Abstract
Background
Familial haemophagocytic lymphohistiocytosis (FHL) is a rare immune deficiency with uncontrolled inflammation; the clinical course usually starts within the first years of life, and is usually fatal unless promptly treated and then cured with haematopoietic stem cell transplant. FHL is caused by genetic mutations resulting in defective cell cytotoxicity; three disease related genes have been identified to date: perforin, Munc13-4 and syntaxin-11. A fourth gene, STXBP2, has been identified very recently as responsible for a defect in Munc18-2 in FHL-5.
Aims
To describe the result of the screening of families with HLH and previously unassigned genetic defects.
Methods
Patients with HLH diagnosed according to current diagnostic criteria, and who lacked mutations in the PRF1, Munc13-4, and STX11 genes were sequenced for mutations in STXBP2. Functional study was performed when material was available.
Results
Among the 28 families investigated, 4 (14%) with biallelic STXBP2 mutations were identified. They originated from Italy, England, Kuwait and Pakistan. The p.Pro477Leu resulting from c.1430C>T, and p.Arg405Gln resulting from the single c.1214G>A nucleotide change are known, while we contribute two novel mutations: p.Glu132Ala resulting from c.395A>C, and p.Gly541Ser, resulting from c.1621G>A. The detrimental effect of the p.Gly541Ser mutation was documented biochemically and functionally in NK and CD8 cells. Additional polymorphisms are also described.
Conclusion
These data expand current knowledge on the genetic heterogeneity of FHL and suggest that patients with FHL5 may have different results in degranulation assays under different conditions.
INTRODUCTION
Haemophagocytic lymphohistiocytosis (HLH) is a genetically heterogeneous disorder characterised by an hyperinflammatory syndrome with fever, hepatosplenomegaly, cytopenia and sometimes central nervous system involvement.1 Bone marrow aspiration is usually performed early during the diagnostic work-up, enabling the identification of haemophagocytosis by activated macrophages. In most cases the natural course of HLH is rapidly fatal within a few weeks, unless appropriate treatment, including corticosteroids, cyclosporine, etoposide, or anti-thymocyte globuline, can obtain transient disease control.2-4 So far, only patients who underwent haematopoietic stem cell transplantation (HSCT) have been cured.5–10
Differential diagnosis of HLH may be difficult.11 To this purpose, diagnostic guidelines for HLH have been established by the Histiocyte Society.12,13 In particular, demonstration of frequent association with common pathogens, together with evidence of impaired natural killer cytotoxic activity, provided the rationale for considering HLH as a selective immune deficiency.14–16 Starting from the original report by Farquhar et al in 1952,17 autosomal recessive inheritance was proposed and then confirmed as the common mode of inheritance for the familial form of HLH (FHLH or FHL).
Previous genetic studies in FHL revealed an extended genetic heterogeneity. A first mapping approach included four consanguineous families of Pakistani origin and identified a 7.8 cm region on chromosome 9q21.3–22, but the underlying defect is still unknown (FHL-1; MIM 267700).18 A second report described linkage of 10 of 17 investigated families to a region on chromosome 10q21–22,19 thus indicating genetic heterogeneity. Mutations in the perforin-1 gene (PRF1) were then described at this locus (FHL-2; MIM 603553).20 About 20–50% of the patients have mutations in PRF1, with a strong geographical bias in the overall frequency.21 Further genetic defects associated with FHL affect proteins involved in transport, membrane fusion or exocytosis of perforin containing lytic granules such as Munc13-4 (FHL-3; MIM 608898) and syntaxin 11 (FHL-4; MIM 603552).22,23
Recently, zur Stadt et al have allocated a novel FHL type, FHL-5 (MIM 613101), to a 1 Mb region on chromosome 19p using high resolution single nucleotide polymorphism (SNP) genotyping in eight unrelated FHL patients from consanguineous families. They have identified mutations in STXBP2, encoding syntaxin binding protein 2 (Munc18-2), a protein involved in the regulation of vesicle transport to the plasma membrane. The 12 patients with FHL-5 originated from Turkey, Saudi Arabia, and Central Europe.24 Almost simultaneously, a similar report was provided by Côte et al.25
In this paper we describe the results of initial screening of STXBP2 mutations among patients with HLH of different geographic origin.
PATIENTS AND METHODS
Patients selection
We selected patients with HLH diagnosed according to current diagnostic criteria13 who lacked PRF1, Munc13-4, and STX11 mutations.
STXBP2 gene analysis
Genomic and mRNA sequences of the STXBP2 gene were retrieved from the National Center for Biotechnology Information (NC_000019.9; NM_006949.2). Genomic DNA was isolated from peripheral blood samples using BioRobot EZ1 Workstation (Qiagen, Milan, Italy). Some samples were retrieved from our DNA library of retrospective patients.1,23,24
To analyse the STXBP2 gene, the 19 coding exons and exon–intron boundaries were amplified and directly sequenced, in both directions, with the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, California, USA). Amplification reactions were performed with 60 ng of DNA, 10 ng of each primer, 200 μM dNTPs, 1× PCR reaction buffer, and 2.5 U Taq polymerase in a final volume of 25 μl; primers are available on request. Sequences obtained using an ABI Prism 3130XL Sequence Detection System (Applied Biosystems) were analysed and compared with the reported gene structure using the dedicated software SeqScape (Applied Biosystems).
In silico analysis
Unknown mutations were tested by bioinformatic facilities. We used three web query tools: SIFT (Sorting Intolerant From Tolerant: http://sift.bii.a-star.edu.sg/), POLYPHEN (prediction of functional effect of human nsSNPs: http://genetics.bwh.harvard.edu/pph/) and The ConSeq Server (http://conseq.tau.ac.il).
SIFT is a sequence homology based tool that predicts whether an amino acid substitution in a protein will have an effect. It is based on the concept that protein evolution is correlated with protein function. Variants that occur at conserved positions are thought to be tolerated less than variants at different positions.
PolyPhen is a computational tool for identification of potentially functional variations. Predictions are based on a combination of phylogenetic, structural and sequence information characterising a substitution and its position in the protein.
The ConSeq Server is a homology based tool for the identification of functionally and structurally important residues in protein sequences. Given a set of homologous proteins in the form of multiple sequence alignment (MSA), the evolutionary rate at each amino acid site in the MSA is calculated.
Degranulation assay
All reagents are from BD Biosciences (Oxford, UK) unless otherwise stated. Peripheral blood mononuclear cells (PBMC) were isolated from heparinised whole blood using a density gradient (Lymphoprep, Axis Shield, Uxbridge, UK). PBMC were stimulated overnight with IL-2 (100 U/ml; Chiron, Ringaskiddy, Ireland) and then incubated with anti-CD107a-FITC (fluorescein isothiocyanate) antibody (1/50) alone or with anti-CD3 (clone CLB-T3/4.E 150 ng/ml; Mast Group Ltd, Bootle, UK) or phytohaemagglutinin (PHA, 6.4 g/ml; Biostat, Stockport, UK) for 2 h at 37°C. Surface markers were stained with anti-CD3-APC, CD56-PE and CD8-PerCP. NK cell degranulation was analysed from the PHA stimulated sample, gating on CD3−CD56+ cells, while T cell degranulation was analysed from the CD3 stimulated sample, gating on CD3+CD8+ cells. Results are reported as CD107a (ie, % CD107a+ cells of stimulated—% CD107a+ cells of unstimulated sample).
PBMC were also cultured in the presence of irradiated 721.221 B-EBV (Epstein–Barr virus) cell line (in the relative proportion of 4:1) and 100 U/ml of IL-2, culture conditions which allow a preferential activation and expansion of NK lymphocytes. Activated lymphocytes were co-cultured with K562 to induce degranulation of NK cells, in the presence of anti-CD107a-PE mAb. Thereafter, the cells were stained and surface expression of CD107a was assessed in the CD3−CD56+ cell fraction, as previously described.26 CTL lines were also generated and cultured as described earlier27 and their degranulation was tested upon activation with plate bound anti-CD3 (1 μg/mL OKT-3) at 5×105 cells per well in a 96 well plate. Cells were incubated for 4 h in FACS buffer in the presence of anti-CD107a to track CD107a cycling, followed by staining for surface CD8 expression.
All samples were analysed by flow cytometry on a BD FACSCalibur.
Cytotoxicity assay
Polyclonal CD8+ T cell were isolated by negative selection (Miltenyi Biotec) from phytohaemagglutinin (PHA) blasts grown in the presence of IL-2, and CTL lines were generated by stimulating cells in vitro with PHA (1 mg/ml), irradiated allogeneic buffy coats and IL-2. CTL lines were cultured in RPMI, 5% human serum, sodium pyruvate, L-glutamine, 2 mercaptoethanol and 100 U/ml rIL-2. Cells were then tested in a 4 h CD3 mediated redirected cytotoxicity assay, using the murine mastocytoma FcγRc+ P815 cell line as targets in the presence of anti-CD3 (0.5 mg/mL UCHT-1). Cytolytic activity was measured by LDH release from targets using the Cytotox96 cytotoxicity assay (Promega, Madison, Wisconsin, USA).
Western blot analysis
Western blot analysis of Munc18-2 protein was performed as previously described.24
Cloning and co-immunoprecipitation analysis
The cloning of the Munc18-2 mutants and co-immunoprecipitation experiments were performed as previously described24.
RESULTS
We have investigated a total of 28 families in which HLH had been diagnosed in one or more subjects, and mutations of PRF1, Munc13-4, and STX11 had been excluded by sequence analysis. Of these families, 17 were of Italian origin, while the others originated from Great Britain (n=4), Turkey (n=2), Rumania, Qatar, Morocco, Kuwait, and Pakistan (n=1).
Four STXBP2 mutations were found in four (14%) families originating from Italy, England, Kuwait and Pakistan (table 1).
Table 1. Presenting features and current status in four patients with haemophagocytic lymphohistiocytosis (HLH) and biallelic STXBP2 gene mutations.
| UPN | 113 | 388 | 392 | 408 |
| Consanguinity | + | + | + | Not known |
| Ethnic origin | Caucasian, Italy | Pakistani | Kuwaiti | Caucasian, UK |
| Mutation | p.Glu132Ala c.395A>C | p.Arg405Gln c.1214G>A | p.Pro477Leu c.1430C>T | p.Gly541Ser c.1621G>A |
| Sex | M | F | F | F |
| Age at diagnosis | 4 years | 4.5 months | 7 months | 7 months |
| Fever | + | + | + | + |
| Splenomegaly | + | + | + | + |
| Haemoglobin (<9 g/dl) | 8.0 | 5.7 | 9.1 | 6.2 |
| Platelets(<100000/mm3) | 23 | 38 | 60 | 11 |
| Neutrophils (<1000/mm3) | Normal | 2.05 | 0.84 | 0.4 |
| Triglycerides (>265 mg/dl) | Normal | 823 | 495 | 600 |
| Fibrinogen (<150 mg/dl) | 75 | 90 | Normal | 110 |
| Haemophagocytosis | + | + | + | + |
| CNS symptoms | + | no | no | no |
| CSF pleocytosis(<5/mm3) | 22 | no | no | ND |
| Degranulation assay | ND | ND | ND | Abnormal* |
| BMT, donor | NP | MUD | MSD | MUD |
| Present status | Deceased of disease at 4.7 years | Deceased 2 years, 8 months, enteropathy post HSCT | Alive 3 years 6 months | Alive 2 years, 8 months |
ND, Not determined; NP, Not performed.
CNS, central nervous system; CSF, cerebrospinal fluid; HSCT, haematopoietic stem cell transplantation; MSD: matched sibling donor; MUD, matched unrelated donor.
Items in bold are comprised among diagnostic criteria for HLH.12
See text for details.
Of these four mutations, two had been reported previously: the Pro477Leu, resulting from the single c.1430C→T nucleotide change,24,25 and Arg405Gln resulting from the single c.1214G→A nucleotide change.24
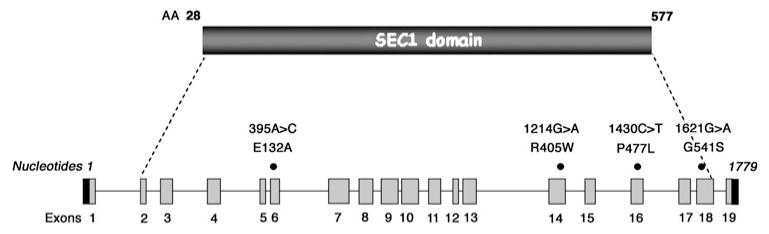
We report two novel mutations (table 1, figure 1). Glu132Ala amino acid change, resulting from the nucleotide change c.395A→C, was observed in UPN 113. The in silico analysis with the three methods suggests that this amino acid is highly conserved and predicted to be critical for protein integrity. This mutation was not found in 120 healthy Caucasian control subjects.
Figure 1. Structure of the STXBP2 gene and location of the observed mutations in four patients with FHL-5.
In vitro co-immunoprecipitation analyses of EGFP tagged Munc18-2 p.Glu132Ala with FLAG-tagged syntaxin 11 demonstrate that Munc18-2 p.Glu132Ala lacks the ability to bind to syntaxin 11 (figure 2).
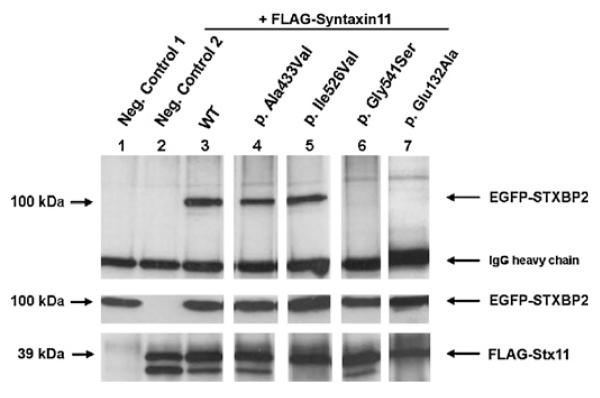
Figure 2. Co-immunoprecipitation analysis.
Upper panel: Neither the negative control 1 (lane 1) without FLAG-Stx11 nor the negative control 2 (lane 2) without EGFP-STXBP2 show unspecific binding to the IP antibody coupled agarose beads. EGFP-STXBP2 WT (lane 3) as well as both EGFP-STXBP2 polymorphic variants (lane 4/lane 5) co-immunoprecipitate with FLAG-Stx11, whereas EGFP-STXBP2 Gly541Ser (lane 6) and Glu132Ala (lane 7) show an impaired bingeing to FLAG-Stx11. The middle panel shows the corresponding input control of the EGFP-STXBP2 proteins. The lower panel presents the IP controls of each approach which reveal approximately equal amounts of FLAG-Stx11 after immunoprecipitation with the capture anti-FLAG-M2 murine IgG1 monoclonal antibody.
The second novel mutation is the Gly541Ser amino acid change, observed in UPN 408, resulting from the nucleotide change c.1621G→A. This mutation was not found in 120 healthy Caucasian control subjects. The Gly541 amino acid is highly conserved and predicted to be a structural residue (The ConSeq Server results).
Biochemical analyses revealed that this mutation disrupts association of Munc 18-2 with syntaxin 11. Using co-immunoprecipitation of EGFP-Munc 18-2 with FLAG-syntaxin 11 we found that syntaxin 11 could co-precipitate Munc 18-2 with the wild-type sequence, while Munc18-2 with the Gly541Ser mutation could not be co-precipitated (figure 2).
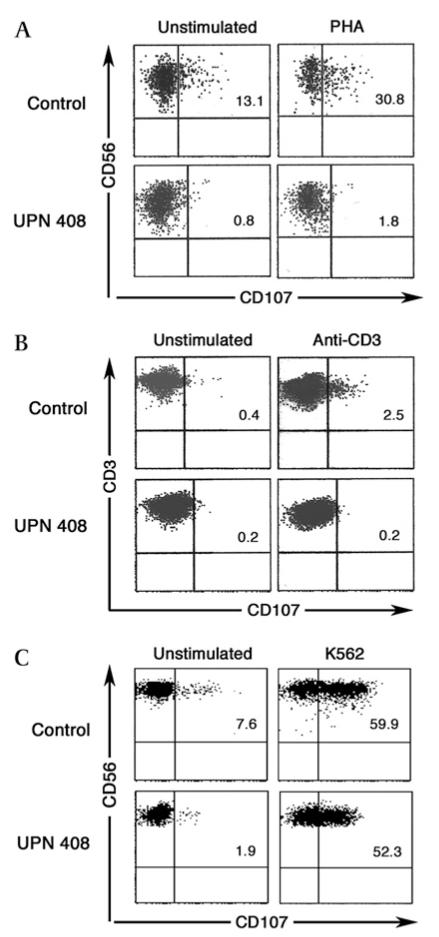
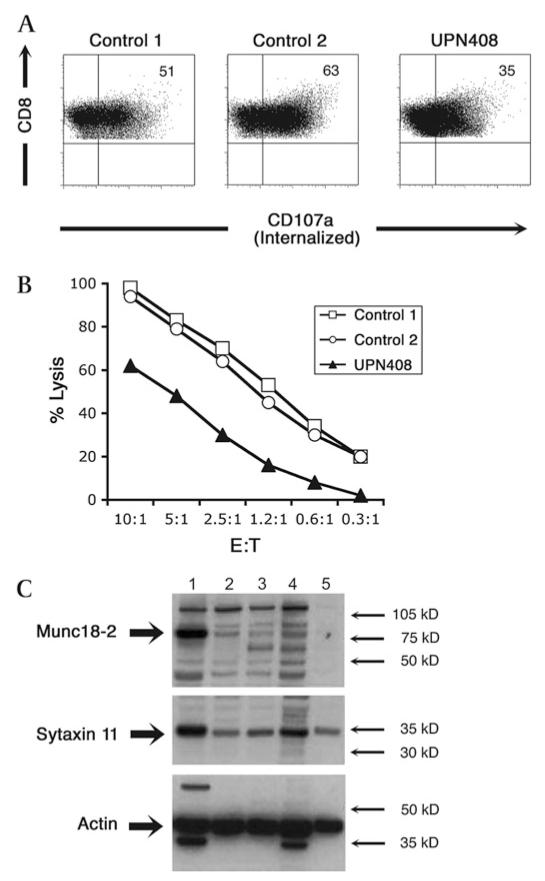
We performed functional assays with lymphocytes derived from UPN 408. The ability of the cells to degranulate upon stimulation was explored using two different types of assay (figure 3). A granule release assay was performed on PBMC cultured for 24 h in IL-2 by using PHA to stimulate NK cells and anti-CD3 antibody to stimulate T cells. The cells from patient UPN 408 gave the following results: minimal release, which was barely above background (≤1% of CD107) level in NK cells (figure 3A) (normal values have a mean±SD of 24.3%±7.1%), and completely absent release in CD3+CD8 cells (figure 3B) (normal values have a mean±SD of 5.3%±3.7%). Lymphocytes were also activated and expanded in culture and the degranulation capacity was tested by CD107a surface expression on NK cells stimulated by K562 cells according to the method originally described by Marcenaro et al26; with this assay, the granule release capacity by UPN 408 appeared to be similar to the healthy control (figure 3C) (normal values have a mean±SD of 53.9%±9). CTL lines from this patient were also tested for degranulation and cytotoxic function (figure 4). CD107a expression was reduced in patient samples (mean=29.5, SE=5.5, n=2) compared with controls (mean=61.4, SE=2.7, n=13) (figure 4A). Patient CTL lines also showed a reduced, although not absent, cell killing capacity (figure 4B). In addition western blotting from cell lysates of cloned CTL or PHA stimulated T cell lines from the patient carrying the Gly541Ser mutation (UPN408, lanes 3–5 figure 4C) showed very low levels of Munc18-2 compared to lysates from healthy donor CTL (lane 1, figure 4C). Lysates from UPN 408 showed a similar level in reduction of Munc18-2 to those of a previously reported patient with a Pro477Leu change23 (lane 2, figure 4C). Levels of syntaxin 11 were reduced in patient samples compared to the control from a healthy donor (figure 4, panel C). These data support a functional interaction between Munc18-2 and syntaxin 11 and show that, in the absence of Munc18-2, syntaxin11 stability is reduced. In addition these results demonstrate that the p.Gly541Ser mutation has a functionally detrimental impact.
Figure 3. Flow cytometry evaluation of CD107 expression of patient UPN 408.
Peripheral blood mononuclear cells (PBMC) from patient and healthy control cultured for 24 h with IL-2 were analysed for CD107 expression by NK cells (gate CD3− CD56+) upon stimulation with phytohaemagglutinin (PHA) (panel A), or by CD3+ CD8+ lymphocytes stimulating with anti-CD3 mAb (panel B). Activated and expanded lymphocytes were analysed for CD107 expression by NK cells upon co-culture with K562 (panel C).
Figure 4. CTL degranulation and cytotoxicity and protein study of patient UPN 408.
Degranulation assay showing CD3 stimulated internalisation of CD107a (panel A), detected with PE-CD107a mAb present throughout the 4 h incubation, from CTL lines from two healthy donors (controls 1 and 2) compared to those from UPN408. Cells were stained for CD8 (y axis) and CD107a-PE (x axis). Cellular cytotoxicity of CTL lines from UPN408 (shaded triangles) compared to those from healthy donors (open symbols) measured using redirected lysis of P815 target cells at different E:T ratios (panel B). Data points were performed in triplicate with standard deviations of <2.5% for all E:T ratios. Western blot of cell lysates from CTL lines (1–4) or phytohaemagglutinin (PHA) blasts (5) from (1) Healthy donor, (2) FHL-5 patient with Pro477Leu mutation (see reference 24), (3–4) UPN408 clones, and (5) UPN408 PHA blasts probed with antibodies to Munc18-2, syntaxin 11 and reprobed with actin to show loading levels (panel C). Molecular weight markers are shown.
We tested 120 healthy controls of Caucasian origin for the substitutions IVS2-7C→T, Thr345Met (c.1034C→T), Ala433Val (c.1298C→T), Ile526Val (c.1576A→G), which all turned out to be frequent neutral polymorphisms. The Thr345 amino acid residue is supposed to be a residue without a predictable functional or structural property (ConSeq results). The p.Ala433Val (c.1298C→T) missense mutation could be detected three times heterozygously in 120 healthy control subjects. In addition co-immunoprecipitation analyses reveal that this mutation does not affect the binding ability of Munc 18-2 to syntaxin 11 (figure 2, lane 4). The Ala433 amino acid turned out to be a variable residue and is not predicted to bear a functional or structural property (The ConSeq Server Results). Due to these results we concluded that this missense mutation is a further neutral Munc 18-2 polymorphic variant, at least in the context concerning the interaction with syntaxin 11. The Ile526Val amino acid exchange is a common SNP of the Munc 18-2 protein (uniprot/Q15833).
We also identified the following silent variants: c.609C→T (p. His203His) and c.1443T→C (p.Asp481Asp).
The geographic and ethnic origin of the four patients with FHL-5 was different, including two Caucasian families one from Italy and one from England, and two Arabic families from Kuwait or Pakistan. Parental consanguinity was documented in three families, not in the fourth—although it originated from an area of England with a high rate of other autosomal diseases due to limited gene pool (island effect).
The clinical and laboratory features of the four patients with STXBP2 mutations fit the diagnostic criteria for HLH (fever, splenomegaly, cytopenia, hypertriglyceridaemia or hypofibrinogenaemia, and haemophagocytosis)12,13 and are summarised in table 1. Their age at onset of the disease was 4 months in one patient, 7 months in two, while one patient had a later onset at 4 years; however, the disease course of this patient with later onset was very aggressive and the child died within 6 months of progressive disease. The remaining 24 patients in whom no STXBP2 mutation was found had a median age of 14 months, with a range of <1 month to 17 years.
Among 451 cases enrolled over more than 20 years in our HLH Registry1 and evaluable for this parameter, the median age at diagnosis was 12 months (range, 1 day to 61 years); 24% were diagnosed within 3 months of age, 39% within 6 months, 63% within 2 years, and 76% within 5 years of age.
All patients received HLH directed therapy, according to current and local regimens. One patient died of reactivation 7 months after the diagnosis, while the remaining three underwent HSCT: two of them are alive and well, while one died of complications.
DISCUSSION
Identification of a genetic marker is of paramount importance in HLH.11,28 This may allow the attending physician to recognise the patient as a candidate for early HSCT from the best available donor, as well as allowing the screening of family donors for HLH. HSCT has been recognised as the only available therapeutic approach with the potential to cure FHL.5–10
Current knowledge on FHL related genes allows the identification of a large proportion of cases. Yet, this proportion is variable according to the different ethnic groups and geographic regions. The only large genotype–phenotype study available so far has been conducted by the Histiocyte Society in FHL-2 and led to identification of significant associations between specific mutations and ethnic groups. Some mutations were found more commonly: c.1122G→A (p.Trp374X), associated with Turkish origin; c.50delT (p.Leu17fsX22) associated with African/African American origin; and c.1090–91delCT (p.Leu364fsX), so far identified only in Japanese patients.21 A similar study is ongoing but has not yet been completed for FHL-3. The contribution of FHL-4 appears to be quite limited so far, with the majority of patients reported from a single geographic area.23,24 Thus, additional genetic defects were expected in order to assign a defect to the remaining families, accounting for a variable proportion included between about 20%—the proportion observed among Italian patients (M Aricò, unpublished data,29,30) and 60–70%—as observed among native German patients—of the total population (U zur Stadt, unpublished data). Current knowledge suggests that the population of patients with an unassigned defect is not clinically distinguishable from those belonging to the currently recognised genetic subgroups (ie, FHL-2 and FHL-3).
Following identification of STXBP2 as a novel FHL related gene, thus becoming FHL-5,24 we decided to screen the series of patients referred to our registry for a genetic study. We found that only a minority of such patients, 14%, belong to the FHL-5 subset of this disease.
The presenting features of these patients appear largely comparable to those included in the remaining subgroups, in particular FHL-2 and FHL-3. Thus, it appears unlikely that FHL-5 may be suspected on the basis of the initial clinical picture.
Overall, a total of 25 patients with FHL-5 are known so far, including the present four. Among the observed mutations, three appear to be more frequent: the c.1430C→T (p.Pro477Leu) mutation, previously reported in seven homozygous patients of Arabian origin24,25 and now in a Kuwaiti family; and the c.12471G→C (p.Val417LeufsX126) mutation, reported in nine patients of different geographic origin, six of them in a homozygous state. Furthermore, the p.Ile232del mutation was reported in two Turkish patients. In addition, we now also report the c.1214G/A (p.Arg405Gln) mutation, which was previously found in a Turkish patient,24 in our patient UPN 388 of Pakistani origin.
We have identified two novel mutations. The c.395A→C (p. Glu132Ala) was found in homozygosity in a consanguineous family from southern Italy, and the Gly541Ser mutation identified in homozygosity in a Caucasian patient of English origin. The pathogenic value of this latter mutation is supported by evidence of defective protein and by defective cytotoxicity by T lymphocytes. Furthermore, both mutations affect the binding of syntaxin 11. A similar impairment in the binding of syntaxin 3 was reported for mutations p.Arg405Pro and p.Gly541Glu by Riento et al.31
Screening of genetic mutations in all the FHL related genes, the gold standard for diagnosing the different subgroups of FHL, is time consuming and not available to most clinical centres. To address this issue, our group has originally introduced the evaluation of expression of CD107 by stimulated cytotoxic cells26; a defect in this function may herald a defect in Munc13-4 protein, as well as in syntaxin11. Defective degranulation impairs the delivery of effector proteins to the target cells, hampering cellular cytotoxicity; the latter is the pathogenic mechanism of FHL in its different subsets. Lack of functional STXBP2 is associated with defective degranulation and defective cellular cytotoxicity, as documented in the present case UPN 408. Yet, compared to patients with FHL-3 due to a defect in Munc13-4, the degranulation defect in patients with FHL-5 may be different. In fact, based on our present finding of a single patient with homozygous Gly541Ser mutation, upon activation these cells may be able to rescue their degranulation capacity, as observed in the classical degranulation assay as described by Marcenaro et al.26 It may be of interest that cells from some patients with FHL-5 may behave normally in the standard CD107 assay performed on activated NK cells, but may show an impaired degranulation capacity when either resting NK24,25 or overnight IL-2 incubated PBMC are tested and, as we describe here, stimulating NK cells with PHA and T lymphocytes by anti-CD3. Long term activated CTL revealed some defect of degranulation. From this point of view, patients with FHL-5 might appear to be closer to patients with FHL-4 (with syntaxin 11 deficiency)32 than to those with FHL-3. These results are consistent with the previous report that degranulation is at least partially restored by culture of cells in IL-2.24,25
In conclusion, mutations of STXBP2 resulting in defective Munc18-2 protein and the clinical HLH phenotype underline the genetic heterogeneity of the disease, although apparently accounting for a minority of cases. Patients with the clinical phenotype, defined by the diagnostic criteria, and unassigned genetic defect are the best candidates for mutation analysis of this gene. Patients with FHL-5 apparently may be not detected by the degranulation assay which is very effective in detecting patients with FHL-3. It is possible that modifications of this method may allow detection of some patients with a more subtle degranulation defect, thus facilitating the screening process. Accumulation of additional cases reported from other groups worldwide might enable definition of its distribution among ethnic groups and genotype–phenotype correlations. Based on current initial knowledge, FHL-5 patients should receive the standard therapy for HLH followed by HSCT. Genetic studies of patients with FHL are still necessary in an attempt to identify additional disease related genes, with the aim of assigning a familial marker to the remaining unassigned patients and families.
Acknowledgements
The authors are grateful to the following sources of funding: European Grant ‘CureHLH’ HEALTH-F2-2008-201461, ‘Antonio Pinzino—Associazione per la Ricerca sulle Sindromi Emofagocitiche (ARSE), ‘Noi per Voi per il Meyer Onlus’; the Fördergemeinschaft Kinderkrebszentrum Hamburg e.V. Some of this work was undertaken at GOSH/UCL Institute of Child Health which received a proportion of funding from the Department of Health’s NIHR Biomedical Research Centres funding scheme. GMG is funded by the Wellcome Trust.
Footnotes
Competing interest None.
Patient consent Not required.
Ethics approval AOU Meyer, Florence Italy.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Aricò M, Janka G, Fischer A, Henter JI, Blanche S, Elinder G, Martinetti M, Rusca MP. Hemophagocytic lymphohistiocytosis. Report of 122 children from the International Registry. FHL study group of the histiocyte society. Leukemia. 1996;10:197–203. [PubMed] [Google Scholar]
- 2.Henter JI, Samuelsson-Horne A, Aricò M, Egeler RM, Elinder G, Filipovich AH, Gadner H, Imashuku S, Komp D, Ladisch S, Webb D, Janka G, Histocyte Society Treatment of hemophagocytic lymphohistiocytosis with HLH-94 immunochemotherapy and bone marrow transplantation. Blood. 2002;100:2367–73. doi: 10.1182/blood-2002-01-0172. [DOI] [PubMed] [Google Scholar]
- 3.Janka GE, Schneider EM. Modern management of children with haemophagocytic lymphohistiocytosis. Br J Haematol. 2004;124:4–14. doi: 10.1046/j.1365-2141.2003.04726.x. [DOI] [PubMed] [Google Scholar]
- 4.Mahlaoui N, Ouachee-Chardin M, de Saint Basile G, Neven B, Picard C, Blanche S, Fischer A. Immunotherapy of familial hemophagocytic lymphohistiocytosis with antithymocyte globulins: a single-center retrospective report of 38 patients. Pediatrics. 2007;120:e622–8. doi: 10.1542/peds.2006-3164. doi:10.1542/peds.2006-3164. [DOI] [PubMed] [Google Scholar]
- 5.Jabado N, de Graeff-Meeder ER, Cavazzana-Calvo M, Haddad E, Le Deist F, Benkerrou M, Dufourcq R, Caillat S, Blanche S, Fischer A. Treatment of familial hemophagocytic lymphohistiocytosis with bone marrow transplantation from HLA genetically nonidentical donors. Blood. 1997;90:4743–8. [PubMed] [Google Scholar]
- 6.Durken M, Horstmann M, Bieling P, Erttmann R, Kabisch H, Loliger C, Schneider EM, Hellwege HH, Kruger W, Kroger N, Zander AR, Janka GE. Improved outcome in haemophagocytic lymphohistiocytosis after bone marrow transplantation from related and unrelated donors: a single-centre experience of 12 patients. Br J Haematol. 1999;106:1052–8. doi: 10.1046/j.1365-2141.1999.01625.x. [DOI] [PubMed] [Google Scholar]
- 7.Horne A, Janka G, Maarten Egeler R, Gadner H, Imashuku S, Ladisch S, Locatelli F, Montgomery SM, Webb D, Winiarski J, Filipovich AH, Henter JI, Histiocyte Society Haematopoietic stem cell transplantation in haemophagocytic lymphohistiocytosis. Br J Haematol. 2005;129:622–30. doi: 10.1111/j.1365-2141.2005.05501.x. [DOI] [PubMed] [Google Scholar]
- 8.Baker KS, Filipovich AH, Gross TG, Grossman WJ, Hale GA, Hayashi RJ, Kamani NR, Kurian S, Kapoor N, Ringdén O, Eapen M. Unrelated donor hematopoietic cell transplantation for hemophagocytic lymphohistiocytosis. Bone Marrow Transplant. 2008;42:175–80. doi: 10.1038/bmt.2008.133. [DOI] [PubMed] [Google Scholar]
- 9.Cooper N, Rao K, Goulden N, Webb D, Amrolia P, Veys P. The use of reduced-intensity stem cell transplantation in haemophagocytic lymphohistiocytosis and Langerhans cell histiocytosis. Bone Marrow Transplant. 2008;42(Suppl 2):S47–50. doi: 10.1038/bmt.2008.283. [DOI] [PubMed] [Google Scholar]
- 10.Cesaro S, Locatelli F, Lanino E, Porta F, Di Maio L, Messina C, Prete A, Ripaldi M, Maximova N, Giorgiani G, Rondelli R, Aricò M, Fagioli F. Hematopoietic stem cell transplantation for hemophagocytic lymphohistiocytosis: a retrospective analysis of data from the Italian Association of Pediatric Hematology Oncology (AIEOP) Haematologica. 2008;93:1694–701. doi: 10.3324/haematol.13142. [DOI] [PubMed] [Google Scholar]
- 11.Aricò M, Allen M, Brusa S, Clementi R, Pende D, Maccario R, Moretta L, Danesino C. Haemophagocytic lymphohistiocytosis: proposal of a diagnostic algorithm based on perforin expression. Br J Haematol. 2002;119:180–8. doi: 10.1046/j.1365-2141.2002.03773.x. [DOI] [PubMed] [Google Scholar]
- 12.Henter JI, Elinder G, Ost A. Diagnostic guidelines for hemophagocytic lymphohistiocytosis. The FHL Study Group of the Histiocyte Society. Semin Oncol. 1991;18:29–33. [PubMed] [Google Scholar]
- 13.Henter JI, Horne A, Aricò M, Egeler RM, Filipovich AH, Imashuku S, Ladisch S, McClain K, Webb D, Winiarski J, Janka G. HLH-2004: Diagnostic and therapeutic guidelines for Hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer. 2007;48:124–31. doi: 10.1002/pbc.21039. [DOI] [PubMed] [Google Scholar]
- 14.Perez N, Virelizier JL, Arenzana-Seisdedos F, Fischer A, Griscelli C. Impaired natural killer activity in lymphohistiocytosis syndrome. J Pediatr. 1984;104:569–73. doi: 10.1016/s0022-3476(84)80549-1. [DOI] [PubMed] [Google Scholar]
- 15.Aricò M, Nespoli L, Maccario R, Montagna D, Bonetti F, Caselli D, Burgio GR. Natural cytotoxicity impairment in familial haemophagocytic lymphohistiocytosis. Arch Dis Child. 1988;63:292–6. doi: 10.1136/adc.63.3.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider EM, Lorenz I, Muller-Rosenberger M, Steinbach G, Kron M, Janka-Schaub GE. Hemophagocytic lymphohistiocytosis is associated with deficiencies of cellular cytolysis but normal expression of transcripts relevant to killer-cell-induced apoptosis. Blood. 2002;100:2891–8. doi: 10.1182/blood-2001-12-0260. [DOI] [PubMed] [Google Scholar]
- 17.Farquhar J, Claireaux A. Familial haemophagocytic reticulosis. Archives of Disease in Childhood. 1952;27:519–25. doi: 10.1136/adc.27.136.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohadi M, Lalloz MR, Sham P, Zhao J, Dearlove AM, Shiach C, Kinsey S, Rhodes M, Layton DM. Localization of a gene for familial hemophagocytic lymphohistiocytosis at chromosome 9q21.3–22 by homozygosity mapping. Am J Hum Genet. 1999;64:165–171. doi: 10.1086/302187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dufurcq-Lagelouse R, Jabado N, Le Deist F, Stéphan JL, Souillet G, Bruin M, Vilmer E, Schneider M, Janka G, Fischer A, de Saint Basile G. Linkage of familial hemophagocytic lymphohistiocytosis to 10q21-22 and evidence for heterogeneity. Am J Hum Genet. 1999;64:172–9. doi: 10.1086/302194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stepp SE, Dufourcq-Lagelouse R, Le Deist F, Bhawan S, Certain S, Mathew PA, Henter JI, Bennett M, Fischer A, de Saint Basile G, Kumar V. Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science. 1999;286:1957–9. doi: 10.1126/science.286.5446.1957. [DOI] [PubMed] [Google Scholar]
- 21.Trizzino A, zur Stadt U, Ueda I, Risma K, Janka G, Ishii E, Beutel K, Sumegi J, Cannella S, Pende D, Mian A, Henter JI, Griffiths G, Santoro A, Filipovich A, Aricò M, Histiocyte Society HLH Study group Genotype-phenotype study of familial haemophagocytic lymphohistiocytosis due to perforin mutations. J Med Genet. 2008;45:15–21. doi: 10.1136/jmg.2007.052670. [DOI] [PubMed] [Google Scholar]
- 22.Feldmann J, Callebaut I, Raposo G, Certain S, Bacq D, Dumont C, Lambert N, Ouachee-Chardin M, Chedeville G, Tamary H, Minard-Colin V, Vilmer E, Blanche S, Le Deist F, Fischer A, de Saint Basile G. Munc13-4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3) Cell. 2003;115:461–73. doi: 10.1016/s0092-8674(03)00855-9. [DOI] [PubMed] [Google Scholar]
- 23.Zur Stadt U, Beutel K, Kolberg S, Schneppenheim R, Kabisch H, Janka G, Hennies HC. Mutation spectrum in children with primary hemophagocytic lymphohistiocytosis: molecular and functional analyses of PRF1, UNC13D, STX11, and RAB27A. Hum Mutat. 2006;27:62–8. doi: 10.1002/humu.20274. [DOI] [PubMed] [Google Scholar]
- 24.zur Stadt U, Rohr J, Seifert W, Koch F, Grieve S, Pagel J, Strauss J, Kasper B, Nürnberg G, Becker C, Maul-Pavicic A, Beutel K, Janka G, Griffiths G, Ehl S, Hennies HC. Familial Hemophagocytic Lymphohistiocytosis Type 5 (FHL-5) Is Caused by Mutations in Munc18-2 and Impaired Binding to Syntaxin 11. Am J Hum Genet. 2009;85:482–92. doi: 10.1016/j.ajhg.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Côte M, Ménager MM, Burgess A, Mahlaoui N, Picard C, Schaffner C, Al-Manjomi F, Al-Harbi M, Alangari A, Le Deist F, Gennery AR, Prince N, Cariou A, Nitschke P, Blank U, El-Ghazali G, Ménasché G, Latour S, Fischer A, de Saint Basile G. Munc18-2 deficiency causes familial hemophagocytic lymphohistiocytosis type 5 and impairs cytotoxic granule exocytosis in patient NK cells. J Clin Invest. 2009:40732. doi: 10.1172/JCI40732. doi: 10.1172/JCI40732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcenaro S, Gallo F, Martini S, Santoro A, Griffiths GM, Aricò M, Moretta L, Pende D. Analysis of natural killer-cell function in familial Hemophagocytic lymphohistiocytosis (FHL): defective CD107a surface expression heralds Munc13-4 defect and discriminates between genetic subtypes of the disease. Blood. 2006;108:2316–23. doi: 10.1182/blood-2006-04-015693. [DOI] [PubMed] [Google Scholar]
- 27.Clark RH, Stinchcombe JC, Day A, Blott E, Booth S, Bossi G, Hamblin T, Davies EG, Griffiths GM. Adaptor protein 3-dependent microtubule-mediated movement of lytic granules to the immunological synapse. Nat Immunol. 2003;4:1111–20. doi: 10.1038/ni1000. [DOI] [PubMed] [Google Scholar]
- 28.Notarangelo LD, Sorensen R. Is it necessary to identify molecular defects in primary immunodeficiency disease? J Allergy Clin. Immunol. 2008;122:1069–73. doi: 10.1016/j.jaci.2008.08.038. [DOI] [PubMed] [Google Scholar]
- 29.Clementi R, zur Stadt U, Savoldi G, Varotto S, Conter V, De Fusco C, Notarangelo LD, Schneider M, Klersy C, Janka G, Danesino C, Aricò M. Six novel mutations in the PRF1 gene in children with haemophagocytic lymphohistiocytosis. J Med Genet. 2001;38:643–6. doi: 10.1136/jmg.38.9.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santoro A, Cannella S, Bossi G, Gallo F, Trizzino A, Pende D, Dieli F, Bruno G, Stinchcombe JC, Micalizzi C, De Fusco C, Danesino C, Moretta L, Notarangelo LD, Griffiths GM, Aricò M. Novel Munc13-4 mutations in children and young adult patients with haemophagocytic lymphohistiocytosis. J Med Genet. 2006;43:953–60. doi: 10.1136/jmg.2006.041863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riento K, Kauppi M, Keranen S, Olkkonen VM. Munc18-2, a functional partner of syntaxin 3, controls apical membrane trafficking in epithelial cells. J Biol Chem. 2000;275:13476–83. doi: 10.1074/jbc.275.18.13476. [DOI] [PubMed] [Google Scholar]
- 32.Bryceson YT, Rudd E, Zheng C, Edner J, Ma D, Wood SM, Bechensteen AG, Boelens JJ, Celkan T, Farah RA, Hultenby K, Winiarski J, Roche PA, Nordenskjöld M, Henter JI, Long EO, Ljunggren HG. Defective cytotoxic lymphocyte degranulation in syntaxin-11 deficient familial hemophagocytic lymphohistiocytosis (FHL4) patients. Blood. 2007;15:1906–15. doi: 10.1182/blood-2007-02-074468. [DOI] [PMC free article] [PubMed] [Google Scholar]