Abstract
Background
Corneal intraepithelial dyskeratosis is an extremely rare condition. The classical form, affecting Native American Haliwa-Saponi tribe members, is called hereditary benign intraepithelial dyskeratosis (HBID). Herein, we present a new form of corneal intraepithelial dyskeratosis for which we identified the causative gene by using deep sequencing technology.
Methods and results
A seven member Caucasian French family with two corneal intraepithelial dyskeratosis affected individuals (6-year-old proband and his mother) was ascertained. The proband presented with bilateral complete corneal opacification and dyskeratosis. Palmoplantar hyperkeratosis and laryngeal dyskeratosis were associated with the phenotype. Histopathology studies of cornea and vocal cord biopsies showed dyskeratotic keratinisation. Quantitative PCR ruled out 4q35 duplication, classically described in HBID cases. Next generation sequencing with mean coverage of 50× using the Illumina Hi Seq and whole exome capture processing was performed. Sequence reads were aligned, and screened for single nucleotide variants and insertion/deletion calls. In-house pipeline filtering analyses and comparisons with available databases were performed. A novel missense mutation M77T was discovered for the gene NLRP1 which maps to chromosome 17p13.2. This was a de novo mutation in the proband’s mother, following segregation in the family, and not found in 738 control DNA samples. NLRP1 expression was determined in adult corneal epithelium. The amino acid change was found to destabilise significantly the protein structure.
Conclusions
We describe a new corneal intraepithelial dyskeratosis and how we identified its causative gene. The NLRP1 gene product is implicated in inflammation, autoimmune disorders, and caspase mediated apoptosis. NLRP1 polymorphisms are associated with various diseases.
INTRODUCTION
Corneal intraepithelial dyskeratosis is an extremely rare condition. The classical form is known as hereditary benign intraepithelial dyskeratosis (HBID), (OMIM 127600, http://www.ncbi.nlm.nih.gov/omim) and is a rare autosomal dominant inherited disorder first described in 1959.1 The condition was initially reported in a Native American Haliwa-Saponi Indian community of North Carolina, USA. To date, several cases have been reported, tracing their ancestry back to North Carolina. 2–7 The origins of isolated cases have been reported in Brazil, Italy, Germany, and China.5,8–11
HBID onset is usually at birth or in early childhood.6,12 Clinical findings include ocular and oral mucosal manifestations. Ocular lesions are typically bilateral, symmetric, and highly variable. The lesions range from the more typical mild perilimbal conjunctival plaques with epibulbar hyperaemia to severe extensive corneo-conjunctival plaques causing vision impairment in rare cases. In 1960, Witkop et al 13 first reported oral lesions characterised by white spongy plaques of the buccal mucosa. The mucosal lesions do not invade underlying tissues and remain localised. No malignant transformation has ever been reported. Histopathologic changes of the oral and ocular lesions are similar 12,14 and include acanthotic epithelium with hyperkeratosis and dyskeratosis.7
Significant progress has been made in genetic disease comprehension in the last decade with the development of next-generation sequencing as a recent evolutionary consequence of the human genome project.15 Next-generation and Sanger sequencing technologies are complementary tools for the discovery of novel genome mutations causing unsolved genetic disorders.16,17 Exome sequencing is particularly amenable to gene identification for monogenic rare diseases.18,19
Herein, we present a new form of corneal intraepithelial dyskeratosis, featuring severe corneal manifestations associated with systemic characteristics. We used whole exome sequencing to identify the gene mutation responsible for this novel corneal disease in a Caucasian French family.
METHODS
Patients and blood samples
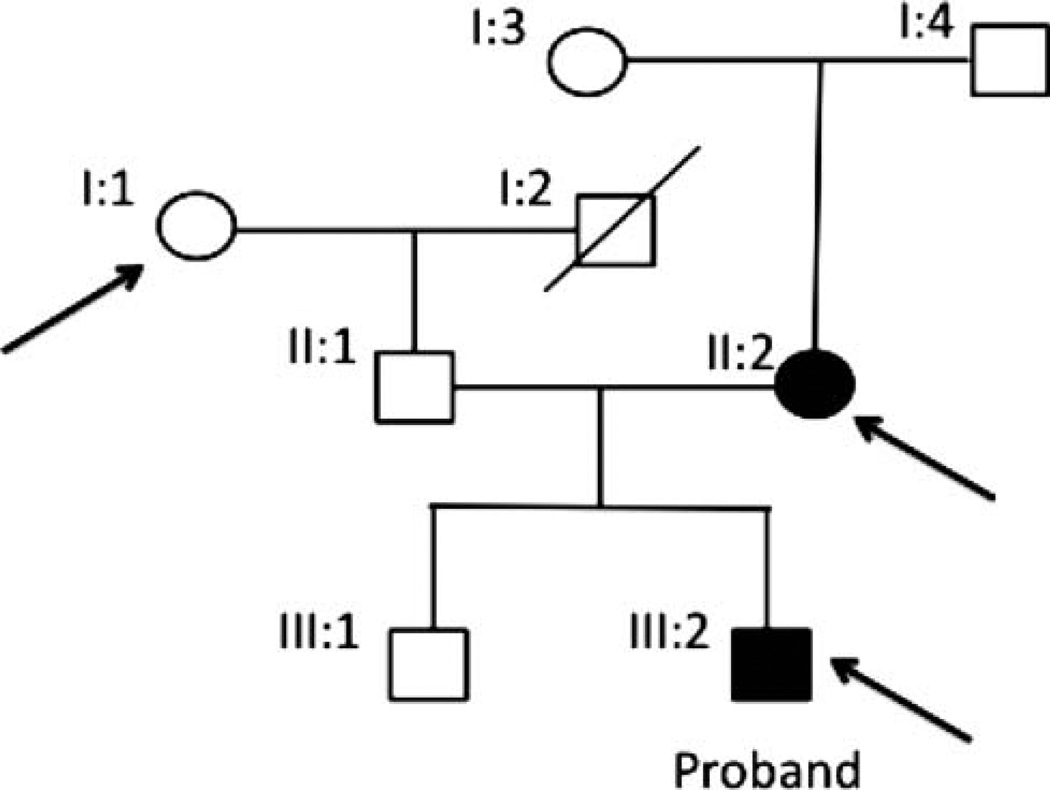
The project was approved by the Purpan Hospital human subjects research study review board entitled ‘Comité de Protection des Personnes Sud-Ouest et Outre-Mer II’. All subjects were ascertained according to the Declaration of Helsinki with informed consent obtained before sample collection. Venous blood was collected from seven French Caucasian family participants. Sample DNA from three individuals were selected for exome sequencing (figure 1): two individuals were affected with corneal dystrophy—the proband (individual III:2) and his mother (individual II:2); and one unaffected individual (I:1) was used as an internal control.
Figure 1.
Corneal dystrophy pedigree. Arrows depict samples that were chosen for exome sequencing: individuals I:1 (unaffected), II:2 (affected) and the proband III:2 (affected).
Total genomic DNA was extracted from blood using AutoPure LS* DNA Extractor and PUREGENE reagents (Gentra Systems Inc, Big Lake, Minnesota, USA), and stored. DNA from 672 ethnically matched Caucasian healthy control participants, with no documented ocular disorder, were commercially purchased from the Centre for Applied Genomics (The Hospital for Sick Children, Toronto, Canada). In addition, we collected blood for DNA extraction from four HBID affected Native American Indian individuals and two unaffected relatives from three different families. These families were cared for by an experienced cornea specialist (NA), and were ascertained with appropriate consent approvals from the Duke University Institutional Review Board.
Sample preparation for pathological analysis
Corneal samples, obtained from the proband during keratoplasty, and vocal cord biopsy were fixed in formalin, paraffin embedded, and 4 µm sections were prepared and counterstained with haematoxylin and eosin. Pathological analysis was performed by an experienced pathologist (BK).
Detection of 4q35 duplication
A multiplex fluorescent PCR based test was performed to amplify polymorphic marker D4S1652 (F-aatccctgggtacattatatttg; R-cagacattctttattctttacctcc) and exon 4 of CFTR gene.20 Reactions were run on a Verity thermocycler (Life Technologies, Saint Aubin, France). Products were loaded on an ABI3130XL genetic analyser and fluorescent intensities were analysed with GeneMapper software (Life Technologies). The quantitative ratio of the areas under the peaks for D4S1652 to the CFTR exon 4 determined the D4S1652 copy number in each individual. Genomic DNA controls were obtained from a healthy control (homozygous) and from the father. The final result was the ratio of the D4S1652 dosage obtained for the patient to the D4S1652 dosage for the normal homozygous control (ie, two-copy control) (see online supplementary figure S1). Tests were repeated three times.
Massively parallel sequencing
Seven micrograms of DNA were submitted to Hudson Alpha Laboratories (Huntsville, Alabama, USA) (http://www.hudsonalpha.org/) for deep sequencing processing to achieve an average coverage of 50× read depth. The libraries were prepared according to manufacturer’s guidelines. The SeqCap EZ Exome Library V.2.0 was used with target coverage of 44.1 megabases, or 1.42% of the human genomic regions of consensus coding sequences (CCDS) exons (Roche NimbleGene, Madison, Michigan, USA). Each captured library was sequenced on an Illumina HiSeq 2000 platform (Illumina, Inc, San Diego, California, USA). Ninety base pair (bp) reads were generated.
Read mapping and quality filtering (see online supplementary figure S2)
First, to align the short paired-end reads, the NCBI GRH37 lite reference genome (ftp://ftp.ncbi.gov/genbank/genomes/Eukaryotes/vertebrates_mammals/Homo_sapiens/GRCh37/special_requests) and the Burrows-Wheeler Aligner tool (BWA)21 (http://biowulf.nih.gov/apps/bwa.html) were used. The Sequence Alignment/Map format, called SAMtools,22 was used to sort the previously performed alignment. The BAM files—binary version of the SAM files—obtained from the two lane runs were merged into a single BAM file.
Statistical summary of the coverage analysis was performed by using custom Perl and R scripts. Only reads that had ≤4 mismatches to the reference were selected and uniquely aligned to the reference; the minimum mapping quality score of these reads was one or more. These data were also supplied by Hudson Alpha Laboratories (Huntsville, Alabama, USA).
The Genome Analyzer Toolkit (GATK V.4333) (http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit)23 was used to process all aligned reads in the merged BAM file. To improve quality score and alignment accuracy, reads containing small insertion/deletions (microindels) were first locally realigned before filtering according to quality constraints defined by the maximum amount of mismatches within a bp window. The GATK framework was used to perform consensus calling and to obtain the raw single nucleotide variants (SNVs), using the GATK ‘Unified Genotyper’ program. Then, SNVs were filtered using the GATK ‘Variant Filtration’ program by considering quality/depth, minimum total read depth (coverage), and occurrence of other SNVs or microindels close to the variant position.
After the above filtering steps, to annotate remaining SNVs, we used information from the University of California Santa Cruz (UCSC) genome annotation database (http://genome.ucsc.edu/)24, including information from dbSNP (v132) (http://www.ncbi.nlm.nih.gov/projects/SNP/),25 1000 Genomes Project (4 August 2010) (http://www.1000genomes.org/),26 Consensus CDS (CCDS) (http://www.ncbi.nlm.nih.gov/Ccdsbrowse.cgi), Ensembl (http://www.ensembl.org/index.html),27 RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq/),28 MirBase (http://www.mirbase.org/), and EntrezGene (http://jura.wi.mit.edu/entrez_gene/).29
The GATK ‘Indel Genotyper V2.0’ program was used to discover microindels that demonstrated at least 5× coverage or were present in more than 3% of the reads. The microindel calls were then filtered based upon their proximity to an exon, minimum microindel call depth (coverage), dbSNP database (v132) appearance, minimum average base quality in a 10 bp window, average mapping quality, and the mismatch rate of the reads showing the microindel. Microindels passing these filters were included in a candidate list and annotated with information from multiple databases, such as CCDS, RefSeq, UCSC, Ensembl, EntrezGene, and MirBase.
With additional filtering steps we removed common variants, defined as all variants present in the dbSNP (v132), Exome Variant Server (http://evs.gs.washington.edu/EVS/), and 1000 Genomes Project. Of the remaining variants, we used Polyphen-2 V.2.1.0 (http://genetics.bwh.harvard.edu/pph2/index.shtml)30 and SIFT (Sorting Intolerant From Tolerant) software (http://sift.bii.a-star.edu.sg/),31 to predict the effect of amino acid changes on protein function, analysing only coding non-synonymous variants or splice site mutations. Only SNVs that were predicted to affect protein function and present in affected individuals were considered in the final candidate variant list. The same analysis was conducted by using annotation results supplied by Hudson Alpha.
PCR and sequence analysis
Once the list of candidate variants were obtained, primers flanking a 250 bp variant region for PCR were designed using the Primer 3 program (http://frodo.wi.mit.edu/primer3). Primers were selected to produce amplification product sizes not to exceed 950 bp.
Genomic DNA of the proband (III:2) and one unaffected family member (II:1) was initially screened for validation of exome results. The DNA of the remaining family members was screened to determine co-segregation with affection status.
To determine if NLRP1 mutations accounted for classic HBID in Native Americans, primers for PCR covering all 17 NLRP1 exons and intron–exon boundaries were designed as described above (see online supplementary table S1). Direct sequencing of NLRP1 of four Native American Indian cases and two unaffected relatives from the three families was performed.
Amplicons were visualised by 2% agarose gel electrophoresis, and then submitted with appropriate primers to EtonBio for PCR purification and sequencing (http://www.EtonBio.com). Base pair calls were made using the Sequencher V.5.0 Software (Gene Codes, Ann Arbor, Michigan, USA). Sequences of affected and unaffected individuals were aligned to a known reference genomic sequence (UCSC Genome Browser GRCh37. p5) and compared for sequence variation.
Allelic frequency in the control population
Following custom Taqman SNP (single nucleotide polymorphism) genotyping assays design guidelines, the TaqMan SNP Genotyping system (Applied Biosystems, Carlsbad, California, USA) was used to genotype DNA from 672 ethnically matched controls for the NLRP1 variant to determine the allelic frequency. For each control sample, allelic call reads were performed using ABI 7900 robotics (Applied Biosystems). Genotype calls were accurately analysed with SDS V.2.4 software (Applied Biosystems). We also checked the presence of the NLRP1 variant in 61 exome sequenced subject DNA samples from other projects.
cDNA ocular tissue expression
NLRP1 expression was studied in both fetal and adult human ocular tissues. Ocular samples were obtained from North Carolina Eye Bank (adults eyes) (http://www.nceyebank.org) and from Advanced Bioscience Resources (24 week fetal eyes) (ABR, Alameda, California, USA). Adult and fetal eyes were placed in RNAlater (Ambion, Austin, Texas, USA) within 6.5 h of death and minutes of procurement, respectively. Eyes, stored on ice, were shipped within 12 h to the lab. Dissection of the whole globes was immediately performed upon arrival. Dissected tissues were snap-frozen and placed at −80°C until RNA extraction. Ambion mirVana Total RNA Extraction Kit per protocol (http://www.ambion.com/techlib/prot/fm_1560.pdf) was used to extract RNA from tissue samples homogenised in Ambion’s lysis buffer using an Omni Bead Ruptor 24 Homogeniser per protocol (http://www.omni-inc.com). Reverse transcription reactions were performed with Invitrogen’s SuperScript III First-Strand Synthesis kit (Invitrogen, Carlsbad, California, USA). NLRP1 primers were designed using the Primer3 program. The reactions were run on an Eppendorf Mastercycler Pro S (Eppendorf, Hamburg, Germany) with a standard touchdown PCR protocol. The NLRP1 primers used were F-atacgaagcctttggggact and R-caccgcttctctcatcacaa. PCR products were run on a 2% agarose electrophoresis gel at 70 V for 35 min. Both primers were run on our internal ocular tissue panel.
Corneal epithelium expression
Corneal epithelia were collected from 10 healthy patients undergoing photorefractive keratectomy procedures performed by the same surgeon (FM). Written consent was obtained from all participants. Samples were assigned a laboratory number and remained anonymous. Collected epithelia were immediately stored in 1.5 ml microtubes in liquid nitrogen until RNA extraction. Total RNA was extracted and further purified using the RNeasy Mini kit and RNase-Free DNase Set (Qiagen, Hilden, Germany), according to the manufacturer’s guidelines. RNA samples were immediately stored at −80°C. Reverse transcription reactions were performed with SuperScript VILO cDNA Synthesis Kit (Invitrogen). The corneal epithelium cDNA was stored at −20°C. PCR conditions were the same as described above.
Protein sequence, phylogenetic trees, and protein structural analyses
Amino acid sequences in FASTA format of each of the 14 members of the NLRP family in humans (NLRP1–NLRP14) were downloaded from UNIPROT.32 Next, finding orthologues for each member of the NLRP family was done using an orthologue search in ANNOTATOR (http://annotator.bii.a-star.edu.sg).33 We created a multiple alignment for all 228 orthologues of the 14 members of the NLRP family by using MAFFT with L-INS-I.34,35 After deleting sequences that had large gaps in Jalview,36 132 remaining sequences were compared. The multiple alignment of these 132 sequences was used to create phylogenetic trees using a maximum likelihood method with gamma distributed rates by using MEGA5.37
For protein structural analysis, we used the nuclear magnetic resonance structure of the human pyrin domain (PDB entry: 1PN5)38 from the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). FoldX (http://foldx.crg.es/)39 was used with prior energy minimisation using the RepairPDB function and five repetitions of the mutation stability changes to predict the SNV effect on protein structure stability.
RESULTS
Clinical and histopathologic findings
The three-generation family comprised two affected members (the proband, III:2, and his mother, II:2) and five unaffected members (figure 1). The proband’s mother (individual II:2) had unilateral keratopathy (right eye), with neovascularisation and complete corneal opacification. She also had a systemic issue of palmoplantar hyperkeratosis, dyshidrosis, and maxillary decalcification associated with alveolysis and tooth loss.
The proband (individual III:2) was born in December 2004. By the age of 2 years, he had presented with corneal dyskeratosis characterised by bilateral inferior corneo-limbal infiltrates, limbal thickening, neovascularisation, and keratin deposits. Despite topical corticotherapy, he developed bilateral centripetal evolution of the infiltrates resulting in bilateral visual impairment.
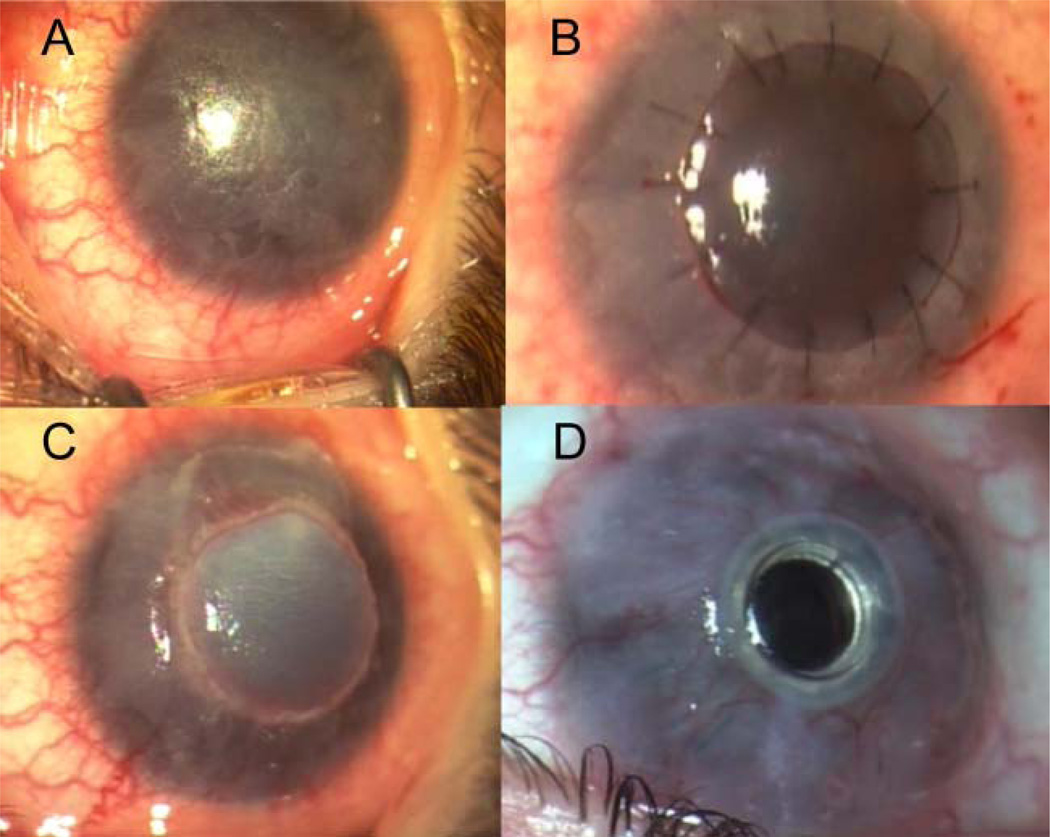
Three years later, the patient was referred to the Toulouse University Hospital (Toulouse, France). Visual acuity of both eyes was limited to hand motions. Clinical examination revealed bilateral corneal thickening, complete corneal opacification with dyskeratosis, and circumferential corneo-limbal neovascularisation (figure 2A). A deep anterior lamellar keratoplasty (DALK) was performed on the right eye (figure 2B). After a brief period of visual improvement, the disease recurred with complete graft opacification 2 months after the DALK (figure 2C). A second DALK was performed on the same eye 3 months after the first, followed by a limbal stem cell graft due to delayed epithelial healing. After developing in the right eye bacterial keratitis, disease recurrence and a third DALK failure 1 year later, the patient obtained a Boston type 1 keratoprosthesis (figure 2D) with cataract surgery and posterior intraocular lens implantation 6 months later. Seven months after keratoprosthesis implantation, an examination revealed visual acuity improvement to 20/200 (Snellen chart) of the operated right eye. The peripheral corneal graft had conjunctival overgrowth and the keratoprosthesis was clear. Keratoplasty and keratoprosthesis surgeries were performed by two anterior segment surgeons (PF and J-LA). The proband had pruritic hyperkeratotic scars, palmoplantar hyperkeratosis, chronic rhinitis, and a raspy voice. His finger and toenails were dystrophic with prominent thickening of the nail beds. He presented with a long philtrum, short neck, bulging chest, and finger joint hypermobility on general examination.
Figure 2.
Preoperative corneal examination of the right eye and postoperative clinical follow-up. Preoperatively, the patient presented with corneal opacification, corneal thickening, and circumferential corneolimbal neovascularisation (A). An initial deep anterior lamellar keratoplasty (DALK) (B) was performed; the disease recurred resulting in corneal graft opacification noted at a 2 month postoperative clinic session (C). After three DALK failures, the patient underwent a type 1 Boston keratoprosthesis procedure (D) approximately 7 months later.
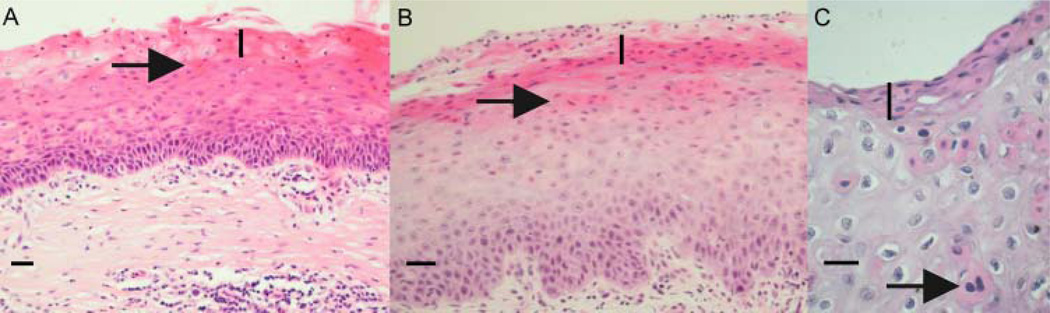
Histopathological findings (figure 3) of the proband’s original excised cornea showed acanthosis, parakeratosis, and dyskeratotic keratinisation affecting most of the corneal epithelium except the basal layers, with absence of Bowman’s layer and a stromal inflammatory infiltrate. Similar epithelial features were found within the second corneal graft.
Figure 3.
Histopathological features of the original cornea sample, the first failed corneal graft, and vocal chord tissue. The original cornea (panel A, horizontal bold scale: 100 µm), first corneal graft (panel B, horizontal bold scale: 60 µm), and vocal chord tissue (panel C, horizontal bold scale: 25 µm) show similar epithelial features: parakeratosis (vertical bold line), dyskeratosis (arrows), and acanthosis. Other histological characteristics are the absence of Bowman’s layer and the presence of a stromal inflammatory infiltrate.
The proband’s vocal cord biopsy showed epithelial features similar to those of the original cornea and corneal graft, demonstrating epithelium hyperplasia with dyskeratosis and parakeratosis (figure 3). These pathological findings are consistent with features described for classic HBID.13
Detection of 4q35 duplication
Normalised ratios of the D4S1652 dosage to the D4S1652 dosage for a normal homozygous control were 0.47 for the father (heterozygous for the allele, one copy control), 1.02 for the mother, and 1.08 for the proband (see online supplementary figure S1). Duplication in chromosome 4q35 commonly present in classic HBID was not evident in the mother (II:2) or in the proband (III:2).6
Genetic analysis
DNA samples from individuals II:2 and III:2—affected mother and son—and from the unaffected grandparent I:1 were exome sequenced. We generated an average of 9.2 gigabases (Gb) of sequence with 64.7× average coverage for each individual. Approximately 96.5% (42.7 Mb in length) of the targeted bases were covered sufficiently to pass variant call thresholds for SNVs and short insertions or deletions (indels). Bases with coverage above 20× represented over 78% of the total sequence data (see online supplementary table S2). For individuals I:1 (internal control), II:2 (proband’s mother) and III:2 (proband), we identified 285 888, 312 274, and 265 330 known SNVs, respectively.
To identify potential pathogenic variants, we focused on novel coding non-synonymous variants, splice acceptor and donor site mutations, and coding indels. Based on the disease rarity and on an autosomal dominant inheritance pattern, we hypothesised that the causing variant was rare and likely to be novel, that the proband (III:2) and the mother (II:2)—both affected—should be heterozygous for the SNV, and that the unaffected individual (I:1) should not carry the SNV.
Filtered exome sequence detected 22 novel non-synonymous SNVs and 10 microindels present in both affected individuals. Sanger sequencing validation of these variants resulted in 95.4% of the SNVs (21/22) and 1 microindel (1/10) to be true, whereas remaining variants were false positive. Sanger sequencing of remaining family members for co-segregation resulted in only one mutation, on NLRP1, where an apparent de novo mutation occurred in the mother, was passed down to her affected child, and thus was not present in the other family member, particularly in the affected mother’s parents, I:3 and I:4.
The change was a de novo mutation in NLRP1 gene exon 1 at position chr17 : 5487048 on GRCh37.p5 (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/chromosomes/) (T to C change on negative strand), and at position 230 on cDNA (GenBank Accession Number NM_033004.3; c.230T>C) (http://www.ncbi.nlm.nih.gov/genbank/), converting codon 77 methionine to threonine. The original amino acid is conserved among multiple species including rhesus monkey, mouse, dog, and elephant (http://genome.ucsc.edu/).
Taqman genotyping of the NLRP1 variant was carried out for 672 unrelated unaffected individuals. No variants were present in our controls and in 61 exome sequenced subject DNAs from other studies as well.
Sanger sequencing of the entire NLRP1 gene (17 exons) in four HBID affected Native American individuals and two unaffected relatives from three different families identified no pathogenic mutations (see online supplementary table S3).
Tissue expression
We examined NLRP1 expression in human ocular (fetal and adult) tissue panel (see online supplementary figure S3). In tissues obtained from dissected whole globes, NLRP1 was expressed in adult cornea, and in the following adult and 24-week fetal tissues: choroid, sclera, cornea, optic nerve, and adult retina and fetal retina/retinal pigment epithelium. Moreover, NLRP1 was expressed in corneal epithelia obtained during photorefractive keratectomy. All bands amplified at the expected product size and Sanger sequencing confirmed the cDNA product.
Protein sequence, phylogenetic tree, and structural analyses
The NLRP1 M77T is predicted to ‘affect protein function’ from SIFT31 mutational analysis using NLRP1 orthologues, human NLRP paralogues, or all aligned NLRP families as input (see online supplementary table S4).
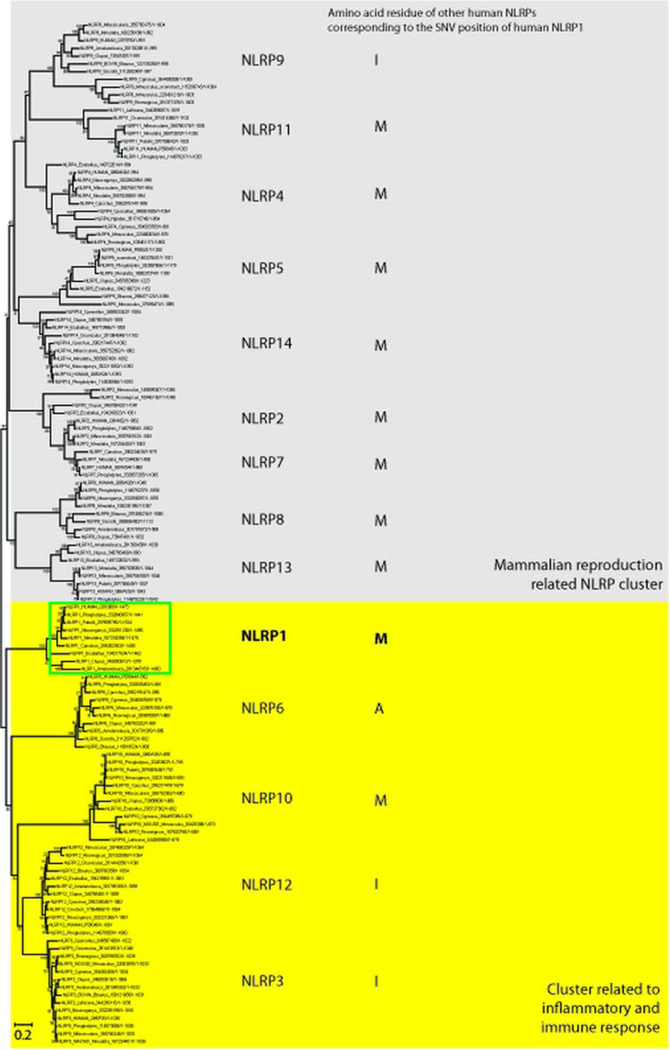
From the human NLRP family multiple alignment, the majority of the human NLRP1 paralogues, that is NLRP2 to NLRP4, and their orthologues have methionine at the NLRP1 SNV position. Methionine at position 77 is conserved in 10 out of the 14 human NLRPs (see online supplementary figure S4).
The phylogenetic tree (figure 4) demonstrates that NLRP1 is in the same cluster as NLRP3, 6, 10, and 12. The remaining NLRP types are in a mammalian reproduction related NLRP cluster including NLRP2, 4, 5, 7, 8, 9, 11, 13 and 14.40
Figure 4.
The phylogenetic tree of the NLRP family. The multiple alignment contained 132 sequences of the NLRP family and was used to create a phylogenetic tree using the maximum likelihood method with gamma distribution. The NLRP1 subtype was found to be in the same cluster with NLRP subtypes 3, 6, 10, and 12 which were related to inflammatory and immune response (highlight in yellow). The other subtypes belong to a mammalian reproduction related cluster (highlight in grey). An amino acid residue of other human NLRPs that is corresponding to the single nucleotide variant position of NLRP1 is also described in the tree.
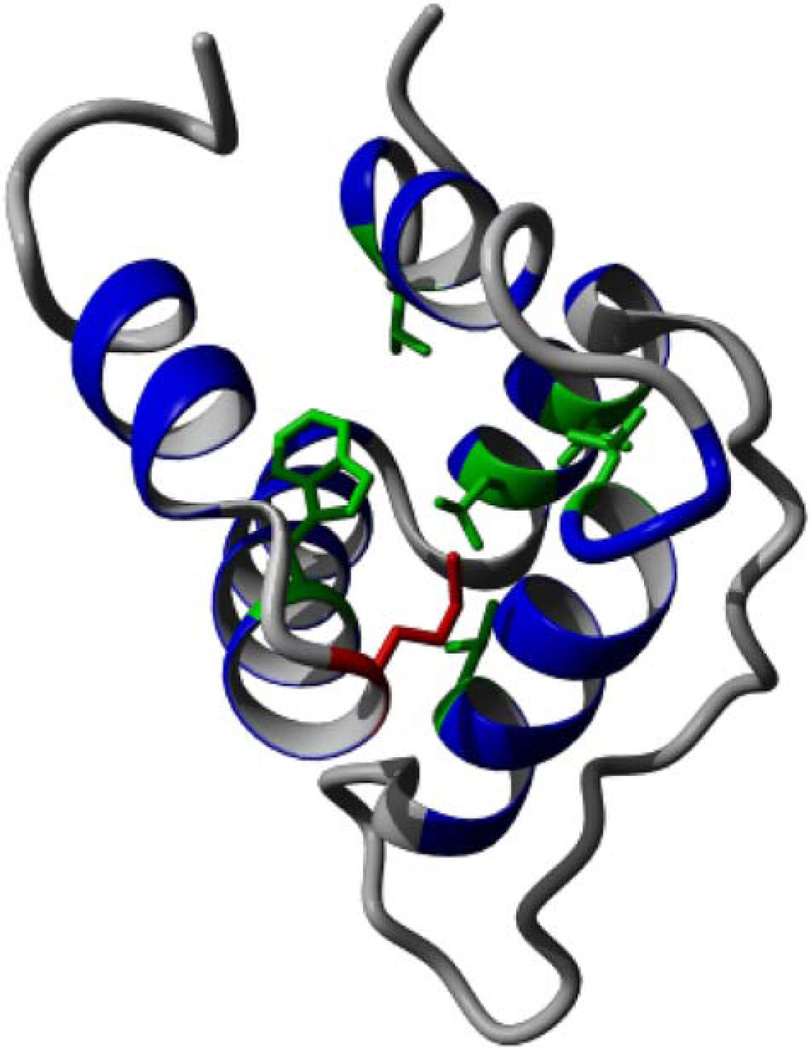
NLRP1 has a pyrin domain in N-terminus (figure 5) consisting of five α helices.38 Methionine 77 is located at the end of the fourth helix.38 By using FoldX,39 which predicts the effect of the SNV on protein structure stability, the average free energy after mutation was predicted to be significantly higher, thus destabilising the protein structure (see online supplementary table S5). Methionine 77 (shown in red in figure 6) is part of the pyrin domain hydrophobic core, together with residues at positions 9, 21, 28, 53, 57, and 74 (shown in green in figure 6),38 indicating its structural importance. Mapping the conservation of the residues from the multiple alignment of all NLRP1 family subtypes to the surface of the structure demonstrated that the SNV region is moderately to highly conserved (see online supplementary figure S5).
Figure 5.

Domain architecture of NLRP1 using UNIPROT information: NLRP1 has a pyrin domain in its N-terminus followed by a central nucleotide -binding and oligomerisation domain (NACHT), six leucine-rich repeats (LRRs), and a caspase recruitment domain (CARD) at the C-terminus.
Figure 6.
Protein structure of the NLRP1 pyrin domain contains five helices with a flexible, disordered loop (PDB ID: 1PN5). The variant, M77, is shown in red and other conserved hydrophobic core residues are shown in green.
DISCUSSION
Intracorneal dyskeratosis is a rare condition. The classical form, called HBID, presents with an autosomal dominant inheritance pattern and clinical, histopathological and genetic characteristics. Most HBID cases have been reported in Native American Haliwa-Saponi Indian tribe members residing in the Halifax and Warren counties in North Carolina.1,3,4,6,7
We report herein a French Caucasian family with corneal histological features similar to the original, classic Native American HBID cases. However, different aspects distinguish our French case from classic HBID cases.1,2 First, the proband has no Native American ancestry. Second, in this report the cornea is widely affected causing vision impairment, whereas in the classic cases, lesions consist generally of white-greyish corneo-conjunctival epithelial plaques2,3 with clear corneal regions, thus rarely impairing visual acuity.2 Thirdly, the onset of the disease is usually at birth or in childhood,6 whereas the proband’s mother (II:2) developed the disease at 27 years of age. Contrary to our family, North Carolina HBID affected patients do not have general involvement, such as pruritic hyperkeratotic scars, palmoplantar hyperkeratosis, chronic rhinitis, vocal cord involvement, and nail thickening.1,2 In addition, among the affected tissues’ epithelial features, there is an absence of Bowman’s layer, which is unusual for HBID.2 Genotypically, our family displayed a de novo mutation event with the mother (II:2), whereas classic North Carolina HBID cases demonstrate 100% penetrance with no skipped generations. The French family did not have chromosome 4q35 duplication, which is associated with the classic form of HBID. Lastly, there were no NLRP1 non-synonymous variants segregating with HBID in our DNA samples from Native American affected cases.
Next generation sequencing is a powerful tool to discover novel mutations, particularly in rare diseases.18,19 Herein, we identified the gene mutation for a novel form of corneal intraepithelial dyskeratosis by analysing exomes of affected and unaffected individuals and using stringent variant filtration criteria. Only one coding non-synonymous variant remained after the filtration strategy. This mutation, located in NLRP1 at position 230 on cDNA (c.230T>C), converting methionine 77 to threonine, was de novo for the mother, and was predicted to affect protein function according to Polyphen-230 and SIFT.31 This sole mutation co-segregated with the disease, was absent from all databases, and was not found in 738 genomes of unrelated unaffected individuals (1476 chromosomes). Moreover, comparative analyses of NLRP1 in other species showed that M77 is highly conserved among primates, mouse, dog and elephant.24
NLRP1 is widely expressed, particularly in epithelial cells. NLRP1 variants are associated with various diseases including coeliac disease, Addison’s disease, type 1 diabetes, Alzheimer’s disease, systemic sclerosis, inflammatory bowel disease, and vitiligo,41–44 but not with Vogt–Koyanagi–Harada disease clinical manifestations.45
NLRP1 maps to chromosome 17p13.2, and belongs to members of the NLR (Nod-like receptors) protein family. As a member of the apoptosis proteins Ced-4 family,38 NLRP1 protein contains a caspase recruitment domain (CARD) and is involved in programmed cell death (http://www.ncbi.nlm.nih.gov/gene/22861) through activation of caspases 3 and 9. Cell apoptosis is induced by NLRP1 overexpression. To date, several NLRP1 transcripts and their encoded isoforms have been described; however their biological functions remain to be investigated.46
Moreover, NLRP1 is involved in the NOD (nucleotide oligomer-isation domain)-like receptor signalling pathway (PATH:hsa04621) (http://www.genome.jp/dbget-bin/www_bget?hsa:22861) and in inflammasome assembly.47 NOD-like receptors are cytoplasmic proteins involved in regulation of inflammation and apoptosis.38,47 Due to evolutionary gene duplication, NOD-like receptors are comprised of 22 family members in human48 and can be divided into three subfamilies based on modular structure: NODs (NOD1-5, CIITA), NLRPs (NLRP1-14), and IPAF subfamily (IPAF/NLRC4, NAIP). All NOD-like receptors share a central nucleotide binding and oligomerisation domain (NACHT) followed by leucine-rich repeats (LRRs) at the C-terminus (with the exception of NLRP10 which lacks these LRRs). The NLRP family is further distinguished by the presence of a pyrin domain at the amino terminus (figure 5), whereas the other NOD-like receptors family members exhibit a caspase activation and recruitment domain (CARD) in the same position.41,48 Interestingly, a rare non-synonymous NLRP1 SNV—A43T (rs145709003)—corresponds to an R42W amino acid change of the pyrin protein, associated with familial Mediterranean fever.38 Moreover, it has been shown that interleukin 1β (IL1β), produced by the inflammasome,47 could participate in normal epithelial growth/differentiation and plays a role in corneal wound healing.49,50 Therefore, the recurrent epithelial and stromal corneal features in our corneal intraepithelial dyskera-tosis case may be a result of misregulation due to chronic inflammatory responses.
Thus, NLRP1 apoptosis and cytokine pathway involvement and its widespread tissue expression—particularly in corneal epithelium—support the plausible causality of this novel NLRP1 SNV for a novel form of corneal intraepithelial dyskeratosis. Functional and animal model studies are needed to confirm NLRP1 involvement in intraepithelial dyskeratosis pathophysiology.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Emilie Tournier and Dr Camille Laurent (Service d’anatomo-pathologie, Purpan Hospital, Toulouse, France) for their help.
Funding This study was supported by the Toulouse Hospital Young Researcher Fellowship, the Fondation pour la Recherche Médicale, Fondation de France (Vincent José Soler) and by a grant from the French Ministry of Health (PHRCN 2007, 0622201) (François Malecaze). Vachiranee Limviphuvadh was supported through A*STAR JCO grant JCOAG04_FG03_2009. This study was also supported by the National Institutes of Health Grant EY014685, by the Lew Wasserman Award from Research To Prevent Blindness Inc, and by the Duke-NUS Signature Research Programs, funded by the Singapore Agency for Science, Technology, and Research and the Singapore Ministry of Health (Steven G Rozen and Terri L Young).
Footnotes
Contributors All the authors contribute to: conception and design (VJS, KNTV), collection of data (VJS, KNTV, SDG, VL, TPK, ESG, PRF, CG, SM-S, FH, CS, BK, NAA, IC, XL, WM, MC, J-LA), data analysis and interpretation (VJS, KNTV, SDG, VL, TPK, SM-S, BK, NAA, IC, XL, PC, SGR, FM, TLY), drafting of the article (VJS, KNTV, SDG, VL, TPK, ESG, PRF, CG, SM-S, FH, CS, BK, NAA, IC, XL, WM, MC, J-LA) or revising it critically for important intellectual content (VJS, KNTV, VL, SM-S, BK, PC, SGR, FM, TLY), and the final approval of the version to be published (all authors).
Competing interests None.
Patient consent Obtained.
Ethics approval Duke University Institutional Review Board (Durham, North Carolina, USA) and the Comité de Protection des Personnes Sud-Ouest et Outre-Mer II (Toulouse, France) and Comité de Protection des Personnes Sud-Ouest et Outre-Mer II (Toulouse, France).
Provenance and peer review Not commissioned; externally peer reviewed.
Web resources
http://www.ncbi.nlm.nih.gov/omim,
http://biowulf.nih.gov/apps/bwa.html,
http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit,
http://www.ncbi.nlm.nih.gov/projects/SNP/,
http://www.ncbi.nlm.nih.gov/Ccdsbrowse.cgi,
http://www.ensembl.org/index.html,
http://www.ncbi.nlm.nih.gov/RefSeq/,
http://jura.wi.mit.edu/entrez_gene/,
http://evs.gs.washington.edu/EVS/,
http://genetics.bwh.harvard.edu/pph2/index.shtml,
http://sift.bii.a-star.edu.sg/,
http://frodo.wi.mit.edu/primer3,
http://www.ambion.com/techlib/prot/fm_1560.pdf,
http://www.uniprot.org/uniprot/Q9C000,
http://annotator.bii.a-star.edu.sg,
http://www.rcsb.org/pdb/home/home.do,
http://hgdownload.cse.ucsc.edu/goldenPath/hg19/chromosomes/,
http://www.ncbi.nlm.nih.gov/genbank/,
REFERENCES
- 1.von Sallmann L, Paton D. Hereditary Dyskeratosis of the Perilimbal Conjunctiva. Trans Am Ophthalmol Soc. 1959;57:53–62. [PMC free article] [PubMed] [Google Scholar]
- 2.Reed JW, Cashwell F, Klintworth GK. Corneal manifestations of hereditary benign intraepithelial dyskeratosis. Arch Ophthalmol. 1979;97:297–300. doi: 10.1001/archopht.1979.01020010149011. [DOI] [PubMed] [Google Scholar]
- 3.Cummings TJ, Dodd LG, Eedes CR, Klintworth GK. Hereditary benign intraepithelial dyskeratosis: an evaluation of diagnostic cytology. Arch Pathol Lab Med. 2008;132:1325–1328. doi: 10.5858/2008-132-1325-HBIDAE. [DOI] [PubMed] [Google Scholar]
- 4.McLean IW, Riddle PJ, Schruggs JH, Jones DB. Hereditary benign intraepithelial dyskeratosis. A report of two cases from Texas. Ophthalmology. 1981;88:164–168. doi: 10.1016/s0161-6420(81)35058-1. [DOI] [PubMed] [Google Scholar]
- 5.Jham BC, Mesquita RA, Aguiar MC, Carmo MA. Hereditary benign intraepithelial dyskeratosis: a new case? J Oral Pathol Med. 2007;36:55–57. doi: 10.1111/j.1600-0714.2006.00456.x. [DOI] [PubMed] [Google Scholar]
- 6.Allingham RR, Seo B, Rampersaud E, Bembe M, Challa P, Liu N, Parrish T, Karolak L, Gilbert J, Pericak-Vance MA, Klintworth GK, Vance JM. A duplication in chromosome 4q35 is associated with hereditary benign intraepithelial dyskeratosis. Am J Hum Genet. 2001;68:491–494. doi: 10.1086/318194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shields CL, Shields JA, Eagle RC., Jr Hereditary benign intraepithelial dyskeratosis. Arch Ophthalmol. 1987;105:422–423. doi: 10.1001/archopht.1987.01060030142045. [DOI] [PubMed] [Google Scholar]
- 8.Baroni A, Palla M, Aiello FS, Ruocco E, Faccenda F, Vozza A, Satriano RA. Hereditary benign intraepithelial dyskeratosis: case report. Int J Dermatol. 2009;48:627–629. doi: 10.1111/j.1365-4632.2009.03893.x. [DOI] [PubMed] [Google Scholar]
- 9.Gombos F, Ruocco V, Satriano RA. (Benign hereditary intraepithelial dyskeratosis. Study of a family nucleus) G Ital Dermatol Venereol. 1986;121:97–101. [PubMed] [Google Scholar]
- 10.Cai R, Zhang C, Chen R, Bi Y, Le Q. Clinicopathological features of a suspected case of hereditary benign intraepithelial dyskeratosis with bilateral corneas involved: a case report and mini review. Cornea. 2011;30:1481–1484. doi: 10.1097/ICO.0b013e31820357e2. [DOI] [PubMed] [Google Scholar]
- 11.Dithmar S, Stulting RD, Grossniklaus HE. (Hereditary benign intraepithelial dyskeratosis) Ophthalmologe. 1998;95:684–686. doi: 10.1007/s003470050335. [DOI] [PubMed] [Google Scholar]
- 12.Haisley-Royster CA, Allingham RR, Klintworth GK, Prose NS. Hereditary benign intraepithelial dyskeratosis: report of two cases with prominent oral lesions. J Am Acad Dermatol. 2001;45:634–636. doi: 10.1067/mjd.2001.116336. [DOI] [PubMed] [Google Scholar]
- 13.Witkop CJ, Jr, Shankle CH, Graham JB, Murray MR, Rucknagel DL, Byerly BH. Hereditary benign intraepithelial dyskeratosis. II. Oral manifestations and hereditary transmission. Arch Pathol. 1960;70:696–711. [PubMed] [Google Scholar]
- 14.von Sallmann L, Paton D. Hereditary benign intraepithelial dyskeratosis. I. Ocular manifestations. Arch Ophthalmol. 1960;63:421–429. [Google Scholar]
- 15.Mardis ER. A decade’s perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 16.Pareek CS, Smoczynski R, Tretyn A. Sequencing technologies and genome sequencing. J Appl Genet. 2011;52:413–435. doi: 10.1007/s13353-011-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 18.Ku CS, Naidoo N, Pawitan Y. Revisiting Mendelian disorders through exome sequencing. Hum Genet. 2011;129:351–370. doi: 10.1007/s00439-011-0964-2. [DOI] [PubMed] [Google Scholar]
- 19.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean M, White MB, Amos J, Gerrard B, Stewart C, Khaw KT, Leppert M. Multiple mutations in highly conserved residues are found in mildly affected cystic fibrosis patients. Cell. 1990;61:863–870. doi: 10.1016/0092-8674(90)90196-l. [DOI] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pedersen JS, Hsu F, Hinrichs AS, Harte RA, Diekhans M, Clawson H, Bejerano G, Barber GP, Baertsch R, Haussler D, Kent WJ. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, Coates G, Fairley S, Fitzgerald S, Fernandez-Banet J, Gordon L, Graf S, Haider S, Hammond M, Holland R, Howe K, Jenkinson A, Johnson N, Kahari A, Keefe D, Keenan S, Kinsella R, Kokocinski F, Kulesha E, Lawson D, Longden I, Megy K, Meidl P, Overduin B, Parker A, Pritchard B, Rios D, Schuster M, Slater G, Smedley D, Spooner W, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wilder S, Zadissa A, Birney E, Cunningham F, Curwen V, Durbin R, Fernandez-Suarez XM, Herrero J, Kasprzyk A, Proctor G, Smith J, Searle S, Flicek P. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.UniProt C. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ooi HS, Kwo CY, Wildpaner M, Sirota FL, Eisenhaber B, Maurer-Stroh S, Wong WC, Schleiffer A, Eisenhaber F, Schneider G. ANNIE: integrated de novo protein sequence annotation. Nucleic Acids Res. 2009;37:W435–W440. doi: 10.1093/nar/gkp254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26:1899–1900. doi: 10.1093/bioinformatics/btq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiller S, Kohl A, Fiorito F, Herrmann T, Wider G, Tschopp J, Grutter MG, Wuthrich K. NMR structure of the apoptosis- and inflammation-related NALP1 pyrin domain. Structure. 2003;11:1199–1205. doi: 10.1016/j.str.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 39.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian X, Pascal G, Monget P. Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evol Biol. 2009;9:202. doi: 10.1186/1471-2148-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zambetti LP, Laudisi F, Licandro G, Ricciardi-Castagnoli P, Mortellaro A. The rhapsody of NLRPs: master players of inflammation … and a lot more. Immunol Res. 2012;53:78–90. doi: 10.1007/s12026-012-8272-z. [DOI] [PubMed] [Google Scholar]
- 42.Spritz RA. Recent progress in the genetics of generalized vitiligo. J Genet Genomics. 2011;38:271–378. doi: 10.1016/j.jgg.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zurawek M, Fichna M, Januszkiewicz-Lewandowska D, Gryczynska M, Fichna P, Nowak J. A coding variant in NLRP1 is associated with autoimmune Addison’s disease. Hum Immunol. 2010;71:530–534. doi: 10.1016/j.humimm.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Dieude P, Guedj M, Wipff J, Ruiz B, Riemekasten G, Airo P, Melchers I, Hachulla E, Cerinic MM, Diot E, Hunzelmann N, Caramaschi P, Sibilia J, Tiev K, Mouthon L, Riccieri V, Cracowski JL, Carpentier PH, Distler J, Amoura Z, Tarner I, Avouac J, Meyer O, Kahan A, Boileau C, Allanore Y. NLRP1 influences the systemic sclerosis phenotype: a new clue for the contribution of innate immunity in systemic sclerosis-related fibrosing alveolitis pathogenesis. Ann Rheum Dis. 2011;70:668–674. doi: 10.1136/ard.2010.131243. [DOI] [PubMed] [Google Scholar]
- 45.Horie Y, Saito W, Kitaichi N, Miura T, Ishida S, Ohno S. Evaluation of NLRP1 gene polymorphisms in Vogt-Koyanagi-Harada disease. Jpn J Ophthalmol. 2011;55:57–61. doi: 10.1007/s10384-010-0887-9. [DOI] [PubMed] [Google Scholar]
- 46.D’Osualdo A, Reed JC. NLRP1, a regulator of innate immunity associated with vitiligo. Pigment Cell Melanoma Res. 2012;25:5–8. doi: 10.1111/j.1755-148X.2011.00942.x. [DOI] [PubMed] [Google Scholar]
- 47.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 48.Proell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS One. 2008;3:e2119. doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li DQ, Tseng SC. Three patterns of cytokine expression potentially involved in epithelial-fibroblast interactions of human ocular surface. J Cell Physiol. 1995;163:61–79. doi: 10.1002/jcp.1041630108. [DOI] [PubMed] [Google Scholar]
- 50.Li DQ, Tseng SC. Differential regulation of cytokine and receptor transcript expression in human corneal and limbal fibroblasts by epidermal growth factor, transforming growth factor-alpha, platelet-derived growth factor B, and interleukin-1 beta. Invest Ophthalmol Vis Sci. 1996;37:2068–2080. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.