Abstract
Senescence is defined as a stable cell growth arrest. Oncogene-induced senescence (OIS) occurs when an activated oncogene is expressed in a normal cell. OIS acts as a bona fide tumor suppressor mechanism by driving stable growth arrest of cancer progenitor cells harboring the initial oncogenic hit. OIS is often characterized by aberrant DNA replication and the associated DNA damage response. Nucleotides, in particular deoxyribonucleotide triphosphates (dNTPs), are necessary for both DNA replication and repair. Imbalanced dNTP pools play a role in a number of human diseases, including during the early stages of cancer development. This review will highlight what is currently known about the role of decreased nucleotide metabolism in OIS, how nucleotide metabolism leads to transformation and tumor progression, and how this pathway can be targeted as a cancer therapeutic by inducing senescence of cancer cells.
1. Introduction
Nucleotides are necessary for a variety of cellular processes. It has been well characterized that imbalances in nucleotide levels lead to a variety of human diseases, including cancer [1–4], immunodeficiency [5, 6], aging [7, 8], kidney diseases [9, 10], gout [6], and a number of mitochondrial pathologies [11, 12].
1.1. Synthesis of nucleotides: The de novo pathway
Nucleotides can be synthesized through either the de novo pathway or the salvage pathway [13]. In the de novo pathway, glucose and glutamine are the main nutrients needed to synthesize nucleotides [14]. Glucose is converted to ribose-5-phosphate during the pentose phosphate pathway, which is used for both purine and pyrimidine synthesis [15]. Glutamine is necessary for supplying nitrogen [16]. Purines and pyrimidines are synthesized in two distinct ways [13, 15, 17]. Purines are made by directly assembling the atoms that comprise the purine ring onto ribose-5-phosphate through 11 steps. This yields inosine monophosphate (IMP), which is further modified to produce adenosine monophosphate (AMP) and guanosine monophosphate (GMP). In contrast, during pyrimidine synthesis, the pyrimidine ring is completed before addition of the ribose-5-phosphate moiety. Pyrimidines are made through a 6-step process, which produces uridine monophosphate (UMP). UMP can then be converted into cytidine triphosphate (CTP). Thymine nucleotides are synthesized after uridine diphosphate (UDP) and cytidine diphosphate (CDP) are reduced, and thymidylate synthase (TS) is necessary for dTTP synthesis [17].
1.2. Synthesis of nucleotides: The salvage pathway
In addition to the de novo pathway, a salvage pathway exists for both purine and pyrimidines [13, 17, 18]. Normal cells undergo turnover and degradation of cellular materials, leading to release of free purines or substrates that compose the pyrimidine ring [17]. These can be converted back into dNTPs by a variety of enzymes in both the cytosol and mitochondria [17, 18]. Interestingly, pyrimidine salvage is more efficient than purine salvage [17].
1.3. Synthesis of deoxyribonucleotides
One particular type of nucleotide, 2’-deoxyribonucleoside 5’-triphosphates (dNTPs), is necessary for both DNA replication and repair [17, 19]. Without the correct levels of dNTPs, cells cannot faithfully replicate either nuclear or mitochondrial DNA, and DNA damage cannot be repaired [7, 20]. The rate-limiting step in dNTP synthesis is reduction of ribonucleoside di- or tri-phosphates (NDPs/NTPs) at the 2’ position of ribose sugar to deoxyribonucleotide-di- or tri-phosphates (dNDPs/dNTPS) by ribonucleotide reductase (RNR) [17, 19]. During reduction of ribonucleosides, RNR is oxidized and then reduced by either thioredoxin or glutathione [19]. Nicotinamide adenine dinucleotide phosphate (NADPH) is the ultimate source of the electrons. RNR reduces all four rNDPs/rNTPs (i.e., ADP/ATP, GDP/GTP, UDP/UTP, and CDP/CTP) [17]. RNR activity is tightly regulated by allosteric regulation and enzyme specificity [19]. RNR is a tetrameric complex consisting of two large catalytic subunits (R1: ribonucleotide reductase M1, RRM1) and two small regulatory subunits (R2: ribonucleotide reductase M2, RRM2; or p53R2/RRM2B) [17, 19]. RRM1 contains both the catalytic site and the allosteric regulatory sites [19]. RRM1 is expressed throughout all phases of the cell cycle [21]. The R2 subunit contains the tyrosyl radical, the site necessary for the reduction reaction [19]. RRM2 is the R2 subunit that controls reduction during S phase of the cell cycle when dNTPs are needed for DNA replication [21]. Therefore, RRM2 expression is rate-limiting for RNR activity [19]. In contrast, p53R2 is involved in supplying dNTPs for DNA repair and mitochondrial DNA synthesis in the G0/G1 phase of the cell cycle [22].
1.4. Senescence
First described in 1961 by Leonard Hayflick and Paul Moorhead, cellular senescence is defined as a stable cell growth arrest [23]. Senescence can be induced by a number of different stimuli, including critically shortened telomeres, activated oncogenes, DNA damage, and some cancer therapeutics [24]. Senescent cells have unique morphological and molecular characteristics [25]. Phenotypically, they are characterized by a large, flat morphology and increased activity of β-galactosidase (termed senescence-associated β-galactosidase or SA-β-gal) [26].
1.5. Oncogene-induced senescence
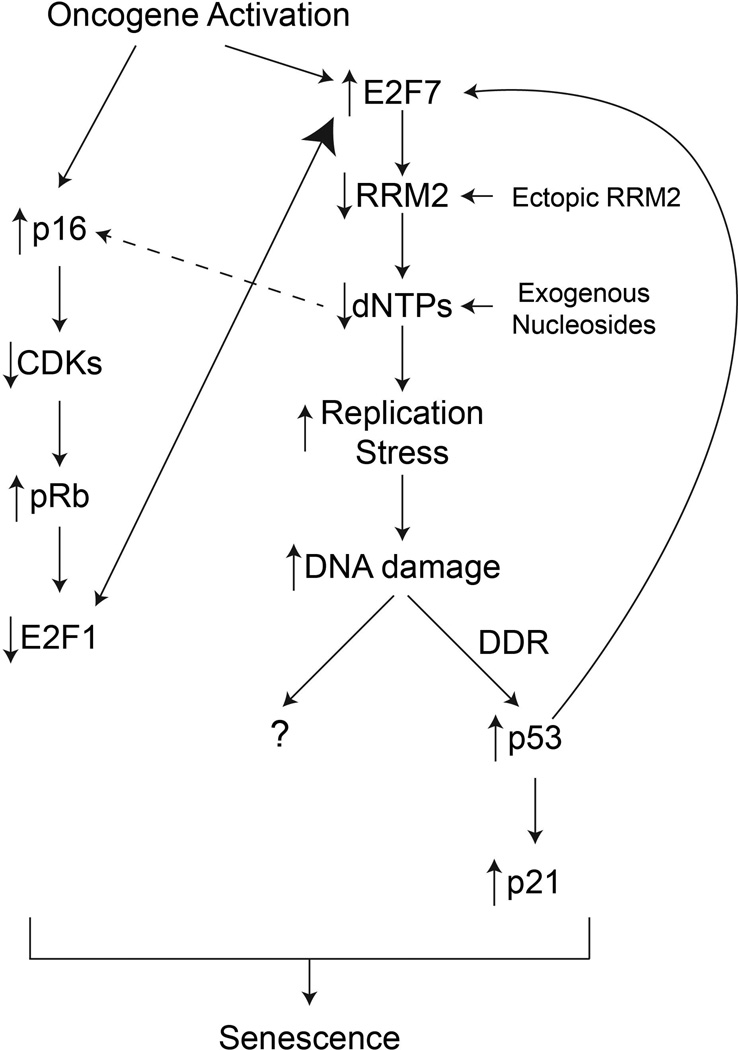
Oncogene-induced senescence (OIS) occurs when an oncogene (such as RAS, BRAF, etc…) becomes activated in a primary (normal) mammalian cell [27]. Paradoxically, expression of an activated oncogene leads to a cell cycle exit and sustained growth arrest [28]. OIS is therefore considered a bona fide tumor suppressor mechanism in vivo [24, 29]. The hallmarks of OIS include DNA replication stress leading to a sustained DNA damage response (DDR) [30, 31] and upregulation of the p53/p21 and p16/pRb pathways (Figure 1), which contribute to the stable growth arrest [32].
Figure 1. Overview of the oncogene-induced senescence pathway.
Oncogene-induced senescence begins with activation of an oncogene in a primary (normal) mammalian cell. This upregulates the repressive E2F7, which inhibits transcription of the RRM2 gene. Inhibition of RRM2 leads to a significant decrease in dNTP levels. This leads to two outcomes: 1) replication stress and accumulation of DNA damage; and 2) upregulation of p16 through an unknown mechanism. Replication stress and DNA damage accumulation activates the DNA damage response (DDR), in particular p53. p53 activation positively feeds back by further increasing E2F7 activity. Sustained activation of p53 induces high expression of p21, which helps facilitate cell cycle arrest. DNA damage can also lead to cell cycle arrest independent of p53 and p21 status. Upregulation of p16 inhibits cyclin-dependent kinases (CDKs), which relieves their inhibition of pRb. pRb can then repress E2F1 activity, which leads to more E2F7 binding to repress RRM2 transcription. Decreased E2F1 also leads to a cell cycle exit through a decrease in transcription of numerous cell cycle-related genes. These pathways all act in concert to establish and maintain the stable senescence-associated cell cycle exit.
This review will focus on the role of nucleotide metabolism as a newly identified pathway in OIS. In addition, we will discuss how changes in nucleotide metabolism can overcome OIS and transform cells. Finally, we will briefly outline how nucleotide metabolism is a diagnostic and prognostic biomarker for cancer and how this pathway could be targeted as a cancer therapeutic by inducing senescence of cancer cells.
2. Role of nucleotide metabolism in oncogene-induced senescence
Nucleotide metabolism and imbalances in dNTP pools have long been known to play a role in a variety of human pathologies [1–4]. However, until recently, there was no specific research to demonstrate that nucleotide metabolism plays a role in OIS. The following sections will review what is currently known about the role of dNTP pools in OIS, including their role in replication stress, the DNA damage response, and execution of senescence pathways after oncogene activation.
2.1. Role of decreased dNTP pools in replication stress during OIS
Senescence induced by activated oncogenes such as RAS is characterized by accumulation of cells in S-phase of the cell cycle [30]. This is due to the induction of replication stress leading to stalled and collapsed replication forks, thereby arresting cells in S-phase [30, 31]. The replication stress leads to activation of either ataxia telangiectasia and Rad3-related protein (ATR) (stalled forks) or ataxia telangiectasia mutated (ATM) (collapsed forks), which effectively activates an intra-S phase checkpoint [33, 34]. Indeed, suppression of S-phase progression is sufficient to block RAS-induced senescence [35]. Early studies showed that a decrease in dNTP pools by hydroxyurea (HU) leads to an S-phase arrest [36, 37]. Recent evidence from our lab suggests that the replication stress observed during OIS is specifically due to a decrease in dNTP levels (Figure 1) [38]. Specifically, we showed a transcriptional decrease in RRM2, whose expression is rate-limiting for rNDP/rNTP reduction to dNDPs/dNTPs [19]. Indeed, ectopic expression of RRM2, which is sufficient to restore cellular dNTP levels, or supplementation with exogenous nucleosides is able to overcome the replication stress observed during OIS (Figure 1) [38]. Interestingly, replication stress due to decreased dNTP pools has also been shown to play a role in longevity in yeast [39]. Additionally, patients with mutations in replication regulators such as the RecQ helicases [e.g., Werner syndrome ATP-dependent helicase (WRN), (Bloom syndrome RecQ helicase-like (BLM), and RecQ Protein-Like 4 (RECQL4)] display replication stress, genomic instability, and accelerated aging phenotypes [40, 41]. Given that cellular senescence is thought to contribute to tissue aging, these data suggest that dNTP pools play a larger role in replication stress during senescence.
2.2. Role of decreased dNTP pools in the activation of the DNA damage response during OIS
In addition to replication stress, accumulation of DNA damage and activation of the DDR are hallmarks of OIS (Figure 1) [30, 31, 42]. Until recently, it was unclear whether replication stress or oxidative stress was the cause of the DNA damage and DDR during OIS. Results from our lab indicate that replication stress induced by a decrease in dNTP pools plays a major role in the accumulation of DNA damage observed during OIS (Figure 1) [38]. Specifically, we show that restoration of cellular dNTP levels by either addition of exogenous nucleosides or ectopic RRM2 expression is sufficient to suppress the DDR. This correlates with rescue of the oncogene-induced DNA replication stress [38]. Another report has corroborated our finding that decreased dNTP levels are the cause of the DNA damage during senescence induced by either oncogenic RAS [43] or c-myc [44]. These authors also found that thymidylate synthase (TS) and the large subunit RRM1 were also downregulated during OIS [43], suggesting a global decrease in the nucleotide metabolic pathway during OIS. However, data from our lab demonstrates that RRM2 downregulation, but not other regulators of the nucleotide metabolic pathway such as p53R2 or RRM1, occurs before the cell cycle exit [38]. This indicates that RRM2 downregulation drives the suppression of nucleotide metabolism observed during OIS, and downregulation of other regulators of the nucleotide metabolic pathway may be simply a consequence of OIS-associated cell growth arrest.
2.3. The role of decreased dNTP pools in the execution of the OIS-associated cell cycle exit
Senescence was initially defined as an irreversible cell cycle exit [45]. In contrast to what has been observed with the reversible effects of HU on the cell cycle [36, 37], cells cannot re-enter the cell cycle after a decrease in dNTP pools due to decreased RRM2 (Figure 1) [38]. Therefore, the consequence of decreased dNTP pools must go beyond replication stress and the associated DDR. It is known that numerous signaling pathways are activated during OIS, in particular the p53/p21 and p16/pRb pathways (Figure 1) [32]. Indeed, as discussed above, the decrease in dNTP pools leads to accumulation of DNA damage and a sustained induction of the DDR [38]. Additionally, the decrease in dNTP pools also leads to elevated expression of the cell cycle regulators p21 and p16 [38], which are both upregulated during OIS [32]. p16 phosphorylates cyclin-dependent kinases (CDKs), which relieves the inhibition of pRb (Figure 1) [46]. pRb inhibits activating E2Fs such as E2F1 [47–50]. Under normal growth conditions, E2F1 contributes to transcription of RRM2 [51]. Interestingly, RRM2 downregulation occurs at the transcriptional level through replacement of the transcriptional activator E2F1 with the transcriptionally repressive E2F7 at the promoter of RRM2 gene (Figure 1) [38]. This reinforces the decrease in dNTP pools and therefore the OIS-associated cell cycle exit. This is consistent with results from a previous report demonstrating a positive feedback loop between p53 and E2F7 [52]. Downregulation of RRM2 activates p53 [38], which then enhances E2F7 expression [52]. In addition, pRb decreases E2F1 activity [49, 50], which may allow for enhanced binding of the repressive E2F7 to the RRM2 promoter. This suggests that these pathways act in concert to maintain the stable cell growth arrest. Interestingly, the decrease in RRM2 seems to be one of the first signals in this feedback loop as it occurs while cells maintain proliferation [38]. Moreover, in RAS-infected cells, ectopic RRM2 or supplementation with exogenous nucleosides can override the activation of p53 (Figure 1). These data all point to the presence of a feedback loop whereby the decrease in RRM2 leads to a further decrease in RRM2 through activation of the p53/E2F7 pathway and pRb pathway. Interestingly, senescence of cancer cells such as melanoma and ovarian cancer cells induced by RRM2 inhibition is independent of both p53 and pRB pathway [38, 53]. However, this correlates with activation of the DDR. This suggests senescence induced by RRM2 inhibition depends upon DNA damage and the associated pathway (Figure 1). Indeed, exogenous nucleosides suppress the DDR and also inhibit senescence induced by RRM2 inhibition. Future studies are warranted to elucidate the downstream pathways activated by DNA damage that mediate senescence and the associated cell growth arrest induced by suppression of nucleotide metabolism.
3. Nucleotide metabolism in cancer
OIS is considered to be a tumor suppressor mechanism in vivo [29]. However, cells that have undergone OIS can accumulate additional oncogenic hits over time, which may lead to senescence bypass and transformation. For instance, benign nevi (moles) that have undergone OIS due to expression of oncogenic BRAF or NRAS can eventually become melanomas [54]. It is important to understand how cells can bypass senescence and become transformed. These observations may allow for better preventative measures or novel cancer therapeutics. The following sections will discuss the role of nucleotide metabolism in transformation, cancer progression, and cancer therapeutics.
3.1 Role of nucleotide metabolism in genomic integrity, senescence bypass, and transformation
We have already discussed the role of decreased dNTP pools in replication stress and the DDR (Section 2.1 and 2.2). It is well known that replication stress can lead to genomic instability [7], which is a hallmark of cancer cells [55, 56]. Indeed, patients with mutations in proteins necessary for genomic integrity (i.e., BRCA1/2, Rad51, etc…) are prone to cancer [57]. Notably, early studies in yeast models demonstrated that decreased dNTP levels lead to increased mutagenesis through an increase in genomic instability [58]. A number of recent publications have attempted to elucidate the mechanism of OIS bypass and transformation. Our lab demonstrated that melanocytes expressing oncogenic BRAF or NRAS can bypass senescence when exogenous nucleosides are supplemented into the cell culture medium (Figure 1) [38]. These data have been corroborated in fibroblasts expressing HRAS [38, 43]. Additionally, we found that either ectopic expression of RRM2 or exogenous nucleoside supplementation could overcome the cell cycle exit in fully senescent fibroblasts [38]. Because these cells maintain oncogene expression, it is easy to surmise that these senescent bypassed cells could become transformed. Indeed, one report demonstrates that decreased dNTP pools in E6/E7 cells is what causes transformation of these cells [59]. Although E6/E7 expression does not induce senescence, this study clearly indicates that low dNTP pools may play a role in the early phases of tumorigenesis.
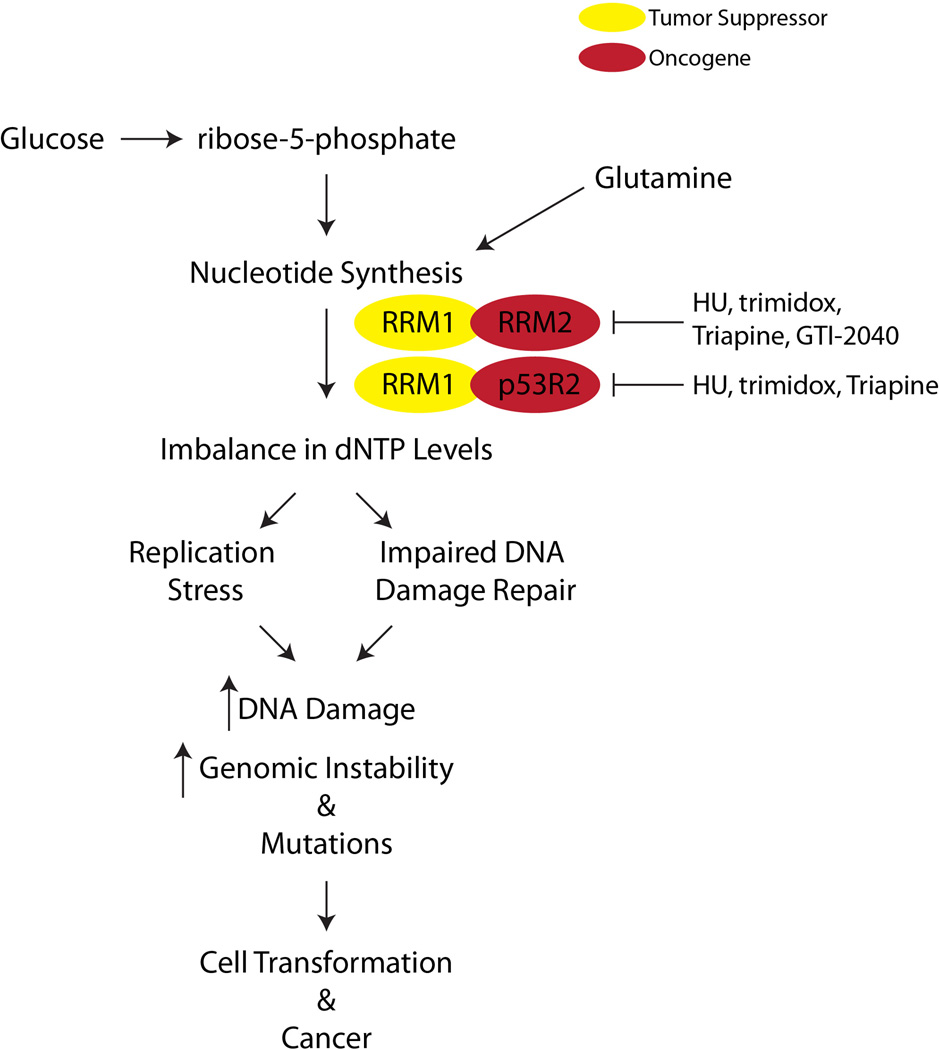
Interestingly, overexpression of RRM2 or p53R2 is tumorigenic by itself (Figure 2) [60]. Increased RRM2 or p53R2 expression is mutagenic in vitro and leads to lung neoplasms in vivo. Notably, RRM2 overexpression induces a higher mutation frequency in vitro compared to p53R2 overexpression. In addition, RRM2 transgenic mice have larger and more malignant lung neoplasms compared to p53R2 transgenic mice. These results indicate that RRM2 may be a more potent oncogene than p53R2. Additionally, another nucleotide metabolic enzyme TS can independently transform cells and lead to tumor formation in vivo [61–63]. In contrast, expression of RRM1 is thought to be tumor suppressive (Figure 2). Indeed, in both in vitro and in vivo models, overexpression of RRM1 can reduce tumor formation, migration, and metastasis [64–66]. These studies indicate that nucleotide metabolism plays an important role in senescence bypass, tumor formation, and progression.
Figure 2. Overview of nucleotide metabolic pathway in cancer.
Synthesis of nucleotides begins with formation of ribose-5-phosphate from glucose. Along with glutamine, which donates the necessary nitrogen, a number of modifications lead to formation of nucleotides. A change in RRM1, RRM2, or p53R2 expression can lead to dNTP pool imbalances. Ultimately, this imbalance leads to replication stress and impaired DNA damage repair, which consequently increases DNA damage, genomic instability, and mutations. This ultimately contributes to cell transformation and cancer. Numerous inhibitors have been developed to inhibit RRM2 and p53R2, including HU (hydroxyurea), trimidox, Triapine (3-AP), and GTI-2040.
3.2 Role of nucleotide metabolism as a cancer biomarker
It is clear that changes in nucleotide metabolism can lead to transformation and tumorigenesis (discussed in Section 3.1). Therefore, many studies have sought to determine whether components of the nucleotide metabolic pathway are either prognostic or diagnostic biomarkers in a variety of cancers.
A number of studies have shown that RRM2 is both a prognostic and diagnostic biomarker. RRM2 has been shown to be a diagnostic biomarker in colon, breast and pancreas [67–69]. Recent studies from our lab have shown that RRM2 expression is both a prognostic and diagnostic biomarker for melanomas with oncogenic BRAF or NRAS [38] and epithelial ovarian cancer (EOC) [53]. In EOC, RRM2 expression positively correlates with the cell proliferation marker Ki67, tumor grade, and stage [53]. We also observed that high RRM2 expression independently predicts a shorter overall survival in both EOC and melanoma patients [53]. [38]. Interestingly, in the same cohort of melanoma patients, RRM1 did not correlate with survival [38]. This underscores the importance of RRM2 in cancer progression.
The role of the other R2 subunit, p53R2, as a biomarker is not as clear. p53R2 has been shown to be diagnostic biomarker for a variety of cancer types, including melanoma [70], non-small cell lung cancer [71], and squamous cell carcinoma [72]. In addition, high p53R2 expression is also a poor prognostic biomarker in non-small cell lung cancer [71] and squamous cell carcinoma [73]. However, some studies have indicated that high p53R2 leads to a better prognosis [74, 75]. Future studies will need to be done on larger patient cohorts to truly determine whether p53R2 expression predicts better or worse patient outcome.
The role of RRM1 in the diagnosis and prognosis of cancer is also unclear. Studies have indicated that RRM1 is a tumor suppressor (Figure 2) [65]; therefore, high RRM1 expression is a predictor of better survival [76–78]. In contrast, other studies have found that high RRM1 expression leads to poor survival [78, 79]. In fact, one study found that depending on what treatment patients received, high RRM1 expression was inconclusive for patient prognosis [80]. Our study in melanoma compared RRM1 and RRM2 expression. We found that RRM2, but not RRM1, predicted better survival in patients with BRAF or NRAS positive tumors [38]. The conflicting results between studies make it hard to fully distinguish whether RRM1 expression is important for the diagnosis and prognosis of cancer patients.
Other enzymes further upstream in the nucleotide biosynthetic pathway have also been shown as important cancer biomarkers. For instance, high expression of thymidylate synthase, which is necessary for dTTP synthesis [17], is a predictor of poor survival in different cancer types [81, 82].
Taken together, these studies demonstrate that nucleotide pool imbalances, and in particular dNTP pool imbalances, are both diagnostic and prognostic biomarkers for a large variety of human cancers.
3.3 Inhibiting nucleotide metabolism for cancer therapeutics
Since nucleotide metabolism plays a role in transformation and tumor progression (discussed in Sections 3.1 and 3.2), inhibition of this pathway has long been considered a therapeutic strategy for cancer. As tumor cells have a higher need for dNTPs [55, 56], many of the anti-tumor therapeutics affecting nucleotide metabolism are aimed at RNR [83–87]. The first class of drugs that were found to inhibit nucleotide metabolism are free radical scavengers, including hydroxyurea [88] and trimidox (Figure 2) [89]. Because these compounds are free radical scavengers, they can inhibit RNR by inactivating the tyrosyl radical on R2 necessary for its reductive capacity [89–94]. Although hydroxyurea was initially found to be a potent anti-neoplastic agent, it has many limitations, including low affinity for RNR and a very short half-life [95, 96]. These limitations have lead to a decrease in the use of hydroxyurea in the clinic.
More recently, iron chelators have been used as a way to target RNR [97] because iron is necessary for formation of the tyrosyl radical center [19]. 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, Triapine) is the best studied iron chelator that inhibits RNR activity (Figure 2) [98]. It is 100-fold more potent than hydroxyurea in both enzyme inhibition and cancer cell growth inhibition. It can inhibit the activity of both RRM2 and p53R2 [98, 99], which disallows for compensation by the other R2 subunit. Triapine as a single agent has generally not been effective as a therapeutic for cancer patients [100, 101]. However, in combination with DNA damage agents such as cisplatin or radiation, Triapine shows some synergistic activity [102, 103], and many clinical trials are still underway (clinicaltrials.gov). One of the main issues with Triapine is the number of side effects, including dyspnea and methemoglobinemia [97, 103, 104], which limits the use of this drug.
Data from both hydroxyurea and Triapine demonstrate that more specific RNR inhibitors are needed to minimize toxic side effects. With the identification of antisense, more recent work has gone into specifically targeting RNR subunits. Currently, an antisense oligonucleotide to RRM2 (GTI-2040) is in clinical trials (clinicaltrials.gov) (Figure 2). Phase II clinical trials with published results have shown little added benefit with GTI-2040 [105–107]. It is possible that p53R2 could compensate for RRM2 when it is inhibited. Additionally, the effect of GTI-2040 has not been fully examined in combination with cisplatin or radiation. This may lead to a synergistic effect similar to combination therapy with Triapine (discussed above). Results from our lab using a short hairpin RNA (shRNA) specific for RRM2 indicate that these cells undergo senescence [38, 53] and not cell death. Senescent cells remain metabolically active [24], which may allow for cellular changes that could overcome senescence. Therefore, finding combination strategies that would kill these senescent cells will be important for future targeting of RNR.
4. Concluding remarks
dNTPs are necessary for both DNA replication and repair. The rate-limiting step in dNTP synthesis is the reduction of rNDPs to dNDPs by RNR. Over the past year, it has become clear that the small subunit of RNR, RRM2, plays a major role in the senescence tumor suppression mechanism. Without downregulation of RRM2, cells with activated oncogenes can bypass senescence, which may ultimately lead to cell transformation and tumorigenesis (Figure 1). Because this enzyme is so important in the establishment of uncontrolled growth in cancer and the maintenance of proliferation, anti-tumor drugs have been developed that target this specific part of the nucleotide metabolic pathway (Figure 2). More work needs to be done to explore new combination strategies by targeting RNR to drive senescence of cancer cells treated with chemotherapeutics or targeted therapies that only induce a transient cell growth arrest. Further, it will be interesting to investigate ways to eliminate senescent cells induced by RNR inhibition via promoting apoptosis. Together, these approaches may ultimately lead to a sustained, long-term response to therapeutics in cancer and enhance survival of cancer patients.
Acknowledgements
This work was supported by a NIH/NCI grant (R01CA160331 to R.Z.), a DoD Ovarian Cancer Academy Award (OC093420 to R.Z.) and an NIH/NCI training grant (T32CA9171-35 to K.M.A.). Support of Core Facilities used in this study was provided by Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute.
List of Abbreviations
- 3-AP
3-aminopyridine-2-carboxaldehyde thiosemicarbazone
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related protein
- AMP/ADP/ATP
adenosine mono-, di-, and tri-phosphate
- BLM
Bloom syndrome RecQ helicase-like
- BRAF
V-raf murine sarcoma viral oncogene homolog B
- BRCA1/2
Breast Cancer 1/2, Early Onset
- CDK
cyclin-dependent kinase
- CMP/CDP/CTP
cytidine mono-, di-, and tri-phosphate
- c-myc
v-myc avian myelocytomatosis viral oncogene homolog
- DNA
deoxyribonucleic acid
- DDR
DNA damage response
- dNDP
deoxyribonucleotide diphosphate
- dNTP
deoxyribonucleotide triphosphate
- E2F1
E2F transcription factor 1
- E2F7
E2F transcription factor 7
- EOC
epithelial ovarian cancer
- GMP/GDP/GTP
guanosine mono-, di-, and tri-phosphate
- HRAS
Harvey Rat Sarcoma Viral Oncogene Homolog
- HU
hydroxyurea
- IMP
inosine monophosphate
- NADPH
Nicotinamide adenine dinucleotide phosphate
- NDP
ribonucleoside diphosphate
- NRAS
neuroblastoma RAS viral (V-Ras) oncogene homolog
- NTP
ribonucleoside triphosphate
- OIS
oncogene-induced senescence
- p53R2/RRM2B
Ribonucleotide Reductase M2 B (TP53 Inducible)
- pRb
retinoblastoma protein
- R1
ribonucleotide reductase subunit 1
- R1/RRM1
ribonucleotide reductase M1
- R2
ribonucleotide reductase subunit 2
- Rad51
Rad51 recombinase
- RAS
rat sarcoma oncogene
- RECQL4
RecQ Protein-Like 4
- RNR
ribonucleotide reductase
- RRM2
ribonucleotide reductase M2
- SA-B-Gal
senescence-associated beta-galactosidase
- shRNA
short hairpin RNA
- TS
thymidylate synthase
- TTP
thymidine triphosphate
- UDP
uridine diphosphate
- UMP
uridine monophosphate
- WRN
Werner syndrome ATP-dependent helicase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp Cell Res. 1989;181:305–316. doi: 10.1016/0014-4827(89)90090-6. [DOI] [PubMed] [Google Scholar]
- 2.Weinberg G, Ullman B, Martin DW., Jr Mutator phenotypes in mammalian cell mutants with distinct biochemical defects and abnormal deoxyribonucleoside triphosphate pools. Proc Natl Acad Sci U S A. 1981;78:2447–2451. doi: 10.1073/pnas.78.4.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chabosseau P, Buhagiar-Labarchede G, Onclercq-Delic R, Lambert S, Debatisse M, Brison O, Amor-Gueret M. Pyrimidine pool imbalance induced by BLM helicase deficiency contributes to genetic instability in Bloom syndrome. Nat Commun. 2011;2:368. doi: 10.1038/ncomms1363. [DOI] [PubMed] [Google Scholar]
- 4.Chang L, Guo R, Huang Q, Yen Y. Chromosomal instability triggered by Rrm2b loss leads to IL-6 secretion and plasmacytic neoplasms. Cell Rep. 2013;3:1389–1397. doi: 10.1016/j.celrep.2013.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ammann AJ. Purine nucleotide imbalance in immunodeficiency disorders. Basic Life Sci. 1985;31:487–502. doi: 10.1007/978-1-4613-2449-2_32. [DOI] [PubMed] [Google Scholar]
- 6.Boss GR, Seegmiller JE. Genetic defects in human purine and pyrimidine metabolism. Annu Rev Genet. 1982;16:297–328. doi: 10.1146/annurev.ge.16.120182.001501. [DOI] [PubMed] [Google Scholar]
- 7.Burhans WC, Weinberger M. DNA replication stress, genome instability and aging. Nucleic Acids Res. 2007;35:7545–7556. doi: 10.1093/nar/gkm1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathews CK. DNA precursor metabolism and genomic stability. Faseb J. 2006;20:1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- 9.Kimura T, Takeda S, Sagiya Y, Gotoh M, Nakamura Y, Arakawa H. Impaired function of p53R2 in Rrm2b-null mice causes severe renal failure through attenuation of dNTP pools. Nat Genet. 2003;34:440–445. doi: 10.1038/ng1212. [DOI] [PubMed] [Google Scholar]
- 10.Dobrovolsky VN, Bucci T, Heflich RH, Desjardins J, Richardson FC. Mice deficient for cytosolic thymidine kinase gene develop fatal kidney disease. Mol Genet Metab. 2003;78:1–10. doi: 10.1016/s1096-7192(02)00224-x. [DOI] [PubMed] [Google Scholar]
- 11.El-Hattab AW, Scaglia F. Mitochondrial DNA depletion syndromes: review and updates of genetic basis, manifestations, and therapeutic options. Neurotherapeutics. 2013;10:186–198. doi: 10.1007/s13311-013-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitceathly RD, Smith C, Fratter C, Alston CL, He L, Craig K, Blakely EL, Evans JC, Taylor J, Shabbir Z, Deschauer M, Pohl U, Roberts ME, Jackson MC, Halfpenny CA, Turnpenny PD, Lunt PW, Hanna MG, Schaefer AM, McFarland R, Horvath R, Chinnery PF, Turnbull DM, Poulton J, Taylor RW, Gorman GS. Adults with RRM2B-related mitochondrial disease have distinct clinical and molecular characteristics. Brain. 2012;135:3392–3403. doi: 10.1093/brain/aws231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blakley RL, Vitols E. The control of nucleotide biosynthesis. Annu Rev Biochem. 1968;37:201–224. doi: 10.1146/annurev.bi.37.070168.001221. [DOI] [PubMed] [Google Scholar]
- 14.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–890. doi: 10.1101/gad.189365.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartman SC, Buchanan JM. Nucleic acids, purines, pyrimidines (nucleotide synthesis) Annu Rev Biochem. 1959;28:365–410. doi: 10.1146/annurev.bi.28.070159.002053. [DOI] [PubMed] [Google Scholar]
- 16.Newsholme P, Procopio J, Lima MM, Pithon-Curi TC, Curi R. Glutamine and glutamate--their central role in cell metabolism and function. Cell Biochem Funct. 2003;21:1–9. doi: 10.1002/cbf.1003. [DOI] [PubMed] [Google Scholar]
- 17.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 18.Murray AW. The biological significance of purine salvage. Annu Rev Biochem. 1971;40:811–826. doi: 10.1146/annurev.bi.40.070171.004115. [DOI] [PubMed] [Google Scholar]
- 19.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 20.Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 21.Engstrom Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- 22.Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281:7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 23.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 24.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 25.Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192:547–556. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yaswen P, Campisi J. Oncogene-induced senescence pathways weave an intricate tapestry. Cell. 2007;128:233–234. doi: 10.1016/j.cell.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Mooi WJ, Peeper DS. Oncogene-induced cell senescence--halting on the road to cancer. N Engl J Med. 2006;355:1037–1046. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 29.Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, Dorken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- 30.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 31.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 32.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 33.Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 2002;12:509–516. doi: 10.1016/s0962-8924(02)02380-2. [DOI] [PubMed] [Google Scholar]
- 34.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, Trotman LC, Cheng K, Varmeh S, Kozma SC, Thomas G, Rosivatz E, Woscholski R, Cognetti F, Scher HI, Pandolfi PP. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010;120:681–693. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bianchi V, Pontis E, Reichard P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J Biol Chem. 1986;261:16037–16042. [PubMed] [Google Scholar]
- 37.Matsumoto M, Rey DA, Cory JG. Effects of cytosine arabinoside and hydroxyurea on the synthesis of deoxyribonucleotides and DNA replication in L1210 cells. Adv Enzyme Regul. 1990;30:47–59. doi: 10.1016/0065-2571(90)90008-p. [DOI] [PubMed] [Google Scholar]
- 38.Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A, Wu H, Wei Z, Wagner SN, Herlyn M, Zhang R. Suppression of Nucleotide Metabolism Underlies the Establishment and Maintenance of Oncogene-Induced Senescence. Cell Rep. 2013 doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinberger M, Feng L, Paul A, Smith DL, Jr, Hontz RD, Smith JS, Vujcic M, Singh KK, Huberman JA, Burhans WC. DNA replication stress is a determinant of chronological lifespan in budding yeast. PLoS One. 2007;2:e748. doi: 10.1371/journal.pone.0000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larizza L, Magnani I, Roversi G. Rothmund-Thomson syndrome and RECQL4 defect: splitting and lumping. Cancer Lett. 2006;232:107–120. doi: 10.1016/j.canlet.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 42.Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mannava S, Moparthy KC, Wheeler LJ, Natarajan V, Zucker SN, Fink EE, Im M, Flanagan S, Burhans WC, Zeitouni NC, Shewach DS, Mathews CK, Nikiforov MA. Depletion of deoxyribonucleotide pools is an endogenous source of DNA damage in cells undergoing oncogene-induced senescence. Am J Pathol. 2013;182:142–151. doi: 10.1016/j.ajpath.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mannava S, Moparthy KC, Wheeler LJ, Leonova KI, Wawrzyniak JA, Bianchi-Smiraglia A, Berman AE, Flanagan S, Shewach DS, Zeitouni NC, Gudkov AV, Mathews CK, Nikiforov MA. Ribonucleotide reductase and thymidylate synthase or exogenous deoxyribonucleosides reduce DNA damage and senescence caused by C-MYC depletion. Aging (Albany NY) 2012;4:917–922. doi: 10.18632/aging.100512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 47.Hiebert SW, Chellappan SP, Horowitz JM, Nevins JR. The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 1992;6:177–185. doi: 10.1101/gad.6.2.177. [DOI] [PubMed] [Google Scholar]
- 48.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 49.Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flemington EK, Speck SH, Kaelin WG., Jr E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc Natl Acad Sci U S A. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang YW, Jones TL, Martin SE, Caplen NJ, Pommier Y. Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. J Biol Chem. 2009;284:18085–18095. doi: 10.1074/jbc.M109.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aksoy O, Chicas A, Zeng T, Zhao Z, McCurrach M, Wang X, Lowe SW. The atypical E2F family member E2F7 couples the p53 and RB pathways during cellular senescence. Genes Dev. 2012;26:1546–1557. doi: 10.1101/gad.196238.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aird KM, Li H, Xin F, Konstantinopoulos PA, Zhang R. Identification of ribonucleotide reductase M2 as a potential target for pro-senescence therapy in epithelial ovarian cancer. Cell Cycle. 2014;13 doi: 10.4161/cc.26953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vredeveld LC, Possik PA, Smit MA, Meissl K, Michaloglou C, Horlings HM, Ajouaou A, Kortman PC, Dankort D, McMahon M, Mooi WJ, Peeper DS. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 2012;26:1055–1069. doi: 10.1101/gad.187252.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 56.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 57.Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 58.Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 2005;19:853–862. doi: 10.1101/gad.329905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu X, Page JL, Surtees JA, Liu H, Lagedrost S, Lu Y, Bronson R, Alani E, Nikitin AY, Weiss RS. Broad overexpression of ribonucleotide reductase genes in mice specifically induces lung neoplasms. Cancer Res. 2008;68:2652–2660. doi: 10.1158/0008-5472.CAN-07-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen M, Rahman L, Voeller D, Kastanos E, Yang SX, Feigenbaum L, Allegra C, Kaye FJ, Steeg P, Zajac-Kaye M. Transgenic expression of human thymidylate synthase accelerates the development of hyperplasia and tumors in the endocrine pancreas. Oncogene. 2007;26:4817–4824. doi: 10.1038/sj.onc.1210273. [DOI] [PubMed] [Google Scholar]
- 62.Rahman L, Voeller D, Rahman M, Lipkowitz S, Allegra C, Barrett JC, Kaye FJ, Zajac-Kaye M. Thymidylate synthase as an oncogene: a novel role for an essential DNA synthesis enzyme. Cancer Cell. 2004;5:341–351. doi: 10.1016/s1535-6108(04)00080-7. [DOI] [PubMed] [Google Scholar]
- 63.Voeller D, Rahman L, Zajac-Kaye M. Elevated levels of thymidylate synthase linked to neoplastic transformation of mammalian cells. Cell Cycle. 2004;3:1005–1007. [PubMed] [Google Scholar]
- 64.Fan H, Huang A, Villegas C, Wright JA. The R1 component of mammalian ribonucleotide reductase has malignancy-suppressing activity as demonstrated by gene transfer experiments. Proc Natl Acad Sci U S A. 1997;94:13181–13186. doi: 10.1073/pnas.94.24.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gautam A, Bepler G. Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res. 2006;66:6497–6502. doi: 10.1158/0008-5472.CAN-05-4462. [DOI] [PubMed] [Google Scholar]
- 66.Gautam A, Li ZR, Bepler G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene. 2003;22:2135–2142. doi: 10.1038/sj.onc.1206232. [DOI] [PubMed] [Google Scholar]
- 67.Liu X, Zhang H, Lai L, Wang X, Loera S, Xue L, He H, Zhang K, Hu S, Huang Y, Nelson RA, Zhou B, Zhou L, Chu P, Zhang S, Zheng S, Yen Y. Ribonucleotide reductase small subunit M2 serves as a prognostic biomarker and predicts poor survival of colorectal cancers. Clin Sci (Lond) 2013;124:567–578. doi: 10.1042/CS20120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fujita H, Ohuchida K, Mizumoto K, Itaba S, Ito T, Nakata K, Yu J, Kayashima T, Souzaki R, Tajiri T, Manabe T, Ohtsuka T, Tanaka M. Gene expression levels as predictive markers of outcome in pancreatic cancer after gemcitabine-based adjuvant chemotherapy. Neoplasia. 2010;12:807–817. doi: 10.1593/neo.10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones DT, Lechertier T, Mitter R, Herbert JM, Bicknell R, Jones JL, Li JL, Buffa F, Harris AL, Hodivala-Dilke K. Gene expression analysis in human breast cancer associated blood vessels. PLoS One. 2012;7:e44294. doi: 10.1371/journal.pone.0044294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsushita S, Ikeda R, Fukushige T, Tajitsu Y, Gunshin K, Okumura H, Ushiyama M, Akiyama S, Kawai K, Takeda Y, Yamada K, Kanekura T. p53R2 is a prognostic factor of melanoma and regulates proliferation and chemosensitivity of melanoma cells. J Dermatol Sci. 2012;68:19–24. doi: 10.1016/j.jdermsci.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 71.Uramoto H, Sugio K, Oyama T, Hanagiri T, Yasumoto K. P53R2, p53 inducible ribonucleotide reductase gene, correlated with tumor progression of non-small cell lung cancer. Anticancer Res. 2006;26:983–988. [PubMed] [Google Scholar]
- 72.Yanamoto S, Kawasaki G, Yoshitomi I, Mizuno A. Expression of p53R2, newly p53 target in oral normal epithelium, epithelial dysplasia and squamous cell carcinoma. Cancer Lett. 2003;190:233–243. doi: 10.1016/s0304-3835(02)00588-8. [DOI] [PubMed] [Google Scholar]
- 73.Okumura H, Natsugoe S, Yokomakura N, Kita Y, Matsumoto M, Uchikado Y, Setoyama T, Owaki T, Ishigami S, Aikou T. Expression of p53R2 is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res. 2006;12:3740–3745. doi: 10.1158/1078-0432.CCR-05-2416. [DOI] [PubMed] [Google Scholar]
- 74.Hsu NY, Wu JY, Liu X, Yen Y, Chen CY, Chou MC, Lee H, Cheng YW. p53R2 expression as a prognostic biomarker in early stage non-small cell lung cancer. Oncol Lett. 2010;1:609–613. doi: 10.3892/ol_00000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu X, Lai L, Wang X, Xue L, Leora S, Wu J, Hu S, Zhang K, Kuo ML, Zhou L, Zhang H, Wang Y, Zhou B, Nelson RA, Zheng S, Zhang S, Chu P, Yen Y. Ribonucleotide reductase small subunit M2B prognoses better survival in colorectal cancer. Cancer Res. 2011;71:3202–3213. doi: 10.1158/0008-5472.CAN-11-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bepler G, Sharma S, Cantor A, Gautam A, Haura E, Simon G, Sharma A, Sommers E, Robinson L. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:1878–1885. doi: 10.1200/JCO.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 77.Pesta M, Kulda V, Fiala O, Safranek J, Topolcan O, Krakorova G, Cerny R, Pesek M. Prognostic significance of ERCC1, RRM1 and BRCA1 in surgically-treated patients with non-small cell lung cancer. Anticancer Res. 2012;32:5003–5010. [PubMed] [Google Scholar]
- 78.Jordheim LP, Seve P, Tredan O, Dumontet C. The ribonucleotide reductase large subunit (RRM1) as a predictive factor in patients with cancer. Lancet Oncol. 2011;12:693–702. doi: 10.1016/S1470-2045(10)70244-8. [DOI] [PubMed] [Google Scholar]
- 79.Wang Q, Liu X, Zhou J, Huang Y, Zhang S, Shen J, Loera S, Yuan X, Chen W, Jin M, Shibata S, Liu Y, Chu P, Wang L, Yen Y. Ribonucleotide reductase large subunit M1 predicts poor survival due to modulation of proliferative and invasive ability of gastric cancer. PLoS One. 2013;8:e70191. doi: 10.1371/journal.pone.0070191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie H, Jiang W, Jiang J, Wang Y, Kim R, Liu X. Predictive and prognostic roles of ribonucleotide reductase M1 in resectable pancreatic adenocarcinoma. Cancer. 2012;119:173–181. doi: 10.1002/cncr.27715. [DOI] [PubMed] [Google Scholar]
- 81.Pestalozzi BC, Peterson HF, Gelber RD, Goldhirsch A, Gusterson BA, Trihia H, Lindtner J, Cortes-Funes H, Simmoncini E, Byrne MJ, Golouh R, Rudenstam CM, Castiglione-Gertsch M, Allegra CJ, Johnston PG. Prognostic importance of thymidylate synthase expression in early breast cancer. J Clin Oncol. 1997;15:1923–1931. doi: 10.1200/JCO.1997.15.5.1923. [DOI] [PubMed] [Google Scholar]
- 82.Allegra CJ, Paik S, Colangelo LH, Parr AL, Kirsch I, Kim G, Klein P, Johnston PG, Wolmark N, Wieand HS. Prognostic value of thymidylate synthase, Ki-67, and p53 in patients with Dukes' B and C colon cancer: a National Cancer Institute-National Surgical Adjuvant Breast and Bowel Project collaborative study. J Clin Oncol. 2003;21:241–250. doi: 10.1200/JCO.2003.05.044. [DOI] [PubMed] [Google Scholar]
- 83.Nocentini G. Ribonucleotide reductase inhibitors: new strategies for cancer chemotherapy. Crit Rev Oncol Hematol. 1996;22:89–126. doi: 10.1016/1040-8428(95)00187-5. [DOI] [PubMed] [Google Scholar]
- 84.Cerqueira NM, Fernandes PA, Ramos MJ. Ribonucleotide reductase: a critical enzyme for cancer chemotherapy and antiviral agents. Recent Pat Anticancer Drug Discov. 2007;2:11–29. doi: 10.2174/157489207779561408. [DOI] [PubMed] [Google Scholar]
- 85.Cerqueira NM, Pereira S, Fernandes PA, Ramos MJ. Overview of ribonucleotide reductase inhibitors: an appealing target in anti-tumour therapy. Curr Med Chem. 2005;12:1283–1294. doi: 10.2174/0929867054020981. [DOI] [PubMed] [Google Scholar]
- 86.Cory JG. Ribonucleotide reductase as a chemotherapeutic target. Adv Enzyme Regul. 1988;27:437–455. doi: 10.1016/0065-2571(88)90030-1. [DOI] [PubMed] [Google Scholar]
- 87.Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets. 2006;6:409–431. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- 88.Madaan K, Kaushik D, Verma T. Hydroxyurea: a key player in cancer chemotherapy. Expert Rev Anticancer Ther. 2011;12:19–29. doi: 10.1586/era.11.175. [DOI] [PubMed] [Google Scholar]
- 89.Szekeres T, Gharehbaghi K, Fritzer M, Woody M, Srivastava A, van't Riet B, Jayaram HN, Elford HL. Biochemical and antitumor activity of trimidox, a new inhibitor of ribonucleotide reductase. Cancer Chemother Pharmacol. 1994;34:63–66. doi: 10.1007/BF00686113. [DOI] [PubMed] [Google Scholar]
- 90.Lassmann G, Thelander L, Graslund A. EPR stopped-flow studies of the reaction of the tyrosyl radical of protein R2 from ribonucleotide reductase with hydroxyurea. Biochem Biophys Res Commun. 1992;188:879–887. doi: 10.1016/0006-291x(92)91138-g. [DOI] [PubMed] [Google Scholar]
- 91.Nigovic B, Kujundzic N, Sankovic K. Electron transfer in N-hydroxyurea complexes with iron(III) Eur J Med Chem. 2005;40:51–55. doi: 10.1016/j.ejmech.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 92.Elford HL. Functional regulation of mammalian ribonucleotide reductase. Adv Enzyme Regul. 1972;10:19–38. doi: 10.1016/0065-2571(72)90004-0. [DOI] [PubMed] [Google Scholar]
- 93.Elford HL, Van't Riet B, Wampler GL, Lin AL, Elford RM. Regulation of ribonucleotide reductase in mammalian cells by chemotherapeutic agents. Adv Enzyme Regul. 1980;19:151–168. doi: 10.1016/0065-2571(81)90014-5. [DOI] [PubMed] [Google Scholar]
- 94.Elford HL, Wampler GL, van't Riet B. New ribonucleotide reductase inhibitors with antineoplastic activity. Cancer Res. 1979;39:844–851. [PubMed] [Google Scholar]
- 95.Beckloff GL, Lerner HJ, Frost D, Russo-Alesi FM, Gitomer S. Hydroxyurea (NSC-32065) in biologic fluids: dose-concentration relationship. Cancer Chemother Rep. 1965;48:57–58. [PubMed] [Google Scholar]
- 96.Gwilt PR, Tracewell WG. Pharmacokinetics and pharmacodynamics of hydroxyurea. Clin Pharmacokinet. 1998;34:347–358. doi: 10.2165/00003088-199834050-00002. [DOI] [PubMed] [Google Scholar]
- 97.Yu Y, Gutierrez E, Kovacevic Z, Saletta F, Obeidy P, Suryo Rahmanto Y, Richardson DR. Iron chelators for the treatment of cancer. Curr Med Chem. 2012;19:2689–2702. doi: 10.2174/092986712800609706. [DOI] [PubMed] [Google Scholar]
- 98.Finch RA, Liu MC, Cory AH, Cory JG, Sartorelli AC. Triapine (3-aminopyridine-2-carboxaldehyde thiosemicarbazone; 3-AP): an inhibitor of ribonucleotide reductase with antineoplastic activity. Adv Enzyme Regul. 1999;39:3–12. doi: 10.1016/s0065-2571(98)00017-x. [DOI] [PubMed] [Google Scholar]
- 99.Finch RA, Liu M, Grill SP, Rose WC, Loomis R, Vasquez KM, Cheng Y, Sartorelli AC. Triapine (3-aminopyridine-2-carboxaldehyde-thiosemicarbazone): A potent inhibitor of ribonucleotide reductase activity with broad spectrum antitumor activity. Biochem Pharmacol. 2000;59:983–991. doi: 10.1016/s0006-2952(99)00419-0. [DOI] [PubMed] [Google Scholar]
- 100.Nutting CM, van Herpen CM, Miah AB, Bhide SA, Machiels JP, Buter J, Kelly C, de Raucourt D, Harrington KJ. Phase II study of 3-AP Triapine in patients with recurrent or metastatic head and neck squamous cell carcinoma. Ann Oncol. 2009;20:1275–1279. doi: 10.1093/annonc/mdn775. [DOI] [PubMed] [Google Scholar]
- 101.Attia S, Kolesar J, Mahoney MR, Pitot HC, Laheru D, Heun J, Huang W, Eickhoff J, Erlichman C, Holen KD. A phase 2 consortium (P2C) trial of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP) for advanced adenocarcinoma of the pancreas. Invest New Drugs. 2008;26:369–379. doi: 10.1007/s10637-008-9123-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kunos CA, Radivoyevitch T, Waggoner S, Debernardo R, Zanotti K, Resnick K, Fusco N, Adams R, Redline R, Faulhaber P, Dowlati A. Radiochemotherapy plus 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, NSC #663249) in advanced-stage cervical and vaginal cancers. Gynecol Oncol. 2013;130:75–80. doi: 10.1016/j.ygyno.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kunos C, Radivoyevitch T, Abdul-Karim FW, Fanning J, Abulafia O, Bonebrake AJ, Usha L. Ribonucleotide reductase inhibition restores platinum-sensitivity in platinum-resistant ovarian cancer: a Gynecologic Oncology Group Study. J Transl Med. 2012;10:79. doi: 10.1186/1479-5876-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kunos CA, Radivoyevitch T, Ingalls ST, Hoppel CL. Management of 3-aminopyridine-2-carboxaldehyde thiosemicarbazone-induced methemoglobinemia. Future Oncol. 2012;8:145–150. doi: 10.2217/fon.11.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Leighl NB, Laurie SA, Chen XE, Ellis P, Shepherd FA, Knox JJ, Goss G, Burkes RL, Pond GR, Dick C, Yen Y, Zwiebel JA, Moore MJ. A phase I/II study of GTI-2040 plus docetaxel as second-line treatment in advanced non-small cell lung cancer: a study of the PMH phase II consortium. J Thorac Oncol. 2009;4:1163–1169. doi: 10.1097/JTO.0b013e3181a949b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sridhar SS, Canil CM, Chi KN, Hotte SJ, Ernst S, Wang L, Chen EX, Juhasz A, Yen Y, Murray P, Zwiebel JA, Moore MJ. A phase II study of the antisense oligonucleotide GTI-2040 plus docetaxel and prednisone as first-line treatment in castration-resistant prostate cancer. Cancer Chemother Pharmacol. 2010;67:927–933. doi: 10.1007/s00280-010-1389-7. [DOI] [PubMed] [Google Scholar]
- 107.Stadler WM, Desai AA, Quinn DI, Bukowski R, Poiesz B, Kardinal CG, Lewis N, Makalinao A, Murray P, Torti FM. A Phase I/II study of GTI-2040 and capecitabine in patients with renal cell carcinoma. Cancer Chemother Pharmacol. 2008;61:689–694. doi: 10.1007/s00280-007-0524-6. [DOI] [PubMed] [Google Scholar]