Abstract
BACKGROUND
Anti-TNF-α therapy has made a significant impact on the treatment of psoriasis. Despite being designed to neutralize TNF-α activity, the mechanism of action of these agents in the resolution of psoriasis remains unclear.
OJECTIVES
To better understand the mechanism of action of etanercept by examining very early changes in the lesional skin of psoriasis patients responding to etanercept.
METHODS
20 chronic plaque psoriasis patients were enrolled and received 50mg etanercept twice weekly. Skin biopsies were obtained before treatment and on days 1, 3, 7 and 14 post-treatment. Skin mRNA expression was analysed by QRT-PCR and microarray; cytokine and phosphoprotein levels were assessed using multiplexed bead arrays.
RESULTS
In etanercept responders, we observed no significant changes in IL-17A, IL-22 and IFN-γ mRNA or protein in the first week of treatment; however, there was a 2.5-fold down-regulation of IL17RC mRNA (p<0.05) after day 1, accompanied by decreased ERK1/2 phosphorylation. Transcriptional analysis revealed genes suppressed by etanercept significantly overlapped with IL-17A-induced genes, and a marked overlap was also observed between the genes suppressed by etanercept and by the anti-IL17A therapy ixekizumab. Finally we show that TNF-α enhances the expression of IL-17RC and shRNA inhibition of IL-17R expression abrogates synergistic gene induction by TNF and IL17A.
CONCLUSIONS
These results suggest that the early responses of psoriasis plaques to etanercept may be due to decreased tissue responsiveness to IL-17A due to suppressed IL17RC expression in keratinocytes, blunting the strong synergy between TNF-α and IL-17, which contributes to the maintenance of psoriasis lesions.
INTRODUCTION
Biologic agents targeting TNF-α have made a significant impact on the treatment of psoriasis, arthritis and Crohn’s disease 1–3. Despite being designed to neutralize TNF-α activity, the mechanism of action of these agents in the resolution of disease remains unclear. Psoriasis vulgaris is a common inflammatory and hyperproliferative skin disease, affecting over 4 million individuals4. The most characteristic feature of psoriasis is the marked hyperproliferation and altered differentiation of epidermal keratinocytes. This epidermal hyperproliferation is now thought to be driven largely by IL-17A, IL-22, IFN-γ and TNF-α-secreting T cells in the skin 5–8. The IL-17 cytokine family members IL-17A, C and F have been shown to be significantly elevated in lesional psoriasis skin 9. These cytokines utilise a family of five receptor subunits (IL-17RA-RE) all of which have been detected in skin 9 with IL-17A and IL-17F using a combination of IL-17RA and IL-17RC for signalling 9,10. The synergistic pro-inflammatory activity of cytokines has recently become a focus of attention 11–14 with an understanding that rather than acting alone, each cytokine is participating in an inflammatory network 15–17 with the possibility that sequestration of one key member of this network could result in the collapse of the network and resolution of inflammation.
We propose that the mechanism of action of etanercept involves dismantling the powerful synergy between TNF-α and IL-17A, reducing IL-17A signalling and the expression of IL-17A-induced chemokines prior to changes in T cell numbers, keratinocyte differentiation and proliferation. To test our hypothesis, we analysed the expression of mRNA, cytokines and phospho-proteins in the lesional skin of chronic plaque psoriasis patients treated with the anti-TNF agent etanercept twice weekly, focusing on the first 2 weeks of treatment before improvements in disease severity were clinically evident. We found that IL-17A, IL-22 and IFN-γ mRNA and protein showed no significant change in the first week of treatment; however, there was a 2.5-fold down-regulation of IL17RC mRNA and decreased activated ERK1/2, a signal transduction element downstream of the IL17 receptor. In vitro, TNF-α increased the expression of IL-17RC mRNA and protein by keratinocytes and shRNA suppression of IL-17RC curtailed synergistic TNF-IL17A responses. Furthermore, analysis of global gene expression revealed that etanercept induced a marked suppression of IL-17A-induced genes in lesional skin and this overlapped with the effects of the anti-IL-17A drug ixekizumab.
These results suggest that the early responses of psoriasis plaques to etanercept may be due in part to diminished tissue responses to IL-17A resulting from decreased expression of one IL-17 receptor subunit (IL-17RC), thus responses to tissue Th17 cytokines are blunted, breaking a potentially self-sustaining cycle contributing to the maintenance of psoriasis lesions.
MATERIALS AND METHODS
Study Population
Twenty individuals with chronic plaque psoriasis were enrolled (age range 18–75 years). Entry criteria included age greater than 18 years and stable plaque-type psoriasis involving at least 10% body surface area. Exclusion criteria included use of systemic psoriasis therapy within 4 weeks, topical therapy within 2 weeks, or severe co-morbid diseases. For 12 weeks, subjects received etanercept (Enbrel) 50mg twice a week subcutaneously. At baseline, 6 mm punch biopsies were obtained under local anaesthesia (lidocaine) from uninvolved skin and a target plaque. Subsequent biopsies were taken on days 1, 3, 7 and 14 of therapy from the same target plaque at least 1cm away from previous biopsy sites. Disease severity was recorded on enrolment and at weeks 1, 2, 4 and 12 using percentage body surface area (BSA) and physician global assessment (PGA) scores. Informed consent was obtained from all subjects, under protocols approved by the Institutional Review Board of the University of Michigan Medical School (HUM00037994). This study was conducted in compliance with good clinical practice and according to the Declaration of Helsinki Principles.
Real Time Quantitative Reverse Transcription PCR (qRT-PCR)
After removal from the skin, biopsies were bisected and snap-frozen in liquid nitrogen and stored at −80°C until use. Biopsies were pulverized with a hammer while still frozen, dissolved in RLT buffer (Qiagen, Chatsworth, CA), homogenized using glass beads (Biospec Products Inc, Bartlesville, OK) and total RNA was isolated (RNeasy Mini kit, Qiagen). 200ng RNA was reverse transcribed (High Capacity cDNA Transcription kit, Applied Biosystems Inc., Foster City, CA) and transcripts quantified using a 7900HT Fast Real-time PCR system (Applied Biosystems) using Taqman primer sets purchased from Applied Biosystems (IL17A, Hs00174383_m1; IL22, Hs00220924_m1; IL17RA, Hs01064648_m1; IL17RC, Hs00994305_m1; IL10RB, Hs00175123_m1; IL22RA1, Hs00222035_m1). All values were normalized to the expression of the housekeeping gene ribosomal protein, large, P0 (RPLP0, Hs99999902_m1).
RNA processing and microarray hybridization
Total RNA was isolated as above from biopsies from 6 individuals, quality checked using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) and transcribed for probe biotinylation using 5μg RNA according to the manufacturer’s protocols (Ambion, Austin, TX). Six samples were obtained from each individual, including two baseline samples (PP and PN), as well as four samples obtained following 1, 3, 7 and 14 days of etanercept therapy, respectively (36 samples in total). Samples were run on Affymetrix Human Genome U133 Plus 2.0 arrays, which include 54,675 probe sets corresponding to 20,184 human genes (Affymetrix, Foster City, CA). CEL image files were obtained for each sample and post-hybridization quality control checks were performed using metrics appropriate for Affymetrix gene chips (i.e., average background, intensity scale factor, % probe sets called present, RNA degradation score, RLE median, RLE IQR, NUSE median and NUSE IQR)18,19. Based upon these analyses, one day 3 PP sample was removed for one patient, and a day 14 PP sample was removed for another, yielding a total of 34 samples upon which further analyses were based. Normalization of these 34 samples was then performed using the Robust Multichip Average (RMA) method 20. The Wilcoxon signed rank test was also used to calculate presence/absence for each of the 54,675 probe set features represented on the Affymetrix array platform21. Prior to further analyses, we removed 19,258 probe sets not expressed above background with respect to at least 5% of the 34 samples. This yielded 35,417 probe sets upon which subsequent analyses were based. Raw microarray data has been deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) and is accessible through GEO Series accession number GSE47751.
Statistics and Gene Set Analyses
Data were tested for normality and statistical significance calculated using Student’s t-test, Mann Whitney or Friedman test as appropriate using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA). To evaluate the effects of etanercept on gene expression, paired moderated t-tests were carried out using the “limma” package developed for the R statistical software 22. Four separate analyses were performed in which lesional expression in treated patients was compared to the baseline lesional sample (i.e., day 1 vs. day 0, day 3 vs. day 0, day 7 vs. day 0, day 14 vs. day 0; n = 5–6 for each comparison). To generate ranked gene lists, results from these analyses were filtered to remove redundant probe sets corresponding the same human gene23. This was done by choosing a representative probe set for each gene, corresponding to the probe set for which the lowest p-value was obtained in the aforementioned test for differential expression. Following this filtering, ranked lists of genes were generated for each comparison, based upon the evidence for differential expression provided by raw p-values. Overlap between these lists and genes decreased in the skin of ixekizumab-treated patients or genes induced by cytokines in vitro was assessed using the hypergeometric distribution24 as recently described 25 (Figure 5). For each analysis, the number of overlapping genes between two ranked genes lists was compared at different depths, with etanercept-decreased genes compared to genes induced by IFN-γ, IL-17, TNF-α or IL-22 in cultured keratinocytes (GEO accessions GSE2737 and GSE36287) or suppressed by ixekizumab (GSE31652) (Figure 5 and S1). This provided an idea of how the number of genes belonging to the intersection region of a Venn diagram would change when the top N genes are selected and compared from two gene lists (with 10 < N < 2000) (Figure 5). In addition to such comparisons, we identified Gene Ontology biological process terms enriched among the 100 genes most strongly decreased by etanercept following 14 days of therapy (i.e., day 14 vs. day 0; Figure S2), and we performed the same analyses with respect to the 100 genes most strongly increased by etanercept (Figure S3). These analyses were generated using the conditional hypergeometric enrichment test procedure implemented in the GOstats package designed for the R statistical software 26.
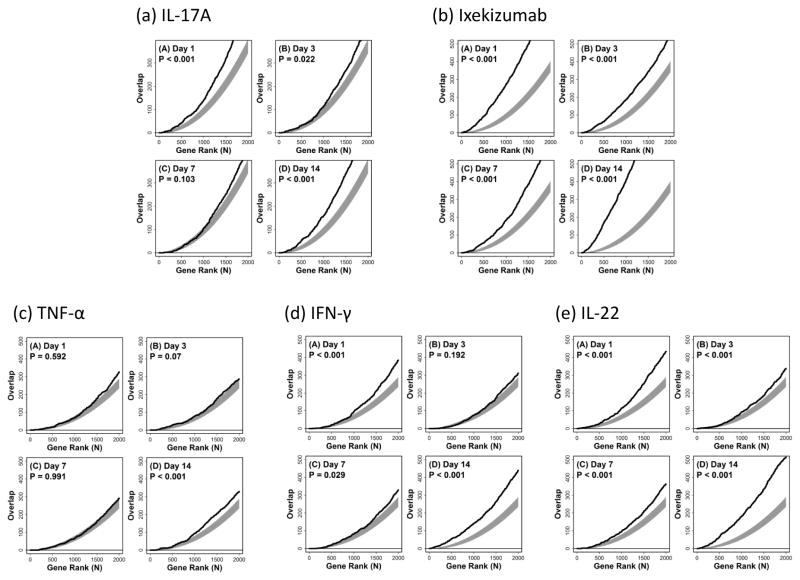
Figure 5. Enbrel treatment leads to suppression of IL-17-induced genes and its effect overlaps with that of the anti-IL-17A biologic LY2439821 (Ixekizumab).
Following RNA extraction, microarray analysis was performed on skin biopsies taken from 6 individuals on days 1, 3, 7 and 14 of etanercept treatment. We compared lists of genes down-regulated by etanercept treatment against a list of IL-17A-induced genes (a) or genes strongly decreased by the IL-17 inhibitor ixekizumab (LY2439821) (b) or genes induced by either TNF-α (c) or IFN-γ (d). The overlap between etanercept-decreased genes and IL-17A-induced genes (a) was significantly larger than expected at days 1, 7 and 14. There is a strong association between etanercept-decreased genes with genes most strongly decreased by the IL-17A inhibitor LY2439821, even after only 1 day of treatment (b). The overlap between etanercept-decreased genes and TNF-α (c) or IFN-γ (d) -induced genes was much less pronounced. In each box, the black line shows the level of overlap among the top n genes taken from each list, whereas the grey region outlines the central 95% of the null distribution under a random sampling model (hypergeometric distribution). Larger than expected overlap (for given n) is present, if the line lies above the grey region, whereas lower than expected overlap (for given n) is present, if the line lies below the grey region. P-values are based upon the overlap between the top 1000 genes from each gene list.
Multiplex bead assays
Biopsies were pulverized while still frozen then suspended in 500μl ice-cold lysis buffer (Bio-Rad, Hercules, CA) and ground in a glass tissue grinder. After 1 freeze-thaw cycle, samples were centrifuged and supernatants collected and diluted to 1mg/ml total protein in PBS. Bead assays were conducted as directed by manufacturer (Bio-Plex, Bio-Rad) using duplicate 50μl samples. Concentrations were determined by 4- or 5-parameter fitting of standard curves.
Keratinocyte culture
Normal human keratinocyte (NHK) cultures were established from sun-protected adult human skin as described 27 in serum-free medium optimized for high-density keratinocyte growth (Medium 154, Invitrogen, Portland, OR). NHKs were used for experiments in the second or third passage. All cells were plated at 5000 cells/cm2 and maintained to 4-days post-confluence. Cultures were then starved of growth factors in unsupplemented M154 for 24 hours before use. NHK were stimulated with recombinant human TNF-α and IL-17A (R&D Systems, Minneapolis, MN) under low calcium (0.1mM) conditions.
IL-17 Receptor knock-down
shIL17RA (shIL17RA-53, Sigma) and shIL17RC (shIL17RC-10, Sigma) pLKO.1-puro vectors (Sigma) were grown in 293FT cells and constructs were tested for knock-down of IL-17RA and IL-17RC expression, respectively as assessed by QRT-PCR and Western blotting using antibodies against IL17RC (NBP1-32481, 1:1000 dilution, Novus, Littleton, CO) and β-actin to control for protein loading (clone AC-74, 1:2500 dilution, Sigma). The shIL17RA-53 and shIL17RC-10 constructs were selected for use in KC stimulation experiments on day 2 post-infection when knock-down was most effective.
RESULTS
Clinical results
On recruitment disease severity was recorded as a median body surface area (BSA) of 35% and median physician global assessment (PGA) score of 3.6. We determined that 18 of 20 subjects were responders to treatment as defined as at least a 25% decrease in BSA by week 12. The mean decrease in BSA score was 64% which was in accord with our previous study where 25 of 30 patients responded to etanercept with a mean BSA score decrease of 62% 28. The decrease in disease severity was significant at weeks 4 and 12 with respect to both BSA and PGA scores (Fig. 1a and b). Analyses of the 2 non-responders were underpowered to detect differences due to small sample size, thus only responder data are presented in this report.
Figure 1. Clinical changes in disease severity were evident after 4 weeks’ of etanercept treatment.
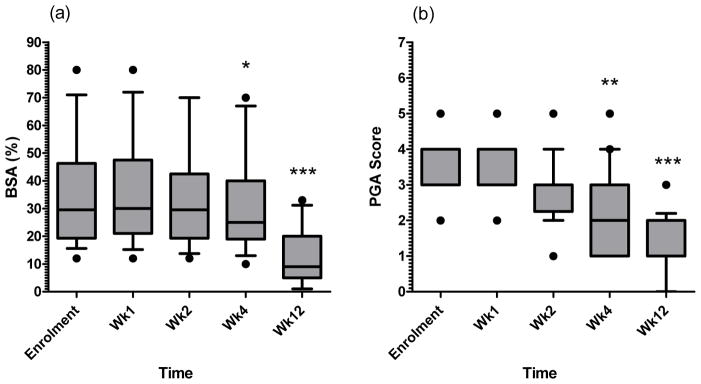
On recruitment the median body surface area (BSA, a) was recorded as 35%. 18/20 subjects responded to treatment with at least a 25% decrease in BSA by week 12, with the mean decrease at 64% (range 26–99%). The 2 subjects with poor responses (0 and 17% reduction in BSA at week 12) were excluded from further analysis as they were considered as non-responders. The decrease in disease severity was significant at week 4 and week 12 with respect to BSA and Physician Global Assessment scores (PGA, b). Boxes extend from the 25th to 75th percentiles showing median as a line, whiskers denote the 10th and 90th percentiles and dots show minimum and maximum values if outside this range. Statistical significance determined using Friedman test (p<0.001, both series) with Dunn’s post-test on individual time points, indicated * p<0.05, ** p<0.01, *** p<0.001, n=18.
Etanercept alters tissue cytokine and receptor expression
Given that our hypothesis is that the mechanism of action of etanercept involves early tissue responses which precede changes in T cell numbers, keratinocyte differentiation and proliferation, we first analysed cytokine mRNA expression by QRT-PCR in the 18 responding patients. In the first 2 weeks of treatment both IL-17A and IL-22 remained significantly elevated compared with uninvolved skin, with IL-17A significantly decreased only at day 14 (p<0.05, Fig. 2a). The IL-17A receptor subunit IL-17RA expression remained unchanged (Fig. 2b), however, IL-17RC expression declined rapidly with significant decrease in expression observed already at day 1 (p=0.0167 overall, Fig. 2c). This preceded any changes in T cell infiltration or human β-defensin 2 expression (Fig. 2g and 2h). In contrast, neither of the receptor subunits for IL-22 were altered in expression (Fig. 2e and f).
Figure 2. Etanercept alters tissue cytokine and receptor mRNA expression in the first 2 weeks of treatment.
Total RNA was extracted from snap-frozen 6mm punch biopsies of responding patients and analysed with qRT-PCR. Transcripts for IL-17A (a) and IL-22 (d) were significantly more abundant in lesional (Day 0) than non-lesional skin (PN). During treatment, mRNA for IL-17RC (c) but not IL-17RA (b) was significantly decreased. Neither IL-22 (d) nor its receptor subunits (e and f) were down-regulated in the first 2 weeks’ treatment. The decrease in IL-17RC preceded any changes in CD3+ T cell infiltration (g) or human β-defensin 2 expression (h). Statistical significance determined with Mann Whitney test (PN vs Day 0) or Friedman test with Dunn’s multiple comparison test (time course) and indicated * p<0.05, ** p<0.01, *** p<0.001. Bars, mean + SEM, n=18.
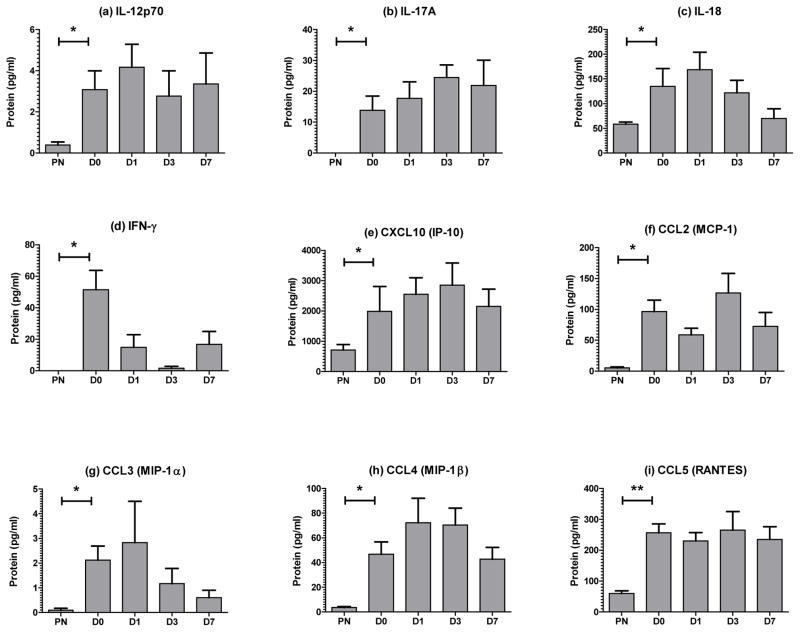
We also analysed tissue cytokine protein levels in the skin biopsies using multiplex bead assays. Levels of IL-12p70, IL-17A, IL-18, IFN-γ, CXCL10 (IP-10), CCL2 (MCP-1), CCL3 (MIP-1α), CCL4 (MIP-1β) and CCL5 (RANTES) were all significantly higher in lesional psoriasis (PP) biopsies than non-lesional skin (PN) at day 0 (Fig. 3). However, of these only IFN-γ (Fig. 3d, p=0.078, day 7) and CCL3 (Fig. 3g, p=0.039, overall) showed a substantial decline in protein expression in the first week of treatment.
Figure 3. Etanercept alters tissue cytokine protein expression in the first 2 weeks of treatment.
6mm punch biopsies were snap-frozen, pulverized and diluted to 1mg/ml total protein before analysis with Bio-Rad human cytokine multiplex kits. IL-12p70 (a), IL-17A (b), IL-18 (c), IFN-γ (d), CXCL-10 (e), CCL2 (f), CCL3 (g), CCL4 (h) and CCL5 (i) protein levels were all determined to be significantly elevated in psoriasis plaques at day 0 (D0) compared with uninvolved skin (PN) from the same individuals (Mann Whitney test). During the first 7 days’ treatment, CCL3 (g, p=0.039) showed a significant decline in protein levels (Friedman test with Dunn’s multiple comparison test). Bars, mean + SEM, n=5, statistical significance indicated as * p<0.05.
Etanercept treatment results in decreased IL-17A signalling
Given that we saw suppressed IL-17RC mRNA expression in the psoriatic lesions, we examined events immediately downstream of cytokine receptors, using multiplex bead array assays to examine the expression of the activated (phosphorylated) forms of several important signal transduction molecules downstream of the IL-17 receptor. We observed a 64% decrease in the level of phospho-ERK1/2 on day 1 compared with pre-treatment; however we detected no changes in the levels of phospho-STAT3 or phospho-Tyk-2 in skin (Fig. 4). Suggesting that in the first week of treatment there are no alterations in the activity of Type 1 interferons (via phospho-STAT2) or IL-22 and IL-6 (and other cytokines that signal via phospho-STAT3) in the skin.
Figure 4. STAT2, STAT3 and ERK1/2 are significantly activated in lesional skin compared to uninvolved skin.
6mm punch skin biopsies were snap-frozen, pulverized and diluted to 1mg/ml total protein before analysis with Bio-Rad human phosphoprotein multiplex kits. Tissue levels of phospho-ERK1/2 (a) and phospho-GSK3a/b (c) were suppressed on days 1 and 7 after treatment with etanercept. Bars, mean fluorescence ± SEM, n=5. Statistical significance determined with paired t-test and indicated as ** p<0.01 and * p<0.05.
Etanercept treatment is accompanied by decreased IL-17A-induced gene expression in skin
Following RNA extraction, microarray analysis was performed on skin biopsies taken from 6 individuals on days 1, 3, 7 and 14 of etanercept treatment. Using publicly available datasets archived on the Gene Expression Omnibus (GEO), specifically GEO series GSE31652 GSE36287 and GSE7216 (Table S1), we compared lists of genes most strongly down-regulated by etanercept treatment against a lists of genes induced by treatment of NHK with IL-17A, TNF-α, IFN-γ, IL-22 and treatment of psoriasis patients with the IL-17 inhibitor ixekizumab. We found a significantly larger than expected overlap between etanercept-decreased genes and those genes induced by IL-17A on days 1, 7 and 14 (Fig. 5a). Next, we compared a list of genes down-regulated by etanercept treatment against those genes decreased by the IL-17 inhibitor ixekizumab (LY2439821). We found a strong association between the genes suppressed by etanercept and ixekizumab, which was evident as soon as day 1 of treatment and remained significant at each time point tested (Fig. 5b). We also examined the overlap between etanercept-decreased genes and lists of keratinocyte genes induced by TNF-α (Fig. 5c), IFN-γ (Fig. 5d) and IL-22 (Fig. 5e) with significant gene list overlaps only on day 1 of treatment for TNF-α and IFN-γ, and days 1, 7 and 14 for IL-22-induced genes. This overlap between the top 1000 genes most strongly decreased by etanercept and the top 1000 genes most strongly altered by IL-17A, TNF-α, IFN-γ, IL-22 and ixekizumab are also depicted in an alternative graphic (Fig. S1). Further, we analysed the genes most strongly altered by etanercept with respect to Gene Ontology (GO) biological processes, i.e., those genes increased or decreased in day 14 lesions as compared to those lesions obtained at baseline (Figs. S2 and S3). Among the top 100 genes most strongly decreased in the day 14 lesions, we detected significant enrichment for GO terms such as response to “type I interferon”, “leukocyte migration” and “regulation of T-cell apoptosis” (FDR < 0.05). These functional terms are consistent with those associated with genes most strongly elevated in psoriasis lesions (as compared to normal uninvolved skin), suggesting that the effect of etanercept is to shift gene expression patterns towards those in normal skin.
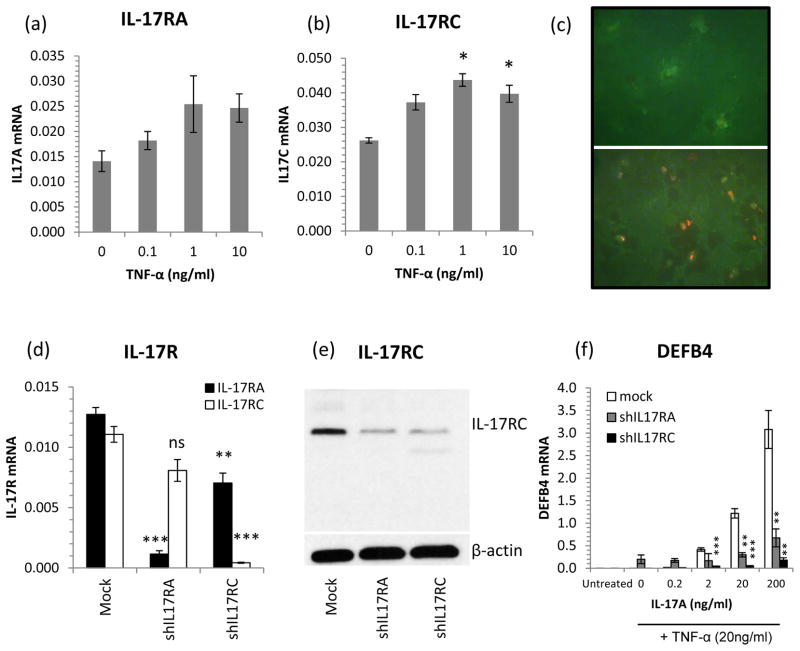
Keratinocyte IL-17RA and IL-17RC are induced by TNF-α and shRNA knock-down of IL-17RA or RC abrogates TNFa-IL-17A synergism
Given that we observed a decrease in the expression of IL-17RC in the skin of etanercept responders, we tested the effects of TNF-α on IL-17 receptor expression by keratinocytes. NHK were treated with 0–10ng/ml TNF-α for 24h and the mRNA expression of the two receptor subunits IL-17RA and IL-17RC was quantified by QRT-PCR. We found that while TNF-α increased the expression of IL-17RA, the effect was more pronounced on IL-17RC (Fig. 6a and b). The increased expression of IL-17RC was confirmed using immunohistochemical detection of IL-17RC in resting and TNF-α-treated NHK. After 24h of TNF-α treatment, strikingly increased immunofluorescence for IL-17RC could be seen (Fig. 6c).
Figure 6. TNF-α induces IL-17R expression by keratinocytes and shRNA knock-down of IL17R expression abolishes synergistic TNF-α – IL-17A gene induction.
NHK treated with TNF-α for 24h significantly increased IL-17RC but not IL-17RA mRNA expression (a and b). Immunocytochemical detection of IL-17RA (green) and IL-17RC (red) protein expression in resting (top) and TNF-α-stimulated (bottom, 10ng/ml, 24h) NHK showing increased IL-17RC protein expression after TNF-α treatment (c). Targeted knock-down of IL-17RA and IL-17RC expression (d, RT-PCR and e, Western blot) lead to significantly decreased DEFB4 responses to IL-17A + TNF-α in IL-17RA (grey bars) and IL-17RC (black bars) deficient cells compared with mock transfected cells (open bars) (f). All bars, mean ± SEM (n=3), statistical significance determined by ANOVA (a and b) or Student’s t-test (d, e and f) and indicated: * p<0.05, ** p<0.01, *** p<0.001.
To show how sequestering TNF-α by etanercept, leading to a decrease in IL-17 receptor expression, could lead to a decrease in the expression of keratinocyte genes induced by the synergistic activity of TNF-α and IL-17A, we selectively knocked down IL-17RA or IL-17RC expression in NHK using short hairpin (sh) RNA (Fig. 6d and e). IL-17RA or IL-17RC mRNA expression was suppressed to 9% and 4% of control levels respectively (Fig. 6d) and Western blotting revealed a strong suppression in IL-17RC protein expression in shRNA treated cells (Fig. 6e). Keratinocytes with suppressed IL-17RA or IL-17RC expression were stimulated with IL-17A in the presence of TNF-α to invoke the synergistic induction of human β-defensin 2 (DEFB4), one of the most up-regulated proteins in lesional psoriasis skin. Knock-down of either receptor subunit, but IL-17RC in particular, lead to a significant suppression of TNF/IL-17A induced DEFB4 mRNA expression (Fig. 6f).
DISCUSSION
The anti-TNF biologics, etanercept (Enbrel), infliximab (Remicade) and adalimumab (Humira) have revolutionised the treatment of chronic skin disease over the last 13 years, with disease remission rates (PASI75) up to 80% 29–32 demonstrating that specific cytokine targeting can be extremely effective in managing this disease, and as such and etanercept is considered a first-line systemic biologic for chronic plaque psoriasis. The anti-TNF biologics typically require a few weeks before significant clinical improvement is noted 29–32 but early studies on etanercept’s mechanism of action in psoriasis have identified a number of changes in immune cell frequencies33 and cytokine and chemokine mRNAs 34 as early as 1 month of treatment, before any meaningful clinical improvement occurred, and more recent studies have revealed biochemical and histological changes as early as 1 week into treatment 35,36. IL-1β and IL-8 expression was shown to be decreased after 1 week 36, and interestingly this occurred in both patients who did and did not respond to etanercept treatment (about 30% of psoriasis patients do not respond well clinically 29). Using gene set enrichment analysis it was further shown that both etanercept responders and non-responders had decreases in TNF-α-induced genes but only responders had decreases in IL-17A-induced gene sets 36. This suggests that the control of IL-17A activity is a crucial part of mechanism of action of etanercept. In that respect, etanercept has been shown to block TNF-α activation of myeloid DC leading to IL-23-driven Th17 responses 35. Moreover, IL-17A and TNF-α have been demonstrated to have additive and synergistic effects directly on keratinocytes 11,28 which may be driven by the stabilisation TNF-α-induced mRNA transcripts by IL-17A 37. Interestingly, the biology behind the poor responses of the group of patients unresponsive to etanercept is unknown and there are still no reliable predictors of clinical response.
IL-17 has been implicated in psoriasis for more than 15 years 14 and interest in this cytokine is now very strong given the promising data from trials of anti-IL-17 biologics in psoriasis 38–40. It is now widely appreciated that cytokines can have strongly synergistic activities 11–14, working in a complex inflammatory network 15–17,41 promoting immune activation, proliferation and inflammation, thus we hypothesized that etanercept acts by dismantling the powerful synergy between TNF-α and IL-17, reducing IL-17 signalling and the expression of IL-17-induced chemokines prior to later changes in T-cell numbers, keratinocyte differentiation and proliferation (Figure 7). To test this idea, we took the approach of examining very early time points in the treatment protocol to detect some of the earliest biochemical changes which precipitate the collapse of the inflammatory circuits that drive the psoriatic hyperplasia.
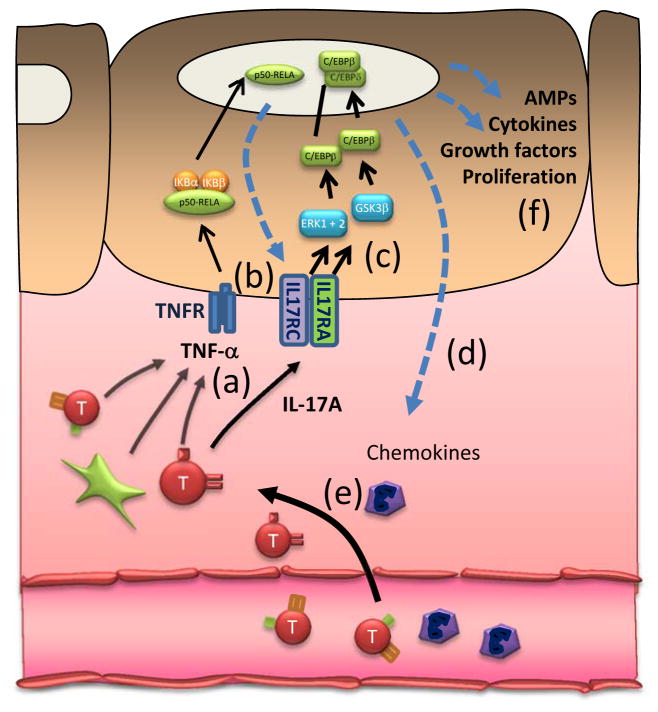
Figure 7. A proposed model of the action of etanercept in psoriasis lesions.
TNF-α and IL-17A are integral parts of a self-sustaining pro-inflammatory cytokine network which maintains psoriatic lesions. We hypothesized that etanercept acts by dismantling the powerful synergy between TNF-α and IL-1711,28,37 (a), by reducing keratinocyte IL-17 receptor expression (b), decreasing IL-17 signalling (c) and decreasing the induction of IL-17-induced chemokines (d), all prior to changes in T cell (red), APC (green) and neutrophil (purple) numbers in skin (e), and keratinocyte differentiation and proliferation (f).
We found that IL-17A, IL-22 and IFN-γ mRNA and protein showed no significant change in the first week of treatment (Fig. 2 and 3); however, there was a 2.5-fold down-regulation of IL17RC mRNA after day 1 (Fig. 2c). In contrast, IL-17RA, and subunits of the IL-22 receptor (IL-22RA and IL-10RB) were unchanged (Fig. 2b, e and f). Examining the activation of pathways downstream of these receptors showed a reduction in the phosphorylation of GSK3a/b and ERK1/2 by 50% and 64% respectively on day 1, suggesting reduced IL17 receptor signalling (Fig. 4). When we examined global gene expression by microarray, we found than etanercept induced a suppression of IL-17A-induced genes (Fig. 5a and S1) which is consistent with the findings of Zaba et al.42 who found that IL-17 pathway genes were strongly down-regulated in etanercept responders. Moreover, this etanercept-suppressed gene set overlapped with those genes suppressed by the anti-IL-17A drug ixekizumab (Fig. 5b). Moreover, TNF-α was shown to increase the expression of IL-17RC mRNA and protein by keratinocytes (Fig. 6a–c) and shRNA inhibition of IL-17RC expression curtailed synergistic TNF-IL17A responses (Fig. 6d–f). Since IL-17A utilises the IL-17RA/RC complex for signalling, the decreased expression of IL-17RC induced by etanercept is likely to lead to decreased keratinocyte activation by IL-17A with subsequent decreases in T cell and APC activity 28,35,36 and an eventual decrease in disease activity resulting in resolution of skin inflammation (Figure 7). Along these lines, the recent data on the efficacy of biologics targeting both IL-17A 38,39 and its IL-17RA 40 receptor chain highlight the crucial role of IL-17A in psoriatic inflammation. Moreover, given the need for IL-17RC in the IL-17R complex, agents targeting IL-17C may also hold the promise of efficacy for psoriasis and Th17-driven inflammatory diseases.
CONCLUSIONS
These results support the view that the early responses of psoriasis plaques to etanercept are due to decreased IL-17A activity, not from lowered Th17 cytokine expression, as the lesions are still rich in IL-17A, IL-22 and activated STAT3, but from diminished tissue responsiveness to IL-17A as a result of decreased expression of an essential subunit of the IL-17A receptor (IL17RC), thus blunting responses to Th17 cytokines and breaking a potentially self-sustaining cycle which contributes to the maintenance of psoriasis lesions.
Supplementary Material
Figure S1: The association between etanercept-altered gene expression and the genes altered by ixekizumab, TNF-α, IFN-γ, and IL-22. We identified the 1000 genes most strongly induced by treatment of cultured KCs with cytokines (IL-17, TNF, IFN-γ, and IL-22) as well as the 1000 genes most strongly repressed in lesional skin of patients treated with ixekizumab. Additionally, we identified the 1000 genes most strongly repressed in lesional skin of patients treated with etanercept following 1, 3, 7 and 14 days of treatment, respectively. The number of genes associated with the intersection of these gene sets was then evaluated (see left margin). Yellow boxes outline the null distribution for the level of overlap expected on the basis of chance (middle 95% of null hypergeometric distribution). P-values listed in the right margin are derived from a test of whether the observed level of overlap is significantly large (i.e., outside of the yellow null region; Fisher’s Exact Test). Table S1 provides a description of source datasets used to define gene sets considered in these analyses.
Figure S2. Gene ontology (GO) biological process terms associated with etanercept-repressed genes. We identified the GO biological process terms most strongly overrepresented among the 100 genes most strongly repressed in lesions of patients following 14 days of etanercept treatment (left margin). For each GO term, the value in parentheses indicates the number of associated etanercept-repressed genes (out of 100). The right margin lists up to three of the etanercept-repressed genes associated with the indicated GO term. All listed terms show statistically significant overrepresentation (P < 0.05; Fisher’s Exact Test). Among the top 100 genes most strongly decreased in the day 14 lesions, we detected significant enrichment for GO terms such as response to “type I interferon”, “leukocyte migration” and “regulation of T-cell apoptosis” (FDR < 0.05). These functional terms are consistent with those associated with genes most strongly elevated in psoriasis lesions (as compared to normal uninvolved skin), suggesting that the effect of etanercept is to shift gene expression patterns towards those in normal skin.
Figure S3. Gene ontology (GO) biological process terms associated with etanercept-induced genes. We identified the GO biological process terms most strongly overrepresented among the 100 genes most strongly induced in lesions of patients following 14 days of etanercept treatment (left margin). For each GO term, the value in parentheses indicates the number of associated etanercept-induced genes (out of 100). The right margin lists up to three of the etanercept-induced genes associated with the indicated GO term. All listed terms show statistically significant overrepresentation (P < 0.05; Fisher’s Exact Test). Among the top 100 genes most strongly increased in day 14 lesions, we detected enrichment for GO terms related to development and differentiation (FDR < 0.05). Such terms are often associated with genes most strongly decreased in psoriasis lesions (as compared to normal uninvolved skin), consistent with the idea that etanercept effectively normalizes the transcriptome, shifting expression patterns towards those of normal skin.
What’s already known about this topic?
TNF-α inhibitors are effective for the treatment of psoriasis yet their mode of action appears to involve more than just inhibition of TNF-α responses.
What does this study add?
The study provides evidence that the early responses of psoriasis plaques to etanercept may be due to diminished tissue responsiveness to IL-17A as a result of the decreased expression of IL17RC, blunting responses to Th17 cytokines and breaking a potentially self-sustaining cycle which contributes to the maintenance of psoriasis lesions.
Acknowledgments
This work was supported in part by Amgen Inc. AJ is supported by The American Skin Association, The Dermatology Foundation, a Babcock Foundation Endowment and NIH K01 AR064765. WRS is funded in part by the American Skin Association Carson Family Research Scholar Award in Psoriasis. JEG is supported by NIH K08 AR060802, the National Psoriasis Foundation, Taubman Medical Research Institute (as the Kenneth and Frances Eisenberg Emerging Scholar) and a Babcock Foundation Endowment.
We thank the research subjects who participated in this study. We also thank Dr. Stefan W. Stoll for kindly providing NHK cultures and Xianying Xing and Marybeth Riblett for excellent technical assistance. Amgen Inc. provided partial funding and etanercept syringes for research purposes. Amgen had no involvement in the design or conduct of the study or in the collection, management, analysis, and interpretation of the data. Amgen was not involved in the preparation or review of the manuscript.
References
- 1.Moreland LW, Schiff MH, Baumgartner SW, et al. Etanercept therapy in rheumatoid arthritis. A randomized, controlled trial. Ann Intern Med. 1999;130:478–86. doi: 10.7326/0003-4819-130-6-199903160-00004. [DOI] [PubMed] [Google Scholar]
- 2.Targan SR, Hanauer SB, van Deventer SJ, et al. A short-term study of chimeric monoclonal antibody cA2 to tumor necrosis factor alpha for Crohn’s disease. Crohn’s Disease cA2 Study Group. N Engl J Med. 1997;337:1029–35. doi: 10.1056/NEJM199710093371502. [DOI] [PubMed] [Google Scholar]
- 3.Mease PJ, Goffe BS, Metz J, et al. Etanercept in the treatment of psoriatic arthritis and psoriasis: a randomised trial. Lancet. 2000;356:385–90. doi: 10.1016/S0140-6736(00)02530-7. [DOI] [PubMed] [Google Scholar]
- 4.Fowler JF, Duh MS, Rovba L, et al. The impact of psoriasis on health care costs and patient work loss. J Am Acad Dermatol. 2008;59:772–80. doi: 10.1016/j.jaad.2008.06.043. [DOI] [PubMed] [Google Scholar]
- 5.Wilson NJ, Boniface K, Chan JR, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950–7. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 6.Hijnen D, Knol EF, Gent YY, et al. CD8(+) T cells in the lesional skin of atopic dermatitis and psoriasis patients are an important source of IFN-gamma, IL-13, IL-17, and IL-22. J Invest Dermatol. 2013;133:973–9. doi: 10.1038/jid.2012.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Res PC, Piskin G, de Boer OJ, et al. Overrepresentation of IL-17A and IL-22 producing CD8 T cells in lesional skin suggests their involvement in the pathogenesis of psoriasis. PLoS One. 2010;5:e14108. doi: 10.1371/journal.pone.0014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eyerich S, Eyerich K, Pennino D, et al. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119:3573–85. doi: 10.1172/JCI40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnston A, Fritz Y, Dawes SM, et al. Keratinocyte overexpression of IL-17C promotes psoriasiform skin inflammation. J Immunol. 2013;190:2252–62. doi: 10.4049/jimmunol.1201505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge D, You Z. Expression of interleukin-17RC protein in normal human tissues. International archives of medicine. 2008;1:19. doi: 10.1186/1755-7682-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiricozzi A, Guttman-Yassky E, Suarez-Farinas M, et al. Integrative responses to IL-17 and TNF-alpha in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol. 2011;131:677–87. doi: 10.1038/jid.2010.340. [DOI] [PubMed] [Google Scholar]
- 12.Kryczek I, Bruce A, Gudjonsson J, et al. Induction of IL-17+ T cell trafficking and development by IFN-gamma: Mechanism and pathological relevance in psoriasis. Journal of Immunology. 2008;181:4733–41. doi: 10.4049/jimmunol.181.7.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston A, Gudjonsson JE, Aphale A, et al. EGFR and IL-1 signaling synergistically promote keratinocyte antimicrobial defenses in a differentiation-dependent manner. J Invest Dermatol. 2011;131:329–37. doi: 10.1038/jid.2010.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teunissen MB, Koomen CW, de Waal Malefyt R, et al. Interleukin-17 and interferon-gamma synergize in the enhancement of proinflammatory cytokine production by human keratinocytes. J Invest Dermatol. 1998;111:645–9. doi: 10.1046/j.1523-1747.1998.00347.x. [DOI] [PubMed] [Google Scholar]
- 15.Gudjonsson JE, Johnston A, Sigmundsdottir H, et al. Immunopathogenic mechanisms in psoriasis. Clinical and experimental immunology. 2004;135:1–8. doi: 10.1111/j.1365-2249.2004.02310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickoloff BJ, Xin H, Nestle FO, et al. The cytokine and chemokine network in psoriasis. Clin Dermatol. 2007;25:568–73. doi: 10.1016/j.clindermatol.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Swindell WR, Xing X, Stuart PE, et al. Heterogeneity of inflammatory and cytokine networks in chronic plaque psoriasis. PLoS One. 2012;7:e34594. doi: 10.1371/journal.pone.0034594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolstad BM, Collin F, Brettschneider J, et al. Quality assessment of Affymetrix GeneChip Data. In: Gentleman R, Carey V, Huber W, et al., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. pp. 33–47. [Google Scholar]
- 19.Popova T, Mennerich D, Weith A, et al. Effect of RNA quality on transcript intensity levels in microarray analysis of human post-mortem brain tissues. BMC Genomics. 2008;9:91. doi: 10.1186/1471-2164-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu WM, Mei R, Di X, et al. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–9. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 23.Stalteri MA, Harrison AP. Interpretation of multiple probe sets mapping to the same gene in Affymetrix GeneChips. BMC bioinformatics. 2007;8:13. doi: 10.1186/1471-2105-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lottaz C, Yang X, Scheid S, et al. OrderedList--a bioconductor package for detecting similarity in ordered gene lists. Bioinformatics. 2006;22:2315–6. doi: 10.1093/bioinformatics/btl385. [DOI] [PubMed] [Google Scholar]
- 25.Johnston A, Xing X, Swindell WR, et al. Susceptibility-associated genetic variation at IL12B enhances Th1 polarization in psoriasis. Human molecular genetics. 2013;22:1807–15. doi: 10.1093/hmg/ddt034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–8. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 27.Elder JT, Fisher GJ, Zhang QY, et al. Retinoic acid receptor gene expression in human skin. J Invest Dermatol. 1991;96:425–33. doi: 10.1111/1523-1747.ep12469889. [DOI] [PubMed] [Google Scholar]
- 28.Wang F, Smith N, Maier L, et al. Etanercept suppresses regenerative hyperplasia in psoriasis by acutely downregulating epidermal expression of interleukin (IL)-19, IL-20 and IL-24. Br J Dermatol. 2012;167:92–102. doi: 10.1111/j.1365-2133.2012.10961.x. [DOI] [PubMed] [Google Scholar]
- 29.Leonardi CL, Powers JL, Matheson RT, et al. Etanercept as monotherapy in patients with psoriasis. N Engl J Med. 2003;349:2014–22. doi: 10.1056/NEJMoa030409. [DOI] [PubMed] [Google Scholar]
- 30.Menter A, Tyring SK, Gordon K, et al. Adalimumab therapy for moderate to severe psoriasis: A randomized, controlled phase III trial. J Am Acad Dermatol. 2008;58:106–15. doi: 10.1016/j.jaad.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Barker J, Hoffmann M, Wozel G, et al. Efficacy and safety of infliximab vs. methotrexate in patients with moderate-to-severe plaque psoriasis: results of an open-label, active-controlled, randomized trial (RESTORE1) Br J Dermatol. 2011;165:1109–17. doi: 10.1111/j.1365-2133.2011.10615.x. [DOI] [PubMed] [Google Scholar]
- 32.Mrowietz U, Kragballe K, Reich K, et al. An assessment of adalimumab efficacy in three Phase III clinical trials using the European Consensus Programme criteria for psoriasis treatment goals. Br J Dermatol. 2013;168:374–80. doi: 10.1111/j.1365-2133.2012.11214.x. [DOI] [PubMed] [Google Scholar]
- 33.Lizzul PF, Aphale A, Malaviya R, et al. Differential expression of phosphorylated NF-kappaB/RelA in normal and psoriatic epidermis and downregulation of NF-kappaB in response to treatment with etanercept. J Invest Dermatol. 2005;124:1275–83. doi: 10.1111/j.0022-202X.2005.23735.x. [DOI] [PubMed] [Google Scholar]
- 34.Gottlieb AB, Chamian F, Masud S, et al. TNF inhibition rapidly down-regulates multiple proinflammatory pathways in psoriasis plaques. J Immunol. 2005;175:2721–9. doi: 10.4049/jimmunol.175.4.2721. [DOI] [PubMed] [Google Scholar]
- 35.Zaba LC, Cardinale I, Gilleaudeau P, et al. Amelioration of epidermal hyperplasia by TNF inhibition is associated with reduced Th17 responses. J Exp Med. 2007;204:3183–94. doi: 10.1084/jem.20071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaba LC, Suarez-Farinas M, Fuentes-Duculan J, et al. Effective treatment of psoriasis with etanercept is linked to suppression of IL-17 signaling, not immediate response TNF genes. J Allergy Clin Immunol. 2009;124:1022–10. e1–395. doi: 10.1016/j.jaci.2009.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartupee J, Liu C, Novotny M, et al. IL-17 enhances chemokine gene expression through mRNA stabilization. J Immunol. 2007;179:4135–41. doi: 10.4049/jimmunol.179.6.4135. [DOI] [PubMed] [Google Scholar]
- 38.Leonardi C, Matheson R, Zachariae C, et al. Anti-interleukin-17 monoclonal antibody ixekizumab in chronic plaque psoriasis. N Engl J Med. 2012;366:1190–9. doi: 10.1056/NEJMoa1109997. [DOI] [PubMed] [Google Scholar]
- 39.Papp KA, Langley RG, Sigurgeirsson B, et al. Efficacy and safety of secukinumab in the treatment of moderate-to-severe plaque psoriasis: a randomized, double-blind, placebo-controlled phase II dose-ranging study. Br J Dermatol. 2013;168:412–21. doi: 10.1111/bjd.12110. [DOI] [PubMed] [Google Scholar]
- 40.Papp KA, Leonardi C, Menter A, et al. Brodalumab, an anti-interleukin-17-receptor antibody for psoriasis. N Engl J Med. 2012;366:1181–9. doi: 10.1056/NEJMoa1109017. [DOI] [PubMed] [Google Scholar]
- 41.Krueger JG, Fretzin S, Suarez-Farinas M, et al. IL-17A is essential for cell activation and inflammatory gene circuits in subjects with psoriasis. J Allergy Clin Immunol. 2012;130:145–54. e9. doi: 10.1016/j.jaci.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaba LC, Fuentes-Duculan J, Eungdamrong NJ, et al. Identification of TNF-related apoptosis-inducing ligand and other molecules that distinguish inflammatory from resident dendritic cells in patients with psoriasis. J Allergy Clin Immunol. 2010;125:1261–8. e9. doi: 10.1016/j.jaci.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: The association between etanercept-altered gene expression and the genes altered by ixekizumab, TNF-α, IFN-γ, and IL-22. We identified the 1000 genes most strongly induced by treatment of cultured KCs with cytokines (IL-17, TNF, IFN-γ, and IL-22) as well as the 1000 genes most strongly repressed in lesional skin of patients treated with ixekizumab. Additionally, we identified the 1000 genes most strongly repressed in lesional skin of patients treated with etanercept following 1, 3, 7 and 14 days of treatment, respectively. The number of genes associated with the intersection of these gene sets was then evaluated (see left margin). Yellow boxes outline the null distribution for the level of overlap expected on the basis of chance (middle 95% of null hypergeometric distribution). P-values listed in the right margin are derived from a test of whether the observed level of overlap is significantly large (i.e., outside of the yellow null region; Fisher’s Exact Test). Table S1 provides a description of source datasets used to define gene sets considered in these analyses.
Figure S2. Gene ontology (GO) biological process terms associated with etanercept-repressed genes. We identified the GO biological process terms most strongly overrepresented among the 100 genes most strongly repressed in lesions of patients following 14 days of etanercept treatment (left margin). For each GO term, the value in parentheses indicates the number of associated etanercept-repressed genes (out of 100). The right margin lists up to three of the etanercept-repressed genes associated with the indicated GO term. All listed terms show statistically significant overrepresentation (P < 0.05; Fisher’s Exact Test). Among the top 100 genes most strongly decreased in the day 14 lesions, we detected significant enrichment for GO terms such as response to “type I interferon”, “leukocyte migration” and “regulation of T-cell apoptosis” (FDR < 0.05). These functional terms are consistent with those associated with genes most strongly elevated in psoriasis lesions (as compared to normal uninvolved skin), suggesting that the effect of etanercept is to shift gene expression patterns towards those in normal skin.
Figure S3. Gene ontology (GO) biological process terms associated with etanercept-induced genes. We identified the GO biological process terms most strongly overrepresented among the 100 genes most strongly induced in lesions of patients following 14 days of etanercept treatment (left margin). For each GO term, the value in parentheses indicates the number of associated etanercept-induced genes (out of 100). The right margin lists up to three of the etanercept-induced genes associated with the indicated GO term. All listed terms show statistically significant overrepresentation (P < 0.05; Fisher’s Exact Test). Among the top 100 genes most strongly increased in day 14 lesions, we detected enrichment for GO terms related to development and differentiation (FDR < 0.05). Such terms are often associated with genes most strongly decreased in psoriasis lesions (as compared to normal uninvolved skin), consistent with the idea that etanercept effectively normalizes the transcriptome, shifting expression patterns towards those of normal skin.