Abstract
Myopia and hyperopia are at opposite ends of the continuum of refraction, the measure of the eye’s ability to focus light, which is an important cause of visual impairment (when aberrant) and is a highly heritable trait. We conducted a genome-wide association study for refractive error in 4,270 individuals from the TwinsUK cohort. We identified SNPs on 15q25 associated with refractive error (rs8027411, P = 7.91 × 10−8). We replicated this association in six adult cohorts of European ancestry with a combined 13,414 individuals (combined P = 2.07 × 10−9). This locus overlaps the transcription initiation site of RASGRF1, which is highly expressed in neurons and retina and has previously been implicated in retinal function and memory consolidation. Rasgrf1−/− mice show a heavier average crystalline lens (P = 0.001). The identification of a susceptibility locus for refractive error on 15q25 will be important in characterizing the molecular mechanism responsible for the most common cause of visual impairment.
Myopia is the most common eye disorder. It is part of the spectrum of refractive error, measured as spherical equivalent, which is the quantitative strength of spectacle lens required to enable images to be focused on the retina of the eye. Clinically relevant myopia or nearsightedness (more severe than −1 diopter) affects 33% of the adult population in the United States1. Its prevalence is increasing in more recent birth cohorts across both developed and less developed countries, particularly in east Asia, where over 80% of Chinese young adults in Singapore and Taiwan are now affected2,3. Apart from the huge economic cost (estimated at $5.5 billion annually spent by adults in the United States alone4), severe refractive error is a important cause of blindness5-7, particularly in the working-age population8. Vision loss is greatest in the 2%–3% of the population with ocular structural changes associated with high myopia (termed ‘pathological myopia’). The World Health Organization has identified the prevention and treatment of myopia as a priority within the current global initiative to eliminate avoidable blindness7. There are currently no practical therapies to prevent the progression of myopia, even though the condition is easily diagnosed in affected children and adolescents.
The mechanism for the development of myopia is not fully understood. Although environment (in particular, education, urbanization, outdoor activity and close work) clearly plays a role in myopia9,10, numerous cross-sectional studies suggest that genetic heritability might be as high as 80%11. No individual genes have been consistently associated with myopia to date, although several candidate loci have been identified through linkage studies.
In order to further explore potential genetic influences of refractive error development in the general population, we conducted a genome-wide association study (GWAS) using the British TwinsUK adult twin registry cohort (see Online Methods for a detailed description of the population and clinical characteristics).
We analyzed a set of 4,270 individuals with complete information on a panel of 2.2 million SNPs, of which 286,481 directly genotyped in all individuals. All of these SNPs passed stringent quality control criteria (Hardy-Weinberg equilibrium P > 0.001, minor allele frequency of at least 0.04 and a genotyping success rate for the SNP of at least 95%). Homogeneity of the population was confirmed by comparison of sample set similarities with each other and with three reference panels from the HapMap Phase 2 data (Supplementary Fig. 1).
The genomic inflation factor (λ) was 1.04 (Supplementary Fig. 2). Multiple SNPs, both directly genotyped and with imputed values, showed genome-wide significance. Association was maximal for rs8027411 (P = 7.91 × 10−8), although association was consistently high across a region of approximately 120 kb on chromosome 15 (Fig. 1 and Table 1). This association was consistent with an additive model, and we found no evidence of dominance for any of these SNPs. In the TwinsUK cohort, each additional copy of the susceptibility allele for the top SNP lowered the refraction by an average of 0.35 diopters (Supplementary Table 1). Individuals carrying a genotype homozygous for the T allele on the rs8027411 locus were significantly more likely than those homozygous for the non-susceptibility allele to have myopia than to have no refractive error (odds ratio (OR) = 1.16, 95% CI 1.02–1.28, P = 0.02; Fig. 2, see Online Methods for definitions).
Figure 1.

Association scatter plot for SNPs in the RASGRF1 promoter region in the TwinsUK discovery cohort. Negative logarithms of the P values for the association of each SNP with spherical equivalence are plotted. The lead SNP is plotted in blue, with the meta-analysis P value for that SNP indicated. Genotyped SNPs are plotted as diamonds and imputed SNPs as circles, with the color indicating the degree of pairwise LD between the lead and neighboring SNPs. Red indicates strong pairwise LD, with r2 ≥ 0.8; orange indicates moderate LD, with 0.5 < r2 < 0.8 yellow indicates weak LD, with 0.2 < r2 < 0.5; and white indicates no LD, with r2 < 0.2. The associated SNPs lie in a 120-kb region of high LD, delineated by two local high-recombination spots at approximately 77,150 kb and 77,270 kb. The recombination rates are shown as light blue line and the locations of the genes with respect to these SNPs are shown as dark green arrows.
Table 1.
Selection of SNPs associated with spherical equivalent measures in six cohorts of European ancestry at 15q25
| SNP | Allele | Discovery Cohort
|
Replication Cohorts
|
Meta-analysis
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TwinsUK
|
RS-I
|
RS-II
|
RS-III
|
ERF
|
1958BC
|
Australian Twins
|
Combined
|
||||||||||||||||||
| Effect | s.e.m. | P | Effect | s.e.m. | P | Effect | s.e.m. | P | Effect | s.e.m. | P | Effect | s.e.m. | P | Effect | s.e.m. | P | Effect | s.e.m. | P | Effect | s.e.m. | P | ||
| rs17175798 | C | −0.34 | 0.07 | 1.38 × 10−7 | –0.13 | 0.05 | 0.01 | –0.02 | 0.08 | 0.9 | –0.30 | 0.08 | 0.000 | –0.16 | 0.07 | 0.02 | –0.19 | 0.19 | 0.04 | 0.06 | 0.08 | 0.4 | –0.15 | 0.026 | 1.99 × 10−9 |
| rs8027411 | T | −0.35 | 0.07 | 7.91 × 10−8 | –0.12 | 0.05 | 0.01 | –0.02 | 0.08 | 0.8 | –0.29 | 0.08 | 0.001 | –0.16 | 0.07 | 0.02 | –0.18 | 0.18 | 0.05 | 0.06 | 0.08 | 0.4 | –0.15 | 0.026 | 2.07 × 10−9 |
| rs8033963 | T | −0.34 | 0.07 | 1.59 × 10−7 | –0.12 | 0.05 | 0.01 | –0.02 | 0.08 | 0.8 | –0.29 | 0.08 | 0.001 | –0.16 | 0.07 | 0.01 | –0.18 | 0.18 | 0.05 | 0.04 | 0.08 | 0.5 | –0.15 | 0.026 | 1.86 × 10−9 |
| rs939658 | G | −0.34 | 0.07 | 2.09 × 10−7 | –0.12 | 0.05 | 0.01 | –0.02 | 0.08 | 0.8 | –0.29 | 0.08 | 0.001 | –0.17 | 0.07 | 0.01 | –0.18 | 0.18 | 0.05 | 0.04 | 0.08 | 0.6 | –0.15 | 0.026 | 1.85 × 10−9 |
| rs939661 | A | −0.33 | 0.07 | 2.73 × 10−7 | –0.11 | 0.05 | 0.01 | –0.01 | 0.08 | 0.9 | –0.28 | 0.08 | 0.001 | –0.17 | 0.07 | 0.01 | –0.13 | 0.13 | 0.16 | –0.01 | 0.08 | 0.9 | –0.15 | 0.026 | 3.85 × 10−9 |
TwinsUK, the Twin Cohort recruited in the UK that served as a discovery GWAS (N = 4,270); RS−I, the first Rotterdam Study cohort (N = 5,328); RS−II, the second Rotterdam Study cohort (N = 2,157); RS−III, the third Rotterdam Study cohort (N = 2,082); ERF, the Erasmus Rucphen Family (N = 2,032); 1958BC, the 1958 British Birth Cohort (N = 1,000); Aus, the Australian Twin Study (N = 1,086). The combined column includes results from the meta-analysis of the discovery and replication cohorts. Effect, β coefficient of linear regression. The effect sizes denote changes in diopters per each additional copy of the reference allele.
Figure 2.
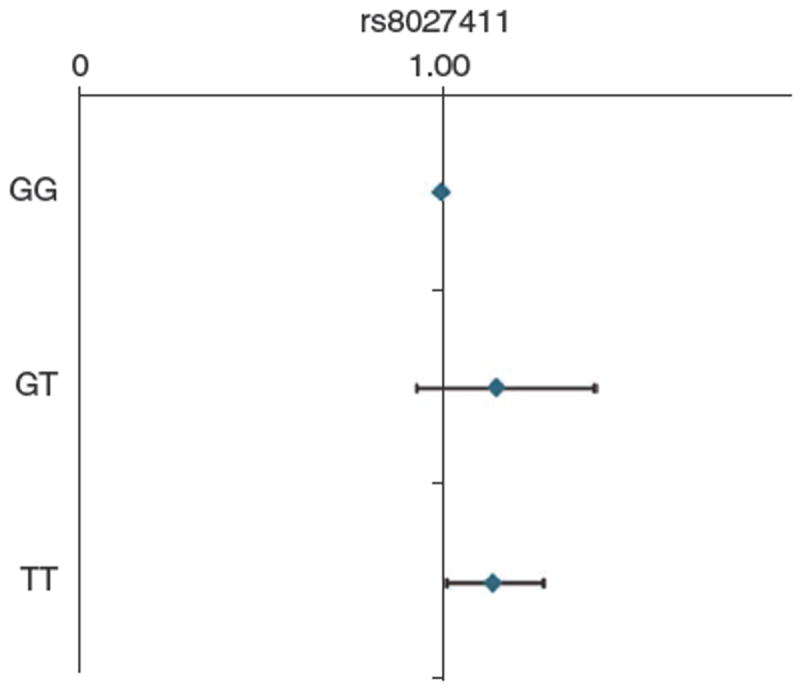
Association of rs8027411 with clinical myopia in the TwinsUK cohort. The values depicted by the diamonds represent ORs. Error bars, 95% CI.
We performed a replication study in 13,414 adults from six independent population-based cohorts of European ancestry: four Dutch cohorts (three within the Rotterdam Study and the Erasmus Rucphen Family Study (ERF), a family-based study in a genetically isolated population in the southwest of The Netherlands), one British cohort (1958 British Birth Cohort) and one Australian cohort (Australian Twin Study). The replication analysis was performed using both observed and imputed genotypes. We observed an association for SNPs at chromosome 15q25 in the Dutch and British replication studies. Although the effect size was higher in the discovery panel (Fig. 3), the effect sizes and directions of the effect were consistent among all the cohorts, with the exception of the Australian cohort. The strongest evidence of association in the meta-analysis was found for rs939658 and rs8027411 (combined P = 1.85 × 10−9 and P = 2.07 × 10−9, respectively), with extra each copy of the myopia susceptibility allele (G and T for the two SNPs, respectively) altering the spherical equivalent by 0.15 diopters (s.e.m. = 0.026) (Table 1). It is of note that the Australian twin group was relatively underpowered (1,086 individuals from 525 families) and substantially younger than the other cohorts. It has been shown that there is evidence of a rapidly increasing prevalence of myopia and, therefore, a cohort effect is possible12.
Figure 3.
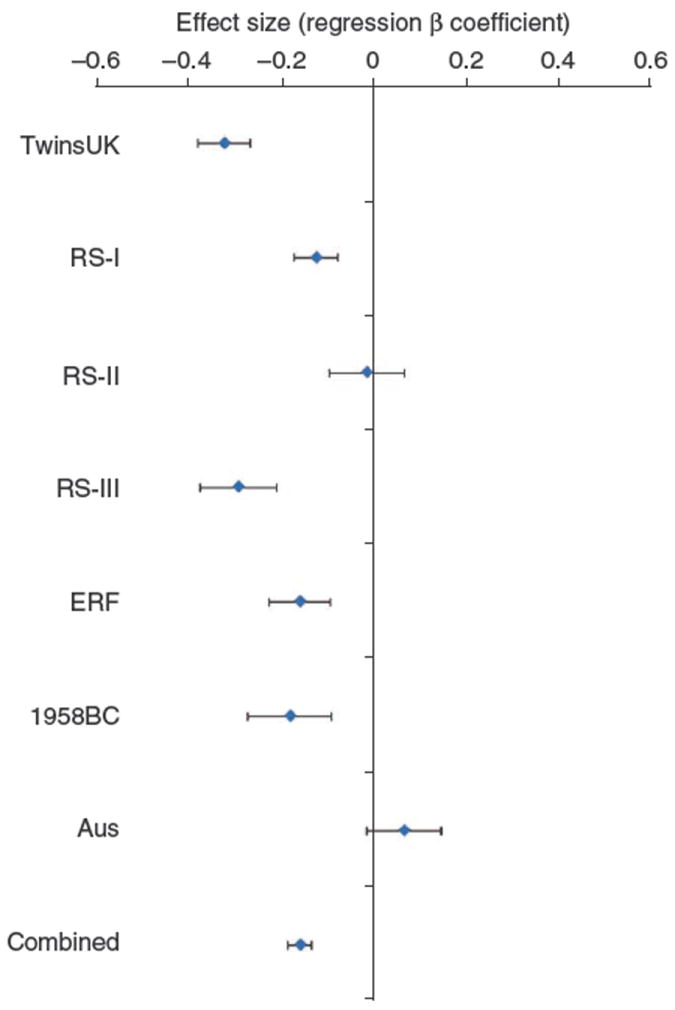
Plot of the effect on refractive error observed for rs8027411 in the seven population panels participating in the study. For each population, the diamond square shows the β linear regression coefficient or the average difference in diopters for each additional copy of the T allele and the bars represent the standard error for the estimate. TwinsUK, the Twin Cohort recruited in London, UK; RS-I, the first Rotterdam Study cohort; RS-II, the second Rotterdam Study cohort; RS-III, the third Rotterdam Study cohort; ERF, the Erasmus Rucphen Family; 1958BC the 1958 British Birth Cohort; Aus, the Australian Twin Study.
All of the identified SNPs associated with the spherical equivalent measures were in high linkage disequilibrium (LD) with one another (Supplementary Fig. 3). This associated region spans about 120 kb on chromosome 15q25 and overlaps the transcription initiation site of RASGRF1 (encoding Ras protein-specific guanine nucleotide-releasing factor 1). This is a large gene spanning 130 kb with 28 known exons and multiple alternative mRNA transcripts. Possible gains or losses in transcription factor–binding sites, because of the DNA polymorphisms within the region of association, are predicted in silico (Supplementary Table 2). The relevance of individual SNPs that potentially influence transcription factor binding and expression of this gene is difficult to assess due to the high LD across the region.
RASGRF1 encodes a protein that is expressed at high levels in neurons and retina in mice13 and that activates Ras14. Our reverse transcription experiments identified high levels of expression of this protein in human retinas (data not shown). In mice, the protein is heavily imprinted in the early developmental phases15,16, but this imprinting fades with time16. RASGRF1 expression is upregulated by stimulation of muscarinic receptors17 and retinoic acid18. Muscarinic inhibitors such as atropine inhibit the development of myopia in animal models and human intervention studies19,20, and choroidal retinoic acid synthesis is altered in animal models of myopia21. RASGRF1 knockdown mouse models have normal brain structure; however, they perform worse than wild-type mice in exercises involving long-term memory22.
Lack of expression of RASGRF1 in knockdown models results in a morphologically normal retina but causes pronounced deficiencies in photoreception and visual sensory processes without any other obvious sensory impairments23. Furthermore, lack of expression of RASGRF1 causes changes in the downstream expression of many genes23, including some genes whose mutations have been reported to cause severe Mendelian vision disorders. Thus, RASGRF1 appears to be involved in the maintenance of normal function of the retina and possibly in the signaling pathways determining myopia.
We further studied the effect of Rasgrf11 on mouse eye parameters. For this purpose, 19 wild-type mice were compared to 16 Rasgrf1 knockdown mice of the same age group (between 3- and 4-months-old, when eye growth has stabilized). We found that the average crystalline lens was significantly heavier in the knockdown mice (P = 0.001). This was independent of the weight of the whole eye, as the ratio of crystalline weight to the total eye weight remained significantly higher in the knockdown mice (P = 0.003; Supplementary Table 3). Axial length was not significantly different between the groups; however, it is known that the measurement of this trait in mice is technically difficult and inaccurate24. In humans, refractive error is mostly due to axial length variations, but refractive error is highly correlated with lens thickness in mice25.
In an accompanying study reported in this issue, C. Klaver and colleagues report a genome-wide association study for refractive error in a Dutch population-based cohort26. Researchers from that study identified a susceptibility locus at genome-wide significant levels on chromosome 15q14. GJD2, also known as Connexin 36, is the gene nearest to the associated chromosome 15q14 locus. In our study, the chromosome 15q14 locus showed suggestive association (P = 6 × 10−7 in the TwinsUK Cohort GWAS), but this locus was not taken forward for inclusion in our replication study (Supplementary Fig. 4).
Both RASGRF1 and GJD2 are involved in synaptic transmission and play important roles in the transmission and processing of visual signals23,27. Animal models of myopia demonstrate that the signal that continues or halts eye growth originates within the retina, and these signals are transmitted through the choroid to stimulate or inhibit scleral remodeling28.
We report, to our knowledge, the first genome-wide association for common myopia and refractive error in multiple European populations, identifying an associated locus upstream of the transcription site of RASGRF1. Rasgrf1 knockdown mice have larger crystalline lenses than wild-type mice, which are likely to result in increased refractive power. RASGRF1 is involved in learning, visual processing and muscarinic signaling pathways, all of which have been associated or implicated with myopia. We believe that identification of this susceptibility locus and the suggestive evidence for involvement of the candidate gene RASGRF1 will greatly improve our understanding of the pathogenesis of myopia.
ONLINE METHODS
Participants and cohorts
The TwinsUK adult twin registry based at St. Thomas’ Hospital in London is a volunteer cohort of over 10,000 twins from the general population. Twins largely volunteered unaware of the eye studies, gave fully informed consent under a protocol reviewed by the St. Thomas’ Hospital Local Research Ethics Committee and underwent non-cyclopleged autorefraction using an ARM-10 autorefractor (Takagi Ltd). Out of the original 4,388 subjects for whom phenotype and genotype information was available, 4,270 subjects (315 males and 3,955 females) with a mean age of 55 years (s.d. = 12; Supplementary Fig. 5a) were included in this study; 118 subjects were excluded after failing quality control. The mean spherical equivalent for both eyes was calculated using the standard formula:
The mean spherical equivalent was −0.4 diopters (s.d. = 2.73 diopters), with a range of −25.12 to +9.4 diopters (Supplementary Fig. 5b). Myopia was defined as the presence of the average spherical equivalent over both eyes ≤−1 diopter, and emmetropia was defined as an average spherical equivalent between −0.5 and 0.5 diopters, inclusive of these values.
A series of three cohorts were derived from the Rotterdam Study (RS). The RS-I was a prospective population-based cohort study comprising 7,983 subjects aged 55 years or older living in Ommoord, a district of Rotterdam, The Netherlands, with baseline examinations that included automated measurement of refractive error in 1991–1993. All participants consented to participation, and the study received ethical approval by the Medical Ethics Committee of the Erasmus University. Complete data on refractive error and from the GWAS were available on 5,328 persons, 99% of whom were of European ancestry. The RS-II included 2,157 new residents of Ommoord with good-quality genotyping data, aged 55 years or older, living in the district since 2000. Baseline examinations took place between 2000 and 2002. The RS-III included 2,082 new residents of Ommoord aged 45 and older living in the district since 2006 and who had DNA samples with good-quality genotyping data. Baseline examinations took place between 2006 and 2009 for RS-III. The fourth Dutch replication study was the Erasmus Rucphen Family (ERF) Study, a family-based study in a genetically isolated population in the southwest area of The Netherlands. This study included 2,032 living descendants from 22 families who had at least six children baptized in the community church between 1880 and 1900, who had good quality genotyping data and were aged 18 years or older. The mean age (s.d.) and mean spherical equivalent (s.d.) were 68.5 (8.6) years and +0.86 (2.45) diopters for RS-I, 64.2 (7.4) years and +0.48 (2.51) diopters for RS-II, and 56.2 (5.8) years and −0.35 (2.62) diopters for RS-III. The mean spherical equivalent (s.d.) for ERF was +0.07 (2.25) diopters and the mean age was 48.5 (14.5) years.
The 1958 British Birth Cohort was a prospective population-based study that initially included 17,000 newborn children whose date of birth was within the first week of March 1958. All participants gave informed written consent to participate in genetic association studies, and the present study was approved by the South East MultiCentre Research Ethics Committee (MREC) and the Oversight Committee for the biomedical examination of the British 1958 Cohort. Biomedical examination protocols were approved by the South East MREC. The phenotype for this cohort was spherical equivalent (mean of both eyes) measured with non-cycloplegic autorefraction (Nikon Retinomax 2). One thousand randomly chosen subjects from this cohort were included in the GWAS. This included 482 males and 518 females, all aged 44–45 years, and the average spherical equivalent (s.d.) in the GWAS was −0.94 (2.01) diopters.
The Australian Twin Eye Study comprised participants examined as part of the Twins Eye Study in Tasmania or the Brisbane Adolescent Twins Study. Ethical approval was obtained from the Royal Victorian Eye and Ear Hospital, the University of Tasmania, the Australian Twin Registry and the Queensland Institute of Medical Research. In all subjects, post-cycloplegic refraction (following instillation of tropicamide 1%) for both eyes was measured using a Humphrey-598 automatic refractor (Carl Zeiss Meditec, Inc.). The mean spherical equivalent of both eyes from all participants 18 years of age or older at the time of examination was used for analysis. This cohort comprised 1,086 individuals from 525 families. The mean age (s.d.) of participants was 27.4 (11.8) years. The mean spherical equivalent (s.d.) was −0.53 (1.64) diopters, with a range of −17.75 to +6.44 diopters.
Genotyping and imputation
Genotyping was carried out using three genotyping platforms from Illumina: the HumanHap 300k Duo for part of the UK Twin Cohort and the HumanHap610-Quad array for the rest of the UK Twin Cohort. The RS-I, RS-II and RS-III studies used the Infinium II HumanHap550 chip v3.0 array to conduct the genotyping, with the exception of 133 of 2,516 subjects in RS-II who were genotyped using the HumanHap610-Quad array. The ERF cohort was genotyped with the Infinium HumanHap300-2 318K array. The Australian cohorts were genotyped on the HumanHap610-Quad array, with part of the sample typed alongside the UK Twin Cohort and the remainder of the sample typed as part of a separate contract with deCODE Genetics. Illumina’s Human1M-Duo chip was used for genotyping of the 1958 British Birth Cohort. Imputation was calculated with reference to HapMap release 22 CEU population data using IMPUTE (see URLs) version 2 for the UK Twin Cohort and the 1958 Birth Cohort. MACH (see URLs) was used for imputation in the Dutch and Australian cohorts.
Statistical analyses
Association analysis was performed using MERLIN in the UK and Australian Twin Cohorts and using PLINK (see URLs) in the Rotterdam cohorts. Ancestry for these individuals was determined initially through self reporting and was verified through principal component decomposition of their ancestry with and without comparison with HapMap phase 2 standard populations. Meta-analysis was carried out using METAL (see URLs).
Laboratory animal handling and eye measurements
In an effort to minimize the number of animals used and their suffering, all procedures with laboratory mice were carried out in accordance with European Union guidelines (directives 86/609/EEC and 2003/65/EC), current Spanish legislation (RD 1201/2005. BOE 252/34367-91, 2005) and the US National Institutes of Health guidelines for the use and care of animals in research. Rasgrf1-null mice were obtained as described previously29. Mice were bred in a pathogen-free facility with water and food ad libitum.
For eye measuring, 16 3–4-month-old Rasgrf1-null mice and 19 wild-type control mice29 were anesthetized with chloral hydrate (5% (wt/vol) in saline, administered intraperitoneally, at 10 ml/g of body weight) and killed by cervical dislocation. After measuring the body weight of each mouse in a precision balance (Sartorius), its eyes were immediately extracted using 7-inch curved tweezers (Lawton), eliminating all rests of the optical nerve, and all vessels and surrounding tissues were cut away. Eye weight was immediately measured in an AX-120 Analytical Weight Balance (Cobos Precision, S.L.). The precision of the balance was 0.001 g. After being weighed, the eye balls were positioned on a glass slide under a microscope (Olympus Provis AX70 photomicroscope, Olympus Optical Co., Ltd.). The coronal and longitudinal axes were captured using an Olympus DP70 digital camera and measured at ×40 magnification using Olympus’ image capture software DP controller, version 1.2.1.108 (Olympus Optical Co., Ltd.).
The lens from each eye was then manually isolated using 7-inch tweezers (Lawton) under a binocular lens (Zeiss, Stemi 2000-C, Carl Zeiss) and measured in an M-220 analytical balance (precision = 0.1 mg) (Denver Instruments GmbH).
Supplementary Material
Acknowledgments
The King’s College London authors acknowledge funding from the Wellcome Trust, the European Union MyEuropia Marie Curie Research Training Network, Guide Dogs for the Blind Association, the European Community’s FP7 (HEALTH-F2-2008-201865-GEFOS), European Network of Genetic and Genomic Epidemiology (ENGAGE) (HEALTH-F4-2007-201413), the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254), US National Institutes of Health (NIH)/National Eye Institute (NEI) grant 1RO1EY018246 and genotyping by the NIH Center for Inherited Disease Research. The study also received support from the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s and St. Thomas’ National Health Service Foundation Trust partnering with King’s College London. We are grateful to the volunteer twins, C. Smoliner and M. Liew, and to R. Metlapally and Felicia Hawthorne in T.L.Y.’s laboratory for RASGRF1 expression studies.
The Rotterdam Study acknowledges Netherlands Organisation of Scientific Research (NWO); the Erasmus Medical Center; Netherlands Organization for the Health Research and Development (ZonMw); UitZicht; Research Institute for Diseases in the Elderly (RIDE); the European Commission Directorate-General XII; the Municipality of Rotterdam; Netherlands Genomics Initiative (NGI); Lijf en Leven; MD Fonds; Henkes; Oogfonds; Stichting Wetenschappelijk Onderzoek Het Oogziekenhuis (SWOO); Swart van Essen; Bevordering van Volkskracht; Blindenhulp; Landelijke Stichting voor Blinden en Slechtzienden (LSBS); Rotterdamse Vereniging voor Blindenbelangen; OOG Foundation; Algemene Nederlandse Vereniging ter Voorkoming van Blindheid (ANVVB); Rotterdam Eye Institute (REI); Laméris Ootech BV; Topcon BV; Heidelberg Engineering; A. Hooghart, C. Brussee, R. Bernaerts-Biskop, P. van Hilten, P. Arp, M. Jhamai, M. Moorhouse, J. Vergeer, M. Verkerk, S. Bervoets and P. van der Spek. E.S., A.F.-M. and L.M. were supported by grants from Instituto de Salud Carlos III (ISCIII) (FIS PS09/01979) and Junta de Castilla y León (JcyL) (SA044A08 and GR93) and received institutional support from Red Temática de Investigación Cooperativa en Cáncer (RTICC) (RD06/0020/000), ISCIII, Spain.
The Australian group acknowledges the National Health and Medical Research Council (NHMRC) for the Australian Twin Registry (ATR) Enabling Grant, Project Grant 350415, Medical Genomics Grant, and the Genetic Cluster Computer (NWO 480-05-003), Clifford Craig Medical Research Trust, Ophthalmic Research Institute of Australia (ORIA), American Health Assistance Foundation (AHAF), Peggy and Leslie Cranbourne Foundation, Foundation for Children and Jack Brockhoff Foundation.
The 1958 British Birth Cohort was funded for biomedical assessment (Medical Research Council), for the GWAS (Wellcome Trust 083478) and analysis at Great Ormond Street Hospital/University College London (UCL). Institute of Child Health and Moorfields Eye Hospital/Institute of Ophthalmology, UCL were each part-funded by NIHR Biomedical Research Centres awards.
Additional acknowledgements are contained within the Supplementary Note.
Footnotes
AUTHOR CONTRIBUTIONS
T.L.Y., D.A.M., T.A. and C.J.H. jointly conceived the project and supervised the work. S.M., Y.J.-L., L.Y.F., P.C.S., N.G.M., P.G.H. and A.M.S. helped with the data analyses. A.F.-M., L.M., A.P. and E.S. performed the animal experiments. A.W.H., J.R.V., M.K.I., C.M.v.D., T.D.S., J.S.R. and C.C.W.K. supervised cohort recruitment, genotyping and analysis in replication cohorts. F.C., S.J.F. and M.C.L. contributed during subject and data collection.
Note: Supplementary information is available on the Nature Genetics website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Vitale S, Ellwein L, Cotch MF, Ferris FL, III, Sperduto R. Prevalence of refractive error in the United States, 1999–2004. Arch Ophthalmol. 2008;126:1111–1119. doi: 10.1001/archopht.126.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan I, Rose K. How genetic is school myopia? Prog Retin Eye Res. 2005;24:1–38. doi: 10.1016/j.preteyeres.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Wu HM, et al. Does education explain ethnic differences in myopia prevalence? A population-based study of young adult males in Singapore. Optom Vis Sci. 2001;78:234–239. doi: 10.1097/00006324-200104000-00012. [DOI] [PubMed] [Google Scholar]
- 4.Rein DB, et al. The economic burden of major adult visual disorders in the United States. Arch Ophthalmol. 2006;124:1754–1760. doi: 10.1001/archopht.124.12.1754. [DOI] [PubMed] [Google Scholar]
- 5.Bamashmus MA, Matlhaga B, Dutton GN. Causes of blindness and visual impairment in the West of Scotland. Eye. 2004;18:257–261. doi: 10.1038/sj.eye.6700606. [DOI] [PubMed] [Google Scholar]
- 6.Evans JR, Fletcher AE, Wormald RP. Causes of visual impairment in people aged 75 years and older in Britain: an add-on study to the MRC Trial of Assessment and Management of Older People in the Community. Br J Ophthalmol. 2004;88:365–370. doi: 10.1136/bjo.2003.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resnikoff S, Pascolini D, Mariotti SP, Pokharel GP. Global magnitude of visual impairment caused by uncorrected refractive errors in 2004. Bull World Health Organ. 2008;86:63–70. doi: 10.2471/BLT.07.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buch H, et al. Prevalence and causes of visual impairment and blindness among 9980 Scandinavian adults: the Copenhagen City Eye Study. Ophthalmology. 2004;111:53–61. doi: 10.1016/j.ophtha.2003.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Saw SM, Katz J, Schein OD, Chew SJ, Chan TK. Epidemiology of myopia. Epidemiol Rev. 1996;18:175–187. doi: 10.1093/oxfordjournals.epirev.a017924. [DOI] [PubMed] [Google Scholar]
- 10.Rose KA, et al. Outdoor activity reduces the prevalence of myopia in children. Ophthalmology. 2008;115:1279–1285. doi: 10.1016/j.ophtha.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 11.Lopes MC, Andrew T, Carbonaro F, Spector TD, Hammond CJ. Estimating heritability and shared environmental effects for refractive error in twin and family studies. Invest Ophthalmol Vis Sci. 2009;50:126–131. doi: 10.1167/iovs.08-2385. [DOI] [PubMed] [Google Scholar]
- 12.Vitale S, Sperduto RD, Ferris FL., III Increased prevalence of myopia in the United States between 1971–1972 and 1999–2004. Arch Ophthalmol. 2009;127:1632–1639. doi: 10.1001/archophthalmol.2009.303. [DOI] [PubMed] [Google Scholar]
- 13.Zippel R, et al. Ras-GRF, the activator of Ras, is expressed preferentially in mature neurons of the central nervous system. Brain Res Mol Brain Res. 1997;48:140–144. doi: 10.1016/s0169-328x(97)00120-4. [DOI] [PubMed] [Google Scholar]
- 14.Raaijmakers JH, Bos JL. Specificity in Ras and Rap signaling. J Biol Chem. 2009;284:10995–10999. doi: 10.1074/jbc.R800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Puente A, et al. Structural characterization of Rasgrf1 and a novel linked imprinted locus. Gene. 2002;291:287–297. doi: 10.1016/s0378-1119(02)00601-7. [DOI] [PubMed] [Google Scholar]
- 16.Yoon BJ, et al. Regulation of DNA methylation of Rasgrf1. Nat Genet. 2002;30:92–96. doi: 10.1038/ng795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattingly RR, Macara IG. Phosphorylation-dependent activation of the Ras-GRF/CDC25Mm exchange factor by muscarinic receptors and G-protein beta gamma subunits. Nature. 1996;382:268–272. doi: 10.1038/382268a0. [DOI] [PubMed] [Google Scholar]
- 18.Tonini R, et al. Expression of Ras-GRF in the SK-N-BE neuroblastoma accelerates retinoic-acid-induced neuronal differentiation and increases the functional expression of the IRK1 potassium channel. Eur J Neurosci. 1999;11:959–966. doi: 10.1046/j.1460-9568.1999.00504.x. [DOI] [PubMed] [Google Scholar]
- 19.Tigges M, et al. Effects of muscarinic cholinergic receptor antagonists on postnatal eye growth of rhesus monkeys. Optom Vis Sci. 1999;76:397–407. doi: 10.1097/00006324-199906000-00020. [DOI] [PubMed] [Google Scholar]
- 20.Tong L, et al. Atropine for the treatment of childhood myopia: effect on myopia progression after cessation of atropine. Ophthalmology. 2009;116:572–579. doi: 10.1016/j.ophtha.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 21.Mertz JR, Wallman J. Choroidal retinoic acid synthesis: a possible mediator between refractive error and compensatory eye growth. Exp Eye Res. 2000;70:519–527. doi: 10.1006/exer.1999.0813. [DOI] [PubMed] [Google Scholar]
- 22.Brambilla R, et al. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390:281–286. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- 23.Fernández-Medarde A, et al. RasGRF1 disruption causes retinal photoreception defects and associated transcriptomic alterations. J Neurochem. 2009;110:641–652. doi: 10.1111/j.1471-4159.2009.06162.x. [DOI] [PubMed] [Google Scholar]
- 24.Schmucker C, Schaeffel F. In vivo biometry in the mouse eye with low coherence interferometry. Vision Res. 2004;44:2445–2456. doi: 10.1016/j.visres.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 25.Zhou X, et al. The development of the refractive status and ocular growth in C57BL/6 mice. Invest Ophthalmol Vis Sci. 2008;49:5208–5214. doi: 10.1167/iovs.07-1545. [DOI] [PubMed] [Google Scholar]
- 26.Solouki A, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q14. Nat Genet. 2010 Sep 12; doi: 10.1038/ng.663. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deans MR, Volgyi B, Goodenough DA, Bloomfield SA, Paul DL. Connexin36 is essential for transmission of rod-mediated visual signals in the mammalian retina. Neuron. 2002;36:703–712. doi: 10.1016/s0896-6273(02)01046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallman J, Winawer J. Homeostasis of eye growth and the question of myopia. Neuron. 2004;43:447–468. doi: 10.1016/j.neuron.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Font de Mora J, et al. Ras-GRF1 signaling is required for normal beta-cell development and glucose homeostasis. EMBO J. 2003;22:3039–3049. doi: 10.1093/emboj/cdg280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


