Abstract
Refractive errors are the most common ocular disorders worldwide and may lead to blindness. Although this trait is highly heritable, identification of susceptibility genes has been challenging. We conducted a genome-wide association study for refractive error in 5,328 individuals from a Dutch population-based study with replication in four independent cohorts (combined 10,280 individuals in the replication stage). We identified a significant association at chromosome 15q14 (rs634990, P = 2.21 × 10−14). The odds ratio of myopia compared to hyperopia for the minor allele (minor allele frequency = 0.47) was 1.41 (95% CI 1.16–1.70) for individuals heterozygous for the allele and 1.83 (95% CI 1.42–2.36) for individuals homozygous for the allele. The associated locus is near two genes that are expressed in the retina, GJD2 and ACTC1, and appears to harbor regulatory elements which may influence transcription of these genes. Our data suggest that common variants at 15q14 influence susceptibility for refractive errors in the general population.
Refractive errors are by far the most common cause of visual impairment in humans1–5. They result from aberrant coordinated effects of the ocular biometric components, most notably of axial length. Elongation of the eye axis leads to myopia (nearsightedness), whereas a shortened axis causes hyperopia (farsightedness). Refractive errors often cause alterations in the anatomical structure of the eye, increasing the risk of clinical complications6. Myopia may lead to ocular morbidity such as glaucoma and retinal detachment, and high myopia in particular can cause posterior staphyloma and macular degeneration7–11. Treatment options for myopia are limited, and myopia is the fifth most common cause of impaired vision and the seventh most common cause of legal blindness worldwide10,11.
The etiology of refractive errors and myopia is complex and largely uncharacterized. The current notion is that eye growth is triggered by a visually evoked signaling cascade that begins in the retina, traverses the choroid and subsequently mediates scleral remodeling. Established risk factors for myopia are education, reading, outdoor exposure and familial predisposition11–14. Familial aggregation studies have quantified a strong genetic basis for myopia; the estimated recurrence risk for siblings of individuals with myopia (λs) varied between 1.5 and 3.0 for low myopia and between 4.9 and 19.8 for high myopia, and heritability estimates of myopia (h2) ranged from 0.60 to 0.90 (ref. 15). Segregation analyses suggested the involvement of multiple genes rather than a single major gene effect11,13,15. In an attempt to identify causal genes, previous linkage mapping studies mainly focused on highly myopic probands with multiple affected relatives and thereby identified at least 20 putative genetic loci11. Replication of these results has been limited, and these candidate genes have been shown to have little to no effect in unselected populations. To our knowledge, no genome-wide association studies (GWAS) for refractive error in the general population have previously been reported.
We performed a GWAS in the population-based Rotterdam Study (RS-I, n = 5,328) for refractive error measured as a quantitative trait. Study design and baseline characteristics are provided in the Online Methods and Supplementary Table 1. The mean spherical equivalent in this older population (> 55 years of age) of European descent was +0.86 (s.d. = 2.45) diopters. Refractive errors occurred in 52% (n = 2,790) of the participants, with measurements ranging from −19 to +10 diopters.
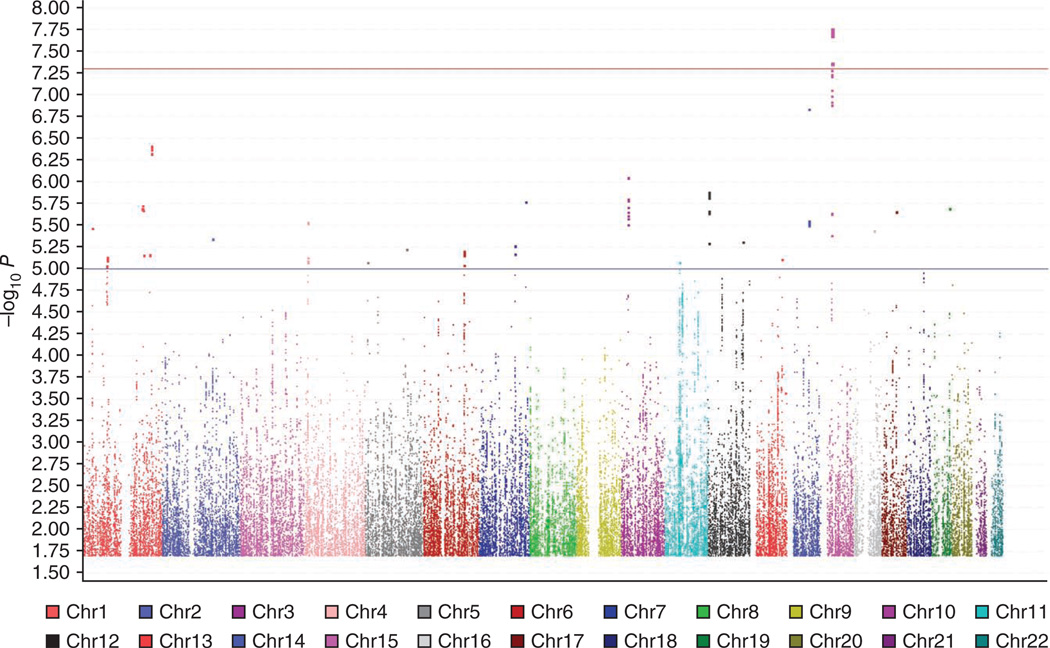
We genotyped the entire cohort using the Illumina HumanHap 550k and 610Q arrays (Online Methods). Genotypes for more than 2.5 million autosomal SNPs were imputed with reference to the HapMap Phase 2 CEU population build 36. Comparison of the observed and expected distributions (for the quantile-quantile plot, see Supplementary Fig. 1) showed modest inflation of the test statistics (genomic control inflation factor (λGC) = 1.054 for RS-I). Using an additive model, we identified a significant association on chromosome 15q14 (rs688220, P = 1.76 × 10−8; Table 1 and Fig. 1). We took forward 31 SNPs spread across four loci on chromosomes 15q14, 14q24, 1q41 and 10p12.3 reaching P < 10−6 (Supplementary Table 2) for further investigation in four independent replication cohorts: RS-II (n = 2,008, λGC = 1.012), RS-III (n = 1,970, λGC = 1.012) and the Erasmus Rucphen Family Study (ERF, n = 2,032, λGC = 1.037) from The Netherlands, and a twin study from the United Kingdom (TwinsUK, n = 4,270, λGC = 1.04). The designs of RS-II and RS-III were population based, whereas those of ERF and TwinsUK were family based. Cohorts were not selected on the basis of a disease phenotype. All studies consisted predominantly of individuals of European ancestry and all used similar protocols to evaluate refractive error (Online Methods and Supplementary Table 2).
Table 1.
Genome-wide association and replication for refractive error at locus 15q14
| Discovery cohort |
Replication |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RS-I (n = 5,328) |
RS-II (n = 2,008) |
RS-III (n = 1,970) |
ERF (n = 2,032) |
TwinsUK (n = 4,270) |
Meta-analysis (n = 15,608) |
||||||||||
| SNP | Position | MA | MAF | β (s.e.m.) |
P | β (s.e.m.) |
P | β (s.e.m.) |
P | β (s.e.m.) |
P | β (s.e.m.) |
P | β (s.e.m.) |
P |
| rs688220 | 32,786,167 | A | 0.45 | −0.27 (0.05) |
1.76 × 10−8 | −0.28 (0.08) |
3.80 × 10−4 | −0.22 (0.08) |
9.27 × 10−3 | −0.03 (0.07) |
6.24 × 10−1 | −0.15 (0.07) |
2.60 × 10−2 | −0.20 (0.0009) |
2.79 × 10−11 |
| rs580839 | 32,786,121 | A | 0.44 | −0.27 (0.05) |
1.89 × 10−8 | −0.27 (0.08) |
4.96 × 10−4 | −0.22 (0.08) |
7.95 × 10−3 | −0.03 (0.07) |
6.34 × 10−1 | −0.16 (0.07) |
1.92 × 10−2 | −0.20 (0.0009) |
2.53 × 10−11 |
| rs619788 | 32,782,398 | A | 0.44 | −0.27 (0.05) |
1.92 × 10−8 | −0.27 (0.08) |
4.94 × 10−4 | −0.22 (0.08) |
7.72 × 10−3 | −0.03 (0.07) |
6.27 × 10−1 | −0.16 (0.07) |
1.85 × 10−2 | −0.20 (0.0009) |
2.53 × 10−11 |
| rs4924134 | 32,781,857 | G | 0.44 | −0.27 (0.05) |
2.04 × 10−8 | −0.27 (0.08) |
4.76 × 10−4 | −0.27 (0.08) |
6.58 × 10−3 | −0.06 (0.07) |
4.10 × 10−1 | −0.16 (0.07) |
1.85 × 10−2 | −0.21 (0.0009) |
1.36 × 10−12 |
| rs560766 | 32,788,234 | A | 0.44 | −0.26 (0.05) |
4.27 × 10−8 | −0.28 (0.08) |
4.54 × 10−4 | −0.21 (0.08) |
1.29 × 10−2 | −0.03 (0.07) |
6.65 × 10−1 | −0.18 (0.07) |
7.68 × 10−3 | −0.20 (0.0009) |
2.49 × 10−11 |
| rs7176510 | 32,786,771 | T | 0.45 | −0.26 (0.05) |
5.16 × 10−8 | −0.28 (0.08) |
5.10 × 10−4 | −0.22 (0.08) |
9.62 × 10−3 | −0.02 (0.07) |
7.51 × 10−1 | −0.16 (0.07) |
1.76 × 10−2 | −0.20 (0.0009) |
6.25 × 10−11 |
| rs7163001 | 32,777,866 | A | 0.44 | −0.26 (0.05) |
5.23 × 10−8 | −0.28 (0.08) |
4.08 × 10−4 | −0.23 (0.08) |
5.89 × 10−3 | −0.07 (0.07) |
3.01 × 10−1 | −0.16 (0.07) |
1.87 × 10−2 | −0.21 (0.0009) |
5.61 × 10−12 |
| rs11073060 | 32,777,143 | A | 0.44 | −0.26 (0.05) |
5.76 × 10−8 | −0.28 (0.08) |
4.05 × 10−4 | −0.23 (0.08) |
5.82 × 10−3 | −0.08 (0.07) |
2.72 × 10−1 | −0.16 (0.07) |
1.91 × 10−2 | −0.21 (0.0009) |
3.65 × 10−12 |
| rs8032019 | 32,778,782 | G | 0.40 | −0.26 (0.05) |
6.09 × 10−8 | −0.28 (0.08) |
5.57 × 10−4 | −0.13 (0.09) |
1.30 × 10−1 | −0.05 (0.07) |
5.12 × 10−1 | −0.16 (0.07) |
1.96 × 10−2 | −0.19 (0.0009) |
3.71 × 10−10 |
| rs685352 | 32,795,627 | G | 0.44 | −0.25 (0.05) |
8.80 × 10−8 | −0.25 (0.08) |
1.28 × 10−3 | −0.19 (0.08) |
1.98 × 10−2 | −0.07 (0.07) |
3.06 × 10−1 | −0.24 (0.07) |
4.43 × 10−4 | −0.21 (0.0009) |
4.19 × 10−12 |
| rs524952 | 32,793,178 | A | 0.47 | −0.25 (0.05) |
1.03 × 10−7 | −0.30 (0.08) |
2.09 × 10−4 | −0.19 (0.08) |
2.56 × 10−2 | −0.06 (0.07) |
4.13 × 10−1 | −0.32 (0.07) |
4.15 × 10−6 | −0.23 (0.0009) |
3.18 × 10−14 |
| rs634990 | 32,793,365 | C | 0.47 | −0.25 (0.05) |
1.03 × 10−7 | −0.30 (0.08) |
2.15 × 10−4 | −0.20 (0.08) |
2.03 × 10−2 | −0.05 (0.07) |
5.11 × 10−1 | −0.33 (0.07) |
2.93 × 10−6 | −0.23 (0.0009) |
2.21 × 10−14 |
| rs11073059 | 32,776,966 | A | 0.44 | −0.25 (0.05) |
1.20 × 10−7 | −0.28 (0.08) |
3.96 × 10−4 | −0.23 (0.08) |
5.83 × 10−3 | −0.08 (0.07) |
2.72 × 10−1 | −0.16 (0.07) |
1.91 × 10−2 | −0.20 (0.0009) |
8.45 × 10−12 |
| rs11073058 | 32,776,918 | T | 0.44 | −0.25 (0.05) |
1.30 × 10−7 | −0.28 (0.08) |
3.93 × 10−4 | −0.23 (0.08) |
5.84 × 10−3 | −0.08 (0.07) |
2.71 × 10−1 | −0.16 (0.07) |
1.90 × 10−2 | −0.20 (0.0009) |
8.45 × 10−12 |
RS-I, Rotterdam Study I; RS-II, Rotterdam Study II; RS-III, Rotterdam Study III; ERF, Erasmus Rucphen Family Study; TwinsUK, the Twin Cohort recruited in London; MA, minor allele; MAF, minor allele frequency; β, effect size on spherical equivalent in diopters.
Figure 1.
Genome-wide signal intensity (Manhattan) plot of the discovery cohort Rotterdam Study-I. The statistical significance values across the 22 autosomes of each SNP’s association with refractive error (measured as spherical equivalent) are plotted as −log10 P values. SNPs with minor allele frequency ≥ 0.01 were included. The blue horizontal line indicates P = 10−5 and the red line indicates P = 5 × 10−8.
Meta-analysis of the combined discovery and replication cohorts showed a significant association between refractive errors and the locus on 15q14 (the most significant association in meta-analysis was at rs634990 with a combined P = 2.21 × 10−14; Table 1). Frequencies of the risk alleles at this region were similar across the studies. The P values were nominally significant (P < 0.05) for the 14 top SNPs in RS-II, RS-III and TwinsUK, and the direction of the effect (regression coefficient β) of the minor alleles was consistent (Table 1). rs634990 accounted for 0.5% of the variance in spherical equivalent.
To determine the effect of this locus on the risk of clinically relevant outcomes, we compared subjects with myopia to those with hyperopia in a logistic regression analysis. We found strong evidence that the C allele of rs634990 conferred a higher risk of myopia than the T allele (Fig. 2). The odds ratio (OR) of low to moderate or high myopia versus low to moderate or high hyperopia was 1.41 (95% CI 1.16–1.70) for heterozygotes and 1.83 (95% CI 1.42–2.36) for homozygotes.
Figure 2.
Forest plot of associations for myopia (spherical equivalent ≤ −3 diopters) versus hyperopia (spherical equivalent ≥ +3 diopters). Forest plot of the estimated per-genotype odds ratio for top SNP rs634990 for the five studies separately and for the meta-analysis of all studies. RS-I, Rotterdam Study I; RS-II, Rotterdam Study II; RS-III, Rotterdam Study III; ERF, Erasmus Rucphen Family Study; TwinsUK, the Twin Cohort recruited in London; OR, odds ratio.
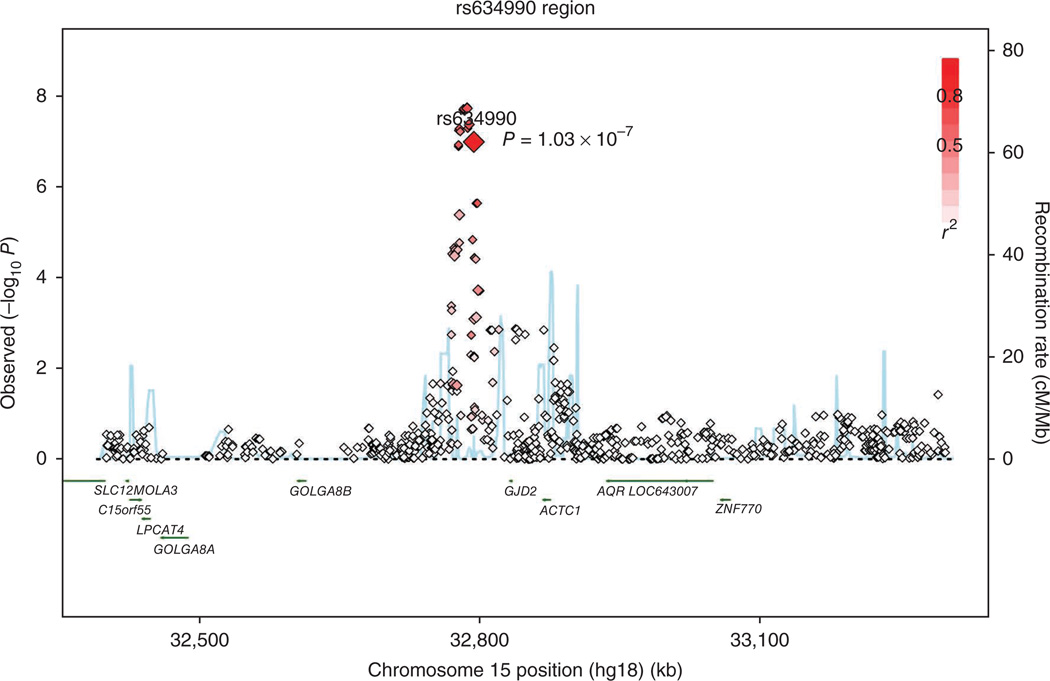
The locus on 15q14 (Fig. 3) is within an intergenic region in the vicinity of the genes GJD2 (39 kb from rs634990 at its 3′ end), ACTC1 (74 kb from rs634990 at its 3′ end) and GOLGA8B (180 kb from rs634990 at its 5′ end). We investigated a potential function for these genes in eye growth development by examining gene expression levels in the retina of postmortem human eyes (Supplementary Table 3) and observed moderate to high expression of GJD2 and ACTC1 and much lower expression of GOLGA8B. GOLGA8B encodes the 67-kDa protein Golgi autoantigen golgin-67, which belongs to a family of Golgi auto-antigens and is localized at the cytoplasmic surface of the Golgi complex16. A specific function of GOLGA8B in the retina has not been reported. ACTC1 encodes the 42-kDa smooth muscle actin cardiac muscle alpha actin 1. The functional role of ACTC1 in the eye is currently unclear, but actins that are similar to it, such as α-SMA, have been shown to be increased in developing myopic eyes17. α-SMA influences the number of contractile myofibroblasts in the sclera and contributes to extracellular matrix remodeling. As these are key factors occurring in eye enlargement, it is intriguing to know whether ACTC1 has these characteristics as well.
Figure 3.
Regional plot at chromosome 15q14. Log10 P values from the discovery cohort RS-I as a function of genomic position (HapMap release 22 build 36). The P value for the top SNP is denoted by the large diamond and P values for other genotyped and imputed SNPs are shown as smaller diamonds. P values for SNPs of unknown type are presented as squares. Superimposed on the plot are gene locations (green) and recombination rates (blue).
The function of GJD2 makes this gene an interesting candidate gene for refractive error. GJD2 encodes the 36-kDa protein Connexin36 (also known as CX36 and gap junction protein delta 2), which is a neuronspecific protein belonging to a multigene family of integral membrane proteins18. CX36 forms gap junction channels between adjacent membranes of neuronal cells, is present in photoreceptors, amacrine and bipolar cells, and plays a critical role in the transmission process of the retinal electric circuitry by enabling intercellular transport of small molecules and ions18–21. Further exploration of GJD2 using Ingenuity analysis (see URLs, Online Methods and Supplementary Fig. 2) alluded to a role for the gene in eye growth regulation as well as in lens fiber maturation in knockdown animals22,23. To identify possible causal variants in this gene, we performed direct sequencing of all exons and intron-exon boundaries of GJD2 in 47 individuals with either high myopia, high hypermetropia or emmetropia. We found neither new mutations nor frequency differences of any variants between groups (Supplementary Table 4), and we conclude that linkage disequilibrium with common functional variants in GJD2 is unlikely to explain the observed association.
The next step was to assess whether associated variants within the intergenic region itself may have functional consequences. We evaluated the expression of SNPs within the associated 15q14 locus in lymphoblastoid cell lines. At least two of the top associated SNPs from the meta-analysis significantly altered expression, suggesting that these may regulate gene expression (rs560766, P = 1.0 × 10−5; rs580839, P = 9.5 × 10−6; Supplementary Table 5). Subsequently, we searched for regulatory elements24,25 in the entire 53-kb locus of highly significantly correlated SNPs using the UCSC Genome Browser and found the predicted presence of seven DNase I hypersensitive sites, six enhancers based on experimentally validated H3 chromatin signatures in Hela and K562 cells24,25, 20 peaks of sequence conservation in alignments of multiple species of placental mammals and 1 insulator site (Supplementary Fig. 3 and ref. 25). Enhancers are known to facilitate transcription of distal genes, and their range of activity is confined by insulators25. Notably, the greatest peak of our association coincided with an insulator site. Precedents of genomic alterations of insulators causing hereditary disease have been previously reported26,27. We speculate that variants or mutations in regulatory elements at 15q14 may lead to illegitimate transcription of genes in the area, for example, of ACTC1 and GJD2.
In GWAS in general, sources of heterogeneity may cause spurious findings. To address this issue and minimize potential biases, we applied genomic control to the cohort-level test statistics in the population cohorts, and we corrected the statistics using the identity-by-descent structure in the family-based cohorts. Three studies, RS-II, RS-III and TwinsUK significantly replicated our initial findings. The fourth study, ERF, showed the same direction of association as the other three studies, albeit a nonsignificant association, and revealed similar risks of myopia for carriers of the risk allele (Fig. 2). Thus, the observed effects of the genetic variants at 15q14 are relatively homogeneous among the five studies, enhancing the credibility of the findings.
In a companion paper in this issue, Christopher Hammond and colleagues report a GWAS for refractive errors in the TwinsUK study28. Researchers in that study found a genome-wide significant association (most significant combined P = 1.85 × 10−9 for rs939658 and P = 2.07 × 10−9 for rs8027411) at a locus on chromosome 15q25, explaining 0.81% of the variance in spherical equivalent measurements. This locus includes the promoter of RASGRF1. This gene is known to be functionally involved in eye development29 and, similar to GJD2, is involved in synaptic transmission of photoreceptor responses30. TwinsUK and RS-I are two of the largest existing refractive error cohorts with GWAS data. Our studies each identified one different genome-wide significant locus, and we both estimated the variation in refractive error explained by these SNPs to be small. The findings of our studies suggest that the genetic variance in refractive error is mostly determined by multiple variants with a low to moderate penetrance, which is similar to the pattern found for traits such as height31.
The mutual replication of the direction and the β coefficient of the effect of variants at 15q14 and 15q25 supports the association of these genomic loci to refractive error and myopia. To unravel the mechanism behind myopia, the next steps should include comprehensive resequencing of the entire associated regions and the flanking genes, validation in cohorts of other ethnicities, functional assays and study of risk modulation by environmental factors. This may help to uncover new pathogenic pathways for refractive errors and may eventually lead to new strategies to reduce the sight-threatening consequences of myopia.
ONLINE METHODS
Participants
Discovery cohort
The Rotterdam Study (RS-I) was a prospective population-based cohort study of 7,983 residents aged 55 years and older living in Ommoord, a suburb of Rotterdam, The Netherlands32. The baseline examination for the ophthalmic portion of the study took place between 1991 and 1993 and included 6,775 persons. Individuals were excluded from the study if they had undergone bilateral cataract surgery, laser refractive procedures or other intra-ocular procedures which might alter refraction. Complete data on refractive error and genome-wide SNPs were available on 5,328 persons, 99% of whom were of European ancestry.
Replication cohorts
The first three replication studies originated from The Netherlands. The first cohort was RS-II, an independent cohort which included 2,157 new participants aged 55 and older living in Ommoord since 2000 (ref. 32) who had good quality genotyping data. Baseline examinations for this cohort took place between 2000 and 2002, and follow-up examination took place from 2004 to 2005. The second replication cohort was RS-III, a study which included 2,082 new participants aged 45 and older living in Ommoord since 2006 who had good quality genotyping data. Baseline examination for this cohort took place between 2006 and 2009. The third replication study was the Erasmus Rucphen Family (ERF) Study, a family-based study in a genetically isolated population in the southwest of The Netherlands. This study included 2,032 living descendants of Erasmus Rucphen aged 18 years and older originating from 22 families who had at least six children baptized in the community church between 1880 and 1900 and who had good quality genotyping data. The fourth replication cohort was derived from the United Kingdom (TwinsUK). This study was an adult twin registry of over 10,000 healthy volunteer twins based at St. Thomas’ Hospital in London. Participants were recruited and phenotyped between 1998 and 2008. A total of 4,270 participants of European descent had complete data on ocular phenotype and genotype33.
As in the discovery cohort, participants in the four replication cohorts were excluded if they had undergone bilateral surgery which inhibited evaluation of the original refractive error.
Measurements of refractive error
All studies used a similar protocol for phenotyping. Participants underwent an ophthalmologic examination which included non-dilated automated measurement of refractive error (RS I–III) and ERF used the Topcon RM-A2000 autorefractor; the TwinsUK cohort used the Humphrey-670 (Humphrey Instruments) from 1998 to 2002 and then the ARM-10 (Takagi Seiko), best-corrected visual acuity and keratometry. Each spherical equivalent was calculated from the standard formula:
spherical equivalent = sphere + (cylinder /2)
In addition to investigating spherical equivalent as a quantitative trait, we stratified spherical equivalent measurements into categories of refractive error to evaluate findings from a clinical viewpoint. Myopia was categorized into low (spherical equivalent from −1.5 to −3 diopters), moderate (spherical equivalent from −3 to −6 diopters) and high (spherical equivalent of −6 diopters or lower). For hyperopia, the categories used were low (spherical equivalent from +1.5 to +3 diopters), moderate (spherical equivalent from +3 to +6 diopters) and high (spherical equivalent of +6 diopters or higher), respectively. We considered spherical equivalent from −1.5 to +1.5 diopters as emmetropia.
Ethics
All measurements in RS-I–III and ERF were conducted after the Medical Ethics Committee of the Erasmus University had approved the study protocols and all participants had given a written informed consent in accordance with the Declaration of Helsinki. In the TwinsUK study, all twins gave fully informed consent under a protocol reviewed by the St. Thomas’ Hospital Local Research Ethics Committee.
Genotyping
Discovery cohort
All persons attending the baseline examination from 1990–1993 consented to genotyping and had DNA extracted from blood leucocytes. Genotyping of autosomal SNPs was performed in persons with high quality extracted DNA (n = 6,449) using the Illumina Infinium II HumanHap550 chip v3.0 array according to the manufacturer’s protocols. Samples with low call rate (<97.5%, n = 209), with excess autosomal heterozygosity (>0.336, n = 21) and with sex-mismatch (n = 36) were excluded, as were outliers identified by the identity-by-state clustering analysis (outliers were defined as being >3 s.d. from population mean, n = 102 or having identity-by-state probabilities >97%, n = 129). 5,974 individuals had good quality genotyping data.
Replication cohorts
In RS-II, the majority of the 2,516 DNA samples were genotyped using the HumanHap550 Duo Arrays; 133 samples (5%) were genotyped using the Human610-Quad Arrays (Illumina). In the RS-III cohort, all DNA samples were genotyped using the Illumina Infinium II HumanHap550 chip v3.0 array. In ERF, DNA was genotyped on one of four different platforms (Illumina 6k, Illumina 318K, Illumina 370K and Affymetrix 250K). Genotyping for the TwinsUK cohort took place in stages; in the first stage, 1,810 individuals were genotyped using Illumina’s HumanHap 300k duo chip and at a later stage, 2,578 persons were genotyped using Illumina’s HumanHap610-Quad chip.
Imputation
The set of genotyped input SNPs used for imputation in each study was selected based on highest quality GWAS data. The call rate was set at >98% in RS-I–III, the minor allele frequency was set at >0.01, and the Hardy-Weinberg P value was set at >10−6. We used the Markov Chain Haplotyping (MACH) package version 1.0.15 software (Rotterdam, The Netherlands; imputed to plus strand of NCBI build 36, HapMap release #22; see URLs) for the analyses. For each imputed SNP, a reliability of imputation was estimated as the ratio of the empirically observed dosage variance to the expected binomial dosage variance (O/E ratio).
Statistical analysis
Discovery cohort
Refractive error measured at baseline as a continuous variable was used as the outcome in the analysis. We calculated the mean spherical equivalent measurements for individuals with measurements available for both eyes and included the spherical equivalent of only one eye if data from the other eye were missing. Linear regression models with a 1 degree of freedom trend test were used to examine the associations between SNPs and spherical equivalent, adjusted for age and gender. Using these linear regression models, we calculated regression coefficients with corresponding 95% CIs. ORs of myopia and hyperopia were calculated with logistic regression analysis, adjusting for age and gender. GWAS analyses were performed using GRIMP34.
We used genomic control to obtain optimal and unbiased results and applied the inverse variance method of each effect size estimated for both autosomal SNPs that were genotyped and imputed in both cohorts. P < 5 × 10−8 was considered genome-wide significant.
Replication analyses
The top SNPs with P <1 × 10−6 from the discovery analysis were examined in the replication cohorts RS-II, RS-III, ERF and TwinsUK using SPSS version 15.0.0 for Windows (SPSS Inc.) and R statistical package version 2.8.1 for Linux. A meta-analysis was performed on all five studies using METAL for Linux.
GRIMP34 was used for the analysis of the population-based replication cohorts. To adjust for family relationships, the GenABEL package35 was used in the ERF study, and Merlin was used in the TwinsUK Study36. SNPs which deviated significantly from Hardy-Weinberg equilibrium (using the threshold of P < 10−6) and SNPs which had minor allele frequency < 0.01 were excluded.
Gene expression data in human eye tissue
Human gene expression data were obtained essentially as described37. In short, postmortem eye bulbs (retinal pigment epithelium was obtained from six donor eyes, choroid was obtained from three donor eyes and photoreceptors were obtained from three donor eyes), provided by the Corneabank Amsterdam, were rapidly frozen using liquid nitrogen. Donors were between 63 and 78 years old and had no known history of eye pathology.
Cryosections were cut from the macula, and we used histology to confirm a normal histological appearance. Retinal pigment epithelium, photoreceptor and choroidal cells were isolated from macular sections using a Laser Microdissection System (PALM). Total RNA was isolated and the mRNA component was amplified, labeled and hybridized to a 44K microarray (Agilent Technologies)38. At least three to six microarrays were performed per tissue. Sample isolation, procedures and expression microarray analysis were carried out according to MIAMI guidelines. As a measure of the level of expression, we sorted all the genes represented on the 44K microarray by increasing their expression, and we calculated the corresponding percentiles (Supplementary Table 3).
Ingenuity database search
We explored the Ingenuity knowledge database using the keyword ‘eye development’ for all genes involved in ‘function or diseases’. This search provided approximately 100 genes, which formed a new network for eye development. We subsequently added GJD2 to the network, and we used the Path Explorer tool to search for possible functional relationships between GDJ2 and these eye development genes in human, mouse, rat and in vitro models (Supplementary Fig. 2a). We continued the search using the keyword ‘eye growth’ for all genes involved in ‘function or diseases’ and investigated functional links between molecules using the connect tool and upstream-downstream analysis (Supplementary Fig. 2b).
Supplementary Material
ACKNOWLEDGMENTS
Major funding of the work performed in The Netherlands came from the Netherlands Organisation of Scientific Research (NWO); Erasmus Medical Center and Erasmus University, Rotterdam, The Netherlands; Netherlands Organization for Health Research and Development (ZonMw); UitZicht; the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); the Municipality of Rotterdam; the Netherlands Genomics Initiative (NGI)/NWO; Center for Medical Systems Biology of NGI; Lijf en Leven; M.D. Fonds; H. Stichting; Oogfonds Nederland; Stichting Nederlands Oogheelkundig Onderzoek; Swart van Essen; Bevordering van Volkskracht; Blindenhulp; Landelijke Stichting voor Blinden en Slechtzienden; Rotterdamse Vereniging voor Blindenbelangen; OOG; Algemene Nederlandse Vereniging ter Voorkoming van Blindheid; the Rotterdam Eye Hospital Research Foundation; Laméris Ootech; Topcon Europe; and Heidelberg Engineering. We thank A. Hooghart, C. Brussee, R. Bernaerts-Biskop, P. van Hilten, P. Arp, M. Jhamai, M. Moorhouse, J. Vergeer, M. Verkerk, S. Bervoets and P. van der Spek for help in execution of the study.
TwinsUK acknowledges the Wellcome Trust, the European Union MyEuropia Marie Curie Research Training Network, Guide Dogs for the Blind Association, the European Community’s Seventh Framework Programme (FP7/2007-2013)/grant agreement HEALTH-F2-2008-201865-GEFOS and (FP7/2007-2013), European Network of Genetic and Genomic Epidemiology (ENGAGE) project grant agreement HEALTH-F4-2007-201413 and the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254), Biotechnology and Biological Sciences Research Council (G20234), Department of Health via US National Institutes of Health, (National Eye Institute grant RO1EY018246), and the Center for Inherited Disease Research. TwinsUK thanks G. Surdulescu, L. Peltonen, P. Deloukas, M. Lathrop, D. Goldstein, A. Palotie and C. Day for help in execution of the study and analyses.
Footnotes
URLs. Ingenuity, http://www.ingenuity.com; MACH, http://www.sph.umich.edu/csg/abecasis/MACH; R, http://www.r-project.org; METAL, http://www.sph.umich.edu/csg/abecasis/metal/.
Accession codes. The relevant expression data are deposited in the GEO database under accession number GSE20191.
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
A.M.S., V.J.M.V. and C.C.W.K. performed analyses and drafted the manuscript. C.M.v.D., B.A.O., F.R., A.G.U., A.H., P.T.V.M.d.J., J.R.V. and C.C.W.K. designed the study and obtained funding. D.D.G.D., L.M.v.K., L.H., W.D.R., M.C., R.K., J.J.M.W.-A., T.G.M.F.G., F.C.C.R. and S.M.A.S. helped in data collection. A.J.M.H.V., M.K.I., N.A., M.S., Y.S.A., A.A.B.B., A.A.L.J.v.O. and A.I. participated in the genetic analyses. P.G.H., T.L.Y., D.A.M., T.D.S. and C.J.H. were responsible for data from the TwinsUK study. M.K.I., R.W.A.M.K., G.v.R., P.G.H., C.J.H., C.M.v.D., A.J.M.H.V., B.A.O., J.R.V. and A.A.B.B. critically reviewed the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Bourne RR, Dineen BP, Ali SM, Noorul Huq DM, Johnson GJ. Prevalence of refractive error in Bangladeshi adults: results of the National Blindness and Low Vision Survey of Bangladesh. Ophthalmology. 2004;111:1150–1160. doi: 10.1016/j.ophtha.2003.09.046. [DOI] [PubMed] [Google Scholar]
- 2.Dandona R, et al. Population-based assessment of refractive error in India: the Andhra Pradesh eye disease study. Clin. Experiment. Ophthalmol. 2002;30:84–93. doi: 10.1046/j.1442-6404.2002.00492.x. [DOI] [PubMed] [Google Scholar]
- 3.Kempen JH, et al. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch. Ophthalmol. 2004;122:495–505. doi: 10.1001/archopht.122.4.495. [DOI] [PubMed] [Google Scholar]
- 4.Sawada A, et al. Refractive errors in an elderly Japanese population: the Tajimi study. Ophthalmology. 2008;115:363–370. doi: 10.1016/j.ophtha.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 5.Vitale S, Ellwein L, Cotch MF, Ferris FL, III, Sperduto R. Prevalence of refractive error in the United States, 1999–2004. Arch. Ophthalmol. 2008;126:1111–1119. doi: 10.1001/archopht.126.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McBrien NA, Gentle A. Role of the sclera in the development and pathological complications of myopia. Prog. Retin. Eye Res. 2003;22:307–338. doi: 10.1016/s1350-9462(02)00063-0. [DOI] [PubMed] [Google Scholar]
- 7.Saw SM, et al. How blinding is pathological myopia? Br. J. Ophthalmol. 2006;90:525–526. doi: 10.1136/bjo.2005.087999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtin BJ, Karlin DB. Axial length measurements and fundus changes of the myopic eye. Am. J. Ophthalmol. 1971;1:42–53. doi: 10.1016/0002-9394(71)91092-0. [DOI] [PubMed] [Google Scholar]
- 9.Saw SM, Gazzard G, Shih-Yen EC, Chua WH. Myopia and associated pathological complications. Ophthalmic Physiol. Opt. 2005;25:381–391. doi: 10.1111/j.1475-1313.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 10.Tano Y, et al. Pathologic myopia: where are we now? Am. J. Ophthalmol. 2002;134:645–660. doi: 10.1016/s0002-9394(02)01883-4. [DOI] [PubMed] [Google Scholar]
- 11.Young TL, et al. Molecular genetics of human myopia: an update. Optom. Vis. Sci. 2009;86:E8–E22. doi: 10.1097/OPX.0b013e3181940655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirani M, et al. Outdoor activity and myopia in Singapore teenage children. Br. J. Ophthalmol. 2009;93:997–1000. doi: 10.1136/bjo.2008.150979. [DOI] [PubMed] [Google Scholar]
- 13.McBrien NA, et al. Myopia: recent advances in molecular studies; prevalence, progression and risk factors; emmetropization; therapies; optical links; peripheral refraction; sclera and ocular growth; signalling cascades; and animal models. Optom. Vis. Sci. 2008 Dec 19; published online. [Google Scholar]
- 14.Saw SM, Hong CY, Chia KS, Stone RA, Tan D. Nearwork and myopia in young children. Lancet. 2001;357:390. doi: 10.1016/S0140-6736(05)71520-8. [DOI] [PubMed] [Google Scholar]
- 15.Young TL, Metlapally R, Shay AE. Complex trait genetics of refractive error. Arch. Ophthalmol. 2007;125:38–48. doi: 10.1001/archopht.125.1.38. [DOI] [PubMed] [Google Scholar]
- 16.Eystathioy T, Jakymiw A, Fujita DJ, Fritzler MJ, Chan EK. Human autoantibodies to a novel Golgi protein golgin-67: high similarity with golgin-95/gm 130 autoantigen. J. Autoimmun. 2000;14:179–187. doi: 10.1006/jaut.1999.0359. [DOI] [PubMed] [Google Scholar]
- 17.Jobling AI, Gentle A, Metlapally R, McGowan BJ, McBrien NA. Regulation of scleral cell contraction by transforming growth factor-beta and stress: competing roles in myopic eye growth. J. Biol. Chem. 2009;284:2072–2079. doi: 10.1074/jbc.M807521200. [DOI] [PubMed] [Google Scholar]
- 18.Kihara AH, et al. Connexin36, an essential element in the rod pathway, is highly expressed in the essentially rodless retina of Gallus gallus. J. Comp. Neurol. 2009;512:651–663. doi: 10.1002/cne.21920. [DOI] [PubMed] [Google Scholar]
- 19.Deans MR, Volgyi B, Goodenough DA, Bloomfield SA, Paul DL. Connexin36 is essential for transmission of rod-mediated visual signals in the mammalian retina. Neuron. 2002;36:703–712. doi: 10.1016/s0896-6273(02)01046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Striedinger K, et al. Loss of connexin36 increases retinal cell vulnerability to secondary cell loss. Eur. J. Neurosci. 2005;22:605–616. doi: 10.1111/j.1460-9568.2005.04228.x. [DOI] [PubMed] [Google Scholar]
- 21.Güldenagel M, et al. Visual transmission deficits in mice with targeted disruption of the gap junction gene connexin36. J. Neurosci. 2001;21:6036–6044. doi: 10.1523/JNEUROSCI.21-16-06036.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rong P, et al. Disruption of Gja8 (a8 connexin) in mice leads to microphthalmia associated with retardation of lens growth and lens fiber maturation. Development. 2002;129:167–174. doi: 10.1242/dev.129.1.167. [DOI] [PubMed] [Google Scholar]
- 23.White TW. Targeted ablation of Connexin50 in mice results in microphthalmia and zonular pulverulent cataracts. J. Cell Biol. 1998;143:815–825. doi: 10.1083/jcb.143.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr. Opin. Genet. Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaloy C, et al. Deletion of WNK1 first intron results in misregulation of both isoforms in renal and extrarenal tissues. Hypertension. 2008;52:1149–1154. doi: 10.1161/HYPERTENSIONAHA.108.120899. [DOI] [PubMed] [Google Scholar]
- 27.Mihaly J, et al. Dissecting the regulatory landscape of theAbd-Bgene of the bithorax complex. Development. 2006;133:2983–2993. doi: 10.1242/dev.02451. [DOI] [PubMed] [Google Scholar]
- 28.Hysi PG, Young TL, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25 which contains theRASGRF1gene. Nat. Genet. 2010 Sep 12; doi: 10.1038/ng.664. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones C, Moses K. Cell-cycle regulation and cell-type specification in the developingDrosophilacompound eye. Semin. Cell Dev. Biol. 2004;15:75–81. doi: 10.1016/j.semcdb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Fernández-Medarde A, et al. RasGRF1 disruption causes retinal photoreception defects and associated transcriptomic alterations. J. Neurochem. 2009;110:641–652. doi: 10.1111/j.1471-4159.2009.06162.x. [DOI] [PubMed] [Google Scholar]
- 31.Lettre G, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hofman A, et al. The Rotterdam Study: 2010 objectives and design update. Eur. J. Epidemiol. 2009;24:553–572. doi: 10.1007/s10654-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spector TD, MacGregor AJ. The St. Thomas’ UK Adult Twin Registry. Twin Res. 2002;5:440–443. doi: 10.1375/136905202320906246. [DOI] [PubMed] [Google Scholar]
- 34.Estrada K, et al. GRIMP: A web- and grid-based tool for high-speed analysis of large-scale genome-wide association using imputed data. Bioinformatics. 2009;25:2750–2752. doi: 10.1093/bioinformatics/btp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 36.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 37.Booij JC, et al. Functional annotation of the human retinal pigment epithelium transcriptome. BMC Genomics. 2009;20:10–164. doi: 10.1186/1471-2164-10-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Soest SS, et al. Comparison of human retinal pigment epithelium gene expression in macula and periphery highlights potential topographic differences in Bruch’s membrane. Mol. Vis. 2007;10:1608–1617. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.