Abstract
Cancer stem cells (CSC) represent a malignant subpopulation of cells in hierarchically organized tumors. They constitute a sub-population of malignant cells within a tumor mass and possess the ability to self-renew giving rise to heterogeneous tumor cell populations with a complex set of differentiated tumor cells. CSC may be the cause of metastasis and therapeutic refractory disease. Because few markers exist to identify and isolate pure CSC, we used cell-based Systematic Evolution of Ligands by EXponential enrichment (cell-SELEX) to create DNA aptamers that can identify novel molecular targets on the surfaces of live CSC. Out of 22 putative DNA sequences, 3 bound to ~90% and 5 bound to ~15% of DU145 prostate cancer cells. The 15% of cells that were positive for the second panel of aptamers expressed high levels of E-cadherin and CD44, had high aldehyde dehydrogenase 1 activity, grew as spheroids under non-adherent culture conditions, and initiated tumors in immune-compromised mice. The discovery of the molecular targets of these aptamers could reveal novel CSC biomarkers.
Introduction
In most cancer cases, patient mortality results from metastasis 1. A new model of cancer has recently emerged based on experiments that measure the ability of isolated subpopulations of tumor cells to initiate tumors 2–6. In this model, a hierarchy of cell types exists within a tumor. This hierarchy is created and sustained by the presence of cancer stem cells (CSC) that divide asymmetrically, giving rise to daughter CSC and highly proliferative progenitor cells. According to this CSC model, metastasis can be understood as the shedding, migration, and engraftment of CSC to distal sites.
One factor influencing the relatively low frequency of metastasis observation, despite the millions of tumor cells that are shed daily from solid tumors, may be the relative rarity of CSC in the primary tumor. Further, CSC may be naturally shielded from drugs through their niche, or they may avoid the cytotoxic effects of drugs by infrequent cell division. Indeed, it has been suggested that CSC may be drug- and radiation resistant 7–10. Although CSC have been identified in hematologic, melanoma, breast, brain, pancreatic, and colon cancers 11–16, specific markers that clearly identify CSC are not available, and most assays are dependent on CSC-enriched cell populations 17. Therefore, the discovery of new markers that clearly identify CSC will help us to understand the nature of metastatic tumor cell populations and, as a result, allow the creation of new avenues to specifically identify these cells for targeted therapies.
There are many methods of biomarker discovery, but only two methods produce molecular probes that can be used to detect the target biomarkers, namely, phage display and Systematic Evolution of Ligands by EXponential enrichment (SELEX). Out of these two methods, SELEX produces molecular probes, termed aptamers, which are single-stranded oligonucleotides that selectively bind to target proteins and other small molecules with high affinity. They can be completely synthesized in vitro and therefore require no animal or bacterial hosts for production. When compared to antibodies, aptamers have many other appealing features, including low molecular weight, easy chemical modification, low toxicity and immunogenicity, and long shelf-life.
Aptamers are selected from libraries of random sequences of synthetic DNA through repetitive binding of these oligonucleotides to target molecules by SELEX 18, 19, an iterative in vitro selection process that generates aptamers with high specificity and affinity to their target molecules. To produce probes for molecular analysis of live tumor cells, we developed a novel method for cell-based aptamer selection called cell-SELEX 20, 21. In cell-SELEX, instead of using a single purified molecule as a target, whole live cells are used as targets to select DNA aptamers. We have applied cell-SELEX to several in vitro systems of human disease, and we have selected aptamers for several types of leukemia cells, small cell lung cancer, liver cancer, colorectal cancer, ovarian cancer and viral-infected cells 20, 22–28. In each case, it has been possible to select a panel of aptamers that bind specifically to the target cells. We have shown that these aptamers can be used to recognize cancer patient samples and bind to tumors in live animals 29, 30. Furthermore, once aptamers have been selected using cell-SELEX, we have shown that biomarker discovery is possible by using aptamer precipitation methods to purify and sequence aptamer protein targets 31, 32.
Based on our previous work, we have hypothesized that a cell-SELEX approach targeting CSC-enriched populations would be able to identify novel markers for CSC by creating DNA-aptamers that bind specifically and with high affinity to the CSC-enriched population. Therefore, combining cell-SELEX with an appropriate human CSC tumor model should result in the development of a system able to detect and identify expressed proteins that are both up-regulated in CSC and functionally involved in specific stages of the tumor growth process.
Prostate cancer has become the most frequently diagnosed cancer and the second leading cause of cancer-related deaths in North American men 33. Surgery and radiation therapy are widely used to treat prostate cancer patients, and although largely successful in early stage disease, these treatment modalities have poor outcomes in advanced metastatic prostate cancer 34, 35. In addition, the prostate-specific antigen test is not clearly associated with reduced mortality from prostate cancer; consequently, there is a great need to discover new markers and diagnostic assays for prostate cancer 36, 37.
There are now several reports suggesting that prostate cancers may originate from CSC 38. In particular, cells from prostate tumors or cell lines that are CD133+/CD44+, CD44+/CD24−, or CD44+/Integrin-α2β1+ have been shown to express embryonic stem cell markers. For example, cells expressing embryonic stem cell markers OCT3/4, Nanog and SOX2 are clonogenic, proliferative, and tumor-initiating 39–43. Recently, we reported the identification of cells expressing these embryonic stem cell markers in patient prostate cancers and human prostate cancer models 44. Using E-cadherin (E-cad) as a marker, these cells could be isolated from prostate cancer cell lines by flow cytometry. E-cad+ cells were exclusively located in colonies demonstrating the classic malignant holoclone morphology. Moreover, these colonies exhibited OCT3/4 nuclear staining and highly expressed both CD44 and integrin-α2β1. We further demonstrated that these E-cad+ cells had characteristics indicative of the CSC phenotype, including high expression of embryonic stem cell markers (SOX2, OCT3/4, Nanog, Klf4 and c-Myc), colony formation, the ability to grow as spheres under non-adherent conditions, and a high degree of tumorigenicity in immunodeficient mice 44. Finally, knockdown of SOX2 or OCT3/4 abolished the tumor-forming ability of these cells 44. Since E-cad could be used reliably as a single marker, to sort prostate CSC we chose to apply cell-SELEX to the E-cad+ subpopulation of the DU145 prostate cancer cell line to generate DNA aptamers for CSC. To this end, we generated two panels of aptamers. The first panel bound to 90% of DU145 cells and could label tumors in live animals. As such, the aptamers in this panel will be useful for detection and diagnostic purposes. In the second panel, aptamers bound to ~15% of DU145 cells. In this case, aptamer binding correlated with high levels of E-cad and CD44 expression, and aptamer-positive cells exhibited high tumorigenicity compared to aptamer-negative cells in immunodeficient mice.
Material and Methods
Cell culture
Human prostate cancer cell lines DU145, PC3, LNCaP and VCaP were obtained from the American Tissue Type Culture Collection and maintained in cell culture medium under the recommended conditions. For cell counting, cells were dissociated by trypsin and stained with 0.04% trypan blue and counted by using heemocytometer.
Design, synthesis and purification of DNA library and primers
The oligonucleotides consisted of a random DNA library containing a segment of randomized sequence of 40 nucleotides (nt) flanked by 19-nt primer hybridization sites at the 5′ and the 3′ ends (5′-ACC TTG GCT GTC GTG TTG T - 40-nt - A GGT CAG TGG TCA GAG CGT -3′), a fluorescein isothiocyanate (FITC)-labeled 5′-forward primer (5′-FITC-ACC TTG GCT GTC GTG TTG T -3′) and a biotinylated 5′-reverse primer (5′-biotin- ACG CTC TGA CCA CTG ACC T -3′). The DNA sequences were synthesized using an ABI3400 DNA/RNA synthesizer (Applied Biosystems) and purified with a ProStar reverse-phase HPLC (Varian) using a C18 column (Econosil, 5U, 250 × 4.6 mm) from Alltech Associates. DNA synthesis reagents were purchased from Glen Research (Sterling, VA). All synthesized sequences were quantified by UV-Vis measurements with a Cary Bio-300 UV spectrometer (Varian).
Flow cytometry and cell sorting
Flow cytometry and cell sorting were performed as previously described44.
SELEX Procedures
Prior to incubation with the target cells, approximately 10nmol of ssDNA library dissolved in 350μL binding buffer (4.5g/L glucose, 5mM MgCl2, 0.1mg/mL yeast tRNA, and 1mg/mL BSA in Dulbecco’s PBS) was denatured by heating at 95°C for 5min and was placed on ice for 10min. This was incubated with the target cells (2×106 for 1st round) in 350μL binding buffer suspension for 1 hour at 4°C. After incubation, the cells were washed three times by centrifugation with washing buffer (4.5g/L glucose, 5mM MgCl2 in Dulbecco’s PBS). The cell-DNA complex was resuspended in 500μL DNAse free water and heated at 95°C for 10min to disrupt the interaction between the DNA and the target on the cell. This was centrifuged at 15,000 rpm to precipitate the cell debris, and the supernatant containing the eluted sequences was recovered.
For the first round of selection, the entire eluted pool was amplified by PCR using the FITC- and biotin-labeled primers. The PCR product was used as a template, and after optimization, preparative PCR was performed to produce more product, which was used to prepare ssDNA. The sense ssDNA strands of the PCR product were separated from the biotinylated antisense ssDNA by alkaline denaturation and affinity purification with streptavidin-coated Sepharose beads. The eluted ssDNA was desalted, quantified and vacuum dried. The ssDNA was resuspended in binding buffer and used to perform the second round of selection. After incubation with target and elution of the binding sequences by heating, the eluted sequences were incubated with the control cells (Ecad−) for 1 hour at 4°C. The unbound sequences were recovered by centrifugation and used to perform PCR.
This procedure was repeated until the 24th round, at which there was significant enrichment for the target cells compared to the control cells. The enriched pools were amplified by PCR with unlabeled primers and subsequently cloned and transformed into Escherichia coli. Positive clones were picked and sequenced at the Genome Sequencing Services Laboratory, University of Florida, to determine the DNA sequences. Based on the sequence analysis and structure prediction, potential aptamer candidates were selected, synthesized, labeled with biotin, and then tested with DU145 cells.
Binding Assays by flow cytometry
Screening potential aptamer candidates and determining their binding affinities were done by flow cytometry. Cells were detached from the culture flask by short-time treatment with 0.05% trypsin in PBS. Dissociated cells were resuspended in culture medium and incubated for 10 hrs on a rocker platform to enable possible regeneration of cell-surface molecules. Cells were then harvested, washed, and resuspended in binding buffer. Then, 100μL of cell suspension containing about 5×105 cells was added to 100μL aptamer solution to a final concentration of 250 nM. After incubation for 30 min at 4°C, the cells were washed and incubated with strepavidin-PE-Cy5.5 or streptavidin-PE conjugate (1:400 optimized dilution) for 10 min at 4°C. The cells were then washed twice with washing buffer, and the binding signal was detected by flow cytometry. The biotin-TD05 (non-binding sequence) was used as a control to set the fluorescence background.
To determine the apparent dissociation constant of the aptamers, parental DU145 cells were used for sequences CSC01, CSC02 and CSC08, while Ecad+ cells were used for CSC13. Cells were incubated with varying concentrations of biotin-aptamer in a 200 μL volume of binding buffer. The cells were washed twice with washing buffer and then incubated with streptavidin-PE. The signal was detected by flow cytometry. Biotin-TD05 was used as a negative control to determine fluorescence background. All binding assays were done in duplicate. The mean fluorescence intensity of the control was subtracted from that of the aptamer with the target cells to determine the specific binding of the labeled aptamer. The equilibrium dissociation constant (Kd) of the aptamer–cell interaction was obtained by fitting the dependence of the intensity of specific binding on the concentration of the aptamers, using the equation . Curve fitting was performed using SigmaPlot (Jandel, San Rafael, CA, USA).
Flow cytometry and FACSorting of aptamer CSC13+ cells
Prostate cancer cells were detached from T150 culture flasks with 0.05% trypsin in PBS and incubated for 10 hrs in fresh medium on a rocker platform to enable possible regeneration of cell-surface molecules. Then, cells were washed, suspended in binding buffer, and incubated separately with CSC13 and non-binding sequence TD05 for 30 mins at 4°C. Cells were washed after incubation and stained with strepavidin-PE-Cy5.5 for 20 mins at 4°C. Cells were resuspended in binding buffer, and PI was added to exclude dead cells. The analysis was done using a FACSCalibur flow cytometer (Becton Dickinson). Live single cells were gated for analysis and sorted (FACSAriaSORP Cell Sorter with Diva 6.1 software, Becton Dickinson). The CSC13 subpopulation (positive and negative) cells were stained with anti-E-cadherin antibody (R&D system), and the activity of aldehyde dehydrogenase-1 (ALDH-1) was assessed. For AldeFluor assay, we used the Aldefluor kit (Stem Cell Technologies), which is designed for optimal identification and isolation of stem cells through specific interaction with human ALDH-1. The experiment followed the manufacturer’s instructions. Briefly, cells obtained from CSC13 sorting by FACS were suspended in ALDFLUOR assay buffer containing ALDH-1 substrate BODIPY aminoacetaldehyde (BAAA, 1μmol/L per 1×106 cells) and incubated for 30 mins at 37°C. As negative control, an aliquot was treated with 50 mmol/L diethylaminobenzaldehyde (DEAB), a specific ALDH-1 inhibitor, for each sample of cells. Using FACScan, the brightly fluorescent ALDH-1-expressing cells were detected in the green fluorescence channel (520–540 nm).
In vivo analysis of enriched cancer stem cells and fluorescent tumor imaging
Tumorigenicity of CSC13+ cells was tested in an immuno-deficient mouse model. Briefly, male C.B-17/IcrHsd-SCID mice were purchased from Harlan Sprague Dawley (Indianapolis, IN). Aptamer CSC13+ and CSC13− DU145 cells were injected subcutaneously into groups of mice (n=5) at a dose of 1×105 cells/mouse. Mice were regularly monitored, and tumor development was observed after four weeks of inoculation. Two positive mice developed tumors, and the tumor size was measured twice a week. Tumor volume was calculated using the formula 0.5ab2, where b is the smaller of the two perpendicular indices. Measurement was stopped when the subcutaneous tumors had reached 15 mm in diameter, and these mice were reserved for whole animal imaging. For tumor imaging, mice were fed with alfalfa free chow (Harlan) for one week before the tumor size endpoint. On the day of imaging, mice were shaved, and 150 μL of 4 μM CSC01-Cy5 conjugate was injected into the tail vein. Mice were then imaged while sedated with isofluorane using a Xenogen IVIS Spectrum Biophotonic Imager (Caliper Life Sciences, Hopkinton, MA). The University of Florida Institutional Animal Care and Use Committee approved all protocols.
Prostate spheroid formation assays
Cancer stem cells under certain growth conditions, such as presence of growth factors and absence of serum, can form spheres. These phenomena were therefore assessed using CSC13 sorted cells. CSC13+ and CSC13− cells were isolated for sorting, and examined the spheroid formation ability. The assay has been previously described44. Briefly, Sorted cells (CSC13+ and CSC13−) from PC3 were grown in suspension culture using ultralow attachment plates (Corning) to generate spheroids. The cells were cultured at a density of 500 cells/ml in serum free DMEM/F12–50/50 supplemented with 20ng/ml EGF (Biosource), 10ng/ml bFGF (Invitrogen), 5μg/ml heparin (Sigma), 2nM Glutamate, 1% penicillin-streptomycin (50 IU penicillin and 50μg/ml streptomycin), 0.2% BSA, 1x B27 without Vitamin A and 1x Insulin-Transferrin-Selenium-A (Gibco). The culture media was changed every 3 to 4 days.
In vitro confocal imaging
DU145 cells were grown on glass-bottom Petri dishes to 80% confluency. Biotinylated aptamers were mixed with streptavidin-Alexa633 in a 1:1 molar ratio. Before imaging, dishes were rinsed with washing buffer and incubated with aptamer-streptavidin solutions for 20 minutes. After a final rinse in washing buffer, cells were imaged using an Olympus FV500-IX81 confocal microscope with 633nm excitation and LP650 nm emission filters.
Statistical analysis
Differences were analyzed using Student’s t-test. p-values of less than 0.05 were considered statistically significant.
Results
Aptamer selection scheme and SELEX progression
In our previous work, E-cad was reported as a surface marker for identifying prostate cancer stem cells in the DU145 and PC3 cell lines 44. The cells expressing high E-cad showed tumorigenic potential, self-renewal ability, and high expression of embryonic stem cell markers (OCT3/4, SOX2, Nanog, c-Myc and Klf4), as well as CD44 and integrin-α2β1, which are reported as surface markers for identifying prostate cancer stem cells. Based on these results, the cell-SELEX methodology was applied to target Ecad+ DU145 cells. To accomplish this, a highly purified E-cad+ cell population was isolated by gating for the top 10% of E-cad+ cells and the bottom 10% E-cad− cells by FACS (Fig. 1a). For each round of selection, approximately 1×106 cells of each subpopulation were sorted. E-cad+ cells were then incubated with the DNA pool, and non-binding DNA was rinsed away. The bound DNA sequences were eluted and incubated with E-cad− cells to subtract DNA sequences binding these control cells. The DNA that bound to E-cad+, but not E-cad−, cells was amplified using PCR and used in the next round of cell-SELEX.
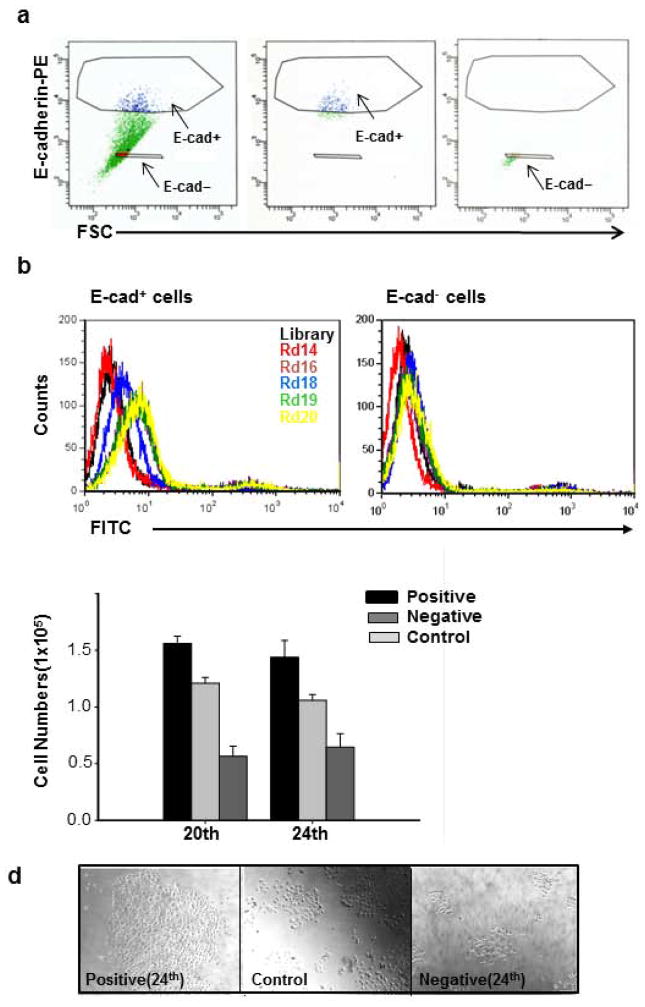
Figure 1.
Cell-SELEX scheme and progression. (a) Flow cytometry dot-plot of E-cadherin-PE staining versus forward scatter signal of DU145 cell line. Gates that select for E-cad+ and E-cad− are shown as polygons (left panel). Unsorted cells are green, E-cad+ are blue, and E-cad− are red. The purity of E-cad+ (middle panel) and E-cad− (right panel) sorted cells was confirmed by post-sorting flow cytometry as shown in the middle and right panels, respectively. (b) Flow cytometry histograms of enriched pools from selection rounds 14 (red), 16, 18 (blue), 19 (green) and 20 (yellow) binding to E-cad+ and E-cad− cells of DU145. A significant increase in intensity is observed for all selection rounds above 15 for E-cad+ cells compared E-cad− cells. The directions of the arrows show increase in fluorescence intensity as selection progressed. (c) DU145 cells positive and negative for the 20th and 24th selection round of enriched pools and unsorted DU145 cells were plated at the same density and allowed to grow for 3 days. The cells positive for the 20th and 24th selection rounds of enriched pools showed higher proliferative capacity compared to negative and unsorted cells. P: positive cells, N: negative cells, C: Control cells. *, P < 0.05 in comparison to negative cells Representative results of two independent experiments are shown. (d) Representaive phase contrast image of cell growth for positive and negative subpopulation for the 24th selection round. Bar = 100 μm.
Starting at the 15th selection round, E-cad+ cells exhibited an observable flow cytometry fluorescence signal when compared to the E-cad− cells. Twenty-four rounds of selection were performed in order ensure the enrichment of the high-affinity aptamers (Fig. 1b). Before determining the exact DNA sequences represented by the DNA-enriched pools, we assessed whether the DNA-enriched pools for selection rounds 20 and 24 were binding to proliferative populations by examining the cell growth. DU145 cells positive and negative for the 20th and 24th enriched pools were sorted by FACS, plated at the same density, and allowed to grow for 3 days. Enriched-pool positive cells showed increased cell growth compared to enriched-pool negative cells and unsorted cells (Fig. 1c and 1d). Since these preliminary data suggested that the enriched pools had targeted a proliferative subpopulation, these pools were cloned and sequenced to identify the specific DNA sequences that were binding to the proliferative cells.
Selectivity and affinity of selected aptamers
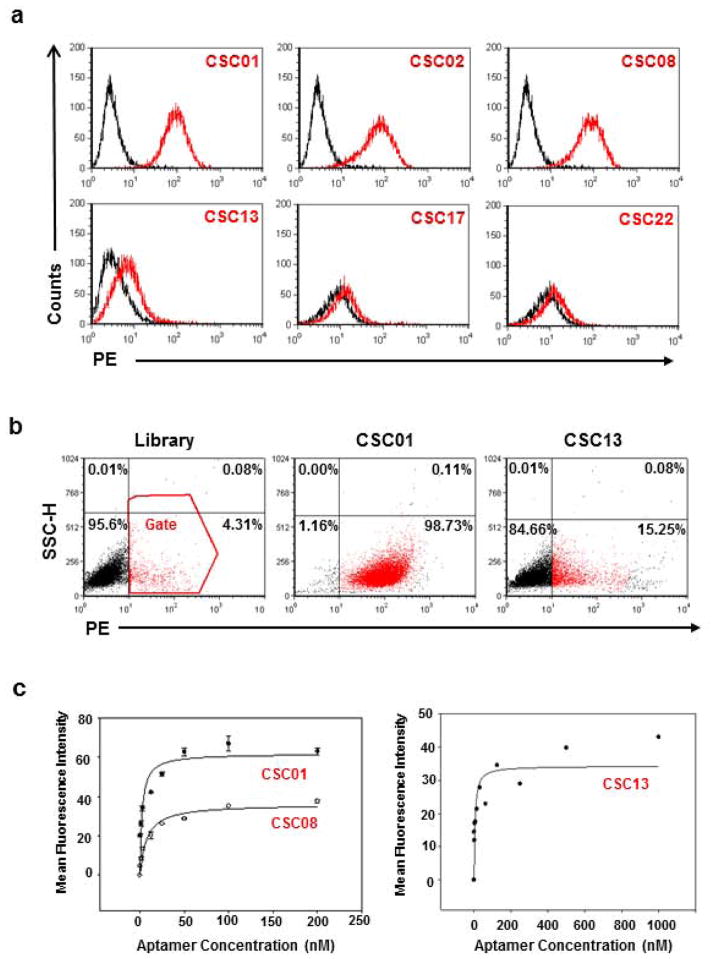
After cloning and sequencing enriched pools for selection rounds 15, 18, 19, 20, 21, and 22, twenty-two putative aptamer sequences were identified and synthesized. These sequences were enumerated as CSC01 through CSC22. Several of these sequences showed consistent binding to all or a part of the DU145 cell line, and the DNA sequences of these aptamers are given in Table 1. A series of flow cytometry-based experiments were performed to confirm that these sequences bound selectively and with high affinity toward E-cad+ DU145 cells. Three aptamers from one family, CSC01, CSC02, and CSC08, bound to over 90% of the DU145 cells, while CSC13, CSC17, and CSC22 bound to less than ~15% of DU145 cells (Fig. 2a and 2b). The side-scatter versus aptamer signal dot-plot analysis is shown in Figure 2b, including the percentage of the cell population of CSC01- and CSC13-positive cells in comparison to the TD05 aptamer background signal. Since TD05 does not bind to DU145 cells, it is the equivalent of an isotype control for these aptamers.
Table 1.
Sequences and Kds of Selected Aptamers for Prostate Cancer Cell line DU145
| Aptamer | Sequence | Kd (nM) |
|---|---|---|
| CSC01 | ACCTTGGCTGTCGTGTTGTAGGTGGTTTGCTGCGGTGGGCTCAAGAAGAAAGCGCAAAGTCAGTGGTCAGAGCGT | 2.2 ± 0.8 |
| CSC02 | ACCTTGGCTGTCGTGTTGTAGTTGGTTTGCTGCGGTGGGCTCAAAAAGAGAGCGCAAAGGTCAGTGGTCAGAGCGT | 7.4 ± 0 2 |
| CSC08 | ACCTTGGCTGTCGTGTTGTGCTCTGAGCCTAGCTTGACCACTTTTCTTTATTCGCTCTGAGGTCAGTGGTCAGAGCGT | 31.3 ± 1.6 |
| CSC13 | ACCTTGGCTGTCGTGTTGTGGGGTGTCGTATCTTTCGTGTCTTATTATTTTCTAGGTGGAGGTCAGTGGTCAGAGCGT | 2.2 ± 1.0 |
| CSC17 | ACCTTGGCTGTCGTGTTGTCACCAGCTCCATACGACACGACCCTCATTCCAACACACAGGTCAGTGGTCAGAGCGT | ND |
| CSC22 | ACCTTGGCTGTCGTGTTGTGTGGGGCTGTGATACTTTACATCTTATTTCTCTAGTGACTAGGTCAGTGGTCAGAGCGT | ND |
Figure 2.
Analysis of potential aptamer candidates in DU145 prostate cancer cell line. (a) Histograms showing initial screening of some potential aptamer candidates. While CSC01, CSC02 and CSC08 bound to ~75% DU145 cells, CSC13, CSC17 and CSC22 bound to less than 20% DU145 cells. (b) Dot-plot showing interaction of aptamer CSC01 and CSC13 with parental DU145 cells. Library (TD05) was used as a control. (c) Using flow cytometry to determine the binding affinity of the FITC-labeled aptamer sequences CSC01, CSC08 and CSC13 to DU145 cells. The nonspecific binding was measured by using FITC-labeled unselected library DNA (TD05). Representative results of three independent experiments are shown.
Aptamers were incubated with various cancer cell lines to determine the selectivity. Although CSC01, CSC02, and CSC08 were not specific to DU145, CSC11 and CSC13 did show some selectivity (Table 2). Dissociation equilibrium constants, Kd, were measured to assess the binding affinity of these aptamers. Kd was measured by titrating the aptamer concentration over 2 to 3 orders of magnitude to generate binding curves and fitting the binding curve data to an equation that represents the ratio of bound receptors to total receptors (see Methods for details). All tested aptamers bound with high affinity, having a Kd < 32 nM. Even though CSC13 bound to a subpopulation of DU145 cells, it bound with high affinity, having a Kd = 2.2 ± 1.0 nM (Fig. 2c).
Table 2.
Selectivity of aptamers to other cell lines
| Cell line/Aptamer | TD05 | CSC01 | CSC02 | CSC08 | CSC11 | CSC13 |
|---|---|---|---|---|---|---|
| Jurkat (leukemia) | − | ++++ | ++ | +++ | − | − |
| Hela (cervical cancer) | − | ++++ | ++++ | ++++ | − | − |
| HCT 116 (colon cancer) | − | ++++ | ++++ | ++++ | − | + |
| HT-29 (colon cancer) | − | ++++ | +++ | ++++ | − | + |
| K562 (leukemia) | − | ++++ | ++++ | ++++ | + | + |
| CEM | − | ++++ | ++++ | ++++ | − | + |
| DU145 at 37°C | − | +++ | ++ | +++ | − | + |
A threshold based on fluorescence intensity of FITC in the flow cytometry analysis was chosen so that 95% of cells incubated with the FITC labeled unselected DNA library would have fluorescence intensity below it. After binding with FITC-labeled aptamer, the percentage of the cells with fluorescence above the set threshold was used to evaluate the binding capacity of the aptamer to the cells: <10% +, 10–35%; ++, 35–60%; +++, 60–85%; ++++, >85%. The final concentration of aptamer in binding buffer is 100 nM.
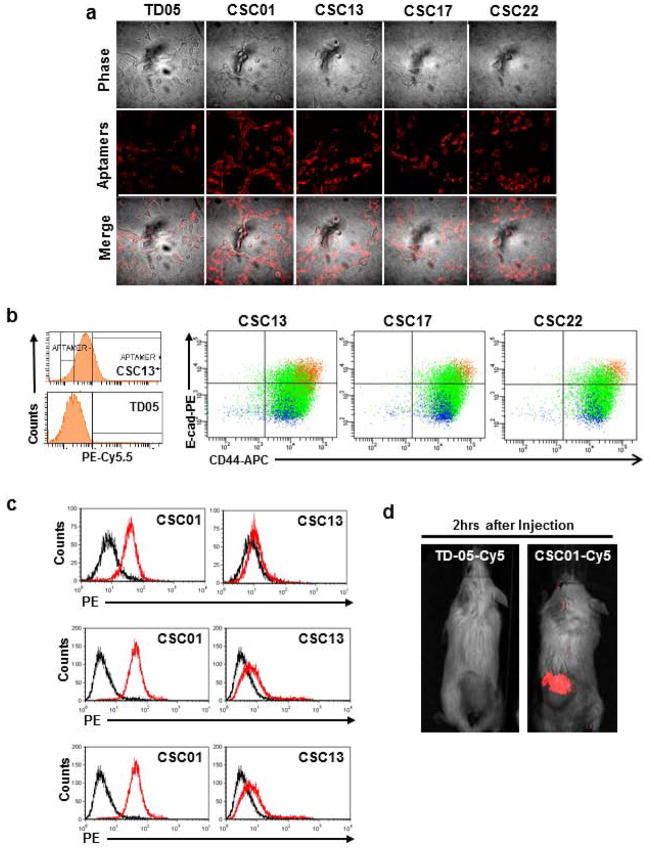
For these aptamers to be useful in vivo, they must bind to the cells at 37°C; therefore, the aptamers were incubated with DU145 cells at 37°C before flow cytometric analysis (Table 2). This analysis showed that CSC01 bound to ~75% of DU145 cells at 37°C and that CSC13 still bound to ~10% of DU145 cells at 37°C (Table 2). Since protein expression profiles can change after releasing cells from their substrate, the binding of these aptamers to live adherent monolayers was also tested using confocal microscopy. Confocal imaging showed bright staining of CSC01 on the surface of DU145 cells (Fig. 3a). CSC13, CSC17, and CSC22 showed increased staining compared to the TD05 non-binding aptamer, and only a subset of cells had bright staining.
Figure 3.
Imaging of aptamer binding to live cell or tumor and immunophenotype of aptamer-positive cells. (a) In vitro confocal imaging of live cells with CSC01, CSC13, CSC17 and CSC22. Monolayer DU145 cells were stained with these aptamers. The staining was visible after 20 mins incubation. (b) Parental DU145 cells were stained with E-cadherin-PE, CD44-APC and aptamers (CSC13, CSC17, or CSC22-PE-Cy5.5). Less than 20% of CSC13+ cells were detected compared to TD05 library control (histogram in left panel). CSC13+, CSC17+ or CSC22+ cells were overlaid with E-cadherin and CD44 double-staining in DU145. Orange color represents aptamer-positive cells, and blue color represents aptamer-negative cells in DU145. (c) Histograms showing the binding pattern of CSC01 and CSC13 in PC3, LNCaP and VCaP. (d) In vivo whole animal imaging of tumor-burdened mice. Either TD05-Cy5 or CSC01-Cy5 was injected into the tail-vein of mice with tumor formed by DU145 cells, and live whole animal imaging was performed. The fluorescent signal above the non-binding aptamer background signal was observed within 2 hours after injection.
We next performed in vivo binding assays to show that CSC01 could be useful for such in vivo applications as targeted drug delivery. CSC01 and TD05 were synthesized with a Cy5 dye conjugated to the 5′ end. Near infrared fluorescent dyes, like Cy5, are useful for in vivo imaging, since tissue autofluorescence is reduced in the near-infrared, and near-infrared light has a better tissue penetration than visible light. Equal doses of CSC01-Cy5 and TD05-Cy5 were injected into the tail-vein of different DU145 tumor-bearing mice, and live whole animal imaging was performed. A fluorescent signal above the TD05-Cy5 aptamer background signal was observed within 2 hours after injection for all mice that had been injected with CSC01-Cy5 (Fig. 3d). These experiments show that our CSC aptamer panel binds selectively and with high affinity to DU145 cells and therefore has the potential for in vivo applications.
Immunophenotype of aptamer-positive cells
Using multi-parameter flow cytometry, we further analyzed the expression level of E-cad and CD44 in the three aptamers that bound to the small subpopulation of DU145 cells: CSC13, CSC17, and CSC22. In these assays, anti-E-cad, anti-CD44, and one of the three aptamers were incubated with cells before flow cytometry analysis. Less than 20% of DU145 cells bound to these aptamers compared to the TD05 aptamer control. Most importantly, the aptamer-positive cells also had high expression of E-cad and CD44 (Fig. 3c). Since our previous work demonstrated that the E-cad+ subpopulation of DU145 cells has stem cell-like characteristics, these flow cytometry data provide evidence that our cell-SELEX scheme has successfully targeted tumor-initiating cells. To test the specificity of these aptamers to other prostate cancer cells, either CSC01 or CSC13 was incubated with PC3, LNCaP and VCaP cell lines. CSC01 and CSC13 had binding patterns similar to PC3, LNCaP and VCaP compared to DU145, with CSC01 binding to ~95% of these cell lines and CSC13 binding to less than 15% of PC3, LNCaP and VCaP cells (Fig. 3c), similar to what was observed in DU145. Since the CSC13 aptamer bound to cells having immunophenotype similar to CSC, as reported in the literature, we assessed the potential of CSC13 to isolate prostate CSC. Based on these findings, CSC01, CSC02, or CSC08 may be useful for prostate cancer detection, while CSC13, CSC17, or CSC22 could be potential probes for detecting prostate cancer stem cells.
In vitro and in vivo characterization of CSC13+ cells
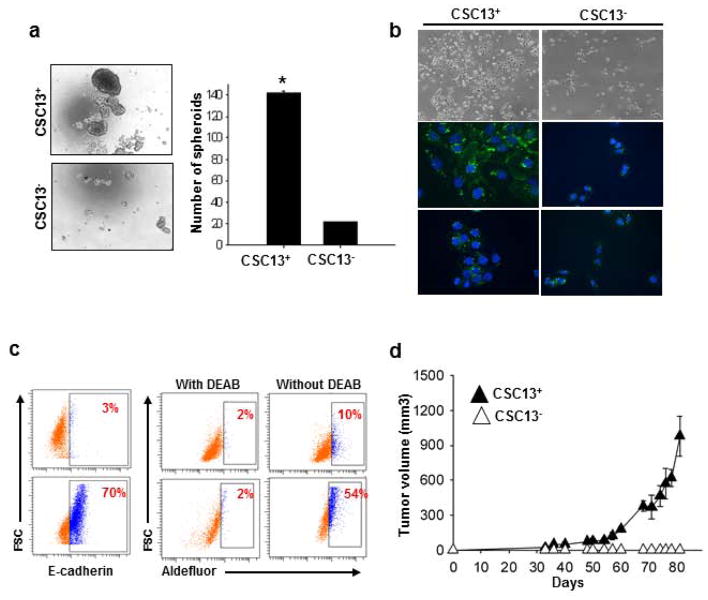
CSC13+ cell populations also expressed high levels of E-cad and CD44, both of which are markers for the CSC population in this cell line. Therefore, it was determined whether CSC13+ cells have cancer stem cell characteristics. One key determinant of stem cells is the capacity for self-renewal. To determine whether CSC13+ cells have high self-renewal capability, CSC13+ and CSC13− DU145 cells were sorted and cultured in prostasphere under non-adherent culture conditions, as previously established for the isolation of prostate cancer stem cells 45. As shown in Figure 4a, CSC13+ DU145 cells showed significantly higher sphere-forming efficiency. Furthermore, sorted CSC13+ and CSC13− cells were cultured for 3 days and immunostained with anti-E-cadherin. As shown Figure 4b, CSC13+ cells produced E-cad+ and E-cad− cells, suggesting that asymmetrical division had occurred. In contrast, CSC13− DU145 cells proliferated slowly and remained negative for E-cadherin. It was further confirmed that sorted CSC13+ and CSC13− cells were equal to E-cad+ and E-cad− cells, respectively, by flow cytometric analysis (Fig. 4c).
Figure 4.
In vitroand in vivo characterization of CSC13+ subpopulation in DU145 cells. Representative results of three independent experiments are shown until otherwise stated: (a) In vitro quantification of prostate cell spheroids formed by CSC13+ and CSC13− cells. The data are represented by the percentage of spheroids formed per 500 seeded cells ± SD. *, P < 0.05 in comparison to CSC13− cells. (b) Immunostaining of CSC13+ (top two panels) and CSC13− (bottom two panels) DU145 cells with E-cadherin antibodies over a 3-day differentiation period. Magnification is 40X for fluorescent cells. The scale bar is 20 μm for fluorescent cells. (c) CSC13+ and CSC13− cells of DU145 were sorted and then stained with E-cadherin-PE antibody; CSC13+ cells expressed high E-cadherin (left panel). CSC13 sorted cells were incubated with Aldefluor in the absence or presence of DEAB, and CSC13+ cells in DU145 also showed high ALDH activity compared to CSC13− cells (right panel). (d) Evaluation of the tumorigenicity of CSC13+ and CSC13− in DU145 cells. CSC13+ and CSC13− DU145 cells (1×105) were sorted by flow cytometry and injected subcutaneously into the backs of mice (5/group); tumors were measured with calipers weekly for the first 3 weeks and then twice a week thereafter. Tumor volume data are reported as the mean ± SD from the five mice that developed tumors. Representative results of two independent experiments are shown.
Recently, aldehyde dehydrogenase 1 (ALDH1) was reported as a marker for malignant colon, prostate, head and neck squamous, lung, and breast cancer stem cells 46–50. Therefore, we analyzed the expression level of ALDH1 between the CSC13+ and CSC13− populations. For the ALDH1 activity assay, CSC13+ DU145 cells were incubated with and without diethylaminobenzaldehyde (DEAB), an ALDH1 inhibitor, followed by the Aldefluor fluorescent assay reagent. ALDH activity correlates with increased fluorescence caused by the cleavage of the Aldefluor reagent. Fifty-four percent of CSC13+ cells were positive for ALDH1 activity compared to only 10% of CSC13− cells (Fig. 4c).
The gold standard in determining CSC is whether the tested cells can preferentially initiate tumor development in animal models. After FACs sorting of CSC13+ and CSC13− DU145 cells, 1×105 cells in PBS were injected subcutaneously into the backs of 5 SCID mice, respectively, and they were observed for tumor development and growth over a period of 4 months (Fig. 4d). Four and a half weeks later, CSC13+ cells yielded tumors with an average volume of 23.0±1.4 mm3 in two mice out of five, whereas CSC13− cells did not produce tumors in any mouse (Fig. 4d). In our second test, CSC13+ cells made tumors in 4 out of five mice, whereas CSC13- cells produced a small tumor mass in only 1 of 5 mice. Therefore, the CSC13+ cells showed higher tumorigenicity compared with the CSC13− cells, implying that the CSC13+ subpopulation could have more tumorigenic property compared with the CSC13− subpopulation. It should be noted that the variation between two independent experiments might be caused by sorting conditions of individual experiments. That is, the formation of one small tumor by CSC13− cells in the second tumorigenicity assay could have resulted from the low efficiency of cell sorting. Also, since aptamer sorting was performed for the first time in this study, optimum conditions should be examined by further study.
Discussion
A deeper molecular level understanding of cancer stem cells would be useful in providing greater insight into the origin of prostate cancer and new therapeutics for prostate cancer. Although it is postulated that cancer stem cells maintain tumor growth, the mechanisms underlying the spread of these cancer stem cells, leading to a distinct metastatic pattern, remain to be elucidated. Although CSC are rare, they are thought to be essential for tumor regrowth and potentially resistant to therapies, making the elimination of CSC essential for long-term success in cancer treatment 4. Therefore, finding molecular probes capable of identifying CSC with specificity is an important challenge. Since previous work44 has shown that DU145 cells expressing E-cad exhibited stem cell-like characteristics, we applied the cell-SELEX methodology to target this CSC population of prostate cancer cells.
In this work, we have shown that the cell-SELEX strategy is powerful enough to produce aptamers which could differentiate between subpopulations of the same cell line; however, the sequencing results were not as simple as our previous cell-SELEX experiments. Typically, 4 to 8 different families of 95% homologous sequences are repeated 20 to 40 times out of 300 clones. However, in this case, there was only one dominant family, with the next highest frequency of 95% homologous sequences repeated between 3 and 6 times out of 300 clones. To circumvent this problem, a total of 1200 clones from rounds 15, 18, 19, 20, 21, and 22 were sequenced. These sequences were then aligned using Sequencher software looking for 70% homology with a 30-base overlap. Following this analysis, 22 putative sequences were selected for further analysis. The difficulty of ascertaining putative aptamer sequences in this system is presumably a reflection of the relative similarity of cell-surface markers between E-cad+ and E-cad− cells. In most of our previous selections involving two cancer cell lines from different tissue origins, about 16 total rounds of selection were required. Even though the enrichment observed up to the twenty-fourth round was only marginal, some aptamers did develop from these pools, such as CSC01, CSC02, and CSC08, and bound to >95% of the cells with high fluorescence intensity. These aptamers may be useful for prostate cancer detection, since they have significant recognition to the four prostate cancer cell lines tested. In addition, since CSC01 bound to live tumors in a xenograft model, these aptamers could be useful for in vivo imaging studies or targeted drug delivery.
Out of the 19 putative aptamer sequences that did not bind the majority of DU145 cells, 5 sequences showed reproducible binding to <15% of the DU145 cell line. Although aptamers CSC11, CSC13, CSC17, CSC21, and CSC22 all showed promising results, we chose to study CSC13 further since it had the highest signal strength and the most reproducible pattern of binding to DU145 cells. To assess the stemness of the cells that were positive for CSC13, we showed that these cells, which also expressed markers for prostate CSC, i.e., CD44, E-cadhigh, and ALDH1, grew as spheres under non-adherent culture conditions and initiated tumors in immunodeficient mice. Based on this evidence, these CSC aptamers may have utility in aptamer-targeted therapeutic platforms, such as siRNA or drug delivery, to generate therapeutics that target CSC in prostate tumors. In the future, the identification of the cell-surface molecular ligands for these aptamers will aid in the biochemical understanding of prostate cancer.
References
- 1.Geiger TR, Peeper DS. Metastasis mechanisms. Biochimica Et Biophysica Acta-Reviews on Cancer. 2009;1796:293–308. doi: 10.1016/j.bbcan.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CHM, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer Stem Cells—Perspectives on Current Status and Future Directions: AACR Workshop on Cancer Stem Cells. Cancer Research. 2006;66:9339–44. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 4.Wicha MS, Liu S, Dontu G. Cancer Stem Cells: An Old Idea—A Paradigm Shift 10.1158/0008-5472. CAN-05-3153. Cancer Research. 2006;66:1883–90. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 5.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–8. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 6.Rosen JM, Jordan CT. The Increasing Complexity of the Cancer Stem Cell Paradigm. Science. 2009;324:1670–3. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirschmann-Jax C, Foster AE, Wulf GG, Goodell MA, Brenner MK. A distinct “side population” of cells in human tumor cells - Implications for tumor biology and therapy. Cell Cycle. 2005;4:203–5. [PubMed] [Google Scholar]
- 8.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 9.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44(+) breast cancer-initiating cells to radiation. Journal of the National Cancer Institute. 2006;98:1777–85. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 10.Li XX, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang XM, Chamness GC, Wong H, Rosen J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. Journal of the National Cancer Institute. 2008;100:672–9. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 11.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature Medicine. 1997;3:730–7. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 12.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 13.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shipitsin M, Campbell LL, Argani P, Werernowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, Sukumar S, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–73. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 15.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 16.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 17.Woodward WA, Sulman EP. Cancer stem cells: markers or biomarkers? Cancer and Metastasis Reviews. 2008;27:459–70. doi: 10.1007/s10555-008-9130-2. [DOI] [PubMed] [Google Scholar]
- 18.Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment - Rna Ligands to Bacteriophage-T4 DNA-Polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 19.Ellington AD, Szostak JW. Invitro Selection of Rna Molecules That Bind Specific Ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 20.Shangguan D, Li Y, Tang ZW, Cao ZHC, Chen HW, Mallikaratchy P, Sefah K, Yang CYJ, Tan WH. Aptamers evolved from live cells as effective molecular probes for cancer study. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:11838–43. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sefah K, Shangguan D, Xiong XL, O’Donoghue MB, Tan WH. Development of DNA aptamers using Cell-SELEX. Nature Protocols. 5:1169–85. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 22.Shangguan DH, Meng L, Cao ZHC, Xiao ZY, Fang XH, Li Y, Cardona D, Witek RP, Liu C, Tan WH. Identification of liver cancer-specific aptamers using whole live cells. Analytical Chemistry. 2008;80:721–8. doi: 10.1021/ac701962v. [DOI] [PubMed] [Google Scholar]
- 23.Tang ZW, Shangguan D, Wang KM, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan WH. Selection of aptamers for molecular recognition and characterization of cancer cells. Analytical Chemistry. 2007;79:4900–7. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 24.Chen HW, Medley CD, Sefah K, Shangguan D, Tang ZW, Meng L, Smith JE, Tan WH. Molecular recognition of small-cell lung cancer cells using aptamers. Chem med chem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sefah K, Tang ZW, Shangguan DH, Chen H, Lopez-Colon D, Li Y, Parekh P, Martin J, Meng L, Phillips JA, Kim YM, Tan WH. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia. 2009;23:235–44. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sefah K, Meng L, Lopez-Colon D, Jimenez E, Liu C, Tan WH. DNA Aptamers as Molecular Probes for Colorectal Cancer Study. Plos One. 2010:5. doi: 10.1371/journal.pone.0014269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Simaeys D, Lopez-Colon D, Sefah K, Sutphen R, Jimenez E, Tan WH. Study of the Molecular Recognition of Aptamers Selected through Ovarian Cancer Cell-SELEX. Plos One. :5. doi: 10.1371/journal.pone.0013770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang ZW, Parekh P, Turner P, Moyer RW, Tan WH. Generating Aptamers for Recognition of Virus-Infected Cells. Clinical Chemistry. 2009;55:813–22. doi: 10.1373/clinchem.2008.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shangguan DH, Cao ZHC, Li Y, Tan WH. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clinical Chemistry. 2007;53:1153–5. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 30.Shi H, Tang ZW, Kim Y, Nie HL, Huang YF, He XX, Deng K, Wang KM, Tan WH. In vivo Fluorescence Imaging of Tumors using Molecular Aptamers Generated by Cell-SELEX. Chemistry-an Asian Journal. 5:2209–13. doi: 10.1002/asia.201000242. [DOI] [PubMed] [Google Scholar]
- 31.Mallikaratchy P, Tang ZW, Kwame S, Meng L, Shangguan DH, Tan WH. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt’s lymphoma cells. Molecular & Cellular Proteomics. 2007;6:2230–8. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Shangguan D, Cao ZH, Meng L, Mallikaratchy P, Sefah K, Wang H, Li Y, Tan WH. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. Journal of Proteome Research. 2008;7:2133–9. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jemal A, Siegel R, Xu JQ, Ward E. Cancer Statistics, 2010. Ca-a Cancer Journal for Clinicians. 60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 34.Coleman RE. Clinical features of metastatic bone disease and risk of skeletal morbidity. Clinical Cancer Research. 2006;12:6243S–9S. doi: 10.1158/1078-0432.CCR-06-0931. [DOI] [PubMed] [Google Scholar]
- 35.Rassweiler J, Schulze M, Teber D, Marrero R, Seemann O, Rumpelt J, Frede T. Laparoscopic radical prostatectomy with the Heilbronn technique: Oncological results in the first 500 patients. Journal of Urology. 2005;173:761–4. doi: 10.1097/01.ju.0000153486.94741.e5. [DOI] [PubMed] [Google Scholar]
- 36.Schroeder FH, Hugosson J, Roobol MJ, Tammela TLJ, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, Denis LJ, Recker F, et al. Screening and Prostate-Cancer Mortality in a Randomized European Study. New England Journal of Medicine. 2009;360:1320–8. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 37.Andriole GL, Grubb RL, Buys SS, Chia D, Church TR, Fouad MN, Gelmann EP, Kvale PA, Reding DJ, Weissfeld JL, Yokochi LA, Crawford ED, et al. Mortality Results from a Randomized Prostate-Cancer Screening Trial. New England Journal of Medicine. 2009;360:1310–9. doi: 10.1056/NEJMoa0810696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang ZA, Shen MM. Revisiting the concept of cancer stem cells in prostate cancer. Oncogene. 2010 doi: 10.1038/onc.2010.530. [DOI] [PubMed] [Google Scholar]
- 39.Dubrovska A, Kim S, Salamone RJ, Walker JR, Maira SM, Garcia-Echeverria C, Schultz PG, Reddy VA. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:268–73. doi: 10.1073/pnas.0810956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG. Hierarchical Organization of Prostate Cancer Cells in Xenograft Tumors: The CD44+α2β1+ Cell Population Is Enriched in Tumor-Initiating Cells. Cancer Research. 2007;67:6796–805. doi: 10.1158/0008-5472.CAN-07-0490. [DOI] [PubMed] [Google Scholar]
- 41.Wei C, Wang GM, Liu YJ, Qian RZ. Cancer stem-like cells in human prostate carcinoma cells DU145 - The seeds of the cell line? Cancer Biology & Therapy. 2007;6:763–8. doi: 10.4161/cbt.6.5.3996. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Chen X, Calhoun-Davis T, Claypool K, Tang DG. PC3 human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Research. 2008;68:1820–5. doi: 10.1158/0008-5472.CAN-07-5878. [DOI] [PubMed] [Google Scholar]
- 43.Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, Coghlan L, Tang DG. Highly purified CD44(+) prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 44.Bae KM, Su Z, Frye C, McClellan S, Allan RW, Andrejewski JT, Kelley V, Jorgensen M, Steindler DA, Vieweg J, Siemann DW. Expression of Pluripotent Stem Cell Reprogramming Factors by Prostate Tumor Initiating Cells. Journal of Urology. 2010;183:2045–53. doi: 10.1016/j.juro.2009.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi XD, Gipp J, Bushman W. Anchorage-independent culture maintains prostate stem cells. Developmental Biology. 2007;312:396–406. doi: 10.1016/j.ydbio.2007.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu SL, Schott A, Hayes D, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde Dehydrogenase 1 Is a Marker for Normal and Malignant Human Colonic Stem Cells (SC) and Tracks SC Overpopulation during Colon Tumorigenesis. Cancer Research. 2009;69:3382–9. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen YC, Chen YW, Hsu HS, Tseng LM, Huang PI, Lu KH, Chen DT, Tai LK, Yung MC, Chang SC, Ku HH, Chiou SH, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochemical and Biophysical Research Communications. 2009;385:307–13. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 49.Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing LX, Wang HJ, Liu ZQ, Su Y, Stass SA, Katz RL. Aldehyde Dehydrogenase 1 Is a Tumor Stem Cell-Associated Marker in Lung Cancer. Molecular Cancer Research. 2009;7:330–8. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burger PE, Gupta R, Xiong X, Ontiveros CS, Salm SN, Moscatelli D, Wilson EL. High Aldehyde Dehydrogenase Activity: A Novel Functional Marker of Murine Prostate Stem/Progenitor Cells. Stem Cells. 2009;27:2220–8. doi: 10.1002/stem.135. [DOI] [PMC free article] [PubMed] [Google Scholar]