Summary
Anaplasma phagocytophilum, which causes granulocytic anaplasmosis in humans and animals, is a tick-transmitted obligate intracellular bacterium that mediates its own uptake into neutrophils and non-phagocytic cells. Invasins of obligate intracellular pathogens are attractive targets for protecting against or curing infection because blocking the internalization step prevents survival of these organisms. The complement of A. phagocytophilum invasins is incompletely defined. Here, we report the significance of a novel A. phagocytophilum invasion protein, AipA. A. phagocytophilum induced aipA expression during transmission feeding of infected ticks on mice. The bacterium upregulated aipA transcription when it transitioned from its non-infectious reticulate cell morphotype to its infectious dense-cored morphotype during infection of HL-60 cells. AipA localized to the bacterial surface and was expressed during in vivo infection. Of the AipA regions predicted to be surface-exposed, only residues 1 to 87 (AipA1–87) were found to be essential for host cell invasion. Recombinant AipA1–87 protein bound to and competitively inhibited A. phagocytophilum infection of mammalian cells. Antiserum specific for AipA1–87, but not other AipA regions, antagonized infection. Additional blocking experiments using peptide-specific antisera narrowed down the AipA invasion domain to residues 9 to 21. An antisera combination targeting AipA1–87 together with two other A. phagocytophilum invasins, OmpA and Asp14, nearly abolished infection of host cells. This study identifies AipA as an A. phagocytophilum surface protein that is critical for infection, demarcates its invasion domain, and establishes a rationale for targeting multiple invasins to protect against granulocytic anaplasmosis.
Introduction
Obligate intracellular bacteria use outer membrane proteins (OMPs) called invasins to enter eukaryotic host cells. Since these organisms are incapable of extracellular survival, infection can be prevented or cured by blocking the internalization step. Thus, it is desirable to identify and characterize invasins of obligate intracellular bacteria, specifically the functional domains that mediate entry into host cells. Anaplasma phagocytophilum is an obligate intracellular bacterium in the order Rickettsiales and family Anaplasmataceae that infects neutrophils to cause granulocytic anaplasmosis in humans and animals. Though primarily an Ixodes spp. tick-borne illness (Truchan et al., 2013), human granulocytic anaplasmosis (HGA) can also be transmitted perinatally, nosocomially, and through blood transfusion (Annen et al., 2012; Carlyon, 2012; Jereb et al., 2012; Alhumaidan et al., 2013). The disease presents as a non-specific febrile illness that can be accompanied by leukopenia, thrombocytopenia, elevated levels of serum transaminases, and increased susceptibility to potentially fatal secondary infections (Truchan et al., 2013). HGA is an emerging infection in the United States, Europe, and Asia (Truchan et al., 2013). The number of reported HGA cases in the United States increased over six-fold from 2003 to 2012, the latest period for which disease reporting statistics are available (Hopkins et al., 2005; Centers for Disease Control and Prevention, 2013).
A. phagocytophilum undergoes a biphasic developmental cycle that begins when an infectious dense-cored (DC) organism binds to and enters its host cell, where it resides within a host cell-derived vacuole. Between 4 and 8 h, the DC develops into the non-infectious reticulate cell (RC) morphotype that subsequently divides by binary fission to yield a bacteria-filled vacuolar inclusion. From 8 to 20 h, the intravacuolar population consists exclusively of replicating RCs. Most RCs transition back into DCs between 28 and 32 h. DCs then exit host cells between 28 to 36 h and initiate the next round of infection (Troese et al., 2009). A. phagocytophilum OMPs that are upregulated during RC-to-DC transition, bacterial exit, and reinfection are attractive candidates to evaluate for both their roles in infection and their prospect as protective antigens.
Given the potential severity of HGA, the limited choices of antibiotics for treating the disease, and the lack of a vaccine, a thorough understanding of A. phagocytophilum cellular invasion is critical. We recently identified OmpA (APH0338) and Asp14 (14-kDa A. phagocytophilum surface protein; APH0248) as being important for A. phagocytophilum entry into mammalian cells (Ojogun et al., 2012; Kahlon et al., 2013). OmpA binds to α2,3-sialic acid of the sialyl Lewis x (sLex) tetrasaccharide that caps P-selectin glycoprotein ligand-1 (PSGL-1) on myeloid cell surfaces. OmpA also recognizes α2,3-sialic acid residues that decorate as yet defined glycoproteins on endothelial cells (Ojogun et al, 2012). The Asp14 receptor is unknown. Evidence implicates involvement of one or more A. phagocytophilum invasins in addition to OmpA and Asp14 in mediating infection (Kahlon et al., 2013). Identifying these invasins and evaluating if targeting them alone or in concert with OmpA and Asp14 can block bacterial entry may foster development of effective prophylaxes against HGA.
A whole genome transcriptional profiling study revealed A. phagocytophilum genes that are upregulated during infection of mammalian versus tick cells (Nelson et al., 2008). Several of these encode putative OMPs, one of which is APH0915. In this study, we show that APH0915, hereafter referred to as AipA (A. phagocytophilum invasion protein A), is important for bacterial entry into mammalian cells and identify its invasion domain. We further demonstrate that a combination of antisera targeting AipA, OmpA, and Asp14 synergistically blocks infection. Our findings not only advance understanding of how A. phagocytophilum employs multiple invasins to promote infection, but also sets the stage for development of a multi-target vaccine that protects against granulocytic anaplasmosis.
Results
A. phagocytophilum differentially expresses AipA during the infectious stage of its biphasic developmental cycle, the transmission bloodmeal of infected ticks, and infection of mammalian hosts
AipA is a 355-amino acid (36.9 kDa) protein and a putative OMP (Nelson et al., 2008). In silico analysis of the AipA amino acid sequence predicted that residues 107 to 127, 136 to 155, and 220 to 243 form transmembrane domains that position residues 1 to 106 and 156 to 219 on the bacterial surface (Figure 1A). Protein BLAST searches revealed that AipA does not display high sequence identity to sequenced proteins of other organisms, including Anaplasmataceae and Rickettsiales members. Because A. phagocytophilum proteins encoded by genes that are upregulated late during the biphasic developmental cycle are important for infection (Huang et al., 2010b; Troese et al., 2011; Mastronunzio et al., 2012), we examined AipA expression throughout the infection cycle in human promyelocytic HL-60 cells. aipA mRNA levels were approximately 10- to 20-fold higher between 30 and 36 hours – time points that correspond to RC-to-DC transition, exit, and reinfection – than between 4 and 26 hours, time points that correspond to conversion to and replication of non-infectious RC organisms (Troese et al., 2009) (Figure 2A). Also, the aipA mRNA level of the DC inoculum was comparable to that detected at 36 h, a time point that correlates with reinfection.
Figure 1. Schematic diagrams of A. phagocytophilum AipA membrane topology and sequence.
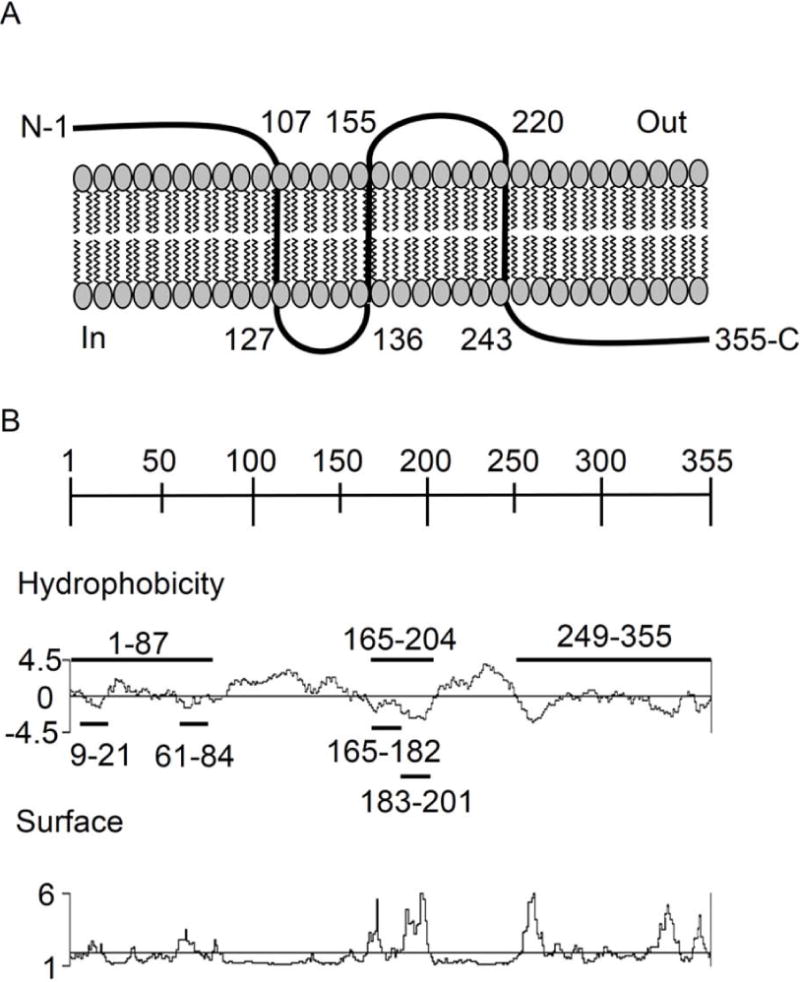
(A) Diagram of the predicted topology of AipA in the A. phagocytophilum outer membrane. N, AipA amino terminus. C, AipA carboxy terminus. Numerical values indicate amino acid coordinates for predicted transmembrane spanning regions. (B) Diagrams of the AipA sequence. The scale indicates 50-amino acid intervals. For the Hydrophobicity diagram, the Kyte-Doolitle algorithm was used to determine hydrophobic (histogram above the axis) and hydrophilic (histogram below axis) regions. For the Surface diagram, the Emini algorithm was used to determine regions that are likely accessible on the surface of the AipA (histogram above the axis) or not (histogram below the axis). The AipA amino acid segments against which antisera were raised are indicated on the Hydrophobicity plot by horizontal lines.
Figure 2. Differential expression profiling of AipA throughout the A. phagocytophilum life cycle.
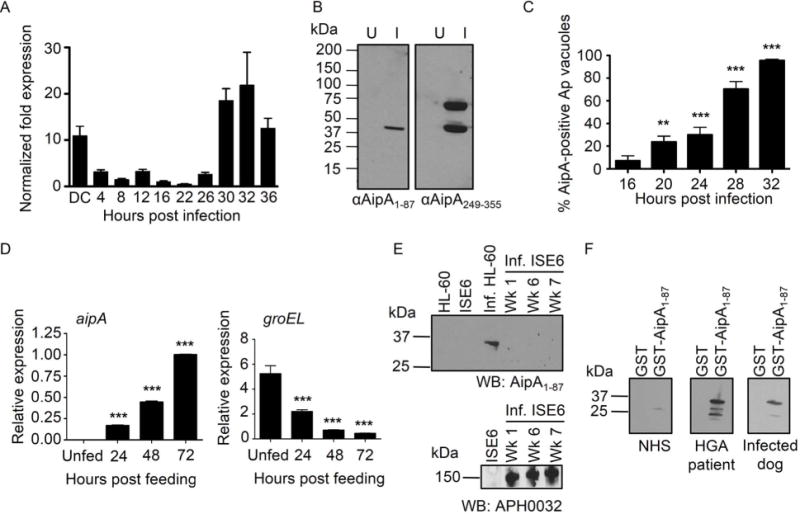
(A) aipA transcriptional profile during A. phagocytophilum infection of HL-60 cells. DC bacteria were incubated with HL-60 cells to establish a synchronous infection. Total RNA isolated from the DC inoculum and infected host cells at several postinfection time points was subjected to reverse transcriptase-quantitative PCR (RT-qPCR). Relative aipA transcript levels were normalized to A. phagocytophilum 16s rRNA gene transcript levels. To determine relative aipA transcription between RC and DC organisms, normalized aipA transcript levels per time point were calculated as the fold change in expression relative to expression at 16 h, a time point at which the entire bacterial population is in the DC morphotype. Data are the means ± standard deviations (SD) for triplicate samples and are representative of two experiments having similar results. (B) Western blot screening of whole-cell lysates of uninfected (U) and A. phagocytophilum infected HL-60 cells (I) using mouse antiserum raised against GST-AipA1–87 (αAipA1–87) and GST-AipA249–355 (αAipA249–355). (C) AipA expression over the course of infection of mammalian host cells. RF/6A cells that had been synchronously infected with A. phagocytophilum (Ap) were screened with antibodies targeting Msp2 (P44) (to denote all A. phagocytophilum inclusions) and AipA viewed by confocal microscopy. Data presented are the mean percentages ± SD of Msp2 (P44)-positive A. phagocytophilum inclusions that were also AipA-positive. At least 100 bacterial inclusions were scored per time point. (D) aipA and groEL expression during transmission feeding of A. phagocytophilum infected ticks on naïve mice. A. phagocytophilum-infected I. scapularis nymphs were allowed to feed on mice for 72 h. Total RNA recovered from unfed and transmission-fed ticks that had been removed at 24, 48, and 72 h postattachment was subjected to RT-qPCR. Relative aipA and groEL transcript levels were normalized to A. phagocytophilum 16S rRNA gene levels. (E) AipA is not expressed during A. phagocytophilum infection of a tick cell line. Western blot analysis of uninfected and A. phagocytophilum infected (Inf.) HL-60 and ISE6 cells using antiserum specific for AipA1–87 or APH0032. The number of weeks (Wk.) during which A. phagocytophilum was maintained in ISE6 cells are indicated. (F) AipA is expressed in vivo and elicits a humoral immune response. Western blot analysis of GST and GST-AipA1–87 screened with sera from an HGA patient and from an A. phagocytophilum infected dog. Results presented in panels B to F are each representative of at two to three independent experiments with similar results. Statistically significant (*, P < 0.05; **, P < 0.005; ***, P < 0.001) values are indicated.
Next, we examined if the differential aipA transcription pattern observed in infected HL-60 cells correlated with differential AipA protein expression. AipA amino acids 1–87 (AipA1–87), 165–204 (AipA165–204) and 249–355 (AipA249–355) contain segments that are hydrophilic and predicted to be accessible on the protein’s surface (Figure 1B). AipA1–87 and AipA165–204 are predicted to be exposed on the bacterial surface, while AipA249–355 is not (Figure 1B). AipA1–87 and AipA249–355 were expressed in Escherichia coli as proteins N-terminally fused to glutathione-S-transferase. Despite numerous attempts, we were unable to express soluble GST-tagged full-length AipA and AipA165–204 (data not shown). Mouse antiserum raised against AipA1–87 and AipA249–355 fusion proteins recognized a band of the expected size in lysates of A. phagocytophilum infected but not uninfected HL-60 cells (Figure 2B). Anti-AipA249–355 recognized an additional band having an apparent mobility that was slightly smaller than 75 kDa, suggesting that AipA may dimerize. Alternatively, anti-AipA249–355 may recognize an epitope that is shared with or is similar in sequence to that of the unknown A. phagocytophilum protein. We used AipA1–87 antibody to screen infected HL-60 cells by immunofluorescence microscopy. Consistent with our transcriptional data, approximately 70% and 96% of the A. phagocytophilum inclusions contained AipA-expressing bacteria at 28 and 32 h, respectively, whereas considerably fewer inclusions were AipA-positive at earlier time points (Figure 2C). These data demonstrate that AipA is both transcriptionally and translationally upregulated during periods when A. phagocytophilum converts to and is in its infectious DC morphotype.
A. phagocytophilum genes that are induced during the tick transmission bloodmeal, such as ompA and asp14, encode proteins that are important for establishing infection in mammalian hosts (Ojogun et al., 2012; Kahlon et al., 2013). Therefore, we examined aipA expression in the salivary glands of A. phagocytophilum infected I. scapularis nymphs over the course of transmission feeding on naïve mice. aipA mRNA was undetectable in unfed infected ticks (Figure 2D). However, aipA transcripts were significantly induced at 24 h of tick feeding and were increasingly expressed through to 72 h. To ensure that A. phagocytophilum transcription was not globally upregulated during tick transmission feeding, we also examined groEL (aph0240) expression. groEL was detected at the highest level in infected unfed nymphs and decreased in expression over the duration of the bloodmeal. Thus, A. phagocytophilum specifically induces select genes, including aipA, as it adapts during the transmission bloodmeal to colonize the mammalian host.
Consistent with a prior report that A. phagocytophilum preferentially expresses aipA during growth in HL-60 cells versus I. scapularis embryo-derived ISE6 cells (Nelson et al., 2008), AipA1–87 antibody failed to detect a protein of the expected size in lysates of ISE6 cells in which A. phagocytophilum had been continually passaged for one, six, or seven weeks (Figure 2E). APH0032, which is an A. phagocytophilum protein that we previously demonstrated to be expressed during infection of ISE6 cells (Huang et al., 2010b), was detected in all infected samples. HGA patient serum and serum from a dog that had been naturally infected with A. phagocytophilum each detected GST-AipA1–87 (Figure 2F), confirming that AipA is expressed and elicits a humoral immune response during A. phagocytophilum infection of humans and dogs. Two additional HGA patient sera recognized GST-AipA1–87 (data not shown).
AipA is an A. phagocytophilum surface protein
To confirm whether AipA localizes to the bacterial outer membrane, we employed confocal microscopy to screen infected RF/6A endothelial cells with AipA1–87 antibody in conjunction with antiserum targeting the A. phagocytophilum major surface protein, Msp2 (P44) (Carlyon, 2012). Both antibodies detected intravacuolar organisms, yielding the ring-like staining pattern on their peripheries that is characteristic for Msp2 (P44) and other confirmed A. phagocytophilum OMPs (Ge et al., 2007; Ojogun et al., 2012; Kahlon et al., 2013) (Figure 3A). Msp2 (P44) signal colocalized with and extended beyond the AipA signal. Next, to determine if immunoaccessible AipA domains are exposed on the bacterial surface, we treated the surfaces of intact, host cell-free DC bacteria with trypsin, an approach that has been used to confirm surface localization of A. phagocytophilum and Chlamydia trachomatis OMPs (Wang et al., 2006; Ojogun et al., 2012; Kahlon et al., 2013). AipA residues 1 to 87 include six lysine and three arginine residues, making this putative surface exposed region susceptible to tryptic digest. If AipA1–87 or portions thereof were exposed on the A. phagocytophilum surface, then incubating intact bacteria with trypsin should result in proteolytic cleavage of this region of the protein, which, in turn, would result in an inability to detect AipA. Trypsin-treated DC organisms were solubilized, Western-blotted, and probed with antiserum specific for AipA1–87 or a confirmed surface-exposed epitope of Asp55 (55-kDa A. phagocytophilum surface protein) (Ge et al., 2007). Blots were also probed with antiserum targeting APH0032, which does not localize to the A. phagocytophilum outer membrane (Huang et al., 2010b). After surface trypsinolysis, APH0032 was detected but AipA and Asp55 were not (Figure 3B). Thus, AipA residues 1 to 87 are exposed on the A. phagocytophilum surface.
Figure 3. AipA is an A. phagocytophilum OMP.
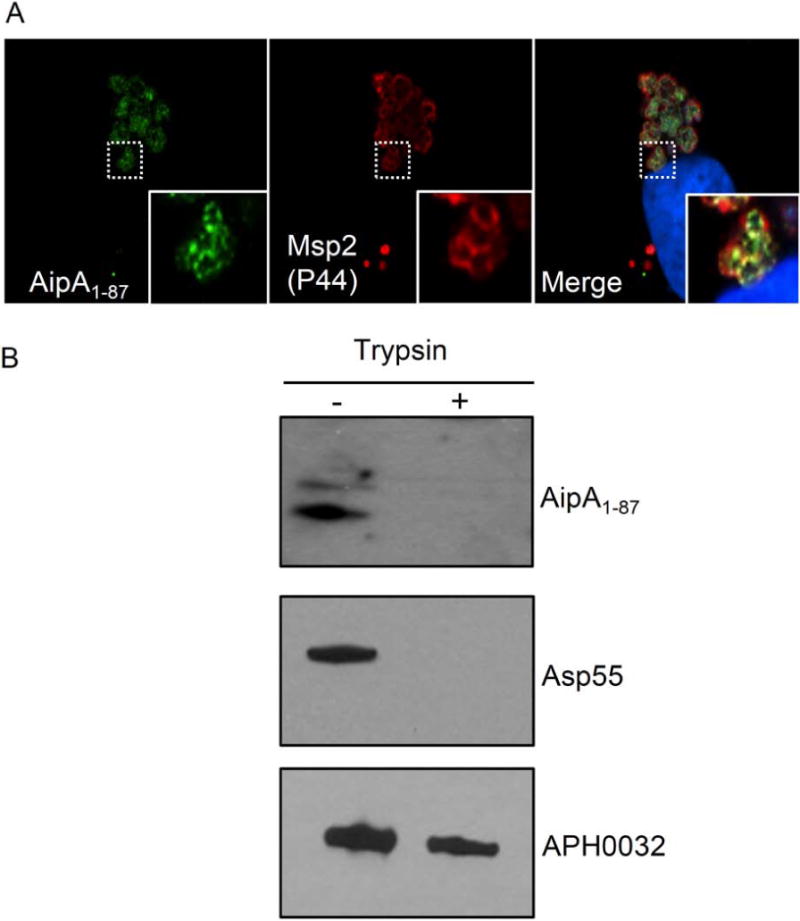
(A) AipA colocalizes with the confirmed OMP, Msp2 (P44). A. phagocytophilum-infected RF/6A cells were fixed and viewed by confocal microscopy to assess immunoreactivity with AipA antiserum (green) in conjunction with Msp2 (P44) antiserum (red). Host cell nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole; blue). The insets demarcated by solid boxes in the lower right corners of each panel are magnified versions of the representative A. phagocytophilum-occupied vacuole that is denoted by the hatched box in each panel. (B) AipA is exposed on the bacterial surface. Intact A. phagocytophilum DC organisms were incubated with trypsin or vehicle control, solubilized, and Western-blotted. Immunoblots were screened with antiserum targeting AipA1–87, Asp55, or APH0032. Data are representative of two experiments with similar results.
GST-AipA requires amino acids 1 to 87 to bind and competitively inhibit A. phagocytophilum infection of mammalian host cells
Since AipA is an exposed surface protein that is induced during key infection stages in the A. phagocytophilum life cycle, we investigated if it facilitates interactions with mammalian host cell surfaces to promote infection. We assessed if GST-tagged AipA1–87 and AipA249–355 bind to RF/6A cells. GST alone served as a negative control. GST antibody detected GST-AipA1–87 that had adhered to the host cells by both immunofluorescence microscopy and flow cytometry (Figure 4, A and B). GST-AipA249–355 bound poorly, at best, and GST alone did not bind to host cells. Based on their differential adhesion capabilities, we rationalized that GST-AipA1–87 but not GST-AipA249–355 would be able to serve as a competitive agonist to inhibit A. phagocytophilum infection. Indeed, preincubating HL-60 and RF/6A cells with GST-AipA1–87 significantly reduced the percentages of infected cells and the number of bacterial inclusions per cell by nearly four-fold relative to incubation with GST alone (Figure 4, C to F). In contrast, GST-AipA249–355 did not inhibit A. phagocytophilum infection of HL-60 cells and reduced infection of RF/6A cells by only a small degree. These data suggest that AipA residues 1 to 87 contain a domain that contributes to A. phagocytophilum invasion of myeloid and endothelial cells.
Figure 4. GST-AipA requires amino acids 1 to 87 to bind and competitively inhibit A. phagocytophilum infection of mammalian host cells.
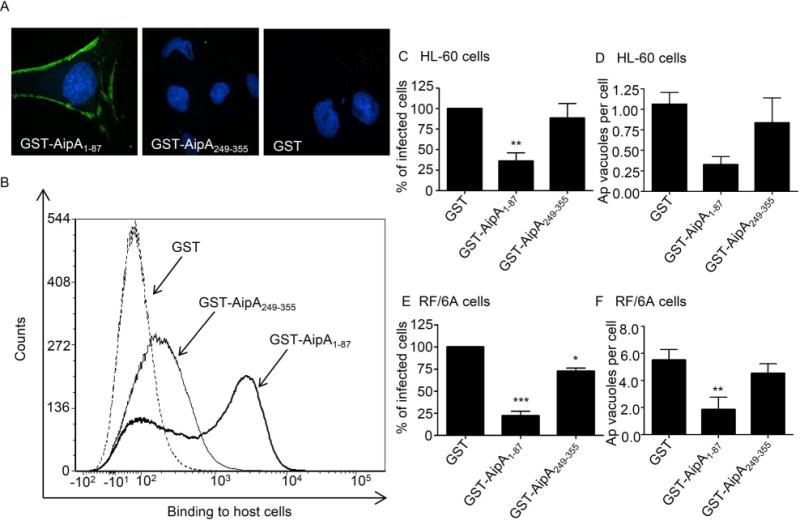
(A and B) GST-AipA1–87 binds to mammalian host cells. RF/6A cells were incubated with GST-AipA1–87, GST-AipA249–355, or GST alone. (A) The host cells were fixed, screened with GST antibody (green), and examined using confocal microscopy. Host cell nuclei were stained with DAPI. Representative merged fluorescent images from three experiments with similar results are shown. (B) Flow cytometric analysis of GST fusion protein binding to RF/6A cells. (C to F) GST-AipA1–87 competitively inhibits A. phagocytophilum infection. HL-60 (C and D) and RF/6A cells (E and F) were incubated with DC bacteria in the presence of GST, GST-AipA1–87, or GST-AipA249–355 for 1 h. Following removal of unbound bacteria, host cells were incubated for 24 h (C and D) or 48 h (E and F) and subsequently examined using confocal microscopy to assess the percentage of infected cells (C and E) or the mean number (± SD) of pathogen-occupied vacuoles per cell (D and F). Results shown are relative to GST-treated host cells and are the means ± SD for three experiments. Statistically significant (*, P < 0.05; **, P < 0.005; ***, P < 0.001) values are indicated.
Antiserum targeting AipA residues 1 to 87 inhibits A. phagocytophilum infection of host cells
Given that AipA amino acids 1 to 87 are exposed on the A. phagocytophilum surface and contribute to infection, we assessed if treating DC organisms with heat-inactivated AipA1–87 antiserum prior to incubating them with HL-60 cells would inhibit infection. OmpA antiserum, for which we previously validated its ability to inhibit A. phagocytophilum infection (Ojogun et al., 2012), was a positive control. Anti-AipA1–87 and anti-OmpA each reduced the number of infected cells and the number of bacterial inclusions per cell by approximately 40% (Figure 5, A and B). In contrast, AipA249–355 antiserum had no effect on A. phagocytophilum infection. Consistent with our published studies of A. phagocytophilum invasins (Ojogun et al., 2012; Kahlon et al., 2013), neither AipA1–87 nor OmpA antiserum inhibited bacterial adhesion to HL-60 cells (data not shown).
Figure 5. Pretreatment of A. phagocytophilum with AipA1–87 antiserum inhibits infection of HL-60 cells but does not alter binding to sLex-capped PSGL-1.
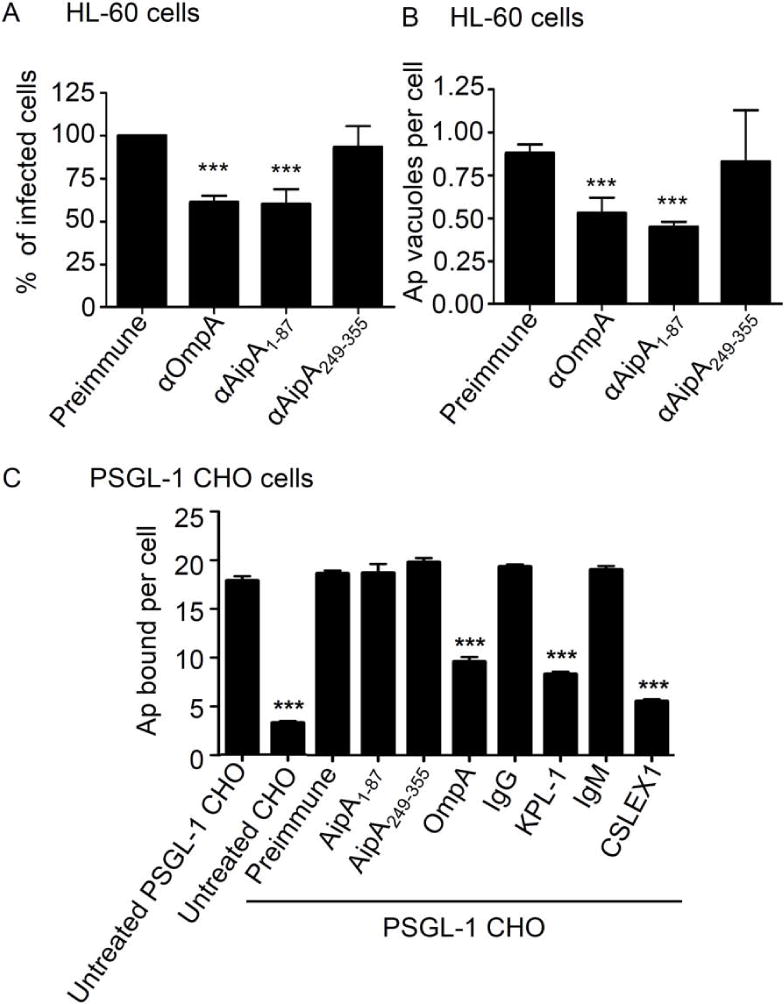
A. phagocytophilum DC organisms were exposed to antiserum targeting AipA1–87, AipA249–355, OmpA, or preimmune serum and then incubated with HL-60 (A and B), PSGL-1 CHO cells, or untransfected CHO cells (C). The infection of HL-60 cells was allowed to proceed for 24 h prior to being assessed, while bacterial binding to PSGL-1 CHO cells was assessed immediately. The mean ± standard deviations of percentages of infected HL-60 cells (A), A. phagocytophilum (Ap) vacuolar inclusions per HL-60 cell (B), and bound DC organisms per PSGL-1 CHO cell or untransfected CHO cell (C) were determined using immunofluorescence microscopy. Additional positive controls for blocking A. phagocytophilum to PSGL-1 CHO cells, besides incubating bacteria with OmpA antiserum, were PSGL-1 CHO cells that had been incubated with PSGL-1 N-terminus blocking antibody KPL-1 or sLex-blocking antibody CSLEX1 prior to the addition of bacteria. Negative controls were PGSL-1 CHO cells that had been incubated with isotype control antibodies prior to the addition of bacteria. Results shown are relative to GST-treated host cells and are the means ± SD for three experiments. Statistically significant (***, P < 0.001) values are indicated.
AipA targets a sLex-capped PSGL-1-independent receptor
sLex-capped PSGL-1 is the only known A. phagocytophilum receptor on myeloid host cells (Herron et al., 2000), and OmpA binds the α2,3-sialic acid determinant of sLex (Ojogun et al., 2012). Since A. phagocytophilum interactions with sLex-capped PSGL-1 involve at least one bacterial surface protein in addition to OmpA (Carlyon et al., 2003; Yago et al., 2003; Reneer et al., 2006; Sarkar et al., 2007; Reneer et al., 2008; Ojogun et al., 2012), we investigated if AipA1–87 antiserum could inhibit bacterial binding to Chinese hamster ovary (CHO) cells transfected to express sLex-capped PSGL-1 (PSGL-1 CHO cells). These cells are excellent models for studying A. phagocytophilum interactions with sLex-capped PSGL-1 as they, but not untransfected CHO cells that do not express the receptor, support bacterial binding (Carlyon et al., 2003; Xia et al., 2003; Yago et al., 2003; Reneer et al., 2006; Sarkar et al., 2007; Reneer et al., 2008; Troese et al., 2009). DC organisms were incubated with AipA1–87 or AipA249–355 antiserum prior to being added to PSGL-1 CHO cells. Bacteria pretreated with preimmune serum were a negative control, whereas bacteria pretreated with OmpA antiserum served as a positive control. Additional positive controls for blocking A. phagocytophilum adhesion were PSGL-1 CHO cells that had been pretreated with KPL-1 or CSLEX1, which are monoclonal antibodies that block the bacterium’s access to the PSGL-1 N-terminus or the α2,3-linked sialic acid determinant of sLex, respectively (Goodman et al., 1999; Herron et al., 2000; Troese et al., 2009). Incubating DC organisms with OmpA antibody and incubating PSGL-1 CHO cells with KPL-1 or CSLEX1 significantly reduced the numbers of bound DC organisms by two- to three-fold (Figure 5C). A. phagocytophilum bound poorly to untransfected CHO cells. Preimmune serum, anti-AipA1–87, and anti-AipA249–355 failed to inhibit bacterial binding to PSGL-1 CHO cells. Therefore, AipA contributes to A. phagocytophilum cellular invasion by interacting with an sLex-capped PSGL-1-independent receptor.
A combination of antisera targeting AipA, OmpA, and Asp14 blocks A. phagocytophilum infection of host cells
A. phagocytophilum infection requires cooperative interactions of multiple invasins with the host cell surface (Truchan et al., 2013). Incubating DC organisms with antiserum targeting full-length OmpA (Ojogun et al., 2012), full-length Asp14 (Kahlon et al., 2013), or AipA1–87 significantly, but only partially, reduced A. phagocytophilum infection of mammalian host cells. We therefore investigated if blocking multiple bacterial-host interactions by treating DC organisms with combinations of AipA1–87, OmpA, and/or Asp14 antisera could improve blocking efficacy. The result was synergistic: whereas antisera combinations targeting two of the three invasins together more effectively inhibited infection than an antiserum targeting an individual protein, the most effective blocking of A. phagocytophilum infection was achieved using antisera against all three invasins (Figure 6). These data demonstrate the potential of simultaneously targeting AipA, OmpA, and Asp14 as an effective means for preventing A. phagocytophilum infection.
Figure 6. A combination of antisera targeting AipA, OmpA, and Asp14 blocks A. phagocytophilum infection of host cells.
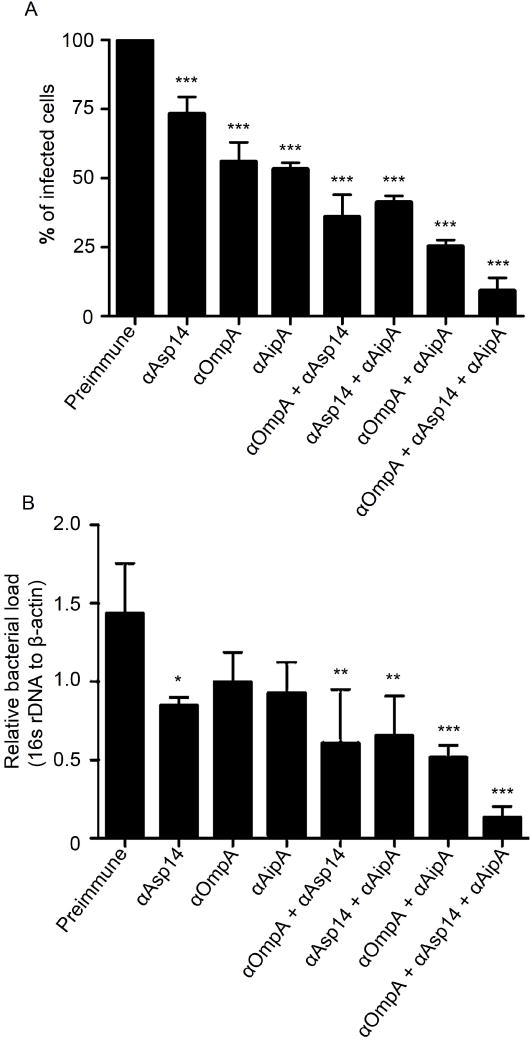
DC bacteria were incubated with preimmune serum or antiserum targeting AipA1–87, OmpA, and/or Asp14 and then incubated with HL-60 cells. (A) The cells were fixed and screened by confocal microscopy to assess the percentage of infected cells. Results shown are relative to host cells that had been treated with preimmune serum and are the means ± SD for three experiments. (B) DNA isolated from the cells was subjected to quantitative PCR analyses. Relative DNA loads of A. phagocytophilum 16s rRNA gene were normalized to DNA loads of the human β-actin gene. Results shown are the means ± SD of triplicate samples and are representative of three independent experiments with similar results. Statistically significant (*, P < 0.05; **, P < 0.005; ***, P < 0.001) values relative to the bacterial load of host cells that had been incubated with preimmune antisera are presented.
The AipA invasion domain is contained within residues 9 to 21
We next sought to pinpoint the AipA invasion domain. Our competitive agonist and antisera blocking studies indicated that this domain lies within residues 1 to 87. Based on hydrophobicity and surface probability analyses (Figure 1B), we rationalized that, of the AipA region of interest, amino acids 9 to 21 and/or 61 to 84 were most likely to facilitate interactions with host cells that promote infection. Accordingly, we generated rabbit antisera against each of these peptides for use in antibody blocking experiments. The AipA region encompassed by residues 165 to 204 (AipA165–204) is predicted to be exposed on the A. phagocytophilum outer membrane, hydrophilic, and accessible on the surface of AipA (Figure 1B). Yet, the contribution of AipA165–204 to infection was unknown due to our inability to express it as a soluble recombinant protein. Therefore, we also raised antisera to peptides corresponding to AipA amino acids 165 to 182 and 183 to 201. Each AipA peptide antiserum recognized endogenous AipA in lysates of A. phagocytophilum infected, but not uninfected HL-60 cells and was specific for the peptide against which it was raised (Figure 7, A and B). Incubating DC organisms with AipA9–21 antiserum reduced infection of HL-60 cells by approximately 47% relative to preimmune control serum, an inhibitory effect that was analogous to the reduction achieved using antiserum against AipA1–87 or OmpA (Figure 7C). Antisera against each of the other three AipA peptides and AipA249–355 had minimal or no inhibitory effect on infection. Thus, the AipA invasion domain is contained within residues 9 to 21.
Figure 7. AipA residues 9–21 are critical for establishing infection in host cells.
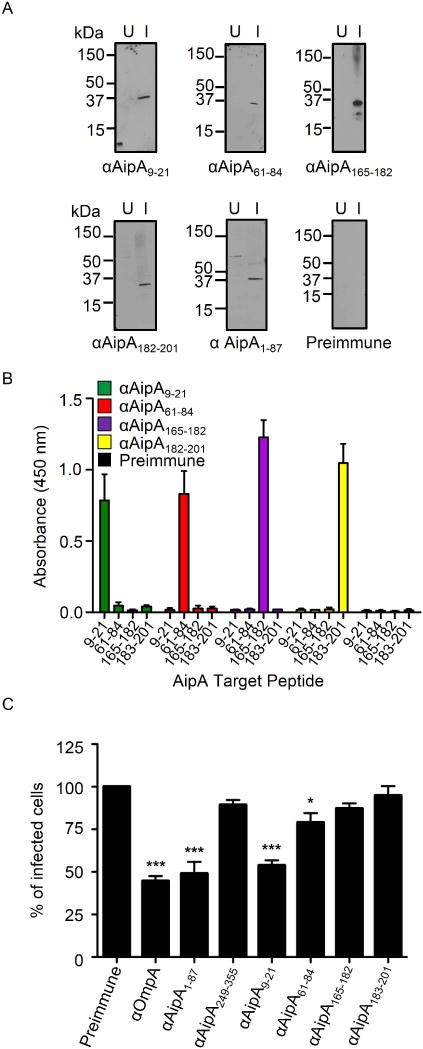
(A) Western blot analyses in which rabbit antiserum targeting AipA9–21, AipA61–84, AipA165–182, AipA183–201, AipA1–87, or preimmune rabbit serum was used to screen whole cell lysates of uninfected (U) and A. phagocytophilum infected HL-60 cells (I). Data are representative of two experiments with similar results. (B) ELISA in which AipA9–21, AipA61–84, AipA165–182, and AipA183–201 antibodies were used to screen wells coated with peptides corresponding to AipA residues 9–21, 61–84, 165–182 and 183–201. Each antiserum only recognized the peptide against which it had been raised. Results shown are the mean (± SD) of triplicate samples. Data are representative of three experiments with similar results. (C) Pretreatment of A. phagocytophilum with AipA9–21 antiserum inhibits infection of HL-60 cells. DC bacteria were pretreated with antiserum specific for AipA9–21, AipA61–84, AipA165–182, AipA183–201, AipA1–87, AipA249–355, OmpA, or preimmune serum for 30 min. Next, the treated bacteria were incubated with HL-60 cells for 60 min. After removal of unbound bacteria, host cells were incubated for 24 h and subsequently examined using Msp2 (P44) antibody and confocal microscopy to assess the percentage of infected cells. Results shown are relative to preimmune serum-treated host cells and are the means ± SD for six experiments. Statistically significant (*, P < 0.05; **, P < 0.005; ***, P < 0.001) values are indicated.
Discussion
Invasins of obligate intracellular pathogens are dualistic: they are essential for bacterial entry into host cells but, as such, they are also “Achilles’ Heels” that can be blocked to prevent infection and pathogen survival. Targeting invasins of arthropod-transmitted pathogens that are induced during the arthropod blood meal would be an attractive approach because of its potential to prevent not only establishment of infection but also disease transmission. Given that A. phagocytophilum infection is predicated on the cooperative actions of multiple bacterial surface-associated invasins (Truchan et al., 2013), most of which are induced during the tick transmission bloodmeal (Ojogun et al., 2012; Kahlon et al., 2013), effective prophylaxis against granulocytic anaplasmosis can potentially be achieved by identifying and targeting these invasins. AipA is an attractive target to include in a multicomponent granulocytic anaplasmosis vaccine. It is an invasin that is induced during tick transmission feeding, is preferentially expressed during the bacterium’s infectious stage, and functions synergistically with OmpA and Asp14 to mediate optimal infection of mammalian host cells.
The exposure of AipA on the infectious DC form surface makes it accessible to blocking antibodies. Indeed, preincubating DC organisms with AipA antiserum significantly reduced infection of HL-60 cells. Pretreatment of DC bacteria with a combination of antibodies targeting AipA, OmpA, and Asp14 nearly abolished infection, whereas pretreatment with antibodies against one or two of the three invasins was less effective. Thus, AipA, OmpA, and Asp14 are collectively critical for infection and targeting all three together blocks infection in vitro. Spotted fever group Rickettsia species, which are in the Order Rickettsiales with A. phagocytophilum, also use multiple surface proteins to promote entry into host cells (Martinez et al., 2004; Cardwell et al., 2009; Chan et al., 2009; Chan et al., 2010; Riley et al., 2010). Moreover, this pathogenic strategy is common among numerous other Gram negative bacterial pathogens, including Chlamydia pneumonia (Molleken et al., 2010; Molleken et al., 2013), Legionella pneumophila (Garduno et al., 1998; Stone et al., 1998; Cirillo et al., 2001; Chang et al., 2005; Vandersmissen et al., 2010; Duncan et al., 2011), Bordetella pertussis (Brennan et al., 1996), Haemophilus influenzae (Jurcisek et al., 2007; Giufre et al., 2008; Chang et al., 2011; Dicko et al., 2011; Jalalvand et al., 2013; Singh et al., 2013) and Leptospira species (Barbosa et al., 2006; Pinne et al., 2010; Verma et al., 2010; Zhang et al., 2012).
GST-AipA is capable of binding to mammalian cells, which suggests that it functions not only as an invasin but also as an adhesin. Yet, AipA antibodies and GST-AipA1–87 each failed to inhibit A. phagocytophilum binding to mammalian cells. In these experiments, the role of AipA as an adhesin was presumably masked by the presence of other adhesins/invasins, such as OmpA and Asp14, on the bacterial surface (Ojogun et al., 2012; Kahlon et al., 2013). The AipA receptor is unknown. However, because AipA antibody failed to inhibit A. phagocytophilum binding to PSGL-1 CHO cells, it can be concluded that AipA recognizes a sLex-capped PSGL-1 independent receptor. AipA and Asp14, which also engages a sLex-capped PSGL-1 independent receptor (Kahlon et al., 2013), complement the sLex-targeting activity of OmpA (Ojogun et al., 2012).
Bacterial genes that are upregulated during transmission feeding of arthropod vectors are critical for various vector-transmitted bacteria to establish infection in their mammalian hosts (Hinnebusch et al., 1996; Perry et al., 1997; Grimm et al., 2004; Tilly et al., 2006). Consistent with these phenomena, aipA is not expressed by A. phagocytophilum during its residence in ISE6 cells or I. scapularis nymphs, but is induced when the bacterium is cultivated in mammalian tissue culture cells and during tick transmission feeding. Furthermore, A. phagocytophilum expresses AipA during infection of humans and dogs. Thus, AipA is dispensable for bacterial colonization of the tick vector, but is important for infecting mammalian hosts. Similar expression profiles have been observed for the other identified invasins OmpA, Asp14, and APH1235 (Mastronunzio et al., 2012; Ojogun et al., 2012; Kahlon et al., 2013). Also, like APH1235 (Troese et al., 2011; Mastronunzio et al., 2012), AipA is pronouncedly upregulated when the bacterium is in the DC stage. In agreement with the invasive role of the DC morphotype, both proteins are important for establishing and/or maintaining infection in mammalian host cells.
How AipA is transported to and associates with the bacterial outer membrane is unclear, as it lacks a canonical signal peptide that would target it for Sec-dependent or twin-arginine secretion. This conundrum is further complicated as AipA is unique to A. phagocytophilum and bears no homology to any known crystal structure. Perhaps AipA is an atypical transmembrane protein or a peripheral membrane protein that is anchored to the bacterial outer membrane via a posttranslational modification. AipA colocalizes with the confirmed outer membrane protein, Msp2 (P44) and functions in concert with OmpA and Asp14, both of which have also been shown to colocalize with Msp2 (P44) (Kahlon et al., 2013; Ojogun et al., 2012). Msp2 (P44) has been proposed to form heteromeric complexes that mediate interactions with host cells (Park et al., 2003). Given that AipA, Asp14, and OmpA synergistically promote A. phagocytophilum infection of host cells, it will be important to determine if they do so as a multimeric invasin complex that includes Msp2 (P44).
The AipA invasion domain lies within residues 9 to 21, which is a hydrophilic region of the protein that is exposed on the bacterial surface. Antiserum targeting this span reduces A. phagocytophilum infection of host cells by a level comparable to that achieved by antiserum targeting the entire surface-exposed N-terminal domain of AipA. The observed inhibition is specific to anti-AipA9–21, as antisera targeting peptides corresponding to all other predicted hydrophilic regions of AipA exhibited no inhibitory effect. Previously, we discovered that the N-terminal ectodomain of OmpA is required for recognition of α2,3-linked sialic acid of sLex (Ojogun et al., 2012) and the Asp14 invasion domain lies within residues 101–124 (Kahlon et al., 2013). Currently, we are further delineating the OmpA and Asp14 invasion domains and confirming whether targeting them in concert with AipA9–21 effectively blocks A. phagocytophilum infection.
Granulocytic anaplasmosis can be debilitating or fatal, and there is no vaccine that protects against the disease. Understanding the A. phagocytophilum invasion mechanism would greatly augment development of novel preventative or therapeutic measures. Here, we have demonstrated that AipA is a promising target because it is an invasin that is markedly expressed at key stages in the A. phagocytophilum infection cycle and is accessible to blocking antibody. Furthermore, we show for the first time that simultaneously targeting multiple A. phagocytophilum invasins effectively blocks infection in vitro. Moving forward, it will be important to surmise the efficacy of AipA, OmpA, and Asp14 as vaccinogens for eliciting protection against A. phagocytophilum infection in vivo.
Experimental Procedures
Cultivation of uninfected and A. phagocytophilum-infected host cell lines
ISE6 cells were kindly provided by Ulrike Munderloh of the University of Minnesota. Human promyelocytic HL-60 cells (CCL-240; American Type Culture Collections (ATCC), Manassas, VA), RF/6A (rhesus monkey choroidal endothelial cells, ATCC CRL-1780), ISE6 cells and A. phagocytophilum (NCH-1 strain) infected HL-60, RF/6A, or ISE6 cells were cultured as previously described (Huang et al., 2012). PSGL-1 CHO cells and untransfected CHO cells, both of which were provided by Rodger McEver of The Oklahoma Medical Research Foundation, were maintained as previously described (Troese et al., 2009).
In silico analyses of AipA
The AipA sequence was assessed for transmembrane domains using the TMpred and TMHMM algorithms (Hofmann et al., 1993; Krogh et al., 2001), each of which yielded highly similar predictions. Results obtained using TMpred are presented in Figure 1. Protean, which is part of the Lasergene software package (version 8.02; DNASTAR, Madison, WI), was used to assess AipA for regions of hydrophobicity and probability of being surface-exposed using the Kyte-Doolittle (Kyte et al., 1982) and Emini (Emini et al., 1985) algorithms, respectively.
Recombinant protein and antiserum production
A PCR amplicon of aipA (aph0915) nucleotides 1 to 261, encoding AipA amino acids 1 to 87, was generated using primers 5′-CACCTTGAGTTTTACAATGTCGAAGTTATCGC-3′ (nucleotides in bold text correspond to a Gateway entry vector-compatible sequence) and 5′-CTATCCTAGCATCCTTCTAGAAGCGGAAG-3′ (underlined nucleotides denote an added stop codon). A PCR product corresponding to aipA nucleotides 745 to 1068, encoding AipA amino acids 249 to 355, was generated using primers 5′-CACCATCTATCAAGGAAATTACGAAGATCGCAAC-3′ and 5′-GAGCAGCATGCTTTA-3′. The amplicons were cloned into the pDest-15 vector (Life Technologies, Carlsbad, CA) downstream of and in frame with the gene encoding GST as described previously (Ojogun et al., 2012). Expression and purification GST-tagged AipA residues 1 to 87 (GST-AipA1–87), 249 to 355 (GST-AipA249–355), and GST alone were performed as previously described (Troese et al., 2011). GST-tagged full-length AipA and AipA amino acids 165 to 204 remained insoluble over a wide range of conditions and thus could not be purified. Generation of murine polyclonal antisera against each GST fusion protein was performed as described previously (Troese et al., 2011). Rabbit polyclonal antisera were raised against synthetic keyhole limpet hemocyanin (KLH)-conjugated peptides corresponding to AipA amino acid residues 9 to 21, 61 to 84, 165 to 182, and 185 to 201 (New England Peptide, Gardner, MA). Specificity of each AipA peptide antiserum for its target peptides was determined by the enzyme-linked immunosorbent assay using the TMB substrate kit (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions.
Differential aipA expression studies
HL-60 cells were synchronously infected with A. phagocytophilum DC organisms (Troese et al., 2009). The infection time course was allowed to proceed for 36 h, a time period that enabled the bacteria to complete their biphasic developmental cycle and initiate a second round of infection (Troese et al., 2009). RNA isolated from aliquots taken every 4 hours was subjected to reverse transcriptase-quantitative PCR (RT-qPCR) as described previously (Troese et al., 2011) using AipA specific primers 5′-CCTCAACTAAAGAAGCGTCATCAAA-3′ and 5′-GTACGGTGTACAAAACGAGGAACA-3′, which targeted nucleotides 179 to 388. Relative aipA transcript levels were normalized to the transcript levels of the A. phagocytophilum 16s rRNA gene (aph1000) using the 2−ΔΔCT method (Livak et al., 2001; Kahlon et al., 2013). To determine if aipA was transcriptionally upregulated in the DC versus RC morphotype, normalized aipA transcript levels were calculated as fold changes in expression relative to expression at 16 h, a time point at which the entire A. phagocytophilum population existed in the RC form (Troese et al., 2009; Mastronunzio et al., 2012). aipA expression during blood meal acquisition by A. phagocytophilum infected nymphs from mice was monitored as described (Mastronunzio et al., 2012) using AipA primers targeting nucleotides 179 to 388. As a control, expression of the A. phagocytophilum groEL gene (aph0240) during tick transmission feeding was monitored using gene-specific primers (Kahlon et al., 2013).
Western blotting and confocal microscopy
Antisera generated in this study and prior studies targeted AipA, APH0032 (Huang et al., 2010b), Asp55 (Ge et al., 2007), and Msp2 (P44) (Troese et al., 2011). Sera from an HGA patient and a dog that had been naturally infected with A. phagocytophilum were previously described (Ojogun et al., 2012) and provided by Dr. Janet Foley of The University of California-Davis, respectively. Western blot analyses (Troese et al., 2011) were performed on uninfected or A. phagocytophilum infected host cells or A. phagocytophilum DC organisms that had been subjected to surface trypsinolysis as described previously (Kahlon et al., 2013). A. phagocytophilum infected host cells were analyzed by spinning disk confocal microscopy as described (Huang et al., 2010a; Huang et al., 2012).
AipA antiserum inhibition of A. phagocytophilum infection
Inhibition of host cell infection by preincubating DC organisms with heat-inactivated polyclonal antiserum targeting GST-AipA1–87, GST-AipA249–355, GST-OmpA (Ojogun, 2012), GST-Asp14 (Kahlon et al., 2013), AipA9–21, AipA61–84, AipA165–182, or AipA183–201 (2 mg mL−1) was assessed as previously described (Ojogun et al., 2012). Serum against GST alone and preimmune serum served as negative controls. In instances where DC bacteria were incubated with combinations of antisera targeting AipA, Asp14, and/or OmpA, each respective antiserum was at a concentration of 2 mg mL−1, and control antiserum was matched accordingly. Following antibody treatment, bacterial adhesion to and infection of HL-60 cells were monitored using spinning disk confocal microscopy (Ojogun et al., 2012).
Binding of GST-AipA to mammalian host cells and competitive inhibition of A. phagocytophilum infection
Mammalian host cells cells were incubated with 4 μM GST, GST-AipA1–87, or GST-AipA249–355 for 1 h at 37°C. Spinning disk confocal microscopy and flow cytometry were used to assess the binding of recombinant proteins to host cells and competitive inhibition of A. phagocytophilum infection as previously described (Ojogun et al., 2012; Kahlon et al., 2013).
Assessment of the relevance of AipA to A. phagocytophilum adherence to PSGL-1 CHO cells
To determine if AipA was important for A. phagocytophilum recognition of sLex-capped PSGL-1, DC organisms were incubated with antiserum targeting AipA1–87, AipA249–355, OmpA (Ojogun et al., 2012), or preimmune control serum as described above. Next, the treated bacteria were incubated with PSGL-1 CHO cells or untransfected CHO cells for 1 h, followed by two rounds of washing with PBS to remove unbound bacteria, and enumeration of bound organisms using spinning disk confocal microscopy as described (Troese et al., 2009). As positive controls for inhibition of bacterial adherence to sLex-capped PSGL-1, PSGL-1 CHO cells were incubated with the PSGL-1 N-terminus-specific antibody, KPL-1 (BD Biosciences, San Jose, CA), or the sLex-specific antibody, CSLEX1 (BD Biosciences) for 30 min prior to the addition of bacteria. Mouse IgG and mouse IgM served as isotype controls for KPL-1 and CSLEX1, respectively.
Statistical analyses
One-way analysis of variance (ANOVA) was performed using the Prism 5.0 software package (Graphpad; San Diego, CA) to assess statistical significance as described (Ojogun et al., 2012). Statistical significance was set at P < 0.05.
Acknowledgments
We thank Yasuko Rikihisa of The Ohio State University for providing antiserum against Asp55, Janet Foley of the University of California, Davis for providing serum from A. phagocytophilum infected dogs, Ulrike Munderloh of the University of Minnesota for providing ISE6 cells, Rodger McEver for providing CHO and PSGL-1 CHO cells, and Andrea Beyer and Hilary Truchan for critical review of this manuscript. This study was supported by NIH grants R01 AI072683 to J.A.C., R01 AI67830 to R.T.M., and R01 AI141440 to E.F. This study was also supported by Financial Assistance Award No. 01-79-14214, which was awarded by the United States Department of Commerce Economic Development Administration to the University of Virginia and sub-contracted to Virginia Commonwealth University (J.A.C. and R.T.M). The Virginia Commonwealth University Flow Cytometry and Imaging Shared Resource Facility is supported in part by funding from NIH-NCI Cancer Center support grant 5P30 CA016059.
References
- Alhumaidan H, Westley B, Esteva C, Berardi V, Young C, Sweeney J. Transfusion-transmitted anaplasmosis from leukoreduced red blood cells. Transfusion. 2013;53:181–186. doi: 10.1111/j.1537-2995.2012.03685.x. [DOI] [PubMed] [Google Scholar]
- Annen K, Friedman K, Eshoa C, Horowitz M, Gottschall J, Straus T. Two cases of transfusion-transmitted Anaplasma phagocytophilum. American journal of clinical pathology. 2012;137:562–565. doi: 10.1309/AJCP4E4VQQQOZIAQ. [DOI] [PubMed] [Google Scholar]
- Barbosa AS, Abreu PA, Neves FO, Atzingen MV, Watanabe MM, Vieira ML, et al. A newly identified leptospiral adhesin mediates attachment to laminin. Infection and immunity. 2006;74:6356–6364. doi: 10.1128/IAI.00460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan MJ, Shahin RD. Pertussis antigens that abrogate bacterial adherence and elicit immunity. American journal of respiratory and critical care medicine. 1996;154:S145–149. doi: 10.1164/ajrccm/154.4_Pt_2.S145. [DOI] [PubMed] [Google Scholar]
- Cardwell MM, Martinez JJ. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infection and immunity. 2009;77:5272–5280. doi: 10.1128/IAI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlyon JA. Establishing intracellular infection: modulation of host cell functions (Anaplasmataceae) In: Palmer GH, Azad A, editors. Intracellular Pathogens II: Rickettsiales. Washington, D. C.: ASM Press; 2012. [Google Scholar]
- Carlyon JA, Akkoyunlu M, Xia L, Yago T, Wang T, Cummings RD, et al. Murine neutrophils require alpha1,3-fucosylation but not PSGL-1 for productive infection with Anaplasma phagocytophilum. Blood. 2003;102:3387–3395. doi: 10.1182/blood-2003-02-0621. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Notice to readers: final 2012 reports of nationally notifiable infectious diseases. MMWR. Morbidity and mortality weekly report. 2013;62:669–682. [PMC free article] [PubMed] [Google Scholar]
- Chan YG, Cardwell MM, Hermanas TM, Uchiyama T, Martinez JJ. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cellular microbiology. 2009;11:629–644. doi: 10.1111/j.1462-5822.2008.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YG, Riley SP, Martinez JJ. Adherence to and invasion of host cells by spotted Fever group rickettsia species. Frontiers in microbiology. 2010;1:139. doi: 10.3389/fmicb.2010.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Kaur R, Michel LV, Casey JR, Pichichero M. Haemophilus influenzae vaccine candidate outer membrane protein P6 is not conserved in all strains. Human vaccines. 2011;7:102–105. doi: 10.4161/hv.7.1.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Kura F, Amemura-Maekawa J, Koizumi N, Watanabe H. Identification of a novel adhesion molecule involved in the virulence of Legionella pneumophila. Infection and immunity. 2005;73:4272–4280. doi: 10.1128/IAI.73.7.4272-4280.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo SL, Bermudez LE, El-Etr SH, Duhamel GE, Cirillo JD. Legionella pneumophila entry gene rtxA is involved in virulence. Infection and immunity. 2001;69:508–517. doi: 10.1128/IAI.69.1.508-517.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicko A, Odusanya OO, Diallo AI, Santara G, Barry A, Dolo A, et al. Primary vaccination with the 10-valent pneumococcal non-typeable Haemophilus influenzae protein D conjugate vaccine (PHiD-CV) in infants in Mali and Nigeria: a randomized controlled trial. BMC public health. 2011;11:882. doi: 10.1186/1471-2458-11-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan C, Prashar A, So J, Tang P, Low DE, Terebiznik M, Guyard C. Lcl of Legionella pneumophila is an immunogenic GAG binding adhesin that promotes interactions with lung epithelial cells and plays a crucial role in biofilm formation. Infection and immunity. 2011;79:2168–2181. doi: 10.1128/IAI.01304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emini EA, Hughes JV, Perlow DS, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garduno RA, Garduno E, Hoffman PS. Surface-associated hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infection and immunity. 1998;66:4602–4610. doi: 10.1128/iai.66.10.4602-4610.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Rikihisa Y. Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. Journal of bacteriology. 2007;189:7819–7828. doi: 10.1128/JB.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giufre M, Carattoli A, Cardines R, Mastrantonio P, Cerquetti M. Variation in expression of HMW1 and HMW2 adhesins in invasive nontypeable Haemophilus influenzae isolates. BMC microbiology. 2008;8:83. doi: 10.1186/1471-2180-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JL, Nelson CM, Klein MB, Hayes SF, Weston BW. Leukocyte infection by the granulocytic ehrlichiosis agent is linked to expression of a selectin ligand. The Journal of clinical investigation. 1999;103:407–412. doi: 10.1172/JCI4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, et al. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:3142–3147. doi: 10.1073/pnas.0306845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron MJ, Nelson CM, Larson J, Snapp KR, Kansas GS, Goodman JL. Intracellular parasitism by the human granulocytic ehrlichiosis bacterium through the P-selectin ligand, PSGL-1. Science. 2000;288:1653–1656. doi: 10.1126/science.288.5471.1653. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BJ, Perry RD, Schwan TG. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science. 1996;273:367–370. doi: 10.1126/science.273.5273.367. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase- A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Hopkins RS, Jajosky RA, Hall PA, Adams DA, Connor FJ, Sharp P, Anderson WJ, Fagan RF, Aponte JJ, Nitschke DA, Worsham CA, Adekoya N, Chang MH. Summary of notifiable diseases—United States, 2003. MMWR Morbity and mortality weekly report. 2005;52:1–85. [PubMed] [Google Scholar]
- Huang B, Hubber A, McDonough JA, Roy CR, Scidmore MA, Carlyon JA. The Anaplasma phagocytophilum-occupied vacuole selectively recruits Rab-GTPases that are predominantly associated with recycling endosomes. Cellular microbiology. 2010a;12:1292–1307. doi: 10.1111/j.1462-5822.2010.01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Troese MJ, Howe D, Ye S, Sims JT, Heinzen RA, et al. Anaplasma phagocytophilum APH_0032 is expressed late during infection and localizes to the pathogen-occupied vacuolar membrane. Microbial pathogenesis. 2010b;49:273–284. doi: 10.1016/j.micpath.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ojogun N, Ragland SA, Carlyon JA. Monoubiquitinated proteins decorate the Anaplasma phagocytophilum-occupied vacuolar membrane. FEMS Immunology and Medical Microbiology. 2012;64:32–41. doi: 10.1111/j.1574-695X.2011.00873.x. [DOI] [PubMed] [Google Scholar]
- Jalalvand F, Su YC, Morgelin M, Brant M, Hallgren O, Westergren-Thorsson G, et al. Haemophilus influenzae protein F mediates binding to laminin and human pulmonary epithelial cells. The Journal of infectious diseases. 2013;207:803–813. doi: 10.1093/infdis/jis754. [DOI] [PubMed] [Google Scholar]
- Jereb M, Pecaver B, Tomazic J, Muzlovic I, Avsic-Zupanc T, Premru-Srsen T, et al. Severe human granulocytic anaplasmosis transmitted by blood transfusion. Emerging infectious diseases. 2012;18:1354–1357. doi: 10.3201/eid1808.120180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurcisek JA, Bookwalter JE, Baker BD, Fernandez S, Novotny LA, Munson RS, Jr, Bakaletz LO. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Molecular microbiology. 2007;65:1288–1299. doi: 10.1111/j.1365-2958.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- Kahlon A, Ojogun N, Ragland SA, Seidman D, Troese MJ, Ottens AK, et al. Anaplasma phagocytophilum Asp14 is an invasin that interacts with mammalian host cells via its C terminus to facilitate infection. Infection and immunity. 2013;81:65–79. doi: 10.1128/IAI.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. Journal of molecular biology. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martinez JJ, Cossart P. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. Journal of cell science. 2004;117:5097–5106. doi: 10.1242/jcs.01382. [DOI] [PubMed] [Google Scholar]
- Mastronunzio JE, Kurscheid S, Fikrig E. Postgenomic analyses reveal development of infectious Anaplasma phagocytophilum during transmission from ticks to mice. Journal of bacteriology. 2012;194:2238–2247. doi: 10.1128/JB.06791-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molleken K, Becker E, Hegemann JH. The Chlamydia pneumoniae invasin protein Pmp21 recruits the EGF receptor for host cell entry. PLoS pathogens. 2013;9:e1003325. doi: 10.1371/journal.ppat.1003325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molleken K, Schmidt E, Hegemann JH. Members of the Pmp protein family of Chlamydia pneumoniae mediate adhesion to human cells via short repetitive peptide motifs. Molecular microbiology. 2010;78:1004–1017. doi: 10.1111/j.1365-2958.2010.07386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CM, Herron MJ, Felsheim RF, Schloeder BR, Grindle SM, Chavez AO, et al. Whole genome transcription profiling of Anaplasma phagocytophilum in human and tick host cells by tiling array analysis. BMC Genomics. 2008;9:364. doi: 10.1186/1471-2164-9-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojogun N, Kahlon A, Ragland SA, Troese MJ, Mastronunzio JE, Walker NJ, et al. Anaplasma phagocytophilum outer membrane protein A interacts with sialylated glycoproteins to promote infection of mammalian host cells. Infection and immunity. 2012;80:3748–3760. doi: 10.1128/IAI.00654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim KJ, Grab DJ, Dumler JS. Anaplasma phagocytophilum major surface protein-2 (Msp2) forms multimeric complexes in the bacterial membrane. FEMS microbiology letters. 2003;227:243–247. doi: 10.1016/S0378-1097(03)00687-6. [DOI] [PubMed] [Google Scholar]
- Perry RD, Fetherston JD. Yersinia pestis--etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinne M, Choy HA, Haake DA. The OmpL37 surface-exposed protein is expressed by pathogenic Leptospira during infection and binds skin and vascular elastin. PLoS neglected tropical diseases. 2010;4:e815. doi: 10.1371/journal.pntd.0000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneer DV, Kearns SA, Yago T, Sims J, Cummings RD, McEver RP, Carlyon JA. Characterization of a sialic acid- and P-selectin glycoprotein ligand-1-independent adhesin activity in the granulocytotropic bacterium Anaplasma phagocytophilum. Cellular microbiology. 2006;8:1972–1984. doi: 10.1111/j.1462-5822.2006.00764.x. [DOI] [PubMed] [Google Scholar]
- Reneer DV, Troese MJ, Huang B, Kearns SA, Carlyon JA. Anaplasma phagocytophilum PSGL-1-independent infection does not require Syk and leads to less efficient AnkA delivery. Cellular microbiology. 2008;10:1827–1838. doi: 10.1111/j.1462-5822.2008.01168.x. [DOI] [PubMed] [Google Scholar]
- Riley SP, Goh KC, Hermanas TM, Cardwell MM, Chan YG, Martinez JJ. The Rickettsia conorii autotransporter protein Sca1 promotes adherence to nonphagocytic mammalian cells. Infection and immunity. 2010;78:1895–1904. doi: 10.1128/IAI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M, Reneer DV, Carlyon JA. Sialyl-Lewis x-independent infection of human myeloid cells by Anaplasma phagocytophilum strains HZ and HGE1. Infection and immunity. 2007;75:5720–5725. doi: 10.1128/IAI.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Al-Jubair T, Morgelin M, Thunnissen MM, Riesbeck K. The unique structure of Haemophilus influenzae protein E reveals multiple binding sites for host factors. Infection and immunity. 2013;81:801–814. doi: 10.1128/IAI.01111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone BJ, Abu Kwaik Y. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infection and immunity. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, et al. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infection and immunity. 2006;74:3554–3564. doi: 10.1128/IAI.01950-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troese MJ, Carlyon JA. Anaplasma phagocytophilum dense-cored organisms mediate cellular adherence through recognition of human P-selectin glycoprotein ligand 1. Infection and immunity. 2009;77:4018–4027. doi: 10.1128/IAI.00527-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troese MJ, Kahlon A, Ragland SA, Ottens AK, Ojogun N, Nelson KT, et al. Proteomic analysis of Anaplasma phagocytophilum during infection of human myeloid cells identifies a protein that is pronouncedly upregulated on the infectious dense-cored cell. Infection and immunity. 2011;79:4696–4707. doi: 10.1128/IAI.05658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchan HK, Seidman D, Carlyon JA. Breaking in and grabbing a meal: Anaplasma phagocytophilum cellular invasion, nutrient acquisition, and promising tools for their study. Microbes and infection / Institut Pasteur. 2013 doi: 10.1016/j.micinf.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandersmissen L, De Buck E, Saels V, Coil DA, Anne J. A Legionella pneumophila collagen-like protein encoded by a gene with a variable number of tandem repeats is involved in the adherence and invasion of host cells. FEMS Microbiol Lett. 2010;306:168–176. doi: 10.1111/j.1574-6968.2010.01951.x. [DOI] [PubMed] [Google Scholar]
- Verma A, Brissette CA, Bowman AA, Shah ST, Zipfel PF, Stevenson B. Leptospiral endostatin-like protein A is a bacterial cell surface receptor for human plasminogen. Infection and immunity. 2010;78:2053–2059. doi: 10.1128/IAI.01282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Berg EA, Feng X, Shen L, Smith T, Costello CE, Zhang YX. Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein science : a publication of the Protein Society. 2006;15:122–134. doi: 10.1110/ps.051616206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Ramachandran V, McDaniel JM, Nguyen KN, Cummings RD, McEver RP. N-terminal residues in murine P-selectin glycoprotein ligand-1 required for binding to murine P-selectin. Blood. 2003;101:552–559. doi: 10.1182/blood-2001-11-0036. [DOI] [PubMed] [Google Scholar]
- Yago T, Leppanen A, Carlyon JA, Akkoyunlu M, Karmakar S, Fikrig E, et al. Structurally distinct requirements for binding of P-selectin glycoprotein ligand-1 and sialyl Lewis x to Anaplasma phagocytophilum and P-selectin. The Journal of biological chemistry. 2003;278:37987–37997. doi: 10.1074/jbc.M305778200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang C, Ojcius DM, Sun D, Zhao J, Lin X, et al. The mammalian cell entry (Mce) protein of pathogenic Leptospira species is responsible for RGD motif-dependent infection of cells and animals. Molecular microbiology. 2012;83:1006–1023. doi: 10.1111/j.1365-2958.2012.07985.x. [DOI] [PubMed] [Google Scholar]


