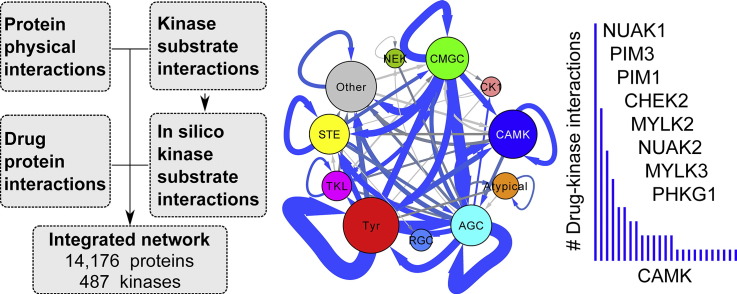
Abstract
Biological matter is organized in functional networks of different natures among which kinase–substrate and protein–protein interactions play an important role. Large public data collections allowed us to compile an important corpus of interaction data around human protein kinases. One of the most interesting observations analyzing this network is that coherence in kinase functional activity relies on kinase substrate interactions primarily and not on which protein complexes are formed around them. Further dissecting the two types of interactions at the level of kinase groups (CMGCs, Tyrosine kinases, etc.) we show a prevalence of intra-group interconnectivity, which we can naturally relate to current scenarios of evolution of biological networks. Tracking publication dates we observe high correlation of kinase interaction research focus with general kinase research. We find a similar bias in the targets of kinase inhibitors that feature high redundancy. Finally, intersecting kinase inhibitor specificity with sets of kinases located at specific positions in the kinase network, we propose alternative options for future therapeutic strategies using these compounds.
Biological significance
Despite its importance for cellular regulation and the fact that protein kinases feature prominent targets of modern therapeutic approaches, the structure and logic of the global, integrated protein phosphorylation network have not been investigated intensively. To focus on the regulatory skeleton of the phosphorylation network, we contemplated a network consisting of kinases, their substrates, and publicly available physical protein interactions. Analysis of this network at multiple levels allowed establishing a series of interesting properties such as prevalence of kinase substrate interactions as opposed to general protein–protein interactions for establishing a holistic control over kinases activities. Kinases controlling many or a few only other kinases, in addition to non-kinases, were distributed in cellular compartments differently. They were also targeted by kinase inhibitors with distinct success rates. Non-kinases tightly regulated by a large number of kinases were involved in biological processes both specific and shared with their regulators while being preferably localized in the nucleus. Collectively, these observations may provide for a new perspective in the elaboration of pharmacological intervention strategies. We complemented our study of kinase interactions with a perspective of how this type of data is generated in comparison with general research about those enzymes. Namely, what was the temporal evolution of the research community attention for interaction versus non-interaction-based kinase experiments.
This article is part of a Special Issue entitled: 20 years of Proteomics in memory of Vitaliano Pallini. Guest Editors: Luca Bini, Juan J. Calvete, Natacha Turck, Denis Hochstrasser and Jean-Charles Sanchez.
Keywords: Bioinformatics, Kinases, Network, System biology, Drugs
Graphical abstract
Highlights
-
•
Comparison of human kinase–protein and kinase–substrate networks
-
•
Prevalence of kinase–substrate interactions
-
•
Relationship with kinase inhibitors
-
•
Potential new therapeutic approaches
-
•
How does kinase interaction research focuses its research.
1. Introduction
The organization of biological matter into functional networks with elements of modularity has been identified as key to warrant the accomplishment of a great variety of biochemical and cellular functions with a limited set of gene products [1–4]. In many instances, molecular networks have been very well studied, such as in intermediate metabolism, protein interactions and phosphorylation. In eukaryotes, and with the added layer of dedicated tyrosine phosphorylation, protein phosphorylation has evolved as a primary intracellular signaling strategy. It bears many intrinsic specificities over other posttranslational modifications, including reversibility, energetic convenience and the ability of changing polarity of protein surfaces, leading to allosteric changes as well as governing protein interaction [5]. As phosphorylation has many biochemical and cellular changes both as input and as outcome, it induces a network that intersects with a plethora of biological processes.
Continuous efforts and methodological developments have greatly augmented our knowledge of how human kinases interact with other proteins or small molecules. More than 11,000 protein–protein physical interactions (PPIs) involving at least one kinase can be retrieved from public databases that compile the individual efforts of a whole community. Different technologies were used to unravel these data with a strong contribution of affinity purification-mass spectrometry (AP-MS) [6–8] and the yeast 2-hybrid (Y2H) system [9,10]. Known kinase interactions were obtained thanks to a multitude of publications, which did not necessarily focus on kinases but nonetheless included some of those enzymes. Recently, dedicated campaigns mapped human protein kinase interactors specifically and on a large-scale. For instance, following a tandem AP–MS approach and using a tetracycline-inducible strep-hemagglutinin tag [7], the protein complexes formed around 32 commonly expressed kinases, i.e. kinases found in most cell types, were mapped in HEK293 cells [11]. Another study employing the same experimental protocol charted the complexes involving CMGC kinases in an unprecedented whole kinase group-wide effort [12].
Since kinases act primarily by regulating other proteins through their enzymatic activity, to map kinase PPIs is not sufficient for understanding their functional relationships. PPIs inform us on their collaboration with other proteins to form protein complexes or molecular machines, whereas kinase–substrate interactions (KSIs) picture what is regulated by their action. Human KSIs have been studied more sparsely than PPIs so far, although data exist from several databases and new large-scale efforts were undertaken. CEASAR, a protein microarray-based strategy unraveled 3,656 KSIs for 289 protein kinases [13], bringing the number of known KSIs to more than 8500. Furthermore, computer-inferred KSIs such as the 75,000 + KSIs predicted by NetworKIN [14], exploiting experimental phospho-proteomic data, known kinase substrate specificity, and kinase physical and genetic interactions, might provide a useful complement after stringent filtering before more experimental KSI are measured.
The regulatory function of kinases has been observed to be deregulated in many diseases, in particular cancer. Accordingly, a large number of kinase inhibitors have been developed by drug discovery laboratories and pharmaceutical companies, which are mostly small molecules or antibodies. To relate kinases with their PPI partners as well as their substrates calls for further annotating such a network with the perturbation entry points available for disease therapy or chemical biology. Several large in vitro kinase screens have provided comprehensive and quantitative drug–protein interaction data (DPIs) for a lot of cancer kinase inhibitors. In this category, 72 inhibitors were screened against 442 kinases by Ambit Biosciences [15] and another set of 178 inhibitors against 300 kinases by the Peterson's group [16]. Furthermore, specialized databases collect DPIs from a broad range of scientific reports, e.g. DrugBank [17] (78 DPIs). Finally, chemical proteomics has emerged as a very interesting, more physiologically correct alternative to in vitro screens, where immobilized compounds serve as bait in affinity purifications to identify kinase inhibitor protein targets in an unbiased and cell type-dependent manner [18,19]. This methodology has been applied successfully to small molecules inhibiting protein kinase activity [20–28].
Altogether, the availability of kinase PPIs, DPIs, as well as KSIs in unprecedented, large numbers created a unique opportunity for assembling a kinase-centered network combining these three kinds of interactions and to perform a global study of kinases in their environment. We hence collected and integrated data from the various sources mentioned above and computed such a network. We started our analysis by examining how protein interaction information was correlated with classical kinase research. Investigating the global topology of PPI and KSI networks we could obtain new insights in how they differ as well as refine previous hypotheses regarding the existence of global kinase communication ways. We finally investigated how existing kinase inhibitors actually cover different classes of kinases and how KSIs might help exploring new therapeutic approaches.
2. Materials and methods
2.1. Statistical analyses and data representation
All statistical analyses were performed with the R system (www.r-project.org). Cytoscape [32] was used to prepare network representations.
2.2. Construction of the network
The list of human kinases with kinase group assignment was downloaded from UniProt web site and was comprised of 508 kinases. Assignment to kinases families (sub-groups) was obtained from kinase.com web site [29]. Binary PPIs were collected from several public repositories: IntAct [30], MINT [31], InnateDB [32], DIP [33], HPRD [34], MatrixDB [35]. Binary PPIs were generated following the spoke model in the case of AP-MS data. PPIs were also obtained from two repositories describing protein complexes: CORUM [36] and NCI PID [37]. We used the matrix model to obtain binary interactions in this case. Experimentally determined KSIs were obtained from IntAct, PhosphoSitePlus [38], and Phospho.ELM [39] databases along with Newman et al. [13] supplementary material. Computational inferences of KSI were retrieved from NetworKIN [14] website. DPIs were compiled from DrugBank [17] (inhibitors of human protein kinases only), in vitro kinase screens [15,16], and several chemical proteomics publications [20,23–25,27].
All interaction datasets downloaded from public databases were obtained from the versions available on the 10th of January 2014. Only human-human interactions derived from physical association methods were retained. UniProtKB/Swiss-Prot [40] human protein sequences were downloaded on the same day and all protein accession codes mapped to Swiss-Prot and updated to the current primary accession code to avoid duplications that can typically account for 2–5% of the proteins when public databases are merged directly. Management of the PPI downloads was operated by a database system implemented in-house using Postgres and Perl.
DPIs and KSIs were downloaded manually and further processed by dedicated Perl and R scripts. Computational KSIs were filtered by comparing with experimental data, see Results and discussion. DPIs were also submitted to filtering. In vitro screen data were imposed a maximum cut-off of 1000 nM (Refs [15,27]) and 50% of remaining activity (Ref. [16]). Chemical proteomics data, except Ref. [27], were taken as filtered in the original publications (Refs [20,23–25]). We further extracted a list of strong interactors by first requiring more stringent thresholds (100 nM for Ref. [15]; 200 nM for Ref. [27]; 25% remaining activity for Ref. [16]; no addition for Refs [11,14–16]). In addition, for each compound in each dataset separately, targets had to be either outliers with the strongest affinity or hits with an affinity not worse than 5 times the most potent target (References [15,16,27]) or 10 times the most potent target (References [20,23–25]). Outliers were defied by the boxplot.stats function of R (default parameters). DPIs from DrugBank were not considered as strong in the absence of affinity measures but we added to the strong target list the primary targets reported in Davis et al. [15] irrespective of the filters above.
2.3. Random networks
We generated two types of random networks to assess the significance of different topological features. A first operation consisted in selecting random nodes from our reference network reduced to PPIs only (Section 3.4). This selection was done to achieve similar node degree distribution and repeated 1000 times. PPI degrees of all kinases in the reference network were extracted and the probability of selecting non-kinase nodes randomly was adjusted to obtain a set of nodes, same number as kinases in the reference network, having the same degree distribution. If kinases with degree d represented x% of the kinases, then the probability to draw a non-kinase node with degree d was set to x%. A second type of random network was generated for evaluating the hierarchical nature of KSI-mediated kinase-kinase interactions (Section 3.5). In this case, we did not generate random selections of sub-networks as above but instead generated 200 full networks whose nodes followed the original degree distribution. This was achieved using the randomNodeGraph function of the graph R package, which implements the algorithm of M.E.J. Newman [41]. Self-connected nodes were removed and a correction factor applied to compensate for the resulting slight reduction in the number of edges.
3. Results
3.1. Reference network
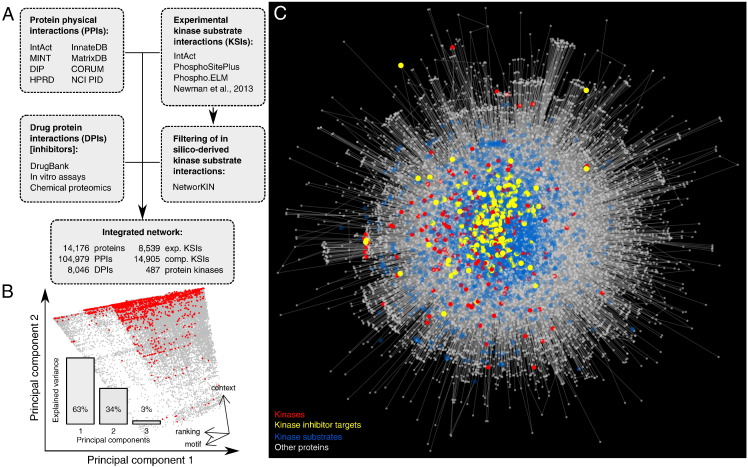
We assembled a comprehensive reference interaction network by incrementally adding the three kinds of interactions we considered in this work (Fig. 1A), starting with all PPIs we retrieved from public sources; see Materials and methods. We eliminated redundancy and PPIs of proteins with themselves that cannot be recognized as real or technical artefacts, e.g. in AP-MS where a bait is almost always detected in its pulldown. A total of 104,964 PPIs linking 13,347 proteins was collected.
Fig. 1.
Construction of a protein kinase-relevant interaction networks. (A) Multiple sources of interaction data were integrated combining protein–protein interactions (PPIs), kinase to substrate interactions (KSIs) and inhibitory drug to kinase interactions (DPIs). (B) To complement experimentally determined KSIs, we included filtered in silico KSIs from NetworKIN. Principal component analysis of the three scores provided by NetworKIN and intersection with experimental KSIs (red) identified a well-defined score range to filter NetworKIN KSIs. Variance explained by the principal components (bottom left) and orientation of the original coordinates in the space spanned by the first 2 principal components (bottom right). (C) The resulting reference network.
In the next step, we added KSIs considering experimental KSIs first. In case an experimental KSI overlapped a PPI, the interaction was considered KSI. We then further proceeded with computational KSIs (compKSIs) obtained from NetworKIN, which makes available a large set of such predictions (75,275 compKSIs). As many new experimental KSIs have been released since NetworKIN inferences were computed, we decided to exploit experimental KSIs to filter computer predictions. NetworKIN compKSIs were associated with three scores (context, motif, and ranking scores) which were potentially correlated. We therefore submitted the whole collection of 75,275 triples to principal component analysis (PCA), a classical statistical procedure to reduce data dimensionality in the presence of redundancy. The result (Fig. 1B) showed that the first two principal components described 97% of data variability (bottom left bar plot). That is, the 2-dimentional projection of the data captured information available in the three NetworKIN scores almost completely. We observed that motif and ranking scores were highly redundant (co-linear in the plot, Fig. 1B bottom right), meaning that the context score and e.g. the motif score were sufficient to describe confidence in compKSIs. Plotting the 943 experimental KSIs overlapping NetworKIN predictions in red, we could clearly observe a rather localized, parallelogram-shaped area. A simple filter could hence be designed: we imposed two independent thresholds to the context and motif scores such that 90% of the experimentally supported compKSIs were above each limit. This filter retained 14,905 compKSIs out of the original 75,275. Altogether, we added 8,539 experimental KSIs, linking 405 kinases to 2,774 substrates, and compKSIs raised these numbers to 23,444 KSIs, linking 409 kinases to 3,941 substrates.
DPIs from distinct sources were filtered separately due to the diversity of the screening methods; see Materials and methods. This resulted in 8,046 DPIs representing clear affinities between 476 kinases and 205 compounds. The reference network we assembled (Fig. 1C) included 14,176 proteins among which 487 kinases.
3.2. Accumulation of knowledge in kinase interaction research
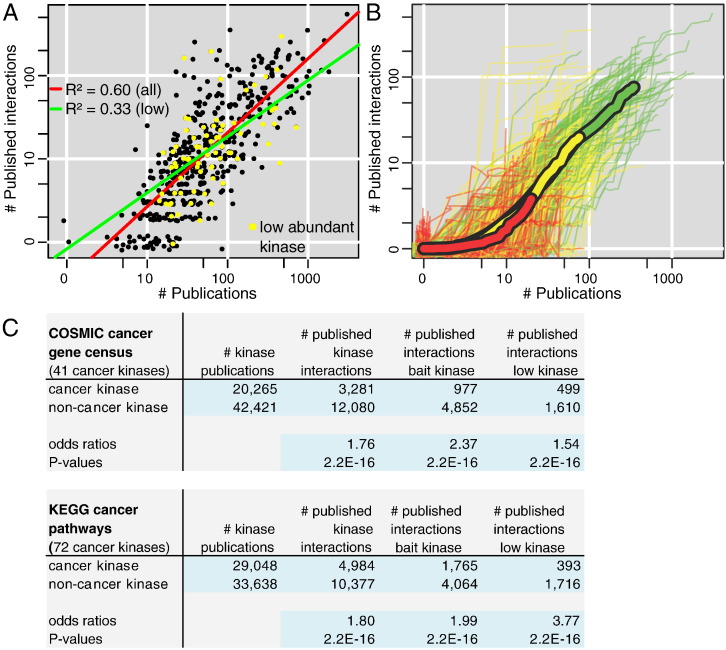
Collecting PPI data from multiple sources, we kept track for each of the PubMed ID (PMID) and the year of publication. In combination with a resource provided by the NCBI that links genes to publications (gene2pubmed, ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/), we could investigate how the community focused its attention to kinases comparing dates of general publications with publications disclosing kinase PPIs from 1975 to 2013. By summing the number of publications of each type for every kinase, we observed a strong correlation between general knowledge and knowledge about interaction partners (Fig. 2A). This means that popular and well-studied kinases were predominantly selected for performing PPI experiments, network biologists aiming at further characterizing “well-known” kinases. This was certainly due to the rather recent emergence of PPI mapping technology, which was naturally directed towards questions already identified as the most important ones instead of embarking in systematic exploration. Interestingly, this trend might change soon as unbiased, systematic studies are emerging [12].
Fig. 2.
An overview of how information accumulates in kinase research from 1975 to 2013. (A) For each kinase we summed the number of publications available (x-axis) and the number of publications disclosing an interaction for this kinase (y-axis). Each dot is thus a kinase and we see a strong correlation (red line) between the two types of knowledge (general and protein interaction). The correlation is preserved but weaker for low abundant and not commonly expressed kinases (yellow dots, green line). (B) Same analysis as in (A) but developing for each kinase a cumulative time course showing how knowledge increases over time (the end point of each curve is a dot in panel A). K-means clustering with three clusters was used to color lines (red, yellow, green). Cluster averages were featured with thick lines. (C) Focus of the community for cancer-related kinases is significantly less pronounced in protein interaction research.
We then wanted to know whether information about kinase PPIs was acquired following different modes such as very intensive early studies followed by no activity afterwards or constant but less intense attention. We therefore considered cumulative time courses counting how many general versus PPI-disclosing publications were released until a certain time point (Fig. 2B). The end-points of those curves were the dots of Fig. 2A (full totals). We clustered the cumulative time courses using the K-means algorithm and found no distinct modes. For instance, K-means applied to the generation of 3 clusters showed that the only differences were in the total amount of information available. The shapes of the cumulative time courses were highly similar otherwise. To apply K-means specifying more clusters did not change this result. We hence concluded that not only the level of attention in kinase PPI research followed closely the one of general kinase research but this relationship remained rather constant over time. It will be interesting to see if further development of PPI mapping technologies will change this in the future.
We asked one further question to our data, which was whether a bias existed at a finer level. Given the prevalent role of kinases in cancer, we classified kinases as cancer kinases and non-cancer kinases. Two different sources were used: the COSMIC cancer gene census [42], which identified 41 kinases; KEGG [43] cancer pathways, which contained 72 kinases (25 shared with COSMIC census). The level of attention to cancer versus non-cancer kinases in general research as opposed to PPI mapping was measured by the total number of publications available for the respective kinase classes. In Fig. 2C (first two columns), one can read that PPI kinase research was much less biased towards cancer than general kinase research. In the case of PPIs, kinase interactions could be measured without choosing a kinase as bait. That is, information was acquired without a kinase being in the original research focus. Consequently, we repeated our analysis only considering publications disclosing kinase interactions where the kinase was the bait and we found the same trend (Fig. 2C, third column).
We finally wanted to assess whether the trend by which kinase PPI data were highly correlated with general information about kinases was not biased by protein abundance. Namely, one could imagine that abundant kinases were easier to analyze and to discover. Therefore, the correlation observed would mostly reflect this practical difficulty. For the purpose of this study, we defined as low abundant or seldom expressed the proteins found in 3 or less of the 11 cell line proteomes recently published by the Mann's group [44], where more than 10,000 proteins were identified in each proteome. We indeed found a lower correlation (R2 = 0.33 instead of 0.60, Fig. 2A) for such proteins, definitely indicating the presence of a bias but the correlation remained clearly positive and significant. The cancer versus non-cancer kinase analysis (Fig. 2C, last column) stayed unchanged.
3.3. How do kinases connect to the human interactome
Fig. 1C visually suggests that kinases (red and yellow) and their substrates (blue) tend to occupy more central positions. We wanted to investigate this global aspect more precisely and considered kinase PPI connectivity, i.e. how many PPIs a protein has with other proteins. Other topological measures exist [45], for instance centrality or the clustering coefficient, but they are usually correlated and for the sake of simplicity it was more convenient to limit ourselves to one measure. Compared to an average human protein, a kinase was more connected with other proteins (Fig. 2A) to participate in more protein complexes most likely. It has been already observed that commonly expressed proteins tend to have more PPIs [46,47]. Here, we defined commonly expressed as detected in all of the 11 proteomes by Geiger et al. [44]. This definition certainly reflected both an important function required by most cells and an abundance level not too low. We could repeat the observation of higher connectivity in our network and further remark that within the kinase family the same trend existed, i.e. common kinases participated in more PPIs (Fig. 2A).
Turning our attention to kinase substrates, we performed a similar analysis and found that they also tended to be more connected with other proteins through PPIs (Fig. 2B). To ensure that this trend would not be caused by the two thirds of computationally derived KSIs we repeated the analysis for experimental KSIs only and observed the same bias. As expected, commonly expressed substrates had even higher connectivity. No correlation was observed between kinase and their substrate PPI connectivity (Fig. 2C, adjusted R2 = 3E− 5; same result for experimental KSI only).
3.4. A kinase–kinase network
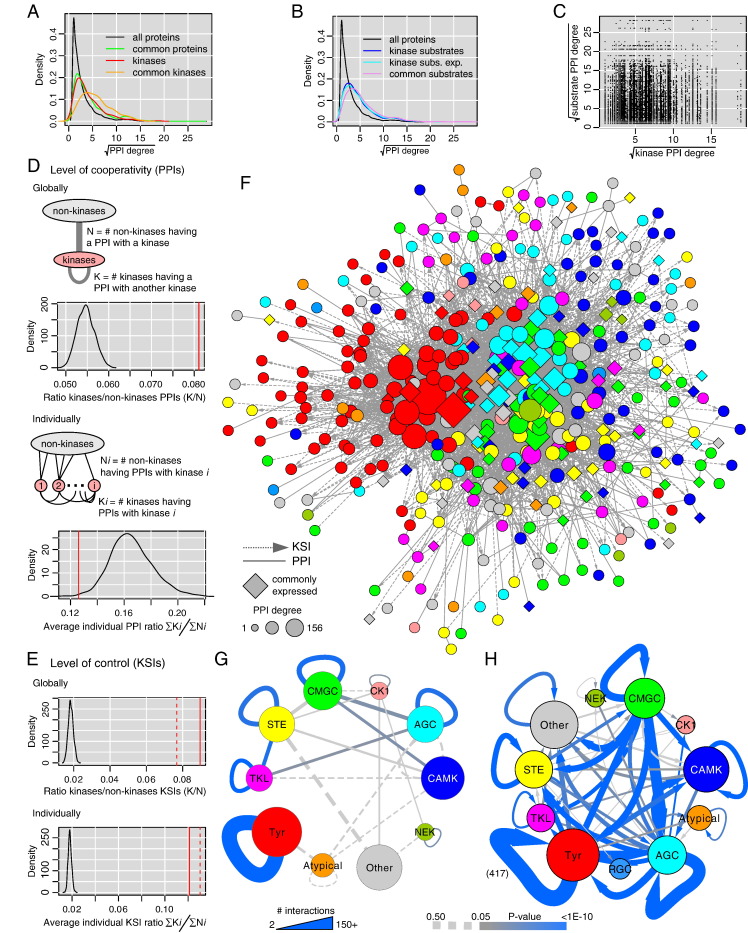
After having considered kinases within the whole human interactome, it was a natural subsequent question to ask our data whether kinases – as a whole – evolved their direct interactions to collaborate with each other (PPIs) and to operate internal control and regulation (KSIs). It has been suggested that through PPIs kinases might establish some significantly dense sub-network in yeast [48] or in human [12] (restricted to CMGC kinase data in the latter case). We hence tried to assess the existence of this phenomenon at the level of the whole kinase family and distinguishing for the first time PPIs and KSIs, which represent very different forms of interaction.
Starting with PPIs, we first adopted a global perspective by marking all the proteins in interaction with one kinase at least. This represents what the whole kinase family can interact with. Among the latter proteins, we counted the numbers of kinases and non-kinases and computed the ratio of these two counts obtaining 0.08. This ratio was close to what was reported for CMGCs only (44/652 = 0.07) [12]. The number of non-kinase proteins in contact with at least one kinase is thus much larger than the number of kinases (1/0.08 = 12.5 times). Nonetheless, 8% was much more than the 2.5% of human genes coding kinases. To properly evaluate whether this ratio was unexpectedly large required careful statistical modeling. As a matter of fact, we saw that kinases had more PPIs than an average human protein, which could artificially bias the observed ratio. Selecting 487 proteins – the number of kinases in our network – randomly that followed the same connectivity distribution as kinases (Materials and methods) we confirmed higher inter-kinase PPI density (Fig. 3D top). To gain deeper insight in the topology of the kinase–kinase PPI network, we repeated a similar analysis but at the level of individual kinases. It turned out that the average ratio kinase versus non-kinase PPIs was not increased (Fig. 3D bottom). This revealed that PPI-mediated collaboration across kinases is not the rule although a certain number of kinases are sufficiently well-connected to other kinases via PPIs to reverse this bias when considering all the kinases as a single entity as we did above in our global analysis. The 10 most PPI-connected kinases were SRC (32 PPIs with other kinases; 228 in total), EGFR (29;357), FYN (25;244), MAP3K7 (21;107),RAF1 (18;82), ERBB2 (18;180), ERBB3 (18;105), LYN (18;80), ABL1 (17;209), and IKBKB (16;102). Among this list LYN, RAF1, ERBB3, MAP3K7 and IKBKB had a very large number of PPIs with other kinases, typically more than 20% of their interactions. For comparison, the more PPI-connected kinases in general were EGFR, IKBKE (341 PPIs), FYN, SRC, ABL1, PLK1 (185), ERBB2, MAP3K3 (178), GSK3B (174), and PRPF4B (173).
Fig. 3.
Connectivity of kinases. (A) Kinases tend to have more PPIs with other proteins than average and this trend is further augmented for commonly expressed kinases that are identified in almost all cell types. (B) Similar observation for kinase substrates. Substrates known from experimental data only have the same bias (cyan). (C) The number of PPIs of a kinase is not correlated with the one of its substrates. (D) Cooperation with other kinases. Globally, the pooled kinase PPIs touch a set of human proteins including 8% of kinases (red line), which is much more than expected by chance (black null distribution). On the contrary, the average ratio of PPIs linking an individual kinase to other kinases versus non-kinases is significantly less than expected by chance. (E) Regulation (control) of other proteins through phosphorylation. Both globally and individually, kinases control other kinases significantly more than expected by chance. (F) Kinase–kinase network linking all the kinases for which we could obtain interaction data (487). Colors associated to each kinase group are given in panels G and H. (G) Analysis of cooperation (PPIs) within and across kinase groups. Since a few interconnections only were significant (P-value < 0.05) we featured edges with at least 3 PPIs and P-value < 0.5 as dashed lines. (H) Same analysis for regulation (KSIs). One self-connection (Tyr to Tyr) was supported by such a large number of KSIs (417) that it was outside the line width scale used to plot the network.
Considering KSIs, we performed similar calculations as above and we found that the part of the proteome globally covered as substrates of kinases was significantly enriched in kinases (Fig. 3E top). Reducing the data to the sole experimentally derived KSIs confirmed this result (same figure). At the individual kinase level, we also found significant over-representation of kinases as substrates of other kinases (Fig. 3E bottom; same result for experimental KSIs). Intra-kinase control or regulation is thus distributed across all kinases and is not the fact of a few global controllers only, a clear difference with PPIs.
The kinase–kinase network combining PPIs and KSIs is depicted in Fig. 3F, where we notice that commonly expressed proteins again seemed to concentrate at the center and kinases of the same groups to be closer to each other. The actual higher connectivity of commonly expressed kinases and substrates is reported in Fig. 3A and B. To evaluate whether inter-group connectivity preference truly exists, we decomposed the interactions (Fig. 3G and H). Taking into account the group sizes, the total number of kinases in the network, and the number of interaction between two groups, or within a single group, we could identify stronger than expected by chance interconnections (hypergeometric test). Most significant PPIs interconnections took place within single groups. In the case of KSIs, we obtained a lot more significant interconnections across two groups although the ones within a group were among the strongest. As we already did, we confirmed this result considering experimental KSIs only (data not shown).
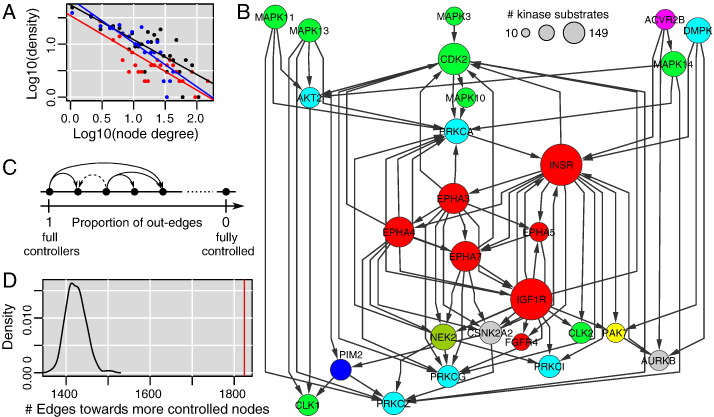
The observation of significant interaction among kinases through KSIs at the level of individual kinases (Fig. 3E bottom) suggested an evolution of KSIs between kinases to globally coordinate the activity of these enzymes. We considered the kinase–kinase network spanned by KSIs only, which we name KSI kinase network, and observed that it had a scale-free, or small world, topology [45,49] (Fig. 4A, black dots) that is commonly observed in biological networks: a few highly-connected hubs and a majority of less connected nodes. Because kinases have a dual role in this network, being either substrates or enzymes, we wanted to explore the existence of a relationship between the network topology and a prevalent enzymatic or substrate role. Selecting kinases that play the role of the enzyme towards other kinases in > 80% of their KSIs, we recognized immediately important signaling cascades (Fig. 4B). We asked our compiled data whether the complete integration of the effect of all signaling and regulation pathways by kinases on themselves would preserve a global structure. The observation that “as enzyme” KSIs and “as substrate” KSIs also induced a scale-free topology (Fig. 4A, red and blue dots) already indicated that a specific structure exists at the functional role level. In order to further ascertain its existence, we compared how kinases that control many other kinases, and are themselves less controlled, connected with other kinases in the real KSI network and in artificial networks with correct topology but random enzymatic/substrate roles. Sorting the kinases from full controllers (always phosphorylating other kinases) to fully controlled (never phosphorylating other kinases) and counting how many times one kinase has kinase substrates that are less controllers than itself (Fig. 4C), we found a significant difference in the real data (Fig. 4D); see also Materials and methods. Kinase controllers thus influence the rest of the kinase activities significantly. Kinases that had less than 5 substrates known (not only kinases) were excluded from the simulation to limit the impact of absent data, which left a KSI network of 357 kinases and 2698 interactions. Identical results were obtained using experimental KSIs only (data not shown).
Fig. 4.
Global organization of the kinase–kinase phosphorylation network. (A) Degree distributions of kinase substrate interactions (black) follow a power-law typical for a scale-free topology; same observation when only considering the number of interactions from a kinase playing the role of the enzyme towards another kinase playing the role of substrate (red) or vice versa (blue). (B) Partial view of the kinase-kinase KSIs involving the kinases that have more than 80% of their interactions towards another kinase (kinase controllers in the text). Edges from and towards kinases not included in this partial network were not drawn. (C) Principle of a statistical test for a directional structure. (D) Bias towards a directional structure.
3.5. Kinases and non-kinases occupying special positions in the phosphorylation network
The demonstrated existence of a global organization with kinases through KSIs called for the selection and inspection of specific sets of kinases, defined by how they were positioned with respect to the rest of the network. For this purpose, we decided to further increase the requirement on data reliability by only considering experimental KSIs and to limit our study to kinases for which at least 10 such KSIs were available, i.e. a rather precise appreciation for their connectivity was possible, which left us with 168 kinases.
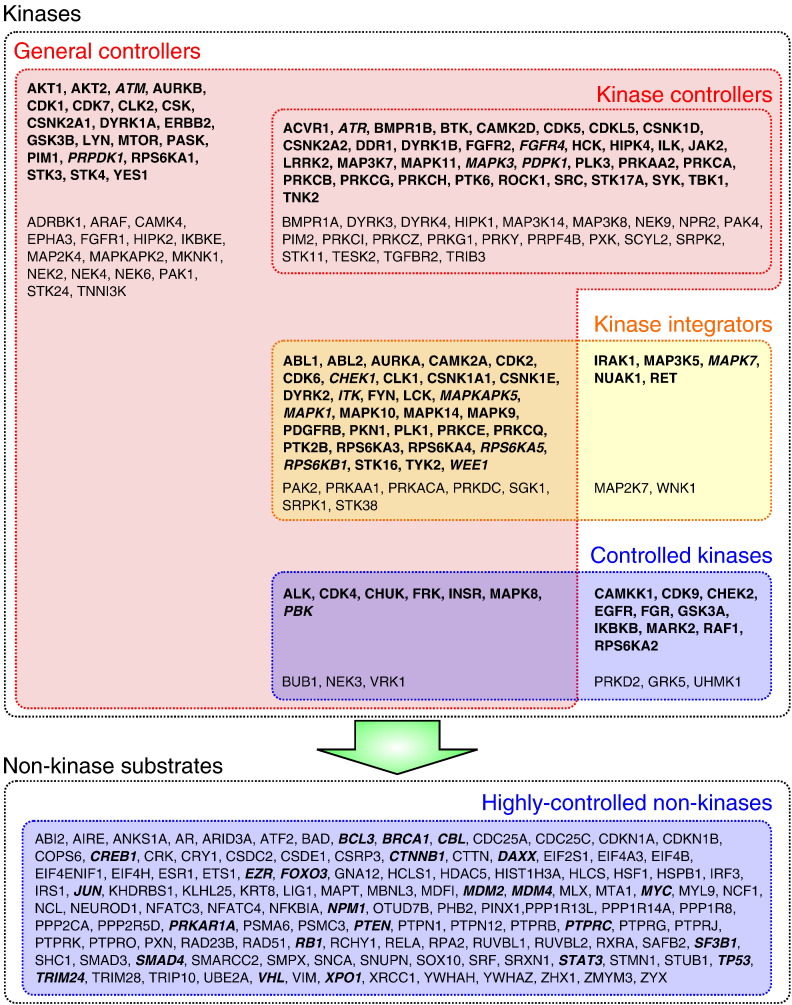
A first set we defined were kinases seldom or never regulated by other kinases (in > 80% of their KSIs they play the role of the enzyme). We identified 143 such proteins and named this set controllers. Then, considering KSIs between kinases, we defined kinase controllers that had > 80% of their KSIs towards other kinases (KSIs towards non-kinase substrates did not count for this second set). We found 57 kinase controllers, which were all included in the controller set (Fig. 5) meaning that kinase controllers are not specialized in this function and hence connect the kinase network to the rest of the interactome. A dual set to the kinase controllers were the 23 controlled kinases that play the role of a substrate in > 80% of their KSIs with other kinases. Some of them were also controllers having a large number of non-kinase substrates (Fig. 5). We further defined a set of 46 kinases that have a comparable number of KSIs as substrates or enzymes (enzymatic role in between 1/3 and 2/3 of their KSIs). We named them kinase integrators to indicate a potential function integrating and propagating multiple signals within the kinase network. As it was the case for the controlled kinases, part of the integrators turned out to be controllers as well (a majority in this case). Highly-controlled non-kinases were defined as the proteins with the largest number of distinct kinases phosphorylating them (top 5%, 122 proteins).
Fig. 5.
Selected kinase sets and highly-regulated non-kinases. Among the kinases, strong drug targets of compound in our data are in bold. We further manually queried PubMed to see if additional literature would indicate the existence of a rather specific inhibitor for more kinases; such positive cases are in bold italic. Non-kinases in bold italic are associated with cancer.
To contrast the different sets we submitted them to GO term [50] analysis. Controllers were localized predominantly in the cytosol, different membranes, Golgi, and also in the nucleus but clearly less than in the cytosol. Kinase controllers and integrators, and controlled kinases were in membranes and the cytosol. Highly-controlled non-kinases were predominantly located in the cytosol though also present in the nucleus (reverse distribution compared to controllers); they were specifically associated with transcription factor complexes and nuclear euchromatin. In terms of function, the four kinase sets associated with all types of kinase activities and signaling cascades with a nice decrease from controllers (strongest) down to controlled kinases (weakest). Controllers and kinase controllers were also enriched in transmembrane receptors, growth factor binding. Highly-controlled non-kinases had a strong and specific association with RNA and DNA binding, and transcription. Immune response and innate immunity pathways were strongly represented in controllers and kinase integrators (TLR signaling). Highly-controlled non-kinases were also involved in immune system processes. Apoptosis and cell proliferation very significantly included the latter non-kinases as well as the controller set. Finally, complementing the preferential nuclear localization and nucleic acid binding specificity of the highly-controlled non-kinases, we found a strongly association with DNA damage signaling and DNA repair.
Compiling the genes annotated to be tumor suppressor or oncogenes in GSEA MSigDB [51], or listed in COSMIC cancer gene census [42] we found an intersection with 24 highly-controlled genes (P < 4E− 14, hypergeometric test).
3.6. Kinase inhibitors, the modulating medical entry points
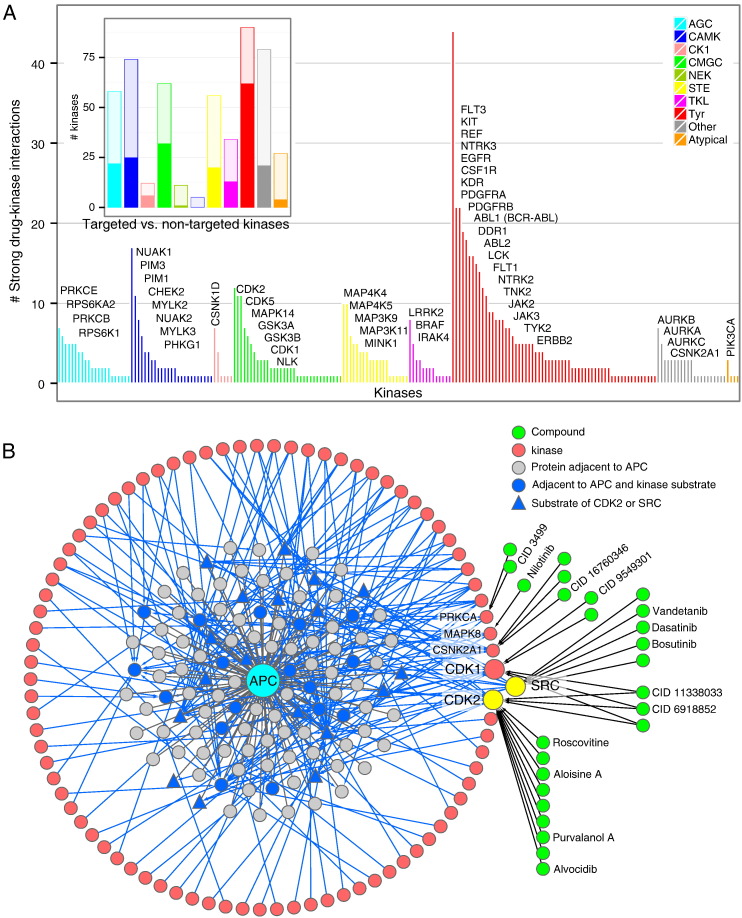
Given the implication of kinases in a large number of disorders [52], numerous kinase inhibitors have been developed to modulate these enzymes as curative agents. Early and extremely successful examples such as imatinib (Gleevec®), aimed at treating chronic myeloid leukemia, were introduced as targeted therapies against a specific kinase (the fusion protein BCR-ABL in this example). Later investigations unraveled the existence of several targets beyond BCR-ABL for this compound, even including a non-kinase (NQO2) [25], and more generally kinase inhibitors tend to be promiscuous compounds [15,20,25]. It is therefore difficult to provide a clear picture of which kinases are specifically and potently targeted by which molecules, separating many tool compounds, approved drugs, and molecules in clinical development or only disclosed in patent applications. For the purpose of this study we preferred to capture trends reflecting the interest of the drug development community and, to a certain extent, the difficulty in targeting certain kinases. Our reference network contained compiled data from large in vitro screens and chemical proteomics that were filtered according to criteria indicating clear inhibition of a kinase by a compound. Here, we needed to define reasonable, more stringent criteria to relate each compound to “strong” targets, i.e. targets for which potency compared to other targets of the same molecule was significantly stronger thus making the compound a likely selective inhibitor for these selected targets (Materials and methods). We obtained a strong target list comprised of 218 kinases interacting with 170 compounds and forming 1261 DPIs. Reporting the number of times a kinase is a strong target for distinct molecules (Fig. 6A) shows a very biased attention of drug development across kinase groups and specific enzymes. A similar figure was reported previously [53].
Fig. 6.
Interface with compounds. (A) Number of compounds for which a kinase is a strong target in the data we compiled. Most frequently targeted kinases in each group are named starting with the most frequent (top left of each list). The inset shows the ratio of (strongly) targeted kinases (filled with color) and untargeted kinases (empty). (B) A conceptual approach to block mutated adenomatosis polyposis coli (APC) activity by disrupting the activity of its PPI partners (gray and blue nodes) through kinase inhibitors targeting kinases phosphorylating such partners. Existence of a compound in our dataset for kinases with at least four substrates among APC partners are indicated by a green node. We indicated a few compounds that have a name or provided the PubChem accession code (CID).
With respect to the four kinase sets we defined previously (Fig. 5) we observed numerous examples of kinases not targeted by any compound suggesting many unexplored therapeutic options for future inhibitor development. Each had a comparable number of strongly targeted kinases but taking into account the multiplicity of compounds strongly targeting each kinases (heights of bars in Fig. 6A), we observed significant differences: controllers that were not kinase controllers were strongly enriched (P < 3.4E− 6, Fisher exact) as well as kinase integrators (P < 1.3E− 8), most of which as controllers. Controlled kinases were not found to be favored drug targets. We further broke down target biases to the kinase family level and identified PDGFR (P < 1.1E− 15), Trk (P < 3.2E− 4), NEK(6.9E− 4), VEGFR (P < 1.3E− 3), Abl (P < 4.9E− 3), and EGFR (P < 5.7E− 3).
To illustrate how a kinase network such as the one we assembled might actually support the implementation of new paradigms in the application kinase inhibitors in cancer therapy, we drafted a strategy to circumvent aberrant behavior of mutated non-kinases. The tumor suppressor adenomatous polyposis coli (APC) has been found to be involved in several tumors when mutated (gastric and colon cancers, medulloblastoma, hepatocellular carcinoma, etc.) [54,55]. As indicated in Fedorov et al. [53] APC is difficult to modulate through small molecules and signaling around this protein could be a preferable target for therapy. From our reference network we collected all PPIs relating APC to its functional context (Wnt and JNK signaling, cell migration) and retrieved 114 proteins, all non-kinases (Fig. 6B). We subsequently found 85 kinases having experimental KSIs with at least one of these proteins (35/144 APC neighbors). The two kinases CDK2 and SRC had the largest number of substrates and covered together 18/144 of the APC partners. One could thus propose to disrupt with maximum efficacy the environment of APC by composing a combined therapy with SRC and CDK2 inhibitors. Dual CDK1 and CDK2 inhibitors would also be an option to explore.
3.7. Discussion and conclusions
We compiled a large corpus of interaction data around human protein kinases and performed an initial characterization of this broad kinase network. Our results are based on available data and clearly may be affected by experimental bias of the underlying technologies. Yet, we feel confident that the general trends observed here are properties of the network too robust to be changed dramatically by acquisition of additional knowledge in the future.
The analysis of how information was acquired in kinase research comparing network biology efforts identifying protein–protein interactions with classical methodology showed that a clear correlation exists between the two (Fig. 2A). Network-orientated work has been so far directed as a complement and this trend was stable over the time period for which we had data, i.e. 1975–2013 (Fig. 2B). A different picture was provided confronting the acquisition of cancer versus non-cancer kinase protein interactions, where we observed a clear departure of kinase PPI mapping from mainstream activity (Fig. 2C). This could be regarded as a contradiction due to the general positive correlation. We believe that this shift was first caused by a difference in the scope of studies realized by the network biology community where authors frequently aim at characterizing an entire pathway or a set of related protein complexes, which causes the inclusion of additional baits, kinases in our case, that are not immediately associated with cancer if cancer was the object of the study. Furthermore, a second, confounding effect is introduced by our partial knowledge of which kinases are cancer-related and to which extent. The boundary between cancer and non-cancer genes remains incompletely characterized and might be more diffuse than previously thought. The increase in maturity of AP-MS and Y2H methods together with the necessary bioinformatics [11,56–58] will surely foster global, unbiased kinase PPI mapping projects eventually covering the dynamical nature of kinase interactions [26]. The contribution of such future data might alleviate the correlation with classical biochemistry letting interaction mapping becoming a driving force in positioning research attention.
Kinases are interacting entities by essence that both establish interactions with other proteins to form complexes, where they provide their enzymatic function, and with their substrates whose activity they modulate. Analyzing the two types of interactions separately and comparing the results unraveled specific properties and notable differences. PPI data suggested that kinases operate the modulation of their substrates by participating in more protein complexes than average human proteins (Fig. 3A), this trend being further increased among kinases commonly expressed by many distinct cell types. This probably reflects an evolution of kinase PPIs that favored the modular reuse of a limited repertoire of enzymatic specificities by creating a multitude of molecular machines, where relevance for a certain cell type, cellular compartment, or condition was contributed by the non-kinase subunits of the complexes, e.g. adaptor or protein bearing binding domains. Substrates themselves tended to be more PPI-connected than average proteins (Fig. 3B), which could implicate another level of evolutionary selection to more tightly modulate multifunctional proteins activities. Absence of correlation between kinase and their substrates PPI degrees (Fig. 3C) excluded a trivial technical bias e.g. caused by protein abundance to be the reason for kinase substrate increased PPIs in our data. We also found that commonly expressed kinases did not have more KSIs thus indicating the existence of cell type-specific kinases, rarely or low expressed, that can drive large biological processes via many substrates upon activation.
Considering kinase to kinase PPIs, other authors raised the hypothesis of existence of a so-called “kinase highway” [12,48]. Although we found a significantly high number of kinase–kinase PPIs by pooling all kinase PPIs as one set, this observation did not hold true at the level of individual kinases (Fig. 3D) meaning that on average a kinase does not bind to more other kinases than expected by chance. We actually made the opposite observation: kinases bound significantly less other kinases than expected by chance. This observation makes sense since the kinase enzymatic function is only required once in a protein complex in general. A few kinases highly PPI-connected to other kinases contributed the global, pooled bias obviously and our data did not support the concept of a PPI-based kinase highway. Turning our attention to KSIs, which logically would be more appropriate to provide the necessary mechanism for creating ways of communications, going beyond individual signaling cascades, we could indeed prove the existence of such a global structure (Figs. 3E and 4).
The evolution of an ancestral kinase domain gave rise to several groups of kinases (CMGCs, tyrosine kinases, CAMKs, etc.), themselves decomposed in families [29]. Identifying significantly increased connectivity within and across kinase groups via PPIs and KSIs, we realized that within group interactions composed an enriched network motif (we could repeat this observation at the finer kinase family level, data not shown). Across group connectivity was not enriched in PPIs but it was in KSIs to a great extent, thus further supporting the importance of the KSI network. The mechanism by which intense interactions within groups emerged could be explained by classical models of protein interaction network evolution, for instance the duplication and divergence (DD) model [59]. Duplicated kinase genes originally preserved their PPI interface with other proteins, which later evolved differentially between the two duplicated genes. Since in many cases the two kinases continued to be involved in the same or related biological processes, evolutionary pressure maintained a high rate of shared PPIs. Obvious examples are provided by kinase families such as CDKs regulating the cell cycle. The same rational can be proposed to explain KSI enrichment within groups – and families – by preserving substrate specificity after gene duplication. The ability to phosphorylate kinases of the same group or family would be acquired by the necessity to coordinate protein activity of related biological processes. This phenomenon could have been favored when the original kinase – before duplication – was able to autophosphorylate thus providing the two kinases obtained after duplication with the capability to cross-phosphorylate.
We decomposed the KSI network in several sets occupying particular positions of its topology (Fig. 5) e.g. distinguishing general controller kinases whose activity was not or seldom modulated by other kinases. We opposed them to the set of non-kinase proteins that were most regulated by kinases (substrates of the largest number of distinct kinases). The general controllers were predominantly located in the cytosol though also present in the nucleus, whereas the highly controlled non-kinases were clearly more present in the nucleus. The general controllers were further enriched in membranes being often transmembrane receptors. A strong association with apoptosis, cell proliferation, and the innate immune system, e.g. TLR signaling, was shared by controllers and highly-controlled proteins. In addition, the latter were specifically involved in RNA and DNA binding, transcription, and DNA damage signaling and repair.
Kinase inhibitors represent one of the major classes of therapeutic agents. They tend to be promiscuous molecules [15,16,25] and despite their multiplicity only part of the kinome has been so far targeted with some specificity and potency (Fig. 6A) [53], leaving open a broad range of options for future research. Similar to kinase interaction research, the drug discovery community has biased its attention to a limited number of kinases identified as important targets without pursuing systematic efforts. In terms of the classes of kinases targeted we found PDGFRs to be the most favored kinase family and, more generally, the general controllers discussed above to be preferred. Interestingly, distinguishing among the general controllers a subset of kinase controllers that had a large number of other kinases as substrates in addition to non-kinases, we showed that successful molecules tended to avoid kinase controllers. We hypothesize that the inhibition of such kinase controllers might induce very deep changes in cells due to a cascade of phosphorylation pattern alterations.
This work may bear interesting stimuli for theoretical evaluations of therapeutic interventions. To start with, controlled kinases and highly-controlled non-kinases could well represent ideal biomarkers that may integrate a variety of signaling states across the kinase network. Strategically chosen, the modification state of a few good such receivers of phosphorylation signaling might have superb diagnostic property. Consequently, they may also offer convenient systems-level readouts for phenotype-based drug discovery campaigns. For what regards targeted therapy, mono- or polypharmacological modulation may well be instructed by the logic of the kinase network. For example, simultaneous inhibition of kinases heavily connected to other kinases may be too prone to catastrophic events common to many cell types, while simultaneous targeting of global controllers that do not regulate other kinases directly may represent perfect therapeutic windows, being more cell–cell context or cell–environment-specific. Furthermore, access to high-resolution, possibly cell type and cell state-dependent KSI maps, would allow the combination of kinase inhibitors in therapies targeting non-kinase mutated proteins by operating intense and surgical disruption of their functional environment as we imagined for APC (Fig. 6B).
One of the most interesting observations of the current analysis is that coherence in the kinase network relies on KSIs primarily and not on PPIs. While experimental bias may play a role, this also indicates that the primary signaling logic involving kinases is intrinsically linked to their enzymatic property and one might wish increased attention of the proteomics community to rapidly complement our knowledge. The accurate measure of phosphorylation sites will not be sufficient. Certainly, more attention to dynamic aspects of protein-protein interaction in the coming years of increased resolution in proteomics experiments will help better model the interface of PPI and KSI networks. Integration with phosphatase specificities and additional regulatory networks induced by other posttranslational modifications such as acetylation, ubiquitination, or SUMOylation, all possibly overlapping [60], will shed light on the many forces contributing to the evolution of kinases sociology.
Conflict of interest
The authors declare no conflict of interest pertaining to this research.
Transparency document
Transparency document.
Transparency document
The Transparency document associated with this article can be found, in the online version.
Acknowledgments
We thank the public databases and repositories which collected all the interaction data we used in this work: IntAct, MINT, InnateDB, DIP, HPRD, MatrixDB, CORUM, NCI PID, PhosphoSitePlus, Phospho.ELM, NetworKIN, and DrugBank. JC was supported by an Austrian Science Fund (FWF) grant no. P 24321-B21.
Footnotes
This article is part of a Special Issue entitled: 20 years of Proteomics in memory of Vitaliano Pallini. Guest Editors: Luca Bini, Juan J. Calvete, Natacha Turck, Denis Hochstrasser and Jean-Charles Sanchez.
Contributor Information
Jacques Colinge, Email: jcolinge@cemm.oeaw.ac.at.
Giulio Superti-Furga, Email: gsuperti@cemm.oeaw.ac.at.
References
- 1.Ravasz E., Somera A.L., Mongru D.A., Oltvai Z.N., Barabasi A.L. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 2.Gavin A.-C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 3.Han J.D., Bertin N., Hao T., Goldberg D.S., Berriz G.F., Zhang L.V. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 4.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 5.Hunter T. Why nature chose phosphate to modify proteins. Philos Trans R Soc Lond B Biol Sci. 2012;367:2513–2516. doi: 10.1098/rstb.2012.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 7.Glatter T., Wepf A., Aebersold R., Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol Syst Biol. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rees J.S., Lowe N., Armean I.M., Roote J., Johnson G., Drummond E. In vivo analysis of proteomes and interactomes using Parallel Affinity Capture (iPAC) coupled to mass spectrometry. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002386. [M110 002386] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 10.Braun P., Tasan M., Dreze M., Barrios-Rodiles M., Lemmens I., Yu H. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varjosalo M., Sacco R., Stukalov A., van Drogen A., Planyavsky M., Hauri S. Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat Methods. 2013;10:307–314. doi: 10.1038/nmeth.2400. [DOI] [PubMed] [Google Scholar]
- 12.Varjosalo M., Keskitalo S., Van Drogen A., Nurkkala H., Vichalkovski A., Aebersold R. The protein interaction landscape of the human CMGC kinase group. Cell Rep. 2013;3:1306–1320. doi: 10.1016/j.celrep.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 13.Newman R.H., Hu J., Rho H.S., Xie Z., Woodard C., Neiswinger J. Construction of human activity-based phosphorylation networks. Mol Syst Biol. 2013;9:655. doi: 10.1038/msb.2013.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linding R., Jensen L.J., Pasculescu A., Olhovsky M., Colwill K., Bork P. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 2008;36:D695–D699. doi: 10.1093/nar/gkm902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis M.I., Hunt J.P., Herrgard S., Ciceri P., Wodicka L.M., Pallares G. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 16.Anastassiadis T., Deacon S.W., Devarajan K., Ma H., Peterson J.R. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wishart D.S., Knox C., Guo A.C., Cheng D., Shrivastava S., Tzur D. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colinge J., Rix U., Bennett K.L., Superti-Furga G. Systems biology analysis of protein–drug interactions. Proteomics Clin Appl. 2012;6:102–116. doi: 10.1002/prca.201100077. [DOI] [PubMed] [Google Scholar]
- 19.Rix U., Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol. 2009;5:616–624. doi: 10.1038/nchembio.216. [DOI] [PubMed] [Google Scholar]
- 20.Rix U., Colinge J., Blatt K., Gridling M., Remsing Rix L.L., Parapatics K. A target-disease network model of second-generation BCR-ABL inhibitor action in Ph + ALL. PLoS One. 2013;8:e77155. doi: 10.1371/journal.pone.0077155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winter G.E., Rix U., Lissat A., Stukalov A., Mullner M.K., Bennett K.L. An integrated chemical biology approach identifies specific vulnerability of Ewing's sarcoma to combined inhibition of Aurora kinases A and B. Mol Cancer Ther. 2011;10:1846–1856. doi: 10.1158/1535-7163.MCT-11-0100. [DOI] [PubMed] [Google Scholar]
- 22.Li J., Rix U., Fang B., Bai Y., Edwards A., Colinge J. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat Chem Biol. 2010;6:291–299. doi: 10.1038/nchembio.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Remsing Rix L.L., Rix U., Colinge J., Hantschel O., Bennett K.L., Stranzl T. Global target profile of the kinase inhibitor bosutinib in primary chronic myeloid leukemia cells. Leukemia. 2009;23:477–485. doi: 10.1038/leu.2008.334. [DOI] [PubMed] [Google Scholar]
- 24.Rix U., Remsing Rix L.L., Terker A.S., Fernbach N.V., Hantschel O., Planyavsky M. A comprehensive target selectivity survey of the BCR-ABL kinase inhibitor INNO-406 by kinase profiling and chemical proteomics in chronic myeloid leukemia cells. Leukemia. 2009;24:44–50. doi: 10.1038/leu.2009.228. [DOI] [PubMed] [Google Scholar]
- 25.Rix U., Hantschel O., Durnberger G., Remsing Rix L.L., Planyavsky M., Fernbach N.V. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 26.Li J., Bennett K., Stukalov A., Fang B., Zhang G., Yoshida T. Perturbation of the mutated EGFR interactome identifies vulnerabilities and resistance mechanisms. Mol Syst Biol. 2013;9:705. doi: 10.1038/msb.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 28.Patricelli M.P., Nomanbhoy T.K., Wu J., Brown H., Zhou D., Zhang J. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem Biol. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 30.Kerrien S., Alam-Faruque Y., Aranda B., Bancarz I., Bridge A., Derow C. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cesareni G., Chatr-aryamontri A., Licata L., Ceol A. Searching the MINT database for protein interaction information. Curr Protoc Bioinformatics. 2008;22:8.5.1–8.5.13. doi: 10.1002/0471250953.bi0805s22. [DOI] [PubMed] [Google Scholar]
- 32.Lynn D.J., Winsor G.L., Chan C., Richard N., Laird M.R., Barsky A. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xenarios I., Salwinski L., Duan X.J., Higney P., Kim S.M., Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chautard E., Fatoux-Ardore M., Ballut L., Thierry-Mieg N., Ricard-Blum S. MatrixDB, the extracellular matrix interaction database. Nucleic Acids Res. 2011;39:D235–D240. doi: 10.1093/nar/gkq830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruepp A., Brauner B., Dunger-Kaltenbach I., Frishman G., Montrone C., Stransky M. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res. 2008;36:D646–D650. doi: 10.1093/nar/gkm936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dinkel H., Chica C., Via A., Gould C.M., Jensen L.J., Gibson T.J. Phospho.ELM: a database of phosphorylation sites–update 2011. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu C.H., Apweiler R., Bairoch A., Da Natale, Barker W.C., Boeckmann B. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187-91. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newman M.E.J. 2002. Random graphs as models of networks. [Google Scholar]
- 42.Forbes S.A., Bindal N., Bamford S., Cole C., Kok C.Y., Beare D. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geiger T., Wehner A., Schaab C., Cox J., Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014050. [M111 014050] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 46.Burkard T.R., Planyavsky M., Kaupe I., Breitwieser F.P., Burckstummer T., Bennett K.L. Initial characterization of the human central proteome. BMC Syst Biol. 2011;5:17. doi: 10.1186/1752-0509-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bossi A., Lehner B. Tissue specificity and the human protein interaction network. Mol Syst Biol. 2009;5:260. doi: 10.1038/msb.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breitkreutz A., Choi H., Sharom J.R., Boucher L., Neduva V., Larsen B. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeong H., Tombor B., Albert R., Oltvai Z.N., Barabasi A.L. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 50.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: tool for the unification of biology. Gene Ontol Consortium Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lahiry P., Torkamani A., Schork N.J., Hegele R.A. Kinase mutations in human disease: interpreting genotype–phenotype relationships. Nat Rev Genet. 2010;11:60–74. doi: 10.1038/nrg2707. [DOI] [PubMed] [Google Scholar]
- 53.Fedorov O., Muller S., Knapp S. The (un)targeted cancer kinome. Nat Chem Biol. 2010;6:166–169. doi: 10.1038/nchembio.297. [DOI] [PubMed] [Google Scholar]
- 54.Wallis Y.L., Morton D.G., McKeown C.M., Macdonald F. Molecular analysis of the APC gene in 205 families: extended genotype-phenotype correlations in FAP and evidence for the role of APC amino acid changes in colorectal cancer predisposition. J Med Genet. 1999;36:14–20. [PMC free article] [PubMed] [Google Scholar]
- 55.Lamlum H., Ilyas M., Rowan A., Clark S., Johnson V., Bell J. The type of somatic mutation at APC in familial adenomatous polyposis is determined by the site of the germline mutation: a new facet to Knudson's ‘two-hit’ hypothesis. Nat Med. 1999;5:1071–1075. doi: 10.1038/12511. [DOI] [PubMed] [Google Scholar]
- 56.Venkatesan K., Rual J.F., Vazquez A., Stelzl U., Lemmens I., Hirozane-Kishikawa T. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi H., Larsen B., Lin Z.Y., Breitkreutz A., Mellacheruvu D., Fermin D. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stukalov A., Superti-Furga G., Colinge J. Deconvolution of targeted protein-protein interaction maps. J Proteome Res. 2012;11:4102–4109. doi: 10.1021/pr300137n. [DOI] [PubMed] [Google Scholar]
- 59.Sun M.G., Kim P.M. Evolution of biological interaction networks: from models to real data. Genome Biol. 2011;12:235. doi: 10.1186/gb-2011-12-12-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beltrao P., Bork P., Krogan N.J., van Noort V. Evolution and functional cross-talk of protein post-translational modifications. Mol Syst Biol. 2013;9:714. doi: 10.1002/msb.201304521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.