Abstract
Background
Somatic embryogenesis is a complex process regulated by numerous factors. The identification of proteins that are differentially expressed during plant development could result in the development of molecular markers of plant metabolism and provide information contributing to the monitoring and understanding of different biological responses. In addition, the identification of molecular markers could lead to the optimization of protocols allowing the use of biotechnology for papaya propagation and reproduction. This work aimed to investigate the effects of polyethylene glycol (PEG) on somatic embryo development and the protein expression profile during somatic embryo maturation in papaya (Carica papaya L.).
Results
The maturation treatment supplemented with 6% PEG (PEG6) resulted in the greatest number of somatic embryos and induced differential protein expression compared with cultures grown under the control treatment. Among 135 spots selected for MS/MS analysis, 76 spots were successfully identified, 38 of which were common to both treatments, while 14 spots were unique to the control treatment, and 24 spots were unique to the PEG6 treatment. The identified proteins were assigned to seven categories or were unclassified. The most representative class of proteins observed in the control treatment was associated with the stress response (25.8%), while those under PEG6 treatment were carbohydrate and energy metabolism (18.4%) and the stress response (18.4%).
Conclusions
The differential expression of three proteins (enolase, esterase and ADH3) induced by PEG6 treatment could play an important role in maturation, and these proteins could be characterized as candidate biomarkers of somatic embryogenesis in papaya.
Keywords: Papaya, Somatic embryogenesis, Protein profile, Mass spectrometry, Two-dimensional electrophoresis
Background
Papaya (Carica papaya L. - Caricaceae) is one of the most commonly cultivated fruit trees in tropical and subtropical regions and one of the most frequently consumed fruits in the world [1]. Since the papaya genome was sequenced, some of the identified features have made papaya an excellent model system for studying tropical plants [2,3], including proteomic studies [4]. A good model system in biology should be easy to grow and maintain, have a strong genetic and genomic foundation and be in a suitable position phylogenetically for comparative studies [2].
A recurrent problem in the cultivation of this species has been seed propagation, which generates heterogeneous plants and results in a reduction of fruit production [5]. One solution for increasing the production of this species involves the cloning of elite cultivars that generate more uniform plants. Somatic embryogenesis, a biotechnological tool, has the potential to achieve high multiplication rates to scale up production.
Somatic embryogenesis was first induced in papaya by Bruijne et al. [6], and many efforts have since been made to optimize the process. These protocols have been hampered due to the numerous variables that must be regulated during the process, including the endogenous levels of phytohormones, proteins and genetic and epigenetic factors [7]. Maturation is a critical process during embryo development for the conversion of somatic embryos into plantlets [8]. This stage is characterized by cellular expansion, differentiation and the accumulation of reserve substances and is therefore a key determinant of successful somatic embryo regeneration and the conversion of somatic embryos into plantlets [9].
Water stress, such as that produced by polyethylene glycol (PEG), has been proposed to be an important factor during the maturation of somatic embryos [10-13] and is potentially able to unleash rapid biochemical changes in the activity of specific proteins during maturation [14]. These high-molecular-weight molecules are not able to pass through the cell wall, which leads to a restriction of water absorption, low turgor pressure and a reduction in the intracellular osmotic potential [10], causing desiccation. PEG has been used as a maturation agent in various species, including Hevea brasiliensis[15], Glycine max[16], Aesculus hippocastanum[17], Prosopis laevigata[18] and C. papaya[19,20].
Proteomic studies in plants have resulted in advances in knowledge with regard to the structure, genetic organization and evolution of plant genomes [21]. Proteomic studies in papaya are still scarce but have been increasing in recent years [4,22-25]. However, no reports currently exist that relate protein levels to the development and control of somatic embryogenesis in papaya.
Many studies have been conducted using this tool to understand the mechanisms that control somatic embryogenesis in various species, such as Vitis vinifera[26], Phoenix dactylifera[27], Cyclamen persicum[28], Theobroma cacao[29], Crocus sativus[30] and Quercus suber[31]. The determination of dynamic changes in protein concentrations during embryonic development could lead to improvements in somatic embryogenesis protocols that would allow the use of biotechnological tools for papaya propagation and breeding. The present study investigated the effects of PEG on somatic embryo development and on the protein expression profile during somatic embryo maturation in papaya.
Results and discussion
Maturation and conversion of somatic embryos
A significant effect of PEG was observed with regard to the maturation of somatic papaya embryos after 42 days of culture (Table 1 and Figure 1). The cultures incubated under the control and PEG6 treatments yielded 15 and 40 cotyledonary somatic embryos per colony (Table 1), respectively. The PEG6 treatment enabled the conversion of the somatic embryos into plantlets with leaves and stems, similar to those with a seed origin (Figure 1).
Table 1.
Number of cotyledonary somatic embryos (per 300 mg of cells initially inoculated), percentage of dry matter (DM) and the protein content in embryogenic cultures of the C. papaya hybrid UENF/CALIMAN01 after 42 days of incubation under the different maturation treatments
| Variables |
PEG (%) |
|||
|---|---|---|---|---|
| 0 | 3 | 6 | 9 | |
|
NSE |
15.0 b* |
20.0 b |
40.0 a |
23.0 b |
|
DM (%) |
11.0 b |
12.9 b |
16.2 a |
17.1 a |
| Protein (mg/g DM) | 50.7 a | 49.8 a | 62.2 a | 2.0 b |
*Means followed by the same letters are not significantly different according to the SNK test (P < 0.05). NSE: C.V.(%) = 26.7, n = 4; DM: C.V.(%) = 9.9, n = 4; Protein (mg/g DM): C.V.(%) = 18.3, n = 3. PEG - polyethylene glycol; DM - dry matter; NSE - number of somatic embryos.
Figure 1.
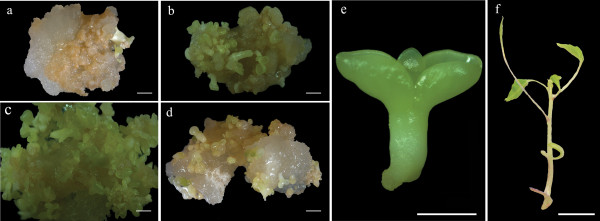
Morphological characteristics of embryogenic cultures of C. papaya subjected to maturation treatments (control) (A); (PEG3) (B); (PEG6) (C) and (PEG9) (D). Morphological characteristics of cotyledonary (mature) somatic embryos (E); and regenerated plantlets (F). PEG: polyethylene glycol; Bars: (A-D): 0.5 mm; (E): 0.2 mm and (F): 15 mm.
There were significant differences in the percentage of dry matter (DM) in the cultures subjected to different concentrations of PEG. The cultures matured under 6 and 9% PEG showed the highest percentages of DM (Table 1). These results indicate that PEG promoted an increase in the DM content of the cultures.
According to the obtained results, the addition of PEG significantly affected the maturation of somatic embryos as well as the protein concentrations and profiles in these embryos during somatic embryogenesis in the C. papaya hybrid UENF/CALIMAN01. PEG6 treatment resulted in the greatest numbers of cotyledonary somatic embryos (Table 1). Similar results were previously described during somatic embryo maturation in this papaya hybrid [20], where the authors observed increased promotion of the maturation of embryogenic cultures when using PEG compared with the control treatment (i.e., without PEG). Koehler et al. [32] found that supplementation with 5% PEG combined with 2 g/L activated charcoal and 5 μM abscisic acid (ABA) improved the quality of somatic embryos and increased the formation of plantlets. These authors concluded that PEG was an important inducer of maturation that aided in the conversion of somatic papaya embryos.
Maturation is a crucial phase during somatic embryogenesis, and the addition of maturation promoters such as PEG, ABA and activated charcoal is crucial for promoting the maturation of somatic embryos and for their conversion into plantlets [33].
PEG is a non-plasmolysing osmoticum that is unable to readily penetrate plant cell walls and produces an in vitro effect similar to that of natural water stress during zygotic embryogenesis [12]. PEG has been shown to enhance the accumulation of storage reserves and increase desiccation tolerance in cotyledonary somatic embryos, and it has also been shown to have a slight impact on endogenous ABA levels in angiosperms and conifers [10-12]. PEG has been reported to act by controlling the expression of several genes involved in embryonic differentiation and the development of apical meristems [13].
During the development of the zygotic embryo, desiccation and the accumulation of reserve substances have been directly related to the differential expression of genes and metabolic pathways, which are critical processes for the successful development and germination of the embryo [9]. The main storage reserves found in the seeds of papaya are lipids (28.8) and proteins (27.8), while the seeds are poor in free monosaccharides, with sucrose being the main sugar present [34]. Somatic embryogenesis, which mimics zygotic embryo development, has been considered as a model for studying zygotic embryogenesis. Studies have been conducted to shed light on embryonic development in many species, such as Citrus sinensis[35], Picea sp. [36,37] and Cyclamen sp. [38].
The maturation of somatic embryos also appears to be highly dependent on stress, such as that induced by PEG. Stress modifies DNA methylation patterns and induces the synthesis of proteins that are essential to embryonic development, enabling embryos to mature [39], as observed in the present work.
Histomorphological sections revealed that cultures subjected to PEG6 treatment presented more cells with embryogenic characteristics. PEG induces a high proportion of small isodiametric cells with dense cytoplasm and large nuclei, organized in meristematic aggregates (Figure 2), as previously described in embryogenic cultures of papaya [40]. Histochemical analysis of these cultures showed that cells with embryogenic characteristics, meristematic aggregates and somatic embryos exhibited an intense reaction to the protein dye CBB (Figure 2). Proteins were visualized in the form of granules in embryos from early stages, as in cotyledonary embryos (Figure 2E, F). In cotyledonary embryos, the proteins were more abundant in the basal region of the embryo (Figure 2F).
Figure 2.
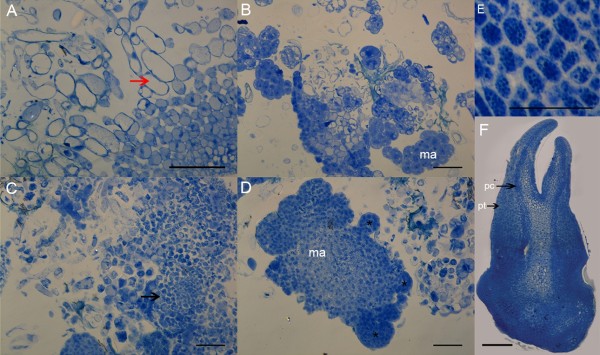
Protein staining with Coomassie brilliant blue R-250 (CBB) in embryogenic cultures of C. papaya after 42 days of incubation in the control (without PEG) and PEG6 (with 6% PEG) maturation treatments. (A-B) Control treatment; (C-D) PEG6 treatment; (E) protein granules in embryogenic cells; (F) cotyledonary somatic embryo under PEG6 treatment. Red arrow: non-embryogenic cells; black arrows: embryogenic cells, ma: meristematic aggregates; Asterisks (*): proembryo; pt: protoderm; pr: procambium. PEG: polyethylene glycol. Bars: 200 μm.
In this context, the increase in protein content observed in cultures incubated under PEG6 treatment is associated with large amounts of protein bodies identified in meristematic cells and somatic embryo tissues, which were more abundant in this treatment. The PEG-induced proteins could be an important factor in somatic embryo development and posterior plantlet conversion. The synthesis of specific proteins, such as storage proteins, is known to be crucial for in vitro morphogenesis, serving as the principal nitrogen source during seed germination [41,42].
Proteomic analysis
Significant differences in protein concentrations were observed between the different treatments (Table 1). The PEG9 treatment induced a reduce in soluble proteins (2.0 mg of proteins/g of DM), and there were no statistically significant differences between control, PEG3 and PEG 6 treatments (Table 1). These results indicate that under the condition of 9% PEG, the stress was so high that cell metabolism was reduced and protein synthesis was affected.Because PEG6 was determined to be the best treatment for the development of mature somatic embryos and led to the highest protein content, the 2-DE profiles of embryogenic cultures from the control and PEG6 treatments were analyzed (Figure 3). The numbers of spots obtained for the control and PEG6 treatments were 422 and 688, respectively, while 217 spots were common to the two treatments.
Figure 3.
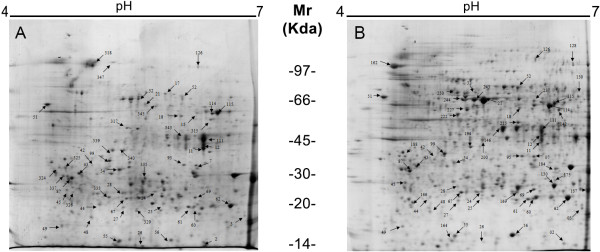
Two-dimensional gel electrophoresis (2-DE) reference maps of embryogenic cultures of the C. papaya after 42 days of incubation in the (A) control (without PEG) and (B) PEG6 (with 6% PEG) maturation treatments. PEG: polyethylene glycol
In addition to being the best treatment for achieving somatic embryo maturation (Table 1), PEG6 induced a change in the 2-DE profile (Figure 3) and number of identified spots compared with the control treatment. The actions of PEG as a maturation agent could be related to the induction of specific proteins associated with the significant dehydration process observed during late embryogenesis, or the PEG-induced changes could be related to the synthesis of storage proteins promoted by water stress [10]. In this context, analyses of molecular and biochemical changes, such as the protein profiles induced by PEG, can provide important information for understanding somatic embryogenesis because proteins play an essential role during somatic embryo histodifferentiation [43]. Additionally, proteins represent the main reserves that accumulate during embryonic development in most species [44].
Of the 135 protein spots selected for MS/MS analysis, 76 were successfully identified, and 38 proteins were common to the two treatments. Among these proteins, 14 were up-regulated, while 3 were down-regulated in the control versus the PEG6 treatment. Additionally, 14 spots were unique to the control treatment, and 24 spots were unique to the PEG6 treatment. Representative graphs of the mean percentage volumes (%vol) of each of these spots are provided in Table 2. In the present study, identification of the proteins was relied essentially on homology search to known sequences of the other plant species because of the poor protein sequence information that is currently available for Carica Papaya at the NCBInr database.
Table 2.
Expressed proteins identified via mass spectrometry in the embryogenic cultures of the C. papaya after 42 days in maturation treatments
| Spot a | Tratament b | Function c | Accession number d | Protein name | Organism | Peptides number e | Coverage (%) f | Score g |
Abundance
h
|
|---|---|---|---|---|---|---|---|---|---|
| Control PEG6 | |||||||||
|
Proteins of higher abundance in PEG6 treatment | |||||||||
| 017 |
Control/PEG6 |
Stress response, glycolysis |
gi|3914394 |
2,3-bisphosphoglycerate-independent phosphoglycerate mutase |
Mesembryanthemum crystallinum |
4 |
8% |
196 |
 |
| 021 |
Control/PEG6 |
Carbohydrate and energy metabolism |
gi|3023685 |
Full = Enolase |
Alnus glutinosa |
3 |
10% |
199 |
 |
| 114 |
Control/PEG6 |
Stress response |
gi|211906436 |
UDP-D-glucose pyrophosphorylase |
Gossypium hirsutum |
4 |
14% |
363 |
 |
| 128 |
PEG6 |
Stress response, Others functions |
gi|151347486 |
Methionine synthase |
Carica papaya |
3 |
5% |
70 |
 |
| 130 |
PEG6 |
Metabolic process |
gi|1743354 |
Aldehyde dehydrogenase (NAD+) |
Nicotiana tabacum |
2 |
4% |
137 |
 |
| 138 |
PEG6 |
Stress response, oxidation-redution process, Carbohydrate and energy metabolism |
gi|15222848 |
Glyceraldehyde 3-phosphate dehydrogenase |
Arabidopsis thaliana |
3 |
15% |
147 |
 |
| 139 |
PEG6 |
Protein metabolism, stress response |
gi|3914394 |
2,3-bisphosphoglycerate-independent phosphoglycerate mutase |
Mesembryanthemum crystallinum |
2 |
3% |
68 |
 |
| 142 |
PEG6 |
Protein metabolism, stress response |
gi|21487 |
Leucine aminopeptidase |
Solanum tuberosum |
1 |
2% |
56 |
 |
| 146 |
PEG6 |
Oxidation-redution process |
gi|29501684 |
Alcohol dehydrogenase 3 |
Petunia x hybrida |
3 |
10% |
136 |
 |
| 162 |
PEG6 |
Protein metabolism, stress response |
gi|38154482 |
Molecular chaperone Hsp90-1 |
Nicotiana benthamiana |
5 |
8% |
189 |
 |
| 163 |
PEG6 |
Others functions |
gi|1707372 |
Ubiquitin-like protein |
Arabidopsis thaliana |
3 |
25% |
119 |
 |
| 165 |
PEG6 |
Unclassified |
gi|20140866 |
Full = Translationally controlled tumor protein homolog |
Cucumis melo |
2 |
13% |
83 |
 |
| 166 |
PEG6 |
Other functions |
gi|295885749 |
Cytokinin oxidase 2 |
Triticum aestivum |
1 |
1% |
46 |
 |
| 169 |
PEG6 |
Fatty acid metabolism, Carbohydrate and energy metabolism |
gi|136057 |
Full = Triosephosphate isomerase, cytosolic |
Coptis japonica |
1 |
3% |
55 |
 |
| 175 |
PEG6 |
Fatty acid metabolism |
gi|147784332 |
Hypothetical protein VITISV_041523 |
Vitis vinifera |
2 |
9% |
69 |
 |
| 184 |
PEG6 |
Metabolic process |
gi|255565327 |
Esterase D, putative |
Ricinus communis |
1 |
6% |
46 |
 |
| 188 |
PEG6 |
Carbohydrate and energy metabolism |
gi|82621108 |
Phosphoglycerate kinase-like |
Solanum tuberosum |
2 |
8% |
212 |
 |
| 194 |
PEG6 |
Oxidation-reduction process, fatty acid metabolism |
gi|2204087 |
Enoyl-ACP reductase |
Arabidopsis thaliana |
2 |
12% |
75 |
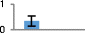 |
| 200 |
PEG6 |
Seed germination, cytoskeleton organization |
gi|32186896 |
Actin |
Gossypium hirsutum |
6 |
22% |
454 |
 |
| 215 |
PEG6 |
Oxidation-redution process, stress response |
gi|449458315 |
PREDICTED: glutamate dehydrogenase 2-like |
Cucumis sativus |
2 |
6% |
95 |
 |
| 222 |
PEG6 |
Others functions |
gi|38564733 |
Initiation factor eIF4A-15 |
Helianthus annuus |
3 |
14% |
190 |
 |
| 227 |
PEG6 |
Carbohydrate and energy metabolism |
gi|3023685 |
Full = Enolase |
Alnus glutinosa |
2 |
8% |
110 |
 |
| 237 |
PEG6 |
Stress response |
gi|169661 |
S-adenosylhomocysteine hydrolase |
Petroselinum crispum |
1 |
8% |
45 |
 |
| 243 |
PEG6 |
Carbohydrate and energy metabolism |
gi|3023685 |
Full = Enolase |
Alnus glutinosa |
2 |
8% |
130 |
 |
| 244 |
PEG6 |
Carbohydrate and energy metabolism |
gi|3023685 |
Full = Enolase |
Alnus glutinosa |
2 |
8% |
110 |
 |
| 250 |
PEG6 |
Unclassified |
gi|462413398 |
Hypothetical protein PRUPE_ppa003850mg |
Prunus persica |
3 |
8% |
130 |
 |
|
Proteins of higher abundance in Control treatment | |||||||||
| 002 |
Control/PEG6 |
Unclassified |
gi|384249809 |
HI0933-like protein |
Coccomyxa subellipsoidea |
1 |
2% |
50 |
 |
| 011 |
Control/PEG6 |
Fatty acid metabolism |
gi|257096376 |
Full = GDSL esterase/lipase |
Carica papaya |
2 |
6% |
82 |
 |
| 012 |
Control/PEG6 |
Fatty acid metabolism |
gi|257096376 |
Full = GDSL esterase/lipase; AltName: Full = CpEST |
Carica papaya |
2 |
6% |
82 |
 |
| 018 |
Control/PEG6 |
Stress response, Carbohydrate and energy metabolism |
gi|414866626 |
TPA: hypothetical protein ZEAMMB73_999129 |
Zea mays |
2 |
6% |
64 |
 |
| 025 |
Control/PEG6 |
Oxidation-redution process |
gi|255555109 |
Flavoprotein wrbA, putative |
Ricinus communis |
1 |
11% |
112 |
 |
| 026 |
Control/PEG6 |
Other functions |
gi|162464222 |
Small GTP binding protein1 |
Zea mays |
2 |
11% |
95 |
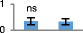 |
| 042 |
Control/PEG6 |
Carbohydrate and energy metabolism |
gi|14423688 |
Full = Enolase 1 |
Hevea brasiliensis |
2 |
8% |
62 |
 |
| 043 |
Control/PEG6 |
Stress response |
gi|2853219 |
Glutathione transferase |
Carica papaya |
5 |
22% |
330 |
 |
| 048 |
Control/PEG6 |
Others functions |
gi|89212810 |
14-3-3-like protein |
Gossypium hirsutum |
4 |
15% |
213 |
 |
| 049 |
Control/PEG6 |
Protein metabolism |
gi|399942 |
Full = Stromal 70 kDa heat shock-related protein |
Pisum sativum |
4 |
9% |
205 |
 |
| 055 |
Control/PEG6 |
Other functions |
gi|162464222 |
Small GTP binding protein1 |
Zea mays |
2 |
11% |
95 |
 |
| 054 |
Control/PEG6 |
Stress response, protein metabolism |
gi|475610277 |
Aspartic proteinase oryzasin-1 |
Aegilops tauschii |
1 |
2% |
60 |
 |
| 111 |
Control/PEG6 |
Fatty acid metabolism |
gi|257096376 |
Full = GDSL esterase/lipase |
Carica papaya |
6 |
18% |
389 |
 |
| 115 |
Control/PEG6 |
Stress response, Carbohydrate and energy metabolism |
gi|7417426 |
UDP-glucose pyrophosphorylase |
Oryza sativa Indica Group |
2 |
6% |
53 |
 |
| 315 |
Control |
Stress response, metabolic process |
gi|32527831 |
UDP-glucose pyrophosphorylase |
Populus tremula x Populus tremuloides |
3 |
9% |
122 |
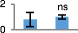 |
| 317 |
Control |
Other functions |
gi|225442221 |
Initiation factor eIF4A-15 |
Vitis vinifera |
4 |
16% |
154 |
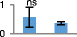 |
| 318 |
Control |
Stress response |
gi|470129411 |
PREDICTED: vicilin-like antimicrobial peptides 2-2-like |
Fragaria vesca subsp. Vesca |
1 |
1% |
49 |
 |
| 324 |
Control |
Protein folding |
gi|166770 |
Heat shock protein 83 |
Arabidopsis thaliana |
1 |
2% |
66 |
 |
| 325 |
Control |
Protein folding |
gi|20559 |
hsp70 (AA 6–651) |
Petunia x hybrid |
2 |
5% |
68 |
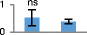 |
| 326 |
Control |
Protein folding |
gi|20559 |
hsp70 (AA 6–651) |
Petunia x hybrid |
2 |
3% |
54 |
 |
| 329 |
Control |
Stress response |
gi|226973436 |
Beta-thioglucoside glucohydrolase |
Carica papaya |
3 |
9% |
165 |
 |
| 333 |
Control |
Oxidation-redution process |
gi|295367043 |
Cinnamoyl alcohol dehydrogenase |
Bambusa multiplex |
5 |
23% |
48 |
 |
| 335 |
Control |
Carbohydrate and enregy metabolism |
gi|359483362 |
PREDICTED: lactoylglutathione lyase |
Vitis vinifera |
3 |
11% |
146 |
 |
| 337 |
Control |
Protein folding |
gi|62433284 |
BiP |
Glycine Max |
2 |
5% |
75 |
 |
| 339 |
Control |
Stress response |
gi|226973436 |
Beta-thioglucoside glucohydrolase |
Prunus pérsica |
3 |
6% |
126 |
 |
| 340 |
Control |
Stress response |
gi|226973436 |
Beta-thioglucoside glucohydrolase |
Carica papaya |
3 |
6% |
108 |
 |
| 343 |
Control |
Fatty acid metabolism |
gi|257096376 |
Full = GDSL esterase/lípase |
Carica papaya |
2 |
6% |
163 |
 |
| 345 |
Control |
Carbohydrate and energy metabolism |
gi|356562473 |
PREDICTED: xylose isomerase-like |
Glycine Max |
3 |
9% |
56 |
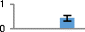 |
| 347 |
Control |
Other functions |
gi|357440579 |
Pentatricopeptide repeat-containing protein |
Medicago truncatula |
7 |
12% |
57 |
 |
|
Proteins expressed similarly in both treatments | |||||||||
| 015 |
Control/PEG6 |
Stress response, metabolic process |
gi|211906436 |
UDP-D-glucose pyrophosphorylase |
Gossypium hirsutum |
5 |
15% |
213 |
 |
| 024 |
Control/PEG6 |
Other functions |
gi|148912162 |
Cytosolic ascorbate peroxidase 1 |
Gossypium hirsutum |
3 |
20% |
221 |
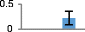 |
| 027 |
Control/PEG6 |
Oxidation-redution process |
gi|449464176 |
PREDICTED: proteasome subunit beta type-3-A-like |
Cucumis sativus |
2 |
15% |
175 |
 |
| 028 |
Control/PEG6 |
Metabolic process |
gi|5669924 |
Soluble inorganic pyrophosphatase |
Populus tremula x Populus tremuloides |
1 |
7% |
37 |
 |
| 032 |
Control/PEG6 |
Stress response |
gi|226973430 |
Beta-thioglucoside glucohydrolase |
Carica papaya |
5 |
13% |
278 |
 |
| 044 |
Control/PEG6 |
Unclassified |
gi|412993224 |
ORF73 |
Bathycoccus prasinos |
1 |
0% |
36 |
 |
| 045 |
Control/PEG6 |
Other functions |
gi|270313547 |
S-adenosylmethionine decarboxylase |
Olea europaea |
1 |
2% |
47 |
 |
| 051 |
Control/PEG6 |
Protein metabolism, Stress response |
gi|233955399 |
Calreticulin |
Carica papaya |
6 |
22% |
323 |
 |
| 052 |
Control/PEG6 |
Oxidation-redution process |
gi|193290660 |
Putative ketol-acid reductoisomerase |
Capsicum annuum |
3 |
10% |
116 |
 |
| 056 |
Control/PEG6 |
Unclassified |
gi|384249809 |
HI0933-like protein |
Coccomyxa subellipsoidea |
1 |
2% |
58 |
 |
| 060 |
Control/PEG6 |
Oxidation-redution process |
gi|449520553 |
PREDICTED: superoxide dismutase [Mn], mitochondrial-like isoform 1 |
Cucumis sativus |
2 |
13% |
204 |
 |
| 061 |
Control/PEG6 |
Oxidation-redution process |
gi|33521626 |
Mn-superoxide dismutase |
Lotus japonicus |
1 |
8% |
65 |
 |
| 062 |
Control/PEG6 |
Unclassified |
gi|384253982 |
Hypothetical protein COCSUDRAFT_39121 |
Coccomyxa subellipsoidea C-169 |
1 |
1% |
44 |
 |
| 067 |
Control/PEG6 |
Unclassified |
gi|460412635 |
PREDICTED: 20 kDa chaperonin, chloroplastic-like |
Solanum lycopersicum |
3 |
12% |
221 |
 |
| 069 |
Control/PEG6 |
Stress response, signaling |
gi|2853219 |
Glutathione transferase |
Carica papaya |
2 |
10% |
41 |
 |
| 087 |
Control/PEG6 |
Others functions |
gi|55375985 |
14-3-3 family protein |
Malus x domestica |
6 |
33% |
504 |
 |
| 095 |
Control/PEG6 |
Stress response |
gi|357514981 |
Annexin-like protein |
Medicago truncatula |
2 |
5% |
57 |
 |
| 099 |
Control/PEG6 |
Strress response |
gi|356531939 |
PREDICTED: putative lactoylglutathione lyase-like |
Glycine Max |
3 |
9% |
152 |
 |
| 126 |
Control/PEG6 |
Metabolic process |
gi|5725356 |
Alpha-D-xylosidase |
Tropaeolum majus |
1 |
1% |
37 |
 |
| 003 |
Control/PEG6 |
Unclassified |
gi|357498189 |
DNA repair and recombination protein PIF1 |
Medicago truncatula |
1 |
2% |
40 |
 |
| 007 | Control/PEG6 | Oxidation-redution process | gi|12322163 | Dormancy related protein, putative | Arabidopsis thaliana | 1 | 3% | 47 |  |
PEG: polyethylene glycol.
aSpot numbers correspond to the numbers indicated in Figure 3.
bMaturation treatments: Control - without PEG; and PEG6 - with 6% PEG.
cFunctional protein classification using Blast2go (http://www.blast2go.com).
dAccession number in the NCBI protein database.
eNumber of unique peptide sequences identified by MASCOT.
fPercentage of predicted protein sequence covered by MASCOT.
gProbability-based MOWSE score from MASCOT software for the hit.
hProtein relative abundance according to the individual spot (%) volume determined using Image Master Platinum v.7 software (GE Healthcare). Change in abundance levels by t-test, *(p ≤ 0.05), **(p ≤ 0.01), ns (not significant).
The identified proteins (Table 2) were assigned to 7 different categories or were unclassified. This classification was performed by comparing the total numbers of proteins in each treatment, including those that were common to the two treatments (Table 2; Figure 4). The most representative class of proteins observed in the control treatment was associated with the stress response (25.8%), while those in the PEG6 treatment were related to carbohydrate and energy metabolism (18.4%) and the stress response (18.4%).
Figure 4.
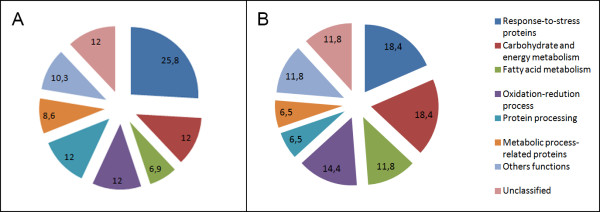
Functional distribution (by Blast2GO) of all 76 proteins identified in the embryogenic cultures of C. papaya after 42 days of incubation in the (A) control without PEG and (B) PEG6 (with 6% PEG) maturation treatments. PEG: polyethylene glycol.
A comparative proteomic analysis between the control and PEG6 treatments was performed in papaya var. UENF/CALIMAN01, and the results are discussed based on the functional classifications of the differentially expressed proteins (Figure 4, Table 2).
Response-to-stress proteins
We identified several stress-related proteins in the embryogenic cultures in both treatments (Figure 4, Table 2). Stress can force cells to switch to an embryogenic state, and recent proteomic studies have strongly emphasized the role of stress proteins during somatic embryogenesis [45]. The similarities of the proteins associated with the stress response identified in the two treatments in the present work indicate that embryogenesis, itself, is a stress inducer that is capable of stimulating various morphogenetic responses, as suggested by Pasternak et al. [46]. These stresses take effect during the regulation of the cell division cycle and reorganize the physiological, metabolic and gene expression patterns that occur during somatic embryo differentiation and development [47].
Glutathione transferase (spots 43 and 69), which was identified in both treatments at similar expression levels, is associated with stress tolerance and is induced by a wide array of biotic and abiotic stresses, including exposure to toxic chemicals, environmental stress and disease [48]. This enzyme was found to be up-regulated in cultures of C. persicum subjected to drought stress caused by transfer from a submerged suspension culture to a semisolid medium [49]. According to our results, the abundance of this protein was similar in the two treatments, indicating that other stresses in addition to water stress can regulate the activity and turnover of this protein during somatic embryo maturation in papaya var. UENF/CALIMAN01.
UDP-glucose pyrophosphorylase (spot 315) was identified in the control treatment, while UDP-D-glucose pyrophosphorylase (spot 15) was found at similar concentrations in the two treatments, and UDP-D-glucose pyrophosphorylase (spot 114) was found at the highest concentration in the PEG6 treatment. These proteins are essential enzymes for the synthesis of sucrose and other nucleotide sugars [50] that are precursors required for cell wall biogenesis [51]. In addition, many proteins involved in cell wall formation during normal development, including actin, are also recruited under stress conditions such as defense-related cell wall-remodeling events [52].
Methionine synthase (spot 128) and S-adenosylhomocysteine hydrolase (spot 237) were unique to the PEG6 treatment. Methionine synthase catalyzes the last step in the plant methionine biosynthesis pathway [53]. It provides methionine for protein synthesis in seeds and is stored throughout the entire process of seed maturation [54]. This protein has also been found in the mature dry seeds of Arabidopsis[55].
Other stress-related proteins identified in the papaya embryogenic cultures have few known functions in plants. For example, 2,3-bisphosphoglycerate-independent phosphoglycerate mutase (spots 17 and 139 ) is involved in plant defense responses, although its function has not yet been fully elucidated [56].
Carbohydrate and energy metabolism proteins
Proteins associated with metabolism and the supply of energy were mainly identified in the PEG6 treatment, rather than the control treatment. These proteins play important roles during plant and embryo development as energy providers, acting in cofactor regeneration and the construction, interconversion and synthesis of metabolites, in addition to playing a role in signaling related to the regulation of various processes [57,58].
Enolase was the most abundant protein identified and was observed in five spots. Three of these spots (227, 243 and 244) were unique to the PEG6 treatment, and two spots (21 and 42) were common to the two treatments, with spot 21 being up-regulated by the PEG treatment (Table 2). Enolase is a ubiquitous enzyme that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate, which is the only dehydration step in the glycolytic pathway, occurring at the end of glycolysis [59]. Enolase was the most frequently detected protein in the seeds of C. persicum, suggesting an important role of this protein in the embryogenesis of this species [60]. Similarly, enolase is the most abundant protein during late somatic embryogenesis in Picea glauca, suggesting that this protein can be used as a marker of embryo maturation [61]. During somatic embryogenesis in Arabidopsis, enolase transcript levels reach a maximum during the torpedo stage [62]. Another spot representing a glycolytic enzyme, corresponding to glyceraldehyde 3-phosphate dehydrogenase (spot 138) was found only in the PEG6 treatment.
Lactoylglutathione lyase proteins were identified in spot 335 in the control treatment and in spot 99 in both treatments. These enzymes are generally associated with cell proliferation [63] and might also be involved in antioxidation processes in tomato species [64].
Proteins involved in fatty acid metabolism
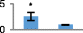
Several proteins identified in the present work play a role in the fatty acid/lipid pathway. Several spots corresponding to GDSL esterase/lipase proteins (spots 11, 12 and 111) were identified in both treatments, being up-regulated in the control treatment, while one spot (343) was unique to the control treatment. Triosephosphate isomerase (spot 169), hypothetical protein VITISV_041523 (spot 175) and enoyl-ACP reductase (spot 194) were unique to the PEG6 treatment.
Papaya seeds are rich in lipids that are used during the germination process [34,65]. The greater number of proteins related to the fatty acid/lipid pathway that was observed in the PEG6 treatment (11.8%) compared with the control treatment (6.9%) (Figure 4) might indicate that water stress, which is inherent to this treatment, could be supporting the synthesis and mobilization of storage lipids during somatic embryogenesis.
Proteins involved in oxidation-reduction processes
Most of the detected oxidation-reduction-related proteins were identified in both treatments, at similar expression levels. One exception was alcohol dehydrogenase 3 (ADH3) (spot 146), which was unique to the PEG6 treatment. ADH3 is involved in regulation of the effects of nitric oxide in the cell and is therefore important for the maintenance of cellular homeostasis [66]. Additionally, ADH3 has been reported to represent a class of enzyme that is closely related to the developmental stages of Vitis rupestris somatic embryos [67]. The other exception was cinnamoyl alcohol dehydrogenase (spot 333), which was unique to the control treatment. This enzyme catalyzes cinnamoyl CoA reductase, a key enzyme in the synthesis of lignin, which acts as a mechanical barrier to various plant pathogens [68].
The cytosolic ascorbate peroxidase 1 protein (spot 24) was observed in both treatments. Ascorbate peroxidases play a role in defense against oxidative stress [69]. This protein functions in the defense against photooxidative stress in Arabidopsis[70]. In the present work, the presence of this protein in both treatments provided evidence of a response to the stress caused by the in vitro culture conditions.
Reactive oxygen species (ROS) play an important role during plant growth and development, as they are extremely reactive in regulating hormonal activity [71]. The activity of ROS is often the first detectable response to biotic or abiotic stress in plants [72]. A large number of studies have shown increases in the levels of ROS during somatic embryo development [73]. ROS play a role as secondary messengers involved in signal transduction, thereby influencing the expression of many genes [74].
This similarity between the two treatments related to the identified proteins and their relative expression levels could be associated with the oxidative stress inherent to the in vitro culture conditions.
Protein processing
Many heat shock proteins (HSPs) were identified during papaya embryogenesis. The HSP70 (spots 325 and 326) and HSP83 (spot 324) proteins were unique to the control treatment, while HSP90 (spot 162) was unique to the PEG6 treatment. The HSP70 protein (spot 49) was observed in both treatments (Table 2).
The HSP70 family consists of chaperone proteins that bind to exposed hydrophobic sequences and maintain the unfolded peptide chain until it folds into the correct three-dimensional conformation [75]. HSP70 proteins are responsible for the translocation and processing of proteins associated with reserve synthesis and mobilization [76], abiotic stress [77] and cellular defense [78]. In addition, HSP70s have been associated with seed development in Dimocarpus longana[79] and somatic embryo development in Vitis vinifera[72] and Picea abies[80].
HSP83 was unique to the control treatment. The function of this protein family is still unknown. However, HSP83 has been detected during the stress response to Ustilago maydis infection in Z. mays[81].
The members of the HSP90 family, which was unique to the PEG6 treatment, are involved in the assembly, maturation, stabilization and activation of proteins that are important as signaling molecules, such as protein kinases, hormone receptors and transcription factors, in eukaryotic cells [82]. They are responsible for refolding denatured proteins and/or folding newly synthesized proteins [83]. These proteins have also been identified during somatic embryogenesis in hybrid larch (Larix × eurolepis) [84] and in the developing seeds of Pinus massoniana[85] and Araucaria angustifolia[86].
BPI (spot 337), which was unique to the control treatment, is a member of the HSP70 family and localizes to the endoplasmic reticulum. It acts in the translocation of proteins through the endoplasmic reticulum and assists in the proper folding and maturation of newly synthesized proteins entering the organelle. Therefore, this protein has been implicated in seed maturation in several species, such as Cucurbita maxima[87], Ocotea catharinensis[88] and Pinus massoniana[85]. However, overexpression of this protein inhibits the accumulation of seed storage proteins in Oryza sativa endosperm cells [89].
A leucine aminopeptidase (spot 142) was found to be unique to the PEG6 treatment. During plant development, proteases exhibit important functions, including the degradation of damaged, misfolded and potentially harmful proteins to provide free amino acids required for the synthesis of new proteins [90]. The observation of this protein only in the PEG6 treatment could be indicative of better control of embryonic development under this treatment.
Metabolic process-related proteins
A high percentage of metabolic process-related proteins was observed in both treatments, and the expression levels of these proteins were similar in most cases (Table 2).
Among these proteins, pyrophosphatase (spot 28), which was common to the two treatments, is a metalloenzyme that catalyzes diverse reactions that are necessary to supply the energy required during various biosynthetic reactions [91]. This protein is associated with stress tolerance [92]. Another protein that was common to both treatments, alpha-D-xylosidase (spot 126), mobilizes xyloglucan during the development of germinated cotyledons in nasturtium (Tropaeolum majus) seeds [93]. Aldehyde dehydrogenase (spot 130) was unique to the PEG6 treatment. Despite being involved in various functions, there is evidence that this protein may play a role in seed desiccation tolerance and vigor [94], as reported in rice [56]. The possible relationship of this protein to desiccation tolerance may explain its expression only in the PEG6 treatment.
The PEG6 treatment also induced the synthesis of the unique protein esterase D (spot 184). This protein is considered a biomarker of competent embryogenic cultures in D. carota[95] and Z. mays[96]. Esterase D is also involved in various cellular functions, such as fruit ripening, abscission, cell expansion, stomatal movement, reproduction and the detoxification of xenobiotics [97].
Proteins related to other functions
Some functional categories contained only a few proteins that were identified in the papaya embryogenic cultures (Table 2). Among these proteins, the initiation factor eIF4A-15 was observed as a unique spot in both treatments (spot 222 and spot 317 in the PEG6 and control treatments, respectively). This protein phosphorylates other proteins involved in translation, allowing protein translation to be coupled with other essential cellular functions, such as the stress response, cell growth and division. Proteins related to changes in the state of chromatin transcription in genes via DNA processing, mRNA translation and post-translational modifications are responsible for regulating the levels of protein expression for several genes [98]. This protein is likely involved in the common, inherent process of somatic embryogenesis in papaya, as it is not dependent on the induction of maturation treatments.
A 14-3-3 protein (spots 48 and 87) that was common to both treatments is an important type of cellular signaling protein that regulates enzymes during primary metabolism and performs other ‘housekeeping’ functions [99]. In addition, this protein participates in the regulation of various biochemical processes during seed development [100].
S-adenosylmethionine decarboxylase (spot 45) was observed in both treatments (Table 2). This protein is a key enzyme involved in the synthesis of polyamines in plants and other organisms [101]. Polyamines play a role in several processes in plants and have been related to the development of somatic embryogenesis [102].
Conclusions
Since the introduction of somatic embryogenesis [103,104], a series of experiments have been performed in an attempt to elucidate the molecular, physiological and biochemical regulation of the morphogenic competence pathways in plant cells. Nevertheless, one key scientific question concerning somatic embryogenic development remains: “How does a single somatic cell become a whole plant?” [105]. The field of proteomics could be at the heart of such research questions, and it has become increasingly important in studies on somatic embryogenic development.
This is the first report describing a proteomic analysis of somatic embryogenesis in papaya. Our results showed that it was possible to detect a pattern of differential protein expression during somatic embryo differentiation and maturation in papaya induced by PEG6 treatment. PEG (6%) treatment promoted increased protein synthesis and induced differential expression of proteins related to carbohydrate and energy metabolism, fatty acid metabolism and oxidation-reduction in embryogenic cultures compared to the control treatment.
We observed that the synthesis of certain proteins induced by PEG treatment, such as enolase, esterase and ADH3, could play an important role in the maturation of somatic papaya embryos. These proteins have been found to be associated with somatic embryo development in other species [60,61,67,106], and esterases are considered important biomarkers of somatic embryos in plants [106]. Additionally, the present study provides a list of PEG-induced proteins that are potential biomarker candidates for future investigations of somatic embryogenic development in papaya.
Materials and methods
Plant material
To induce somatic embryogenesis in papaya, immature zygotic embryos were isolated from mature papaya seeds of the hybrid UENF/CALIMAN01 and used as explants. Immature fruits were kindly provided by the Caliman Agricola S/A company, which is located in the city of Linhares, Espírito Santo (ES), Brazil (19° 23’S and 40° 4’W).
Induction and multiplication of embryogenic cultures
The induction of embryogenic cultures was performed according to Heringer et al. [20]. Briefly, immature fruits were disinfected for 2 min in 70% ethanol and for 30 min in 50% commercial bleach, followed by three washes with distilled, autoclaved water. Immature seeds were subsequently obtained and sorted in a laminar flow cabinet, and immature embryos were isolated for use as explants. The immature embryos were inoculated into tubes (25 × 150 mm) containing 10 mL of MS culture medium [107] (Phytotechnology Lab, Shawnee Mission, KS, USA) supplemented with 3% sucrose (Vetec, São Paulo, Brazil), 20 μM 2,4-dichlorophenoxyacetic acid (2,4-D) (Sigma-Aldrich, St. Louis, USA) and 2.0 g/L Phytagel (Sigma-Aldrich). The pH of the culture medium was adjusted to 5.8 before the Phytagel was added. The culture medium was sterilized via autoclaving at 121°C for 15 min. After inoculation, the tubes with the explants were incubated in the dark at 25°C (±2°C) for 42 days. The induced embryogenic cultures were then isolated and subcultured in a culture medium with the same composition. Four subcultures were then obtained at intervals of 21 days prior to maturation to perform multiplication of embryogenic cultures.
Maturation of embryogenic cultures
After the multiplication phase, the cultures were transferred to maturation treatments. Three colonies, each with a fresh mass (FM) of 300 mg, were inoculated into Petri dishes (90 × 15 mm) containing 20 mL of MS culture medium supplemented with myo-inositol (Merck KGaA, Darmstadt, Germany) (0.005%), Phytagel (2.0 g/L), sucrose (3%) and PEG 3350 (Sigma-Aldrich) at concentrations of 0, 3, 6 and 9%, hereafter referred to as the control, PEG3, PEG6 and PEG9 treatments, respectively. The pH of the culture medium was adjusted to 5.8 before the Phytagel was added. The culture medium was sterilized by autoclaving at 121°C for 15 min. The cultures were incubated in a growth chamber at 25 ± 1°C in the dark for the first 7 days, after which they were subjected to a 16 h light (60 μmol/m2 s1) photoperiod. Four repetitions were performed, with a total of 12 colonies per treatment. After 42 days of cultivation, the number of cotyledonary somatic embryos (mature somatic embryos) per colony was evaluated. Samples of 300 mg (FM) were dried in an oven at 70°C for 48 h for dry matter (DM) determination. Samples of 300 mg FM were also stored at -20°C for proteomic analysis.
Preparation for microscopy
The samples were fixed in an aqueous solution containing glutaraldehyde (2.5%) and formaldehyde (4.0%) diluted in sodium cacodylate buffer (0.1 M), pH 7.3, at room temperature for 24 h. After fixation, the samples were dehydrated in a graded ethanol series and embedded in historesin (Leica, Wetzlar, Germany) [108]. Sections with a thickness of 3 μm were cut with a microtome (Leica) and fixed on slides by heating.
The samples were dehydrated with periodic acid and stained via the periodic acid-Schiff reaction. Protein bodies were stained with Coomassie brilliant blue R-250 (CBB) [109]. The samples were observed under an Axioplan light microscope (Carl Zeiss, Jena, Germany) equipped with an Axiocam MRC5 digital camera (Carl Zeiss), and the images were analyzed using AxioVisionLE version 4.8 software (Carl Zeiss).
Protein extraction
Protein extracts were prepared in biological triplicate (300 mg FM) for each maturation treatment. Soluble proteins were extracted according to the method described by Santa-Catarina et al. [110]. The buffer-soluble proteins were extracted with phosphate buffer (pH 7.5) containing 50 mM sodium phosphate dibasic (Vetec), 10 mM 2-mercaptoethanol (Sigma-Aldrich) and 1 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich). The supernatants were transferred to clear microtubes, and the proteins were precipitated on ice for 30 min with 10% trichloroacetic acid (Merck). The pellet was washed three times with cold acetone (Merck), and the proteins were then resuspended and concentrated in 0.5 mL of a urea/thiourea buffer (7 M urea, 2 M thiourea, 1% dithiothreitol (DTT), 2% Triton X-100, 0.5% pharmalyte (all from GE Healthcare, Freiburg, Germany) and 1 mM PMSF), to which a 0.5% immobilized pH gradient (IPG) buffer (pH 4–7) (GE Healthcare) was added. The protein concentration was determined using the 2-D Quant Kit (GE Healthcare).
Two-dimensional gel electrophoresis (2-DE) and spot matching
The 2-DE analyses were performed in the protein extracts from the embryogenic cultures from both the control (the treatment that resulted in the fewest mature embryos) and PEG6 (the treatment resulting in the greatest number of somatic embryos) treatments to compare these two conditions. Embryogenic cultures were used, once the isolation of somatic embryos in different developmental stages was unfeasible, due not have sufficient material for the extraction of the amount of protein required for the initial stages of development.
Aliquots of the samples containing 500 μg of protein were used for 2-DE. Prior to loading onto 18 cm IPG strips (pH 4 – 7), a sufficient volume of rehydration buffer (7 M urea, 2 M thiourea, 2% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) (GE Healthcare), 0.5% IPG buffer, pH 4–7, 1% DTT and 0.002% bromophenol blue (Sigma-Aldrich)) was added to the sample aliquots to achieve a final volume of 375 μL. After 12 h in the rehydration gel, isoelectric focusing was performed using an IPGphor II apparatus (GE Healthcare) for a total of 35 kVh at 20°C. The IPG strips were then subjected to reduction and alkylation through 2 × 15 min incubations, using a buffer (50 mM Tris–HCl (GE Healthcare), 6 M urea, 30% glycerol (Sigma-Aldrich), 2% sodium dodecyl sulfate (SDS) (Sigma-Aldrich) and 0.002% bromophenol blue) containing 125 mM DTT for the first incubation and 125 mM iodoacetamide (GE Healthcare) for the second incubation. Then, the strips were applied to the top of a 12% polyacrylamide gel. The second dimension of electrophoresis was performed at 25 mA per gel using a Protean II apparatus (Bio-Rad, Hercules, USA), and the gels were stained with Coomassie (0.1% Coomassie brilliant blue G250, 1.2% ortho-phosphoric acid (85%) and 10% ammonium sulfate) according to Neuhoff et al. [111].
The Coomassie-stained 2-DE gels obtained for each of the three biological samples were analyzed using Image Master Platinum v.7 software (GE Healthcare). The authenticity and the outline of each protein spot was validated via visual inspection and edited when necessary. The identification and selection of the differentially expressed proteins were achieved through comparative analysis of the gels, and the volumes of individual spots were obtained following the program’s instructions. To eliminate gel-to-gel variations, the individual spot volume in each gel was normalized relative to the total valid spot volume, expressing the protein abundance as the relative volume (%vol), and the values obtained for the control and PEG6 treatments were compared using Student’s t-test. The 135 most abundant proteins present in three biological replicates, 21 of which were unique to the control, while 47 were unique to PEG6, and 67 were common to both treatments, were selected for MS/MS identification.
In-gel digestion and tandem mass spectrometry (MS/MS) analysis
Protein identifications were obtained through in-gel digestion and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF/TOF)-MS/MS. The protein spots were manually excised from the 2-DE gels and washed with water. The spots were then selected according to the methodology proposed by Balbuena et al. [86] and digested with trypsin (Promega, Madison, WI, USA). The resulting peptides were concentrated and desalted using C18 Zip Tips (Millipore). The final solutions of extracted peptides were stored at -20°C until MS/MS analysis.
Mass spectrometry analysis was performed according to Rocha et al. [112] and Gandra et al. [113]. Briefly, 0.3 μL of the sample solution was mixed with an equal volume of a saturated matrix solution [10 mg/mL α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich) in 50% acetonitrile (Merck)/0.1% trifluoroacetic acid (Sigma-Aldrich)] on a target plate and allowed to dry at room temperature. Raw data for protein identification were obtained using an AB SCIEX TOF-TOF 5800 mass spectrometer (Applied Biosystems, North Warrington, Cheshire, United Kingdom). External calibration was performed in MS mode using a mixture of four peptides: des-Arg1-bradykinin (m/z 904.468); angiotensin I (m/z 1296.685); Glu1-fibrinopeptide B (m/z 1570.677); and ACTH (18–39) (m/z 2465.199) (all from Applied Biosystems). The MS/MS spectra were externally calibrated using the known product ion masses observed in the MS/MS spectrum of angiotensin I.
Database searches
All MS/MS samples were analyzed using Mascot (Matrix Science, http://www.matrixscience.com). Mascot was set up to search the NCBInr database (selected for Viridiplantae) assuming the use of the digestion enzyme trypsin, with up to two missed cleavages. Mascot was searched with a product ion mass tolerance of 0.30 Da and a precursor ion tolerance of 0.60 Da. Methionine oxidation and the iodoacetamide derivative of cysteine were specified as variable and fixed modifications, respectively. Only significant hits, as defined through MASCOT probability analysis (P < 0.05), and peptides identified with individual ion scores greater than 30 were considered.
Functional classification of the proteins identified by Mascot was performed using the program Blast2go (http://www.blast2go.com).
Abbreviations
2,4-D: 2,4-dichlorophenoxyacetic acid; 2-DE: Two-dimensional gel electrophoresis; ABA: Abscisic acid; ADH3: Alcohol dehydrogenase 3; CHAPS: 3-[(3-cholamidopropyl) dimethylammonio]-1- propanesulfonate; DTT: Dithiothreitol; FM: Fresh mass; IPG: Immobilized pH gradient; HSP: Heat shock proteins; MALDI-TOF/TOF: Matrix-assisted laser desorption/ionization time-of-flight; MS: Murashige and Skoog; MS/MS: Tandem mass spectrometry; NCBI: National Center for Biotechnology Information; PEG: Polyethylene glycol; PEG3: Polyethylene glycol 3%; PEG6: Polyethylene glycol 6%; PEG9: Polyethylene glycol 9%; PMSF: Phenylmethylsulfonyl fluoride; SDS: Sodium dodecyl sulfate.
Competing interests
There are no competing interests in this study.
Authors’ contributions
EMV, CSC and VS conceived the study and designed the experiments. EMV, VS, ASH and CSC wrote the manuscript. ASH conducted two-dimensional gel electrophoresis and spot matching. ATSF, MNC, TB and JEAP were responsible for the in-gel digestion and tandem mass spectrometry (MS/MS) analysis and database management. All authors read and approved the final manuscript.
Contributor Information
Ellen de Moura Vale, Email: ellenmoura27@gmail.com.
Angelo Schuabb Heringer, Email: angeloheringer@gmail.com.
Tatiana Barroso, Email: tatianabarroso@hotmail.com.
André Teixeira da Silva Ferreira, Email: atsferreira@ioc.fiocruz.br.
Monique Nunes da Costa, Email: mnqcosta@gmail.com.
Jonas Enrique Aguilar Perales, Email: jperales@ioc.fiocruz.br.
Claudete Santa-Catarina, Email: claudete@uenf.br.
Vanildo Silveira, Email: vanildo@uenf.br.
Acknowledgements
We thank the Carlos Chagas Filho Foundation for Research Support in the State of Rio de Janeiro (FAPERJ), the Coordination for the Improvement of Higher Education Personnel (CAPES) and the National Council for Scientific and Technological Development (CNPq) for financial support and/or providing scholarships to the students. The authors thank Caliman Agrícola S.A. for kindly providing the fruits.
References
- FAOSTAT - Crop Production. http://faostat.fao.org [ http://faostat.fao.org/]
- Aryal R, Ming R. Sex determination in flowering plants: papaya as a model system. Plant Sci. 2014;217–218:56–62. doi: 10.1016/j.plantsci.2013.10.018. [DOI] [PubMed] [Google Scholar]
- Ming R, Yu Q, Blas A, Chen C, Na J-K, Moore P. In: Genomics of Tropical Crop Plants. Volume 1. Moore P, Ming R, editor. New York: Springer New York; 2008. Genomics of Papaya a Common Source of Vitamins in the Tropics; pp. 405–420. Plant Genetics and Genomics: Crops and Models. [Google Scholar]
- Rodrigues SP, Ventura JA, Aguilar C, Nakayasu ES, Almeida IC, Fernandes PMB, Zingali RB. Proteomic analysis of papaya (Carica papaya L.) displaying typical sticky disease symptoms. Proteomics. 2011;11:2592–2602. doi: 10.1002/pmic.201000757. [DOI] [PubMed] [Google Scholar]
- Bhattacharya J, Khuspe SS. In vitro and in vivo germination of papaya (Carica papaya L.) seeds. Sci Hortic. 2001;91:39–49. [Google Scholar]
- Ed B, Ed L, Rv R. Actions of hormones and embryoid formation in callus cultures of Carica papaya. Int Symp Crop Prot, Fytopharmacie en Fytiatrie Rijkslandsbouwhoogeskool Medelingen. 1974;39:637–645. [Google Scholar]
- Joshi R, Kumar P. Regulation of somatic embryogenesis in crops: a review. Agric Rev. 2013;34:1–20. [Google Scholar]
- Marquez-Martin B, Sesmero R, Quesada MA, Pliego-Alfaro F, Sanchez-Romero C. Water relations in culture media influence maturation of avocado somatic embryos. J Plant Physiol. 2011;168:2028–2034. doi: 10.1016/j.jplph.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Mishra S, Sanyal I, Amla DV. Changes in protein pattern during different developmental stages of somatic embryos in chickpea. Biol Plant. 2012;56:613–619. [Google Scholar]
- Misra S, Attree SM, Leal I, Fowke LC. Effect of abscisic-acid, osmoticum, and desiccation on synthesis of storage proteins during the development of white spruce somatic embryos. Ann Bot. 1993;71:11–22. [Google Scholar]
- Attree SM, Fowke LC. Embryogeny of gymnosperms - advances in synthetic seed technology of conifers. Plant Cell Tiss Org Cult. 1993;35:1–35. [Google Scholar]
- Kong L, Attree SM, Fowke LC. Effects of polyethylene glycol and methylglyoxal bis(guanylhydrazone) on endogenous polyamine levels and somatic embryo maturation in white spruce (Picea glauca) Plant Sci. 1998;133:211–220. [Google Scholar]
- Stasolla C, Yeung EC. Recent advances in conifer somatic embryogenesis: improving somatic embryo quality. Plant Cell Tiss Org Cult. 2003;74:15–35. [Google Scholar]
- Rance IM, Tian WZ, Mathews H, Dekochko A, Beachy RN, Fauquet C. Partial desiccation of mature embryo-derived calli, a simple treatment that dramatically enhances the regeneration ability of indica rice. Plant Cell Rep. 1994;13:647–651. doi: 10.1007/BF00232938. [DOI] [PubMed] [Google Scholar]
- Linossier L, Veisseire P, Cailloux F, Coudret A. Effects of abscisic acid and high concentrations of PEG on Hevea brasiliensis somatic embryos development. Plant Sci. 1997;124:183–191. [Google Scholar]
- Walker DR, Parrott WA. Effect of polyethylene glycol and sugar alcohols on soybean somatic embryo germination and conversion. Plant Cell Tiss Org Cult. 2001;64:55–62. [Google Scholar]
- Troch V, Werbrouck S, Geelen D, Van Labeke M-C. Optimization of horse chestnut (Aesculus hippocastanum L.) somatic embryo conversion. Plant Cell Tiss Org Cult. 2009;98:115–123. [Google Scholar]
- Buendia-Gonzalez L, Estrada-Zuniga ME, Orozco-Villafuerte J, Cruz-Sosa F, Vernon-Carter EJ. Somatic embryogenesis of the heavy metal accumulator Prosopis laevigata. Plant Cell Tiss Org Cult. 2012;108:287–296. [Google Scholar]
- Mishra M, Shukla N, Chandra R. In: II International Symposium on Papaya Volume 851. Kumar N, Soorianathasundaram K, Jeyakumar P, editor. Madurai, India: Acta Horticulturae; 2010. Role of Polyethylene Glycol in Maturation and Germination of Transformed Somatic Embryos of Papaya (Carica Papaya L.) [Google Scholar]
- Heringer AS, Vale EM, Barroso T, Santa-Catarina C, Silveira V. Polyethylene glycol effects on somatic embryogenesis of papaya hybrid UENF/CALIMAN 01 seeds. Theor Exp Plant Pysiol. 2013;25:116–124. [Google Scholar]
- Agrawal GK, Sarkar A, Righetti PG, Pedreschi R, Carpentier S, Wang T, Barkla BJ, Kohli A, Ndimba BK, Bykova NV, Rampitsch C, Zolla L, Rafudeen MS, Cramer R, Bindschedler LV, Tsakirpaloglou N, Ndimba RJ, Farrant JM, Renaut J, Job D, Kikuchi S, Rakwal R. A decade of plant proteomics and mass spectrometry: translation of technical advancements to food security and safety issues. Mass Spectrom Rev. 2013;32:335–365. doi: 10.1002/mas.21365. [DOI] [PubMed] [Google Scholar]
- Rodrigues SP, Da Cunha M, Ventura JA, Bueno Fernandes PM. Effects of the Papaya meleira virus on papaya latex structure and composition. Plant Cell Rep. 2009;28:861–871. doi: 10.1007/s00299-009-0673-7. [DOI] [PubMed] [Google Scholar]
- Rodrigues SP, Ventura JA, Aguilar C, Nakayasu ES, Choi H, Sobreira TJ, Nohara LL, Wermelinger LS, Almeida IC, Zingali RB, Fernandes PM. Label-free quantitative proteomics reveals differentially regulated proteins in the latex of sticky diseased Carica papaya L. plants. J Proteomics. 2012;75:3191–3198. doi: 10.1016/j.jprot.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel Huerta-Ocampo J, Alberto Osuna-Castro J, Jareth Lino-Lopez G, Barrera-Pacheco A, Mendoza-Hernandez G, De Leon-Rodriguez A, de la Rosa AP B. Proteomic analysis of differentially accumulated proteins during ripening and in response to 1-MCP in papaya fruit. J Proteomics. 2012;75:2160–2169. doi: 10.1016/j.jprot.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Nogueira SB, Labate CA, Gozzo FC, Pilau EJ, Lajolo FM, Nascimento JR O d. Proteomic analysis of papaya fruit ripening using 2DE-DIGE. J Proteomics. 2012;75:1428–1439. doi: 10.1016/j.jprot.2011.11.015. [DOI] [PubMed] [Google Scholar]
- Marsoni M, Bracale M, Espen L, Prinsi B, Negri AS, Vannini C. Proteomic analysis of somatic embryogenesis in Vitis vinifera. Plant Cell Rep. 2008;27:347–356. doi: 10.1007/s00299-007-0438-0. [DOI] [PubMed] [Google Scholar]
- Sghaier-Hammami B, Drira N, Jorrin-Novo JV. Comparative 2-DE proteomic analysis of date palm (Phoenix dactylifera L.) somatic and zygotic embryos. J Proteomics. 2009;73:161–177. doi: 10.1016/j.jprot.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Bian F, Zheng C, Qu F, Gong X, You C. Proteomic analysis of somatic embryogenesis in Cyclamen persicum Mill. Plant Mol Biol Report. 2010;28:22–31. [Google Scholar]
- Noah AM, Niemenak N, Sunderhaus S, Haase C, Omokolo DN, Winkelmann T, Braun H-P. Comparative proteomic analysis of early somatic and zygotic embryogenesis in Theobroma cacao L. J Proteomics. 2013;78:123–133. doi: 10.1016/j.jprot.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Sharifi G, Ebrahimzadeh H, Ghareyazie B, Gharechahi J, Vatankhah E. Identification of differentially accumulated proteins associated with embryogenic and non-embryogenic calli in saffron (Crocus sativus L.) Proteome Sci. 2012;10:3. doi: 10.1186/1477-5956-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Garay A, Lopez JA, Camafeita E, Bueno MA, Pintos B. Proteomic perspective of Quercus suber somatic embryogenesis. J Proteomics. 2013;93:314–325. doi: 10.1016/j.jprot.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Koehler AD, Carvalho CR, Abreu IS, Clarindo WR. Somatic embryogenesis from leaf explants of hermaphrodite Carica papaya: a new approach for clonal propagation. Afr J Biotechnol. 2013;12:2386–2391. [Google Scholar]
- Calic-Dragosavac D, Zdravkovi-Kora S, Bohanec B, Radojevi L, Vinterhalter B, Stevovi S, Cingel A, Savi J. Effect of activated charcoal, abscisic acid and polyethylene glycol on maturation, germination and conversion of Aesculus hippocastanum androgenic embryos. Afr J Biotechnol. 2010;9:3786–3793. [Google Scholar]
- Marfo EK, Oke OL, Afolabi OA. Chemical composition of papaya (Carica papaya) seeds. Food Chem. 1986;22:259–266. [Google Scholar]
- Pan Z, Guan R, Zhu S, Deng X. Proteomic analysis of somatic embryogenesis in valencia sweet orange (Citrus sinensis Osbeck) Plant Cell Rep. 2009;28:281–289. doi: 10.1007/s00299-008-0633-7. [DOI] [PubMed] [Google Scholar]
- Iraqi D, Tremblay FM. The role of sucrose during maturation of black spruce (Picea mariana) and white spruce (Picea glauca) somatic embryos. Physiol Plant. 2001;111:381–388. doi: 10.1034/j.1399-3054.2001.1110316.x. [DOI] [PubMed] [Google Scholar]
- Rode C, Lindhorst K, Braun H-P, Winkelmann T. From callus to embryo: a proteomic view on the development and maturation of somatic embryos in Cyclamen persicum. Planta. 2012;235:995–1011. doi: 10.1007/s00425-011-1554-1. [DOI] [PubMed] [Google Scholar]
- Winkelmann T, Heintz D, Van Dorsselaer A, Serek M, Braun H-P. Proteomic analyses of somatic and zygotic embryos of Cyclamen persicum Mill. reveal new insights into seed and germination physiology. Planta. 2006;224:508–519. doi: 10.1007/s00425-006-0238-8. [DOI] [PubMed] [Google Scholar]
- Smulders MJM, Klerk GJ. Epigenetics in plant tissue culture. Plant Growth Regul. 2011;63:137–146. [Google Scholar]
- Fernando JA, Melo M, Soares MKM, Appezzato-da-Glória B. Anatomy of somatic embryogenesis in Carica papaya L. Braz Arch Biol Technol. 2001;44:247–255. [Google Scholar]
- Gonçalves JFC, Lima RBS, Fernandes AV, Borges EEL, Buckeridge MS. Physiological and biochemical characterization of the assai palm (Euterpe oleracea Mart.) during seed germination and seedling growth under aerobic and anaerobic conditions. Revista Árvore. 2010;34:1045–4053. [Google Scholar]
- Rocha D, Vieira L, Tanaka F, Silva L, Otoni W. Somatic embryogenesis of a wild passion fruit species Passiflora cincinnata Masters: histocytological and histochemical evidences. Protoplasma. 2012;249:747–758. doi: 10.1007/s00709-011-0318-x. [DOI] [PubMed] [Google Scholar]
- Cangahuala-Inocente GC, Steiner N, Maldonado SB, Guerra MP. Patterns of protein and carbohydrate accumulation during somatic embryogenesis of Acca sellowiana. Pesq Agrop Brasileira. 2009;44:217–224. [Google Scholar]
- Bewley JD, Black M. Seeds: Physiology of Development and Germination. New York: Plenum Press; 1994. [Google Scholar]
- Takac T, Pechan T, Samaj J. Differential proteomics of plant development. J Proteomics. 2011;74:577–588. doi: 10.1016/j.jprot.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Pasternak TP, Prinsen E, Ayaydin F, Miskolczi P, Potters G, Asard H, Van Onckelen HA, Dudits D, Feher A. The role of auxin, pH, and stress in the activation of embryogenic cell division in leaf protoplast-derived cells of alfalfa. Plant Physiol. 2002;129:1807–1819. doi: 10.1104/pp.000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feher A, Pasternak TP, Dudits D. Transition of somatic plant cells to an embryogenic state. Plant Cell Tiss Org Cult. 2003;74:201–228. [Google Scholar]
- Frova C. The plant glutathione transferase gene family: genomic structure, functions, expression and evolution. Physiol Plant. 2003;119:469–479. [Google Scholar]
- Hoenemann C, Ambold J, Hohe A. Gene expression of a putative glutathione S-transferase is responsive to abiotic stress in embryogenic cell cultures of Cyclamen persicum. Electron J Biotechnol. 2012;15:6. [Google Scholar]
- Winter H, Huber SC. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Biochem Mol Biol. 2000;35:253–289. doi: 10.1080/10409230008984165. [DOI] [PubMed] [Google Scholar]
- Gibeaut DM. Nucleotide sugars and glycosyltransferases for synthesis of cell wall matrix polysaccharides. Plant Physiol Biochem. 2000;38:69–80. [Google Scholar]
- Bosch M, Mayer C-D, Cookson A, Donnison IS. Identification of genes involved in cell wall biogenesis in grasses by differential gene expression profiling of elongating and non-elongating maize internodes. J Exp Bot. 2011;62:3545–3561. doi: 10.1093/jxb/err045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquea F, Arce-Johnson P. Identification of genes expressed during early somatic embryogenesis in Pinus radiata. Plant Physiol Biochem. 2008;46:559–568. doi: 10.1016/j.plaphy.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Domoney C, Duc G, Ellis THN, Ferrandiz C, Firnhaber C, Gallardo K, Hofer J, Kopka J, Kuster H, Madueno F, Munier-Jolain NG, Mayer K, Thompson R, Udvardi M, Salon C. Genetic and genomic analysis of legume flowers and seeds. Curr Opin Plant Biol. 2006;9:133–141. doi: 10.1016/j.pbi.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D. Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiol Plant. 2002;116:238–247. doi: 10.1034/j.1399-3054.2002.1160214.x. [DOI] [PubMed] [Google Scholar]
- Shin J-H, Kim S-R, An G. Rice aldehyde dehydrogenase7 is needed for seed maturation and viability. Plant Physiol. 2009;149:905–915. doi: 10.1104/pp.108.130716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. Combined networks regulating seed maturation. Trends Plant Sci. 2007;12:294–300. doi: 10.1016/j.tplants.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Xu SB, Li T, Deng ZY, Chong K, Xue Y, Wang T. Dynamic proteomic analysis reveals a switch between central carbon metabolism and alcoholic fermentation in rice filling grains. Plant Physiol. 2008;148:908–925. doi: 10.1104/pp.108.125633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderstraeten D, Rodriguespousada RA, Goodman HM, Vanmontagu M. Plant enolase - gene structure, expression, and evolution. Plant Cell. 1991;3:719–735. doi: 10.1105/tpc.3.7.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode C, Gallien S, Heintz D, Van Dorsselaer A, Braun H-P, Winkelmann T. Enolases: storage compounds in seeds? Evidence from a proteomic comparison of zygotic and somatic embryos of Cyclamen persicum Mill. Plant Mol Biol. 2011;75:305–319. doi: 10.1007/s11103-010-9729-x. [DOI] [PubMed] [Google Scholar]
- Lippert D, Zhuang J, Ralph S, Ellis DE, Gilbert M, Olafson R, Ritland K, Ellis B, Douglas CJ, Bohlmann J. Proteome analysis of early somatic embryogenesis in Picea glauca. Proteomics. 2005;5:461–473. doi: 10.1002/pmic.200400986. [DOI] [PubMed] [Google Scholar]
- Andriotis VME, Kruger NJ, Pike MJ, Smith AM. Plastidial glycolysis in developing Arabidopsis embryos. New Phytol. 2010;185:649–662. doi: 10.1111/j.1469-8137.2009.03113.x. [DOI] [PubMed] [Google Scholar]
- Paulus C, Kollner B, Jacobsen HJ. Physiological and biochemical-characterization of glyoxalase-I, a general marker for cell-proliferation, from a soybean cell-suspension. Planta. 1993;189:561–566. doi: 10.1007/BF00198220. [DOI] [PubMed] [Google Scholar]
- Sun W, Xu X, Zhu H, Liu A, Liu L, Li J, Hua X. Comparative transcriptomic profiling of a salt-tolerant wild tomato species and a salt-sensitive tomato cultivar. Plant Cell Physiol. 2010;51:997–1006. doi: 10.1093/pcp/pcq056. [DOI] [PubMed] [Google Scholar]
- Puangsri I, Abdulkarim SM, Ghazali HM. Properties of Carica papaya L. (papaya) seed oil following extractions using solvent and aqueous enzymatic methods. J Food Lipids. 2005;12:62–76. [Google Scholar]
- Leterrier M, Chaki M, Airaki M, Valderrama R, Palma JM, Barroso JB, Corpas FJ. Function of S-nitrosoglutathione reductase (GSNOR) in plant development and under biotic/abiotic stress. Plant Signal Behav. 2011;6:789–793. doi: 10.4161/psb.6.6.15161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli L, Scienza A, Villa P, Deponti P, Gianazza E. Enzyme markers for somatic embryogenesis in Vitis. J Plant Physiol. 1993;141:476–481. [Google Scholar]
- Prasad NK, Vindal V, Kumar V, Kabra A, Phogat N, Kumar M. Structural and docking studies of Leucaena leucocephala Cinnamoyl CoA reductase. J Mol Model. 2011;17:533–541. doi: 10.1007/s00894-010-0744-2. [DOI] [PubMed] [Google Scholar]
- Morita S, Nakatani S, Koshiba T, Masumura T, Ogihara Y, Tanaka K. Differential expression of two cytosolic ascorbate peroxidase and two superoxide dismutase genes in response to abiotic stress in rice. Rice Sci. 2011;18:157–166. [Google Scholar]
- Davletova S, Rizhsky L, Liang HJ, Zhong SQ, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell. 2005;17:268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-S, Huang TY, Bokoch GM. Reactive oxygen species regulate a slingshot-cofilin activation pathway. Mol Biol Cell. 2009;20:2650–2660. doi: 10.1091/mbc.E09-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ma H, Chen S, Ji M, Perl A, Kovacs L, Chen S. Stress response proteins’ differential expression in embryogenic and non-embryogenic callus of Vitis vinifera L. cv. Cabernet Sauvignon - a proteomic approach. Plant Sci. 2009;177:103–113. [Google Scholar]
- Zavattieri MA, Frederico AM, Lima M, Sabino R, Arnholdt-Schmitt B. Induction of somatic embryogenesis as an example of stress-related plant reactions. Electron J Biotechnol. 2010;13:12. [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT. Hydrogen peroxide and nitric oxide as signalling molecules in plants. J Exp Bot. 2002;53:1237–1247. [PubMed] [Google Scholar]
- Wang WX, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Forward BS, Osusky M, Misra S. The douglas-fir BiP promoter is functional in Arabidopsis and responds to wounding. Planta. 2002;215:569–576. doi: 10.1007/s00425-002-0775-8. [DOI] [PubMed] [Google Scholar]
- Kosová K, Vítámvás P, Prášil IT, Renaut J. Plant proteome changes under abiotic stress - Contribution of proteomics studies to understanding plant stress response. J Proteomics. 2011;74:1301–1322. doi: 10.1016/j.jprot.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Imin N, Nizamidin M, Daniher D, Nolan KE, Rose RJ, Rolfe BG. Proteomic analysis of somatic embryogenesis in Medicago truncatula. Explant cultures grown under 6-benzylaminopurine and 1-naphthaleneacetic acid treatments. Plant Physiol. 2005;137:1250–1260. doi: 10.1104/pp.104.055277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Weng Z-x, Cheng C-L, Liu H, Liang W-y, Jiang J-m, Chen W. Identification and analysis of differentially expressed proteins during cotyledon embryo stage in longan. Sci Hortic. 2010;126:426–433. [Google Scholar]
- Businge E, Bygdell J, Wingsle G, Moritz T, Egertsdotter U. The effect of carbohydrates and osmoticum on storage reserve accumulation and germination of Norway spruce somatic embryos. Physiol Plant. 2013;149:273–285. doi: 10.1111/ppl.12039. [DOI] [PubMed] [Google Scholar]
- Pegoraro C, Mertz L, Maia L, Rombaldi C, Oliveira A. Importance of heat shock proteins in maize. J Crop Sci Biotechnol. 2011;14:85–95. [Google Scholar]
- Kadota Y, Shirasu K. The HSP90 complex of plants. Biochimica Et Biophys Acta-Mol Cell Res. 1823;2012:689–697. doi: 10.1016/j.bbamcr.2011.09.016. [DOI] [PubMed] [Google Scholar]
- Yamada K, Fukao Y, Hayashi M, Fukazawa M, Suzuki I, Nishimura M. Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J Biol Chem. 2007;282:37794–37804. doi: 10.1074/jbc.M707168200. [DOI] [PubMed] [Google Scholar]
- Teyssier C, Maury S, Beaufour M, Grondin C, Delaunay A, Le Mette C, Ader K, Cadene M, Label P, Lelu-Walter MA. In search of markers for somatic embryo maturation in hybrid larch (Larix x eurolepis): global DNA methylation and proteomic analyses. Physiol Plant. 2013. doi:10.1111/ppl.12081. [DOI] [PubMed]
- Zhen Y, Zhao ZZ, Zheng RH, Shi J. Proteomic analysis of early seed development in Pinus massoniana L. Plant Physiol Biochem. 2012;54:97–104. doi: 10.1016/j.plaphy.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Balbuena TS, Silveira V, Junqueira M, Dias LLC, Santa-Catarina C, Shevchenko A, Floh EIS. Changes in the 2-DE protein profile during zygotic embryogenesis in the Brazilian Pine (Araucaria angustifolia) J Proteomics. 2009;72:337–352. doi: 10.1016/j.jprot.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Hatano K, Shimada T, Hiraiwa N, Nishimura M, HaraNishimura I. A rapid increase in the level of binding protein (BiP) is accompanied by synthesis and degradation of storage proteins in pumpkin cotyledons. Plant Cell Physiol. 1997;38:344–351. doi: 10.1093/oxfordjournals.pcp.a029172. [DOI] [PubMed] [Google Scholar]
- Dias LLC, Balbuena TS, Silveira V, Santa-Catarina C, Shevchenko A, Floh EIS. Two-dimensional gel electrophoretic protein profile analysis during seed development of Ocotea catharinensis: a recalcitrant seed species. Braz J Plant Physiol. 2010;22:23–33. [Google Scholar]
- Yasuda H, Hirose S, Kawakatsu T, Wakasa Y, Takaiwa F. Overexpression of BiP has inhibitory effects on the accumulation of seed storage proteins in endosperm cells of rice. Plant Cell Physiol. 2009;50:1532–1543. doi: 10.1093/pcp/pcp098. [DOI] [PubMed] [Google Scholar]
- Pawlowski TA. Proteome analysis of Norway maple (Acer platanoides L.) seeds dormancy breaking and germination: influence of abscisic and gibberellic acids. BMC Plant Biol. 2009;9:48. doi: 10.1186/1471-2229-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari R, Dubey RS. Effect of nickel toxicity on the alteration of phosphate pool and the suppressing activity of phosphorolytic enzymes in germinating seeds and growing seedlings of rice. Int J Plant Physiol Biochem. 2011;3:50–59. [Google Scholar]
- Geigenberger P, Hajirezaei M, Geiger M, Deiting U, Sonnewald U, Stitt M. Overexpression of pyrophosphatase leads to increased sucrose degradation and starch synthesis, increased activities of enzymes for sucrose-starch interconversions, and increased levels of nucleotides in growing potato tubers. Planta. 1998;205:428–437. doi: 10.1007/s004250050340. [DOI] [PubMed] [Google Scholar]
- Crombie HJ, Chengappa S, Jarman C, Sidebottom C, Reid JSG. Molecular characterisation of a xyloglucan oligosaccharide-acting alpha-D-xylosidase from nasturtium (Tropaeolum majus L.) cotyledons that resembles plant ‘apoplastic’ alpha-D-glucosidases. Planta. 2002;214:406–413. doi: 10.1007/s004250100631. [DOI] [PubMed] [Google Scholar]
- Leprince O, Buitink J. Desiccation tolerance: from genomics to the field. Plant Sci. 2010;179:554–564. [Google Scholar]
- Chibbar RN, Shyluk J, Georges F, Mallard CS, Constabel F. Esterase isozymes as markers of somatic embryogenesis in cultured carrot cells. J Plant Physiol. 1988;133:367–370. [Google Scholar]
- Rao KV, Suprasanna P, Reddy GM. Biochemical-changes in embryogenic and nonembryogenic calli of Zea mays L. Plant Sci. 1990;66:127–130. [Google Scholar]
- Subramani T, Chandrashekharaiah KS, Swamy NR, Murthy KRS. Purification and characterization of carboxylesterase from the seeds of Jatropha curcas. Protein J. 2012;31:120–128. doi: 10.1007/s10930-011-9380-7. [DOI] [PubMed] [Google Scholar]
- Pierrat OA, Mikitova V, Bush MS, Browning KS, Doonan JH. Control of protein translation by phosphorylation of the mRNA 5′-cap-binding complex. Biochem Soc Trans. 2007;35:1634–1637. doi: 10.1042/BST0351634. [DOI] [PubMed] [Google Scholar]
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ. A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol. 2005;137:1397–1419. doi: 10.1104/pp.104.056614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek KN, Graham K, Agrawal GK, Thelen JJ. The 14-3-3 isoforms chi and epsilon differentially bind client proteins from developing Arabidopsis seed. J Proteome Res. 2011;10:4076–4087. doi: 10.1021/pr200263m. [DOI] [PubMed] [Google Scholar]
- Hanfrey C, Franceschetti M, Mayer MJ, Illingworth C, Elliott K, Collier M, Thompson B, Perry B, Michael AJ. Translational regulation of the plant S-adenosylmethionine decarboxylase. Biochem Soc Trans. 2003;31:424–427. doi: 10.1042/bst0310424. [DOI] [PubMed] [Google Scholar]
- Silveira V, Vita A, Macedo A, Dias M, Floh E, Santa-Catarina C. Morphological and polyamine content changes in embryogenic and non-embryogenic callus of sugarcane. Plant Cell Tiss Organ Cult. 2013;114:351–364. [Google Scholar]
- Steward FC, Marion OM, Smith J. Growth and organized development of cultured cells. I. growth and division of freely suspended cells. Am J Bot. 1958;45:693–703. [Google Scholar]
- Reinert J. Morphogenese und ihre kontrolle an gewebekulturen aus carotten. Naturwissenschaften. 1958;45:344–345. [Google Scholar]
- Vogel G. How does a single somatic cell become a whole plant. Science. 2005;309:86–86. doi: 10.1126/science.309.5731.86. [DOI] [PubMed] [Google Scholar]
- Mihaljevic S, Radic S, Bauer N, Garic R, Mihaljevic B, Horvat G, Leljak-Levanic D, Jelaska S. Ammonium-related metabolic changes affect somatic embryogenesis in pumpkin (Cucurbita pepo L.) J Plant Physiol. 2011;168:1943–1951. doi: 10.1016/j.jplph.2011.05.025. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Arnold W, Mitrenga D, Mayersbach HV. Freeze-drying and embedding in glycol methacrylate - results of histochemical reactions. Acta Histochem. 1975;54:271–277. [PubMed] [Google Scholar]
- Gahan PB. Plant Histochemistry and Cytochemistry: An Introduction. Orlando: Academic Press; 1984. [Google Scholar]
- Santa-Catarina C, Silveira V, Balbuena TS, Viana AM, Estelita MEM, Handro W, Floh EIS. IAA, ABA, polyamines and free amino acids associated with zygotic embryo development of Ocotea catharinensis. Plant Growth Regul. 2006;49:237–247. [Google Scholar]
- Neuhoff V, Stamm R, Eibl H. Clear background and highly sensitive protein staining with coomassie blue dyes in polyacrylamide gels - a systematic analysis. Electrophoresis. 1985;6:427–448. [Google Scholar]
- Rocha SLG, Neves-Ferreira AGC, Trugilho MRO, Chapeaurouge A, Leon IR, Valente RH, Domont GB, Perales J. Crotalid snake venom subproteomes unraveled by the antiophidic protein DM43. J Proteome Res. 2009;8:2351–2360. doi: 10.1021/pr800977s. [DOI] [PubMed] [Google Scholar]
- Gandra PG, Valente RH, Perales J, Pacheco AG, Macedo DV. Proteomic analysis of rat skeletal muscle submitted to one bout of incremental exercise. Scand J Med Sci Sports. 2012;22:207–216. doi: 10.1111/j.1600-0838.2010.01235.x. [DOI] [PubMed] [Google Scholar]


