methods for Standard Errors, Confidence Intervals, and Other Measures of Statistical Accuracy. Statistical Science. 1986;1:54. [Google Scholar]
- 50.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999 Dec;20:3551. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 51.Radulovic D, Jelveh S, Ryu S, Hamilton TG, Foss E, Mao Y, Emili A. Informatics platform for global proteomic profiling and biomarker discovery using liquid chromatography-tandem mass spectrometry. Molecular & cellular proteomics: MCP. 2004 Oct;3:984. doi: 10.1074/mcp.M400061-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Li XJ, Yi EC, Kemp CJ, Zhang H, Aebersold R. A software suite for the generation and comparison of peptide arrays from sets of data collected by liquid chromatography-mass spectrometry. Molecular & cellular proteomics: MCP. 2005 Sep;4:1328. doi: 10.1074/mcp.M500141-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Brusniak MY, Bodenmiller B, Campbell D, Cooke K, Eddes J, Garbutt A, Lau H, Letarte S, Mueller LN, Sharma V, Vitek O, Zhang N, Aebersold R, Watts JD. Corra: Computational framework and tools for LC-MS discovery and targeted mass spectrometry-based proteomics. BMC bioinformatics. 2008;9:542. doi: 10.1186/1471-2105-9-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005 Aug;5:3226. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 55.States DJ, Omenn GS, Blackwell TW, Fermin D, Eng J, Speicher DW, Hanash SM. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nature biotechnology. 2006 Mar;24:333. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 56.Deutsch EW, Eng JK, Zhang H, King NL, Nesvizhskii AI, Lin B, Lee H, Yi EC, Ossola R, Aebersold R. Human Plasma PeptideAtlas. Proteomics. 2005 Aug;5:3497. doi: 10.1002/pmic.200500160. [DOI] [PubMed] [Google Scholar]
- 57.Polanski M, Anderson NL. A list of candidate cancer biomarkers for targeted proteomics. Biomarker insights. 2007;1:1. [PMC free article] [PubMed] [Google Scholar]
- 58.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology. 2001 Jan 19;305:567. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 59.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of molecular biology. 2004 Jul 16;340:783. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 60.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein engineering, design & selection: PEDS. 2004 Apr;17:349. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 61.Sidak Z. Rectangular confidence regions for the means of multivariate normal distributions. Journal of the American Statistical Association. 1967;62:626. [Google Scholar]
- 62.Lehmann EL. Nonparametrics Statistical Methods Based on Ranks. Springer; New York: 2006. [Google Scholar]
- 63.Rao PV. Statistical Research Methods in the Life Sciences. Duxbury Press; New York: 1998. [Google Scholar]
- 64.Mercaldo ND, Lau KF, Zhou XH. Confidence intervals for predictive values with an emphasis to case-control studies. Statistics in medicine. 2007 May 10;26:2170. doi: 10.1002/sim.2677. [DOI] [PubMed] [Google Scholar]
- 65.Reiter L, Rinner O, Picotti P, Huttenhain R, Beck M, Brusniak MY, Hengartner MO, Aebersold R. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nature methods. 2011 May;8:430. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 66.Kearney P, Butler H, Eng K, Hugo P. Protein identification and Peptide expression resolver: harmonizing protein identification with protein expression data. Journal of proteome research. 2008 Jan;7:234. doi: 10.1021/pr0705439. [DOI] [PubMed] [Google Scholar]
- 67.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003 Aug 5;100:9440. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pepe MS. The Statistical Evaluation of Medical Tests for Classification and Prediction. Oxford University Press; 2003. p. 220. [Google Scholar]
- 69.Farrah T, Deutsch EW, Omenn GS, Campbell DS, Sun Z, Bletz JA, Mallick P, Katz JE, Malmstrom J, Ossola R, Watts JD, Lin B, Zhang H, Moritz RL, Aebersold R. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Molecular & cellular proteomics: MCP. 2011 Sep;10:M110 006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X, Ender P, Mitchell M, Wells C. Lesson 3 Logistic Regression Diagnostics. [accessed on June 6, 2013];UCLA: Statistical Consulting Group. http://www.ats.ucla.edu/stat/stata/webbooks/logistic/chapter3/statalog3.htm.
- 71.Holland BS, Copenhaver MD. An improved sequentially rejective Bonferroni test procedure. Biometrics. 1987;43:417. Correction in Biometrics 43, 737, 1987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


