Abstract
In Saccharomyces cerevisiae, the glutamate dehydrogenase (GDH) enzymes play a pivotal role in glutamate biosynthesis and nitrogen assimilation. It has been proposed that, in GDH-deficient yeast, either the proline utilization (PUT) or the glutamine synthetase-glutamate synthase (GS/GOGAT) pathway serves as the alternative pathway for glutamate production and nitrogen assimilation to the exclusion of the other. Using a gdh-null mutant (gdh1Δ2Δ3Δ), this ambiguity was addressed using a combination of growth studies and pathway-specific enzyme assays on a variety of nitrogen sources (ammonia, glutamine, proline and urea). The GDH-null mutant was viable on all nitrogen sources tested, confirming that alternate pathways for nitrogen assimilation exist in the gdh-null strain. Enzyme assays point to GS/GOGAT as the primary alternative pathway on the preferred nitrogen sources ammonia and glutamine, whereas growth on proline required both the PUT and GS/GOGAT pathways. In contrast, growth on glucose-urea media elicited a decrease in GOGAT activity along with an increase in activity of the PUT pathway specific enzyme Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH). Together, these results suggest the alternative pathway for nitrogen assimilation in strains lacking the preferred GDH-dependent route is nitrogen source dependent and that neither GS/GOGAT nor PUT serves as the sole compensatory pathway.
Keywords: Yeast, Saccharomyces cerevisiae, glutamate dehydrogenase, nitrogen assimilation
1. Introduction
Like many other microorganisms, Saccharomyces cerevisiae is able to utilize a wide array of nitrogen-containing compounds as its sole source of nitrogen (Magasanik and Kaiser, 2002). In order to utilize diverse nitrogen sources, S. cerevisiae uses two strategies, both of which converge on the amino acid glutamate. This is achieved either through the assimilation of the metabolic by-product ammonia into α-ketoglutarate or by catabolism of precursor molecules to yield glutamate directly. Glutamate produced by these routes can then react with another molecule of α-ketoglutarate to form glutamine. The two amino acids that are formed—glutamate and glutamine—are responsible for 85% and 15% of total cellular nitrogen, respectively (Cooper, 1982). The primary enzyme responsible for generating glutamate in vivo is glutamate dehydrogenase (GDH) (Avendano et al., 1997; De Luna et al., 2001; Miller and Magasanik, 1990). The S. cerevisiae genome contains three GDH genes encoding for enzymes with different functional roles. Two of the genes, GDH and GDH3, encode NADPH-dependent enzymes that catalyze the reductive amination of α-ketoglutarate to form glutamate, whereas the other, GDH2, encodes an NAD+-dependent enzyme that catalyzes the reverse reaction (DeLuna et al., 2001). Although both Gdh1p and Gdh3p catalyze the formation of glutamate, Gdh1p is the preferred biosynthetic enzyme under all conditions (DeLuna et al., 2001). The third GDH gene, GDH2, plays an equally important role in nitrogen metabolism, where it has been shown to be the primary source of ammonia generation in S. cerevisiae (Miller and Magasanik, 1990).
Although previous research posits a central role for GDH in glutamate metabolism and nitrogen assimilation, it does not constitute the sole pathway for these processes (Figure 1) (Avendano et al., 1997; De Luna et al., 2001,Miller and Magasanik, 1990). In fact, at least two other pathways have been shown to exist in S. cerevisiae for the synthesis of glutamate and assimilation of nitrogen—viz. the proline utilization (PUT) and glutamine synthetase-glutamate synthase pathways (GS/GOGAT). Proline gains entry into the cells via either the general amino acid permease, Gap1p, or the proline-specific permease, Put4p (Magasanik and Kaiser, 2002). The enzymes that constitute the PUT pathway are proline oxidase (Put1p) and Δ1-pyrroline-5-carboxylate dehydrogenase (Put2p), which are encoded by the PUT and PUT genes, respectively (Brandriss and Magasanik, 1979; Wanduragala et al., 2010). In laboratory strains of S. cerevisiae, both the PUT pathway and permease genes (Gap1p and Put4p) are tightly controlled by nitrogen catabolite repression (NCR) due to the status of proline as a non-preferred nitrogen source (ter Schure et al., 2000). The second alternative pathway for glutamate production, GS/GOGAT, consists of two enzymes acting consecutively to catalyze: (1) the ATP-dependent amidation of glutamate using free ammonia (GS) and (2) the reductive amination of α-ketoglutarate using the amide nitrogen of glutamine (GOGAT) (Figure 1) (Magasanik and Kaiser, 2002; ter Schure et al., 2000). Ultimately, this sequence of reactions yields two molecules of glutamate for every one that enters the pathway.
Figure 1.
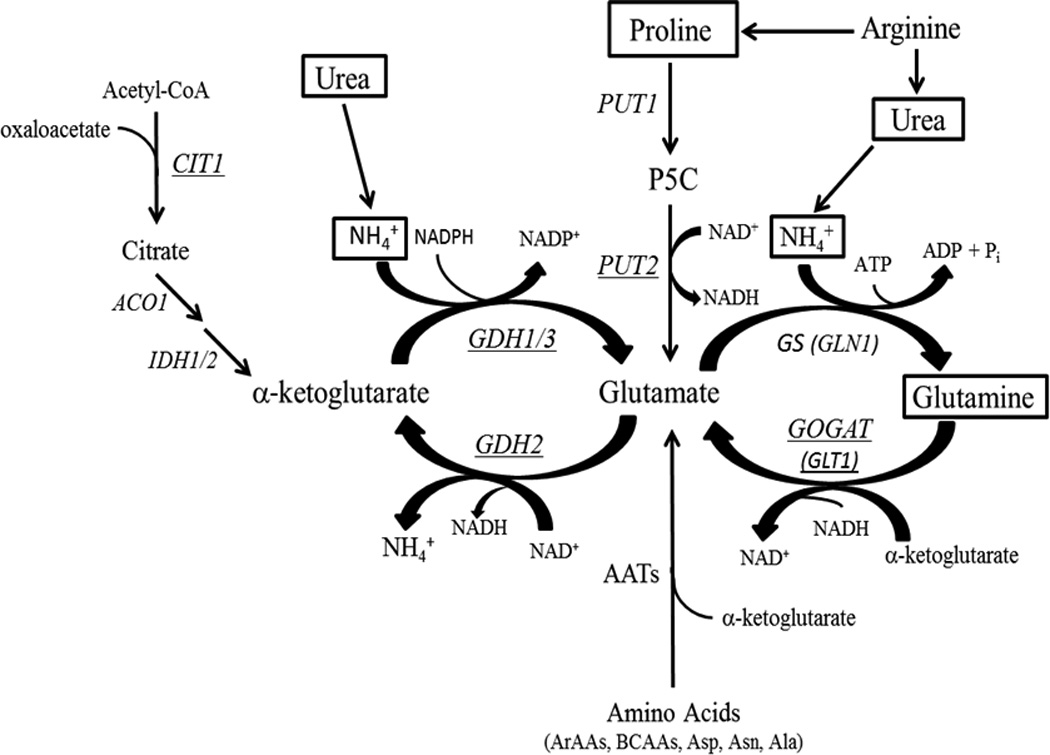
Relevant pathways for glutamate metabolism and nitrogen assimilation in S. cerevisiae. The three primary pathways for glutamate synthesis and nitrogen assimilation are detailed including the two pathways (GS/GOGAT and PUT) tested in this study for up-regulation in response to abrogation of GDH activity. Molecules surrounded by a box denote that it was used as a nitrogen source in this study and genes encoding for enzymes that were assayed are underlined. Co-factors utilized in enzyme assays are also listed. Abbreviations: P5C = Δ1-pyrroline-5-carboxylate; AATs = amino acid transaminases; ArAAs = aromatic amino acids; BCAAs = branched-chain amino acids; BCKA = branched chain keto acids. Figure was adapted from Magasanik, 2002.
Using mutagenesis on a glutamate auxotroph stalled in the citric acid cycle at aconitase (in their nomenclature, glt1-1), Lundgren et al. showed that the PUT pathway could serve as a source of glutamate, as the put2-glt1-1mutant they isolated was no longer able to utilize proline as a nitrogen source (Lundgren et al., 1972). In contrast with the results from Lundgren et al., Avendano and colleagues asserted that the GS/GOGAT pathway, in combination with the GDHs, represents the sole pathway for glutamate biosynthesis in S. cerevisiae (Avendano et al., 1997). In order to address this apparent discrepancy, a gdh-null mutant was created and subjected to experiments exploring the relative contribution of the PUT and GS/GOGAT pathways to glutamate biosynthesis and nitrogen assimilation following adaptation to growth on various nitrogen sources. This approach allowed for an unprecedented exploration of nitrogen assimilation in S. cerevisiae and enabled both the clarification previous findings and the discovery of previously unappreciated mechanisms for nitrogen assimilation in this organism
2. Materials and Methods
2.1. Construction of GDH mutants
The yeast strains used in these studies and their genotypes are listed in Table 1. The strains were obtained using a combination of traditional and molecular genetic techniques. Briefly, the single mutants (i.e. gdh1Δ, gdh2Δ and gdh3Δ) were crossed with a wild-type strain (SEY6211) and mutants with novel genetic markers were isolated following sporulation, tetrad dissection and genotyping (Sherman, 2002).
Table 1.
Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| SEY6211 | ade2 trp1 ura3 leu2 his3 suc2 | M. Yaffe |
| BY4742 (YSC1049) | his3 leu2 lys2 ura3 | Research Genetics |
| gdh1Δ (YOR375c) | his3 leu2 lys2 ura3 gdh1Δ::kanMX4 | Research Geneticsa, This studyb |
| gdh2Δ (YDL215c) | his3 leu2 lys2 ura3 gdh2Δ::kanMX4 | Research Geneticsa, This studyb |
| gdh3Δ (YAL062w) | his3 leu2 lys2 ura3 gdh3Δ::kanMX4 | Research Geneticsa, This studyb |
| gdh1Δ2Δ (PTY84) | his3 leu2 lys2 trp1 ura3 gdh1Δ::TRP1 gdh2Δ::URA3 | This study |
| gdh1Δ3Δ (ASY18A) | his3 leu2 trp1 ura3 gdh1Δ::TRP1 gdh3Δ::kanMX4 | This study |
| gdh2Δ3Δ (ASY19A) | his3 leu2 trp1 ura3 gdh2Δ::URA3 gdh3Δ::kanMX4 | This study |
| gdh1Δ2Δ3Δ (ASY20A) | his3 leu2 trp1 ura3 gdh1Δ::TRP1 gdh2Δ::URA3 gdh3Δ::kanMX4 | This study |
Now available from Open Biosystems (Thermo Scientific).
Variations of these strains with novel markers were generated as explained in methods.
The disrupted genes, gdh1Δ::TRP1and gdh2Δ::URA3, were introduced into gdh single mutants via linearization and homologous recombination from the YEp352-gdh1Δ::TRP1 and pUC18-gdh2Δ::URA3plasmids, respectively. GDH1and GDH2 were amplified from genomic DNA using restriction site (underlined)-flagged primer pairs GDH1-BamF (5’-CAGGATCCACGATTGGCTG-GATAAGAGGA-3’) / GDH1-PstR (5’-TGTCTGCAGTGCCAAATTGGACGAGTAAG-3’) and GDH2-XbaF (5’-GCTCTAGATCGACATCAACACTGACAAGC-3’) / GDH2-KpnR (5’-GGGGTACCAAC-CTGTTTCAATGCTGCCT-3’), respectively (Integrated DNA Technologies). Genes amplified from genomic DNA were restriction digested and ligated into the YEp352 (Hill et al., 1986) and pUC18 (Stratagene; now Agilent Technologies) plasmids using restriction enzyme pairs BamHI-PstI and XbaI-KpnI, respectively. The TRP1 gene was excised from the PJJ281 (Jones and Prakash, 1990) plasmid using restriction enzymes SalI and BglII and ligated into the YEp352-GDH plasmid to create YEp352-gdh1Δ::TRP1. URA3 was amplified from genomic DNA using primers URA3 F-AvaI (5’-CCCTCGAGCGGCATCAGAGCAGATTGTA-3’) and URA3 R-ClaI (5’-CCATCGATCGT-TGGAGTCCACGTTCTTT-3’) (Integrated DNA Technologies) and inserted between the AvaI and ClaI sites in the pUC18-GDH plasmid. Linear fragments were isolated using restriction enzyme pairs PacI-KpnI (gdh1Δ::TRP1) and XbaI-KpnI (gdh2Δ::URA3), gel purified and introduced into the appropriate single and double mutants via homologous recombination using the YEASTMAKER Yeast Transformation System 2 (ClonTech). Following recombination, the mutants were selected for on the appropriate drop out media. Genotypes were confirmed by assessing the growth phenotypes for nutritional markers and through PCR analysis of GDH alleles (data not shown). All restriction enzymes were purchased from New England Biolabs.
2.2. Media and Growth
Minimal Media (Y-Min) contained 0.67% yeast nitrogen base without amino acids or ammonium sulfate, 2% glucose and the appropriate amino acid supplements (Table 1) (Sherman, 2002). The nitrogen sources were added at either 0.2% for ammonium sulfate or 0.1% for glutamine, proline or urea (Miller and Magasanik, 1991). All media reagents were obtained from either Difco or Sigma chemical.
Doubling times were determined for cells during logarithmic growth. Cells pre-grown overnight in Y-Min supplemented with ammonia and the appropriate amino acids were diluted in Y-Min plus the nitrogen source being tested to an optical density of 0.05–0.2 at 600 nm (OD600). Following a lag-phase of 7–8 hours, OD600 readings were recorded every hour for 6–8 hours. At each step, cells were at 30°C with shaking (200 rpm).
2.3. Enzyme Assays
For the determination of enzyme activities, culture conditions were identical to those used in the experiments. Crude lysates were prepared from cultures in late-log phase as described previously (Trotter et al., 2005). Aliquots were stored at −80° C and thawed on the day of the assay. Protein in the crude lysates were determined prior to enzyme assays using the BCA (bicinchoninic acid) method (Sigma Chemical). All chemicals were purchased from either Sigma Chemical or Dow Chemical.
GDH activities were measured by monitoring the oxidation of NADPH (Gdh1/3p) or NADH (Gdh2p) spectrophotometrically according the method of Doherty (Doherty, 1970). Citrate synthase were measured by following the reaction of coenzyme A and 5,5’-dithiobis-(2-nitro-benzoate) (DTNB) at 412 nm in a reaction mixture containing 1 mM DTNB, 10 mM acetyl-CoA, 10 mM that was initiated by adding ~0.06 mg of protein from crude lysate (Srere, 1969).
Glutamate synthase (GOGAT) and Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH) activities measured by monitoring either the oxidation (GOGAT) or reduction (P5CDH) of NAD(H) at 340. GOGAT assays were performed in 1-mL reaction volumes (100 mM potassium phosphate, pH 7.8; 0.2 mM NADH; 10 mM glutamine; ~0.2–0.4 mg protein from crude lysates) and initiated by the addition of α-ketoglutarate (10 mM) (Roon et al., 1974). DL-Δ1-Pyrroline-5-carboxylate (P5C) was synthesized to the method of Williams and Frank and was adjusted to a pH of 7.0 prior to use in assays (Williams and Frank, 1975). P5CDH assays were carried out in 1-mL reaction volumes (50 mM tris(hydroxylmethyl)methyl-3-aminopropanesulfonic acid (TAPS), pH 9.0; 0.8 µM P5C, pH 7.0; 0.2 mM NAD+) and initiated by the addition of between 0.2–0.4 mg of protein from crude lysate (Brandriss and Magasanik, 1979; Williams and Frank, 1975).
3. Results
3.1. Strain Creation and Confirmation
Although the isolation and characterization of several GDH mutants have been reported elsewhere, the gdh2Δ3Δ and gdh1Δ2Δ3Δ strains have yet to be described (Avendano et al., 1997; De Luna et al., 2001; Miller and Magasanik, 1990). For this study, the full complement of double and triple mutants was isolated using standard genetic techniques (see Methods). Confirmation of mutant strains was accomplished via PCR amplification of GDH alleles and growth phenotype for auxotrophic markers (data not shown). To further test the integrity of the isolated strains, NADP-GDH (Gdh1p and Gdh3p) and NAD-GDH (Gdh2p) activities were determined (Table 2). The lack of detectable NADP-GDH activity in the gdh1-deficient strains corroborates well with both our PCR findings and other studies showing the primacy of Gdh1p over Gdh3p in strains grown on glucose (Avendano et al., 1997; De Luna et al., 2001). While significant levels of NAD-GDH activity were not observed for the gdh2Δ, gdh1Δ2Δ or gdh1Δ2Δ3Δ strains, the gdh2Δ3Δ mutant exhibited consistently elevated activity when grown on glucose-ammonia media. It has been previously reported that the GDH2 allele is subject to glucose repression, which can be relieved when strains are grown on either glucose-limiting media or non-fermentable carbon sources (Coschigano et al., 1991). To further test for NAD-GDH activity in putative gdh mutants, wild-type and mutant strains were grown on the non-fermentable carbon source raffinose, which has previously been used to maximize Gdh2p activity (Tang et al., 2011). As expected, when raffinose was substituted for glucose, the putative gdh2 mutants showed no detectable NAD-GDH activity as compared to the wild-type strain, which exhibited a four-fold increase in activity (see Table 2, c). Taken together, these results confirm the isolation of the full complement of GDH mutants.
Table 2.
Strains and doubling times on various nitrogen sources
| Strain | Relative Growtha | NADP-GDH Sp. Activityb (Gdh1p & Gdh3p) |
NAD-GDH Sp. Activityb (Gdh2p) |
|||
|---|---|---|---|---|---|---|
| NH3 | Glutamine | Proline | Urea | |||
| BY4742 | 1.00 ± 0.01 | 1.00 ± 0.01 | 1.00 ± 0.03 | 1.01 ± 0.07 | 88.4 ± 4.1 | 7.7 ± 1.7 |
| gdh1Δ | 0.64 ± 0.05e | 0.79 ± 0.03f | 0.74 ± 0.01e | 0.73 ± 0.04d | ND | 19.8 ± 3.9 |
| gdh2Δ | 0.93 ± 0.05 | 1.02 ± 0.03 | 0.79 ± 0.03e | 0.99 ± 0.07 | 87.9 ± 12.0 | ND |
| gdh3Δ | 0.95 ± 0.01 | 0.95 ± 0.03 | 0.95 ± 0.05 | 1.00 ± 0.05 | 95.0 ± 11.9 | 9.0 ± 0.9 |
| gdh1Δ2Δ | 0.56 ± 0.03f | 0.51 ± 0.07f | 0.80 ± 0.03f | 0.79 ± 0.04 | ND | 2.1 ± 1.7c |
| gdh1Δ3Δ | 0.58 ± 0.01f | 0.39 ± 0.03f | 0.84 ± 0.01e | 0.77 ± 0.03d | ND | 9.9 ± 4.3 |
| gdh2Δ3Δ | 1.06 ± 0.05 | 1.27 ± 0.04e | 0.86 ± 0.01e | 1.21 ± 0.02d | 83.5 ± 8.1 | 4.4 ± 0.7c |
| gdh1Δ2Δ3Δ | 0.67 ± 0.02e | 0.83 ± 0.01f | 0.87 ± 0.03e | 0.83 ± 0.02d | ND | 1.8 ± 1.7c |
Doubling times calculated for cultures during logarithmic growth on various nitrogen sources as described in methods. Values are shown relative to the doubling-time of BY4742 (i.e. 2.3, 2.1, 4.5 and 3.6 hours with NH3, Glutamine, Proline and Urea as nitrogen sources, respectively). Average of 4 separate experiments ± SEM.
NAD(P)-GDH activities were determined on Y-Min + NH3 media as described in methods. Values are expressed in nmol/min/mg of protein. Average of three separate experiments ± SEM. ND, not detectable.
No detectable NAD-GDH activity as compared to wild-type (~33 µmol/min/mg of protein) when grown on raffinose as a carbon source (n=1, data not shown).
P < 0.05 by t-test as compared to BY4742 for the same nitrogen source.
P < 0.01 by t-test as compared to BY4742 for the same nitrogen source.
P < 0.001 by t-test as compared to BY4742 for the same nitrogen source.
3.2. Experimental Conditions and Growth Data
Four nitrogen sources were selected to explore the differential contributions of the PUT and GS/GOGAT pathways to glutamate biosynthesis and nitrogen assimilation in adapted gdh-deficient yeast. As a result of nitrogen catabolite repression (NCR) in S. cerevisiae, pathways for utilization of non-preferred sources of nitrogen—e.g. proline—are down-regulated in the presence of the preferred nitrogen sources ammonia, glutamine and asparagine (ter Schure et al., 2000). In addition to NCR status, nitrogen sources in S. cerevisiae differ in their mode of utilization proceeding either through an ammonium intermediate (Ai) or via direct conversion to glutamate (Glu) (Magasanik and Kaiser, 2002; ter Schure et al., 2000). Four nitrogen sources were chosen corresponding to the four possible combinations of NCR-status and pathways for utilization: (i) ammonia – Ai/repressing; (ii) glutamine – Glu/repressing; (iii) urea – Ai/de-repressing; (iv) proline – Glu/de-repressing (Magasanik and Kaiser, 2002; ter Schure et al., 2000). Figure 1details the relevant pathways for the utilization of these nitrogen sources including the enzymes whose activities were assayed in this study.
Upon transfer and adaptation to fresh medium containing any of the four nitrogen sources, all strains exhibited a lag-phase between 7–8 hours (data not shown). The gdh1-deficient strains had doubling times 56–67% as compared to wild-type when grown on glucose-ammonia media (Table 2). When the gdh1-deficient strains were grown on glutamine as the sole nitrogen source, low growth rates were observed (39–51% of wild-type) with the exception of gdh1Δ and gdh1Δ2Δ3Δ, which exhibited slightly higher (79% and 83% of wild-type) growth rates (Table 2). Growth of the gdh2Δ3Δ strain on glucose-glutamine media was approximately 130% of wild-type (Table 2). Rates of growth for the GDH mutants when urea was utilized as the sole nitrogen source yielded a pattern analogous to that seen on glucose-glutamine media—i.e. a reduction of growth rate in gdh1-deficient strains and an elevation in the gdh2Δ3Δ mutant (Table 2). In contrast, growth on proline yielded a reduction in growth rate to approximately 80% for all mutants with the exception of gdh3Δ. For strains exhibiting severely attenuated growth rates (i.e. those below 50% of WT), growth phenotypes were confirmed using spot assays of serial dilutions (data not shown). Growth phenotypes for the remaining mutants were undiscerable due to the assay’s lack of resolution. Because the elevated growth rate of the gdh2Δ3Δ mutant in glucose-glutamine and glucose-urea media was unexpected, this strain was chosen for inclusion in subsequent experiments.
3.3. Enzyme Assays
Because glutamate metabolism and nitrogen assimilation are intricately linked with the citric acid cycle (CAC; or tricarboxylic acid, TCA, cycle) through the intermediate α-ketoglutarate, the activity of the enzyme citrate synthase (encoded by the gene CIT1) was measured as a proxy for CAC function in GDH-deficient mutants. In the wild-type strain, citrate synthase activities were similar for nitrogen sources that either converge on glutamate (30.9 and 48.7 µmol/min/mg-protein for glutamine and proline, respectively) or proceed through an ammonium intermediate (58.0 and 75.7 µmol/min/mg-protein for ammonia and urea, respectively). On nitrogen sources where the gdh2Δ3Δ strain exhibited enhanced growth rates—i.e. glutamine and urea—citrate synthase activities followed a similar pattern in which the gdh2Δ3Δ exhibited slightly up-regulated and the triple-mutant exhibited either no change or slightly down-regulated activity compared to wild-type (p < 0.05 for both the up-regulations in gdh2Δ3Δ and the down-regulation of the triple-mutant on urea; see Figure 2). While citrate synthase activities showed an analogous pattern on the aforementioned nitrogen sources, the pattern for proline grown cells differed greatly. Although an increase in citrate synthase activity for the gdh2Δ3Δ strain was also observed on proline, the up-regulation was far more pronounced (2.5-fold increase as compared to wild-type, p < 0.01; Figure 2). Additionally, an approximately 2-fold elevation in citrate synthase activity was observed for the gdh1Δ2Δ3Δ strain grown on glucose-proline media (relative to WT, p < 0.01; Figure 2).
Figure 2.
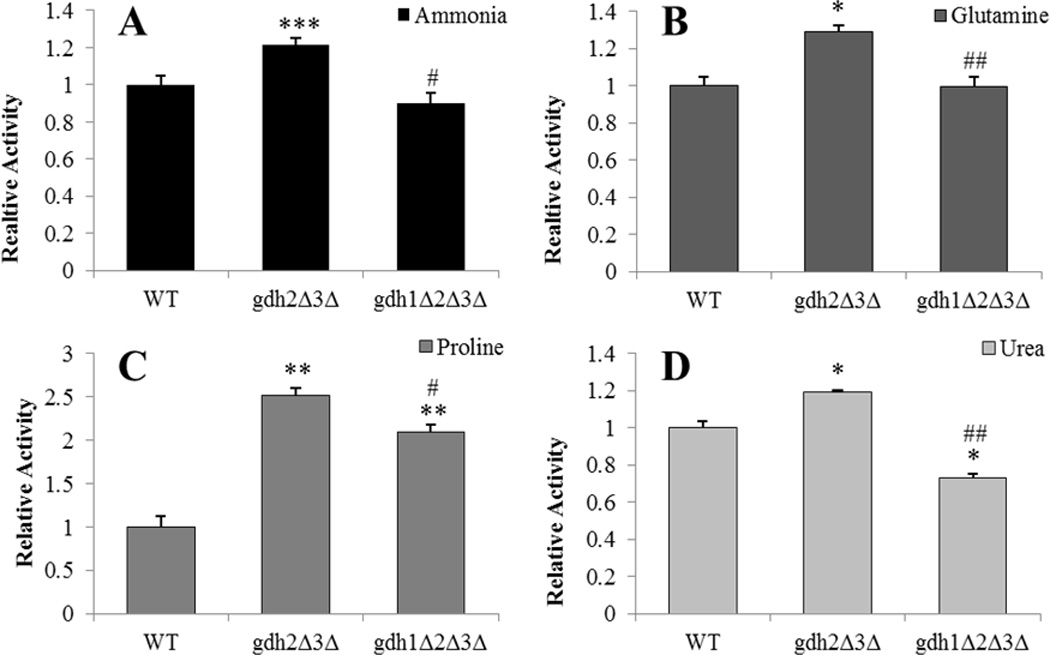
Citrate synthase activities in gdh2Δ3Δ and gdh1Δ2Δ3Δ mutants. Enzyme activities were determined following growth on various nitrogen sources (A–D) and results expressed as the ratio of mutant to wild-type (BY4742) activity ± SEM from three separate experiments. Citrate synthase activities in the wild-type strain were 58.0, 30.9, 48.7 and 75.6 µmol/min-mg for ammonia, glutamine, proline and urea grown cells, respectively. * p < 0.05, ** p < 0.and *** p < 0.0between wild-type and strain indicated by paired t-test. # P < 0.05 and ## P < 0.01 between double and triple-mutant strains by paired t-test.
Previous work in S. cerevisiae has suggested that either the GS/GOGAT or the PUT pathway is responsible for glutamate production in strains deficient in gdh activity (Avendano et al., 1997; Valenzuela et al., 1998). In order to assess the relative contributions of these pathways, activities of the Put2p (P5CDH) and Glt1p (GOGAT) enzymes were determined for the gdh2Δ3Δ and gdh1Δ 2Δ3Δ strains following adaptation to various nitrogen sources. P5CDH activities in the gdh2Δ3Δ mutant were slightly decreased when either proline or urea were supplied as the sole nitrogen source (Figures 5 & 6; ~0.8-fold compared to wild-type, p < 0.08 and p < 0.05 for proline and urea, respectively). When the gdh1Δ2Δ3Δ strain was grown in the presence of either ammonia or urea, an increase in P5CDH activity was observed (Figures 3 & 6; ~1.6-fold and ~1.2-fold increase compared to WT for ammonia and urea, respectively, p < 0.05). P5CDH activities for both mutants on the remaining nitrogen sources remained indistinguishable from wild-type. Analysis of GOGAT activity in the gdh1Δ2Δ3Δ strain showed a significant up-regulation when ammonia, proline or glutamine were utilized as the sole nitrogen source (between 1.4–1.8-fold relative to wild-type, p < 0.08, 0.05 and 0.01 for ammonia, proline and glutamine, respectively; Figures 3–Figure 5). Up-regulation of GOGAT activity was also observed for the gdh2Δ3Δ mutant grown on glucose-ammonia media (~1.6-fold compared to wild-type, p < 0.08; Figure 3B). On the remaining nitrogen source, urea, a decrease in GOGAT activity of 40–60% relative to wild-type was observed for the gdh2Δ3Δ and gdh1Δ2Δ3Δ strains (p < 0.05; Figure 6).
Figure 5.
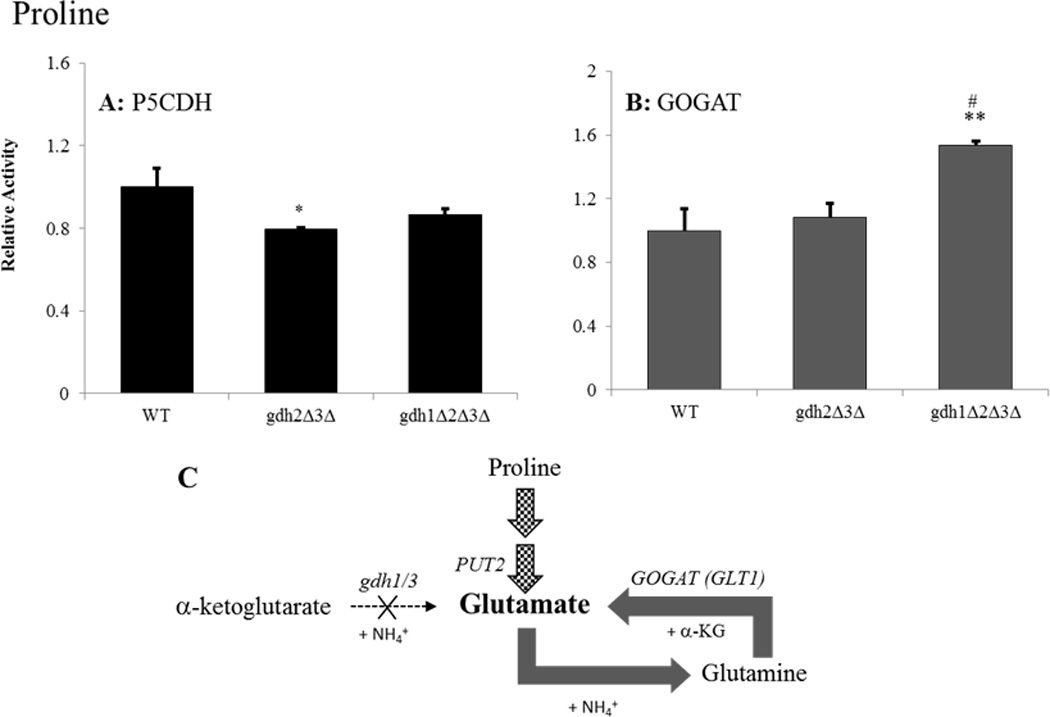
Both the PUT and GS/GOGAT pathways contribute to glutamate biosynthesis and nitrogen assimilation in gdh-null grown on glucose-proline media. Enzyme activities were determined for (A) Δ1-pyrroline-5-carboxylate dehydrogenase (Put2p) or (B) glutamate synthase (GOGAT/Glt1p) following adaptation to growth in glucose-proline media. Results are expressed as the ratio of mutant to wild-type (BY4742) activity ± SEM from three separate experiments. Activities in the wild-type strain were 97.1 and 11.8 nmol/min-mg for Put2p and GOGAT, respectively. (C) Graphical summary of findings for the triple mutant. Gene designations are indicated in italics for each reaction along with the appropriate co-factors. Bolded arrows indicate pathway up-regulation relative to the wild-type strain. Checkered arrows indicated an absolute increase in Put2p activity compared to the same strain grown on glucose-ammonia (83.9 vs. 23.5 nmol/min-mg) * P < 0.05 and ** P < 0.008 between wild-type and strain indicated by paired t-test. # P < 0.05 between double and triple-mutants by paired t-test.
Figure 6.
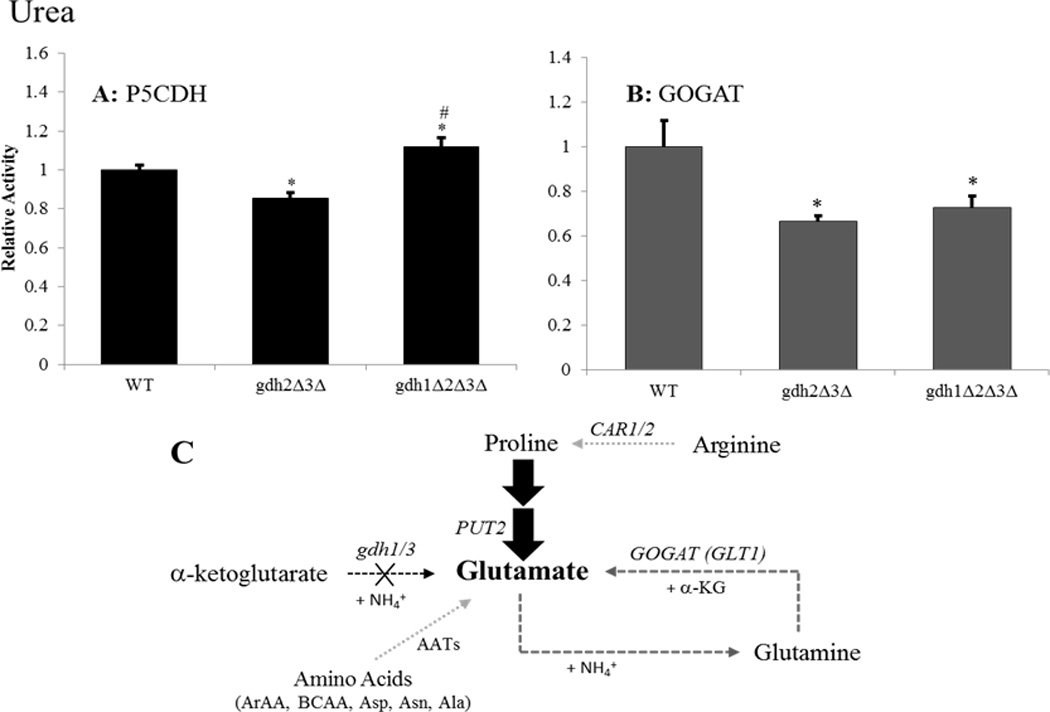
The PUT pathway may represent an important source of glutamate in the gdh-null mutant grown on glucose-urea. Enzyme activities were determined for (A) Δ1-pyrroline-5-carboxylate dehydrogenase (Put2p) or (B) glutamate synthase (GOGAT/Glt1p) following adaptation to growth in glucose-urea media. Results are expressed as the ratio of mutant to wild-type (BY4742) activity ± SEM from three separate experiments. Activities in the wild-type strain were 24.1 and 21.9 nmol/min-mg for Put2p and GOGAT, respectively. (C) Graphical summary of findings for the triple mutant. Gene designations are indicated in italics for each reaction along with the appropriate co-factors. Bolded arrows indicate pathway up-regulation whereas dashed arrows indicated pathway down-regulation relative to the wild-type strain. Potential alternative sources of glutamate are indicated by light gray arrows (Brandriss and Magasanik, 1980; Eden et al., 1996; García-Campusano et al., 2009; Iraqui et al., 1998; Kispal et al., 1996; Morin et al., 1992; Urrestarazu et al., 1998). Abbreviations: AATs = amino acid transaminases; ArAAs = aromatic amino acids; BCAAs = branched-chain amino acids; BCKA = branched chain keto acids. * P < 0.05 between wild-type and strain indicated by paired t-test. # P < 0.05 between double and triple-mutants by paired t-test.
Figure 3.
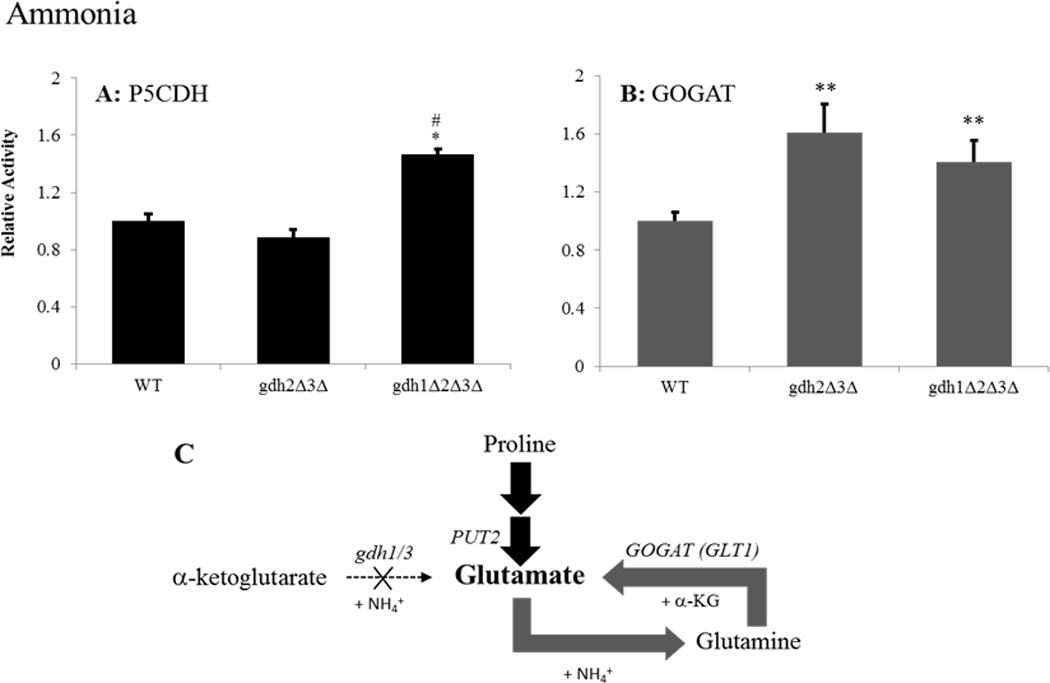
Both the PUT and GS/GOGAT pathways contribute to glutamate biosynthesis and nitrogen assimilation in gdh-null grown on glucose-ammonia media. Enzyme activities were determined for (A) Δ1-pyrroline-5-carboxylate dehydrogenase (Put2p) or (B) glutamate synthase (GOGAT/Glt1p) following adaptation to growth in glucose-ammonia media. Results are expressed as the ratio of mutant to wild-type (BY4742) activity ± SEM from three separate experiments. Activities in the wild-type strain were 17.and 7.nmol/min-mg for Put2p and GOGAT, respectively. (C) Graphical summary of findings for the triple mutant. Gene designations are indicated in italics for each reaction along with the appropriate co-factors. Bolded arrows indicate pathway up-regulated relative to the wild-type strain. * P < 0.05 and **P < 0.08 between wild-type and strain indicated by paired t-test. # P < 0.01 between double and triple-mutants by paired t-test.
4. Discussion
In S. cerevisiae, the amino acid glutamate plays an integral role in nitrogen metabolism, where it is responsible for 85% of total cellular nitrogen (Cooper, 1982; ter Schure et al., 2000). The ability of glutamate to serve as progenitor of 85% of total cellular nitrogen is due, in large part, to the action of glutamate dehydrogenase (GDH), which catalyzes the direct assimilation of free ammonia via reductive amination of α-ketoglutarate (ter Schure et al., 2000). The S. cerevisiae genome contains three GDH genes encoding for enzymes that favor either the assimilation—Gdh1p & Gdh3p—or release—Gdh2p—of ammonia (see Figure 1) (Cooper, 1982; ter Schure et al., 2000). Although previous studies report that Gdh1p serves as the main conduit for nitrogen assimilation and glutamate biosynthesis in vivo, gdh1Δ strains are viable and do not exhibit glutamate auxotrophy suggesting that alternative pathways exist (DeLuna et al., 2001). While the proline utilization (PUT) and glutamine synthetase-glutamate synthase (GS/GOGAT) pathways have been previously identified for their ability to facilitate glutamate biosynthesis in gdh-deficient yeast, questions remain as to the relative contribution of these pathways in mutant strains grown on various nitrogen sources (Avendano et al., 1997; Valenzuela et al., 1998). A gdh-null mutant was created. The strain was able to adapt and grow on various nitrogen sources which allowed the examination of the relative contributions of the PUT and GS/GOGAT pathways to glutamate biosynthesis and nitrogen assimilation in the absence of the favored GDH pathway.
Although previous studies examining the growth characteristics of GDH mutants have primarily used ammonia as the sole nitrogen source, data for wild-type growth on the remaining nitrogen sources have been documented (Miller and Magasanik, 1991). Doubling times for the wild-type strain used in these studies, BY4742, correspond well with these previous results (Table 2, a). When ammonia was supplied as the sole nitrogen source, gdh1-deficient strains exhibited decreased growth rates in the range of 47–63% compared to wild-type, whereas loss of either GDH2 or GDH3 resulted in little reduction in growth rate; these rates largely paralleled those reported previously (DeLuna et al., 2001; Miller and Magasanik, 1990). In a previous study, the growth rate of the gdh1Δ mutant on glucose-glutamine media was found to be 56% of the wild-type rate (Miller and Magasanik, 1991), and our results are generally similar. The elevated growth rate of the gdh2Δ3Δ mutant grown on glucose-glutamine or glucose-urea media, however, was unexpected. While this strain was expected to grow at a near wild-type rate—as seen in the gdh2Δ and gdh3Δ single mutants—a persistent elevation in growth rate was not predicted. In glucose-limiting conditions, the concentration of α-ketoglutarate decreases leading to a switch in preference from GDH to the GS/GOGAT pathway for nitrogen assimilation (Magasanik, 2003; Roon et al., 1974). The preference for the GS/GOGAT pathway in response to low intracellular α-ketoglutarate levels has been proposed to result from a more efficient use of glucose by this pathway as well as the intrinsic properties of the GOGAT enzyme—viz. a three-fold greater affinity for α-ketoglutarate and the irreversibility of the reaction it catalyzes (Magasanik, 2003). Our data suggest that this pathway may be operative in the gdh2Δ3Δ mutant, as the inability of this strain to regenerate α-ketoglutarate through the Gdh2p catalyzed reaction may have led to an analogous drop in intracellular α-ketoglutarate, thus ensuring the use of the more efficient GS/GOGAT pathway. While the observed increase in citrate synthase activity on glucose-glutamine and glucose-urea provides indirect evidence of decreased α-ketoglutarate concentrations in the gdh2Δ3Δ mutant, this explanation is weakened by the absence of a growth-rate enhancement in the gdh2Δ mutant. A possible explanation for this discrepancy is that the Gdh3p isoform may catalyze the oxidative deamination of glutamate to α-ketoglutarate in strains lacking Gdh2p activity. While there is no literature precedent for this alternative activity of the Gdh3p isoform, our growth data for the gdh2Δ3Δ mutant grown on glucose-glutamine or glucose-urea media suggest Gdh3p may compensate for loss of Gdh2p activity under certain physiological conditions.
Previously, Avendano et al. have shown that yeast lacking both NADP-GDH and GOGAT enzymatic activity (gdh1Δgdh3Δglt1Δ) are strict glutamate auxotrophs (Avendano et al., 1997) arguing for the importance of the GS/GOGAT pathway in gdh-deficient yeast. As expected, our data show an increase in GOGAT activity for the gdh-null mutant grown on glucose-ammonia media (Figure 3A) (Avendano et al., 1997; Valenzuela et al., 1998). Interestingly, despite the repressing activity of ammonia on the PUT pathway (ter Schure et al., 2000; Magasanik and Kaiser, 2002), a 1.6-fold increase in P5CDH activity was also observed, which suggests that the GS/GOGAT pathway is not solely responsible for glutamate production under these conditions (Figure 3B). When the gdh-null mutant was grown on glucose-glutamine media, glutamate production was derived primarily from the GS/GOGAT pathway as evidenced by a nearly 2-fold increase in GOGAT activity relative to the wild-type strain grown on this nitrogen source (Figure 4B). Together, these results suggest that the GS/GOGAT pathway serves as the primary, but not sole, route for glutamate production in gdh-null yeast adapted to the preferred nitrogen sources ammonia and glutamine (Figures 3C and 4C).
Figure 4.
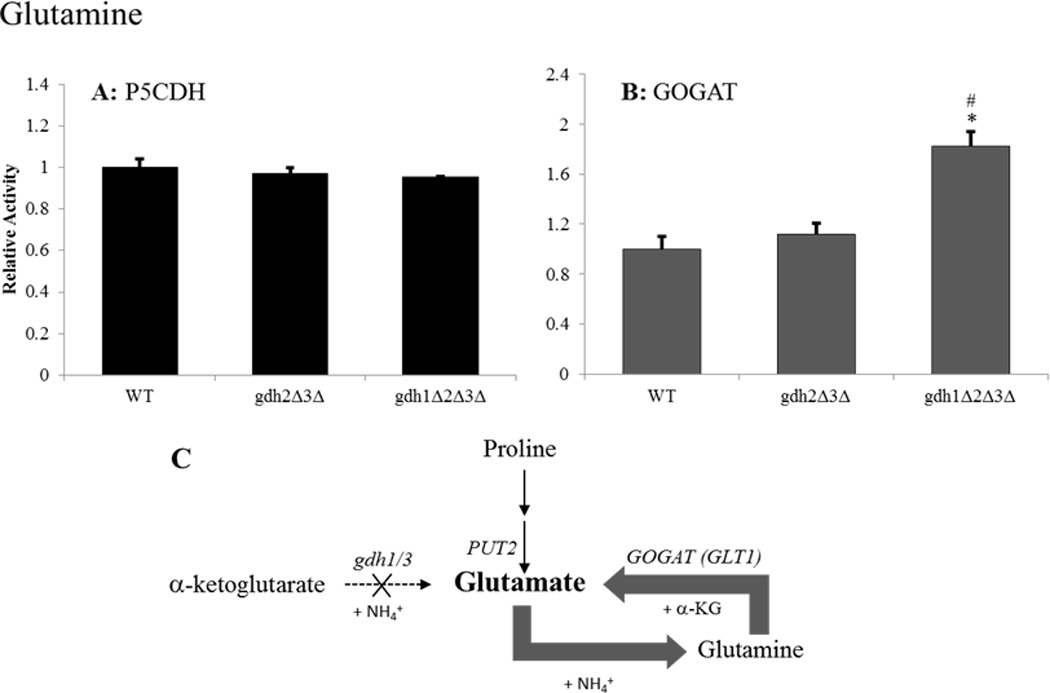
GS/GOGAT is the primary pathway for glutamate biosynthesis and nitrogen assimilation in gdh-null grown on glucose-glutamine media. Enzyme activities were determined for (A) Δ1-pyrroline-5-carboxylate dehydrogenase (Put2p) or (B) glutamate synthase (GOGAT/Glt1p) following adaptation to growth in glucose-glutamine media. Results are expressed as the ratio of mutant to wild-type (BY4742) activity ± SEM from three separate experiments. Activities in the wild-type strain were 21.and 5.2 nmol/min-mg for Put2p and GOGAT, respectively. (C) Graphical summary of findings for the triple mutant. Gene designations are indicated in italics for each reaction along with the appropriate co-factors. Bolded arrows indicate pathway up-regulation whereas regular weighted arrows indicate no observed up-regulation relative to the wild-type strain. * P < 0.01 between wild-type and strain indicated by paired t-test. # P < 0.05 between double and triple-mutants by paired t-test.
While an up-regulation in P5CDH activity compared to the wild-type strain was not observed for the gdh-null mutant grown on glucose-proline media, the absolute level of P5CDH activity in the null mutant was significantly elevated on this nitrogen source (83.9 vs. 23.5 nmol/min-mg for glucose-proline and glucose-ammonia grown cells, respectively). Elevated activity of the P5CDH enzyme argues for the importance of the PUT pathway in gdh-null yeast grown on glucose-proline media. Yet, the observed increases in both citrate synthase and GOGAT activities suggest that alternative pathways may become important when levels of glutamate are significantly depleted. In a previous report, Valenzuela and co-workers observed a slight up-regulation in GOGAT activity when the gdh1Δ mutant was grown on glucose-proline media (~1.3-fold) (Valenzuela et al., 1998). Our results indicate that elimination of the GDH pathway yields an approximately 1.6-fold up-regulation in GOGAT activity on glucose-proline media (Figure 5B). Because citrate synthase catalyzes the rate-limiting step in the CAC, the nearly 2-fold increase in its activity observed for the gdh-null mutant grown on glucose-proline media is evidence of enhanced CAC function (Figure 2C) (Kim et al., 1986). Enhanced CAC activity would be expected to result in elevated levels of α-ketoglutarate, which could then be utilized by the GS/GOGAT pathway to synthesize an additional molecule of glutamate from the one already derived from breakdown of proline. Taken together, these results demonstrate that, in yeast lacking the preferred GDH pathway, the GS/GOGAT and PUT pathways may act in a coordinated manner to ensure that glutamate concentrations do not reach prohibitively low levels (Figure 5C).
In contrast to cells grown in the presence of ammonia, glutamine or proline as sole nitrogen sources, growth of the gdh-null mutant on glucose-urea media elicited a 40% decrease in GOGAT activity (Figure 6B). Utilization of urea as a nitrogen source requires the action of the bi-functional enzyme Dur1,2p, whose catalytic cycle involves the carboxylation of urea followed by hydrolysis of the resulting urea-1-carboxylate intermediate to yield two equivalents of carbon dioxide and ammonia (Cooper et al., 1980; Cooper, 1982; ter Schure et al., 2000). In the absence of NADP-GDH activity, assimilation of urea-derived ammonia was expected to proceed via the action of glutamine synthetase (GS) (see Figure 1) (Minehart and Magasanik, 1992). However, assimilation of urea-derived ammonia via this route would require glutamate as a substrate for the GS catalyzed reaction. This requirement would necessitate the acquisition of glutamate from alternative sources in gdh-null yeast. Our results indicate that the PUT pathway, which exhibits a small up-regulation in the gdh-null mutant (Figure 6A), may serve as an important source of the glutamate required for GS-dependent assimilation of urea-derived ammonia in gdh-null yeast. While the PUT pathway represents a plausible source of glutamate in this strain, we cannot rule out the possibility of other sources. In particular, transaminase enzymes involved in the degradation of various amino acids could represent an unappreciated source of glutamate (see Figure 6C). Further experimentation will be needed to confirm these hypotheses.
In this report, we have described the characterization of a newly isolated gdh1Δ2Δ3Δ triple mutant and its application towards assessing the relative contribution of the PUT and GS/GOGAT pathways to glutamate production and nitrogen assimilation following adaptation on various nitrogen sources—an issue where previous research had yielded discrepant results (Avendano et al., 1997; Valenzuela et al., 1998). Analysis of pathway-specific enzyme activities in the gdh-null strain adapted to various nitrogen sources suggests that the GS/GOGAT pathway is primarily responsible for glutamate production when yeast strains are grown on the preferred nitrogen sources ammonia or glutamine. In contrast, utilization of the non-preferred nitrogen source proline appears to require a combination of both the PUT and GS/GOGAT pathways. Finally, utilization of urea as sole nitrogen source resulted in a down-regulation in GOGAT activity with a concomitant increase in P5CDH activity. This result suggests that the PUT pathway serves as an important source of glutamate needed for GS/GOGAT-dependent assimilation of nitrogen on this nitrogen source. Taken together, these results establish that both the PUT and GS/GOGAT pathways play important—and often complementary—roles in glutamate and nitrogen metabolism in S. cerevisiae.
Acknowledgements
The authors thank Dr. Dennis Voelker (National Jewish Center, Denver) for sharing yeast strains, Mr. Matthew Collinson-Pautz for assisting with cloning, Mr. Brett Bernatowicz for performing citrate synthase assays, Mr. Joshua Rathrod for technical assistance and Dr. Greg Domski for his careful reading of the manuscript. This work was supported by an Academic Research Enhancement Award from the National Institutes of Health (#R15-GM069372 to P.J.T).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avendano A, De Luna A, Olivera H, Valenzuela L, Gonzalez A. GDH3 encodes a glutamate dehydrogenase isozyme, a previously unrecognized route for glutamate biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1997;179(17):5594–5597. doi: 10.1128/jb.179.17.5594-5597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss MC, Magasanik B. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J Bacteriol. 1979;140(2):498–503. doi: 10.1128/jb.140.2.498-503.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandriss MC, Magasanik B. Proline: an essential intermediate in arginine degradation in Saccharomyces cerevisiae. J Bacteriol. 1980;143:1403–1410. doi: 10.1128/jb.143.3.1403-1410.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG, Lam C, Turoscy V. Structural analysis of the dur loci in S cerevisiae: two domains of a single multifunctional gene. Genetics. 1980;94:555–580. doi: 10.1093/genetics/94.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG. Nitrogen metabolism in Saccharomyces cerevisiae. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. pp. 39–99. [Google Scholar]
- Coschigano PW, Miller SM, Magasanik B. Physiological and genetic analysis of the carbon regulation of the NAD-dependent glutamate dehydrogenase of Saccharomyces cerevisiae. Mol Cell Biol. 1991;11(9):4455–4465. doi: 10.1128/mcb.11.9.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuna A, Avendano A, Riego L, Gonzalez A. NADP-glutamate dehydrogenase isoenzymes of Saccharomyces cerevisiae. J Biol Chem. 2001;276(47):43775–43783. doi: 10.1074/jbc.M107986200. [DOI] [PubMed] [Google Scholar]
- Doherty D. L-glutamate dehydrogenases (yeast) Method Enzymol. 1970;17(1):850–856. [Google Scholar]
- Eden A, Simchen G, Benvenisty N. Two yeast homologs of ECA39, a target for c-myc regulation, code for cytosolic and mitochondrial branched-chain amino acid transaminases. J Biol Chem. 1996;271:20242–20245. doi: 10.1074/jbc.271.34.20242. [DOI] [PubMed] [Google Scholar]
- García-Campusano F, Anaya VH, Robledo-Arratia L, Quezada H, Hernández H, Riego L, González A. ALT1-encoded alanine aminotransferase plays a central role in the metabolism of alanine in Saccharomyces cerevisiae. Can J Microbiol. 2009;55(4):368–374. doi: 10.1139/w08-150. [DOI] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Iraqui I, Vissers S, Cartiaux M, Urrestarazu A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol Gen Genet. 1998;257(2):238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- Jones JS, Prakash L. Yeast Saccharomyces cerevisiae selectable makers in pUC18 polylinkers. Yeast. 1990;6:363–366. doi: 10.1002/yea.320060502. [DOI] [PubMed] [Google Scholar]
- Kim KS, Rosenkrantz MS, Guarente L. Saccharomyces cerevisiae contains two functional citrate synthase genes. Mol Cell Biol. 1986;6:1936–1942. doi: 10.1128/mcb.6.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal G, Steiner H, Court DA, Rolinski B, Lill R. Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J Biol Chem. 1996;271:24458–24464. doi: 10.1074/jbc.271.40.24458. [DOI] [PubMed] [Google Scholar]
- Lundgren DW, Ogur M, Yuen S. The isolation and characterization of a Saccharomyces mutant deficient in Δ1-pyrroline-5-carboxylate dehydrogenase activity. Biochim Biophys Acta. 1972;286:360–362. doi: 10.1016/0304-4165(72)90271-1. [DOI] [PubMed] [Google Scholar]
- Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002;290:1–18. doi: 10.1016/s0378-1119(02)00558-9. [DOI] [PubMed] [Google Scholar]
- Magasanik B. Ammonia assimilation by Saccharomyces cerevisiae. Eukaryot Cell. 2003;2(5):827–829. doi: 10.1128/EC.2.5.827-829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SM, Magasanik B. Role of NAD-linked glutamate dehydrogenase in nitrogen metabolism in Saccharomyces cerevisiae. J Bacteriol. 1990;172(9):4927–4935. doi: 10.1128/jb.172.9.4927-4935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SM, Magasanik B. Role of the complex upstream region of the GDH2 gene in nitrogen regulation of the NAD-linked glutamate dehydrogenase in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11(12):6229–6247. doi: 10.1128/mcb.11.12.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minehart PL, Magasanik B. Sequence of the GLN1 gene of Saccharomyces cerevisiae: role of the upstream region in regulation of glutamine synthetase expression. J Bacteriol. 1992;174:1828–1836. doi: 10.1128/jb.174.6.1828-1836.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin PJ, Subramanian GS, Gilmore TD. AAT1, a gene encoding a mitochondrial aspartate aminotransferase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1992;1171(2):211–214. doi: 10.1016/0167-4781(92)90124-i. [DOI] [PubMed] [Google Scholar]
- Roon RJ, Even HL, Larimore F. Glutamate synthase: properties of the reduced nicotinamide adenine dinucleotide-dependent enzyme from Saccharomyces cerevisiae. J Bacteriol. 1974;118:89–95. doi: 10.1128/jb.118.1.89-95.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Schure EG, van Riel AAW, Verrips CT. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2000;24:67–83. doi: 10.1111/j.1574-6976.2000.tb00533.x. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;352:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Srere PA. Citrate synthase: [EC 4.1.3.7 citrate oxaloacetate-lyase (CoA-acetylating)] Method Enzymol. 1969;13:3–11. [Google Scholar]
- Tang Y, Sieg A, Trotter PJ. (13)C-metabolic enrichment of glutamate in glutamate dehydrogenase mutants of Saccharomyces cerevisiae. Microbiol Res. 2011;166:521–530. doi: 10.1016/j.micres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter PJ, Adamson AL, Ghrist AC, Rowe L, Scott LR, Sherman MP, Stites NC, Sun Y, Twiah-Boateng MA, Tibbetts AS, Wadington MC, West AC. Mitochondrial transporters involved in oleic acid utilization and glutamate metabolism in yeast. Arch Biochem Biophys. 2005;442:21–32. doi: 10.1016/j.abb.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Urrestarazu A, Vissers S, Iraqui I, Grenson M. Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination. Mol Gen Genet. 1998;257(2):230–237. doi: 10.1007/s004380050643. [DOI] [PubMed] [Google Scholar]
- Valenzuela L, Ballario P, Aranda C, Filetici P, Gonzalez A. Regulation of expression of GLT1, the gene encoding glutamate synthase in Saccharomyces cerevisiae. J Bacteriol. 1998;180(14):3533–3540. doi: 10.1128/jb.180.14.3533-3540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanduragala S, Sanyal N, Liang X, Becker DF. Purification and characterization of Put1p from Saccharomyces cerevisiae. Arch Biochem Biophys. 2010;498(2):136–142. doi: 10.1016/j.abb.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams I, Frank L. Improved chemical synthesis and enzymatic assay of Δ1-pyrroline-5-carboxylic acid. Anal Biochem. 1975;64:85–97. doi: 10.1016/0003-2697(75)90408-x. [DOI] [PubMed] [Google Scholar]


