Abstract
Purpose
miRNAs regulate the expression of genes in both normal physiology and disease. While miRNAs have been demonstrated to play a pivotal role in aspects of cancer biology, these reports have generally focused on the regulation of single genes. Such single-gene approaches have significant limitations, relying on miRNA expression levels and heuristic predictions of mRNA binding sites. This results in only circumstantial evidence of miRNA-target interaction and typically leads to large numbers of false positive predictions.
Methods
Here, we used a genome-wide approach (High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation, or HITS-CLIP) to define direct miRNA-mRNA interactions in three breast cancer subtypes (estrogen receptor positive, Her2 amplified and triple negative).
Results
Focusing on steroid receptor signaling, we identified two novel regulators of the ER pathway (miR-9-5p and miR-193a/b-3p), which together target multiple genes involved in ER signaling. Moreover, this approach enabled the definition of miR-9-5p as a global regulator of steroid receptor signaling in breast cancer.
Conclusions
We show that miRNA targets and networks defined by HITS-CLIP under physiologic conditions are predictive of patient outcomes and provide global insight into miRNA regulation in breast cancer.
INTRODUCTION
Breast cancer is the most common malignancy and the second most common cause of cancer related mortality among women in the United States. Despite recent advances in our understanding of breast cancer biology, estrogen receptor (ER) expression remains the best predictive and prognostic factor for therapy and patient risk stratification [1]. Over 70% of breast cancers express ER, and its expression generally portends a favorable prognosis. However, there is wide variability in clinical outcomes among patients with ER+ breast cancer, underscoring gaps in our understanding of ER+ breast cancer biology [2].
MicroRNAs (miRNAs) are a class of small non-coding RNAs with well-defined regulatory roles in normal physiology and diseases such as cancer, including breast cancer [3, 4]. Conventionally, their study has been based on defining single miRNA-mRNA target interactions using a combination of miRNA expression arrays and bioinformatic predictions of binding to the 3’ untranslated regions (UTRs) of transcripts [5, 6]. This approach is typically followed by miRNA overexpression and inhibition experiments in the relevant cell type. Such approaches often use transient transfection of miRNAs, raising their expression to non-physiologic levels, resulting in substantial off target effect [7]. In addition, they present only circumstantial evidence for the regulation of a particular target by a given miRNA, providing no evidence of direct binding, and typically do not allow the study of global interactions [8, 9].
In order to capture miRNA-mRNA interactions under physiological conditions in breast cancer, we used High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) [10, 11]. The miRNA-mRNA interaction within the RNA-induced silencing complex (RISC) occurs within the folds of the Argonaute (Ago) protein. Hence cross-linking of RNA to adjacent protein moieties (Argonaute in this case) by ultraviolet (UV) light allows for stringent isolation of the miRNA-mRNA-Ago complexes by immunoprecipitation. RNA isolated from the Ago-miRNA-mRNA complexes is analyzed with massively parallel DNA sequencing to determine the miRNA-mRNA interactome.
We performed HITS-CLIP on three well characterized breast cancer cell lines that represent ER+ (MCF7; ~60–70% of all patients), Her2+ (BT474; ~20% of all patients) and triple negative disease (MDA-MB-231; ~15–20% of all patients). We used these data to identify individual miRNA-mRNA interactions, including those previously defined in the literature as well as novel interactions not previously identified. Specifically, we focused on miRNAs predicted to regulate ER signaling. Multiple downstream targets of ER were found to be targets of miRNAs, suggesting that miRNAs regulate single targets as well as groups of genes representing signaling networks. Finally, we tested the relevance of our observations in the biology of ER+ breast cancer using gene expression profiles of primary breast cancer tissue coupled with patient survival data. This integrated approach enabled us to uncover novel miRNA-mRNA interactions and a program of hormone regulation that directs steroid receptor expression and differentiation, which has direct relevance to tumor biology and patient outcomes.
MATERIALS AND METHODS
Cell culture, transfection and treatment assays
The human breast cancer cell lines T47D and MCF7 were originally obtained from lafa Keydar [12], and Sam Brooks, the Michigan Cancer Foundation, respectively. Cells were maintained in MEM supplemented with 5% fetal bovine serum (FBS) and penicillin / streptomycin. BT474, and MDA-MB-231 cells were obtained from ATCC (Manassas, VA) and maintained in DMEM medium supplemented with 5% FBS and penicillin / streptomycin. HEK-293T cell line was obtained from ATCC (Manassas, VA) and maintained in DMEM with 10% FBS. Cell line authenticity was confirmed by short tandem repeat analyses in the University of Colorado DNA Sequencing Core Laboratory [13].
Plasmids, viral constructs and DNA cloning
Luciferase reporter vectors were created by cloning of the miRNA Responsive Element (MRE) sequences into the XhoI-NotI restriction site of psi-Check2 vector (Promega, Madison, WI) as previously reported [14]. Primers that were used for individual experiments are detailed in Supplementary methods section. Retroviral constructs for stable overexpression of miR-193 and miR-9 were constructed as previously reported [15].
HITS-CLIP protocol for Argonaute protein
HITS-CLIP was performed per Chi, et al’s original protocol adapted with minor modifications (Supplemental Figure 1A, for full protocol see [16]).
Luciferase assays
Luciferase reporter vectors were made by cloning the 3’ UTR or miRNA responsive element (MRE), into the XhoI-NotI restriction sites of psiCheck2 vector (Promega, Madison, WI) as was reported previously (primers used are detailed in supplemental methods section). HEK-293T cells were plated at approximately 40% confluence (50 000 cells per well) in 24 well plates. Sixteen hours later the psi-Check2 vectors and miR mimic / control mimic were co-transfected using Lipofectamine 2000 as per manufacturer’s protocol. Cells were harvested by passive lysis and assayed for firefly and renilla luciferase activity using Dual Luciferase Reporter Assay kit on a Glomax luminometer (Promega) following manufacturer’s instructions. The student’s t-test was used to determine significance.
Western Blotting
Cell lysates were quantitated for protein content, and approximately 25 mg lysates were resolved by standard SDS-PAGE and transferred to PVDF membrane. Blots were probed by antibodies against ER, (Cell Signaling Technology, Danvers, MA), and an appropriate HRP-conjugated secondary antibody. Blots were then stripped and reprobed for alpha tubulin (Sigma-Aldrich, St. Louis, MO) or beta actin (Applied Biological Materials, Richmond, BC, Canada) to ensure equal protein loading. The blots were then visualized using enhanced chemoluminescence using the Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE).
Immunocytochemistry
Cells were fixed onto glass coverslips in ice-cold 70% acetone/ 30% methanol for 5 min and stained by immunocytochemistry (ICC) with antibodies to TFF1, NCOA3 (Cell Signaling Technology, Danvers, MA). Secondary antibodies were anti-mouse and anti-rabbit Alexa Fluors 488 (green). Nuclei of cells were counterstained with DAPI. A Nikon E600 microscope (Nikon Corporation, Tokyo, Japan) was used for photography. Images were shot in black and white using ImagePro software (Media Cybernetics, Rockville, MD) and merged in Adobe Photoshop CS3 (Adobe Systems, San Jose, CA).
Cell Growth Assay
Growth effects were determined using the sulforhodamine B assay [17] after treatment with either either vehicle alone (0.1% ethanol), 4-hydroxytamoxifen (100nM) or 17-beta estradiol (10nM).
Next generation sequencing data analysis
First, raw reads from next generation sequencing were processed by alignment (Novoalign, Selangor, Malaysia) to the human genome 19 (hg19, UCSC). Aligned reads were filtered to exclude those with significant similarity to the 3’ adapter sequence. Statistics of aligned reads were generated using custom Python scripts. Peaks were determined using MACS [18]. Summits of peaks were determined by ascertaining the highest point of the peaks, and a peak footprint was determined by incorporating a 30 bp sequence up and downstream from the summit. Seed sequences of annotated mature human miRNAs (positions 2–7 and 2–8) were generated from mirbase-18. Seeds were mapped to consensus peaks and their interactions were visualized using Cytoscape (http://www.cytoscape.org/). Scatter plots and coefficient of determination were generated using R statistical packages (corrplot function) and Python-based data analysis toolkit Pandas. Custom software packages developed for this project are available for download and use at https://github.com/jayhesselberth/peaktools.
Analysis of microarray data and clinical correlates and network analysis
Publicly available normalized datasets were downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo/, data sets GSE22219, GSE22220) [19] and TCGA (http://tcga-data.nci.nih.gov/tcga/; BRCA Hi-Seq mRNA and Hi-Seq miRNA datasets) [20]. Individual miRNA expression levels were divided into high and low based on their median within the entire data set. Survival package within R was used for plotting of patient outcomes based on individual miRNA used for supervised outcomes analysis. Kaplan Meyer plots were generated with R software.
RESULTS
HITS-CLIP defines miRNA-mRNA targets in breast cancer subtypes
We hypothesized that miRNAs contribute to subtype specific regulation of cellular functions in breast cancer. We used Ago-HITS-CLIP to determine miRNA targets in three major subtypes of breast cancer: ER+, Her2+ and triple negative[10]. Each subtype was represented by a well-characterized breast cancer cell line (MCF7, BT474 and MDA-MB-231, respectively) [21]. Our objectives were to define miRNA-mRNA interactions in each breast cancer subtype, to identify specific miRNA-mRNA targeting events unique to and shared between each subtype, and to characterize clinically relevant targets related to the response of cancer cells to hormonal cues. The above cell lines also have a wealth of genome-wide datasets including mRNA and miRNA expression profiles, which we have incorporated in this study.
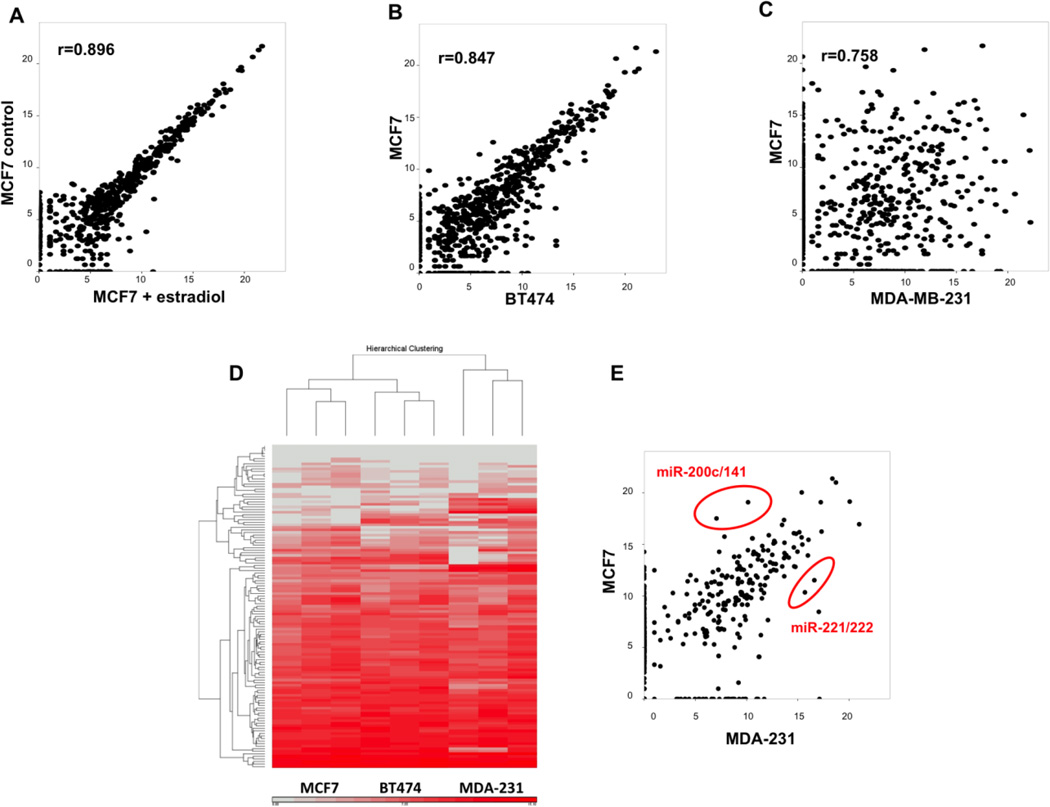
Ago-HITS-CLIP was performed by isolating UV cross-linked RISC-miRNA-mRNA complexes using immunoprecipitation, and analyzing isolated miRNA and mRNA fragments by next-generation DNA sequencing as described [10] (Supplemental Figure 1A). To ensure reproducibility, we performed at least three biological replicates for each cell line, and two biological replicates were performed for both ER positive cells lines stimulated with 17-beta estradiol for 24 hours. A typical experiment yielded about 20 million alignable reads (or 30 million total) matching the human genome reference sequence (hg19) (detailed statistics shown in Supplemental Table 1A and Supplemental Table 1B). Aligned reads were annotated as deriving from regions corresponding to mRNA, miRNA or other RNA (e.g. ribosomal RNA, intergenic RNA and non-coding RNA). The reads were then distributed between 3’ and 5’ untranslated regions (UTR), coding sequences (CDS) and intergenic sequences. MiRNA reads represented on average 4% of the total read counts for each of the experiments (Supplemental Figure 1B). These results are in agreement with previous studies that used Ago-HITS-CLIP [22]. We assessed the technical reproducibility of this approach by calculating the coefficient of determination for biological replicates (R2 greater than 0.9). The coefficient of determination between combined MCF7 samples with and without estrogen treatment showed a slightly higher variability suggesting a small change in the miRNA-mRNA interactome in response to estrogen exposure (Figure 1A). Higher variability was evident in comparisons between MCF7 (ER+, PR+, Her2-) and BT474 (ER+, PR+, Her2+) cell lines (Figure 1B) and maximum variability was evident between MCF7 and the triple negative cell line MDA-MB-231 (Figure 1C). This variability suggests that significant differences in the miRNA-mRNA interactome exist between biological subsets of breast cancer.
Figure 1. Ago HITS-CLIP analysis of breast cancer cells.
A. Correlation of consensus peaks between combined MCF7 and 17-beta estradiol treated MCF7 cells. Coefficient of determination (R2) calculated by Corrplot package. B. Correlation of consensus peaks between MCF7 and BT474 cells. C. Correlation of consensus peaks between MCF7 and MDA-MB-231 cells. D. Hierarchical clustering of miRNA expression from three biological replicates for each breast cancer subtype. MicroRNA read numbers were normalized to total aligned reads and analyzed with Partek genomics suite to generate the heat map. Fifty highly expressed miRNAs in each of the cell type were included in the analysis. E. Correlation of expression levels of individual miRNAs as measured by HITS-CLIP between MCF7 and MDA-MB-231 cells.
Given that HITS-CLIP libraries are prepared with two serial PCR amplification reactions, significant quantitative biases are anticipated in the sequencing results. Next generation sequencing based RNA quantification (RNA-Seq) is known to be very quantitative, and HITS-CLIP relies on similar methodology for library construction [23]. We used hierarchical clustering to determine whether qualitative assumptions about sequencing data would hold true in HITS-CLIP datasets. Both miRNA and mRNA peaks in a particular sample were normalized to total alignable reads in that sample and clustered in an unsupervised fashion using Partek software. When miRNA reads were analyzed (Figure 1D), biological replicates were observed to cluster together. Ago-mRNA peaks did not cluster similarly (data not shown). A potential explanation for this divergence between Ago-miRNA and Ago-mRNA peaks is that the mRNA fragments isolated after RNase digestion are of variable length with significant sample to sample variation, while the mature miRNAs are not affected by RNase digestion, leading to variability in their amplicon abundance. The observation that Ago-mRNA peaks are not quantitative needs to be considered while predicting and validating targets of miRNA. The most abundant miRNAs for each cell line are listed in Supplemental Table 2. These results are again in general agreement with miRNA quantifications reported by other investigators, using microarray or miRNA-Seq techniques. Importantly, our data sets also demonstrated previously noted difference between ER positive and negative breast cancer, such as the differential expression of miR-200c/141 and miR-221/222 between MCF7 and MDA-MB-231 cell lines [24, 25] (Figure 1E).
Global regulation of genes in breast cancer under physiologic conditions
We hypothesized that miRNA-mRNA interactions contribute to regulatory networks that define biological differences between breast cancer subtypes. To test this, we constructed a miRNA-mRNA interactome for each breast cancer subtype using Cytoscape software [26]. Individual nodes in this interactome were defined by the miRNAs or mRNAs that they were predicted to bind. Further hypothesizing that such networks might be best observed in highly abundant miRNAs, we started with analysis of 10 most abundant miRNAs by breast cancer subtype found in our data set (Supplemental Table 2). Based on seed-pairing predictions we generated a list of target mRNAs for these miRNAs. We then used Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com) to organize the gene set and visualize its contribution to cellular activity. The 10 most abundant miRNAs in each cell line were predicted to regulate several basic cellular mechanisms such as metabolism, proliferation and DNA synthesis, (Supplemental Figure 2). As expected, in both MCF7 and BT474 cells, the estrogen receptor pathway was among the most overrepresented networks targeted by the top 10 most abundant miRNAs (p<0.001), with high overlap of target genes among both cell lines (Table 1). In contrast to ER+ cell lines, the total number of miRNA-regulated genes was considerably lower in triple negative breast cancer cells (3413 for MDA-MB-231, compared to 9 978 and 10 785 for MCF7 and BT474, respectively). The top regulated pathways in the ER negative cells were cell proliferation and cell death, with far fewer ER regulated genes appearing in the target set (Table 1). This observation is in agreement with prior data describing dysregulation of miRNA biogenesis in triple negative disease [27]. Our data shows that the ER pathway is regulated by highly expressed miRNAs in both ER+ cell lines.
Table 1.
ER Targets with CLIP peaks containing seed sites for the top 10 expressed miRNAs in each cell line. Targets appearing in both ER+ lines are bolded and targets appearing in ER+ and ER- cell lines are underlined.
| Cell Line | ER Targets |
|---|---|
| BT474 | AHNAK, CEBPA, CLEC2D, CYP1B1, DDIT4, EDN1, EFNA1, ESR1, FAM102A, FOXA1, GATA3, GPAM, GREB1, HMGCR, HOXC6, IGF1R, INHBB, LDLR, MAP2K7, MDM2, MSX2, NRCAM, PDCD4, PGR, PRKCD, PRLR, PTEN, SLC7A2 TNPO1, TP53, TXNIP, VIM, YPEL3 |
| MCF7 | CCND1, CCNG2, CDKN1A, CELSR2, CLEC2D, CREBBP, CRKL, CYP1B1, DDIT4, ESR1, FOSL2, FOXA1, GATA3 ICOSLG, IGF1R, NOTCH3, PGR, PRLR, SLC7A2, SLC7A5, TXNIP |
| MDA-MB-231 | CAV2, CRKL, CYP1B1, DDIT4, SERPINE1, TXNIP |
miRNA regulation of the endocrine signaling axis
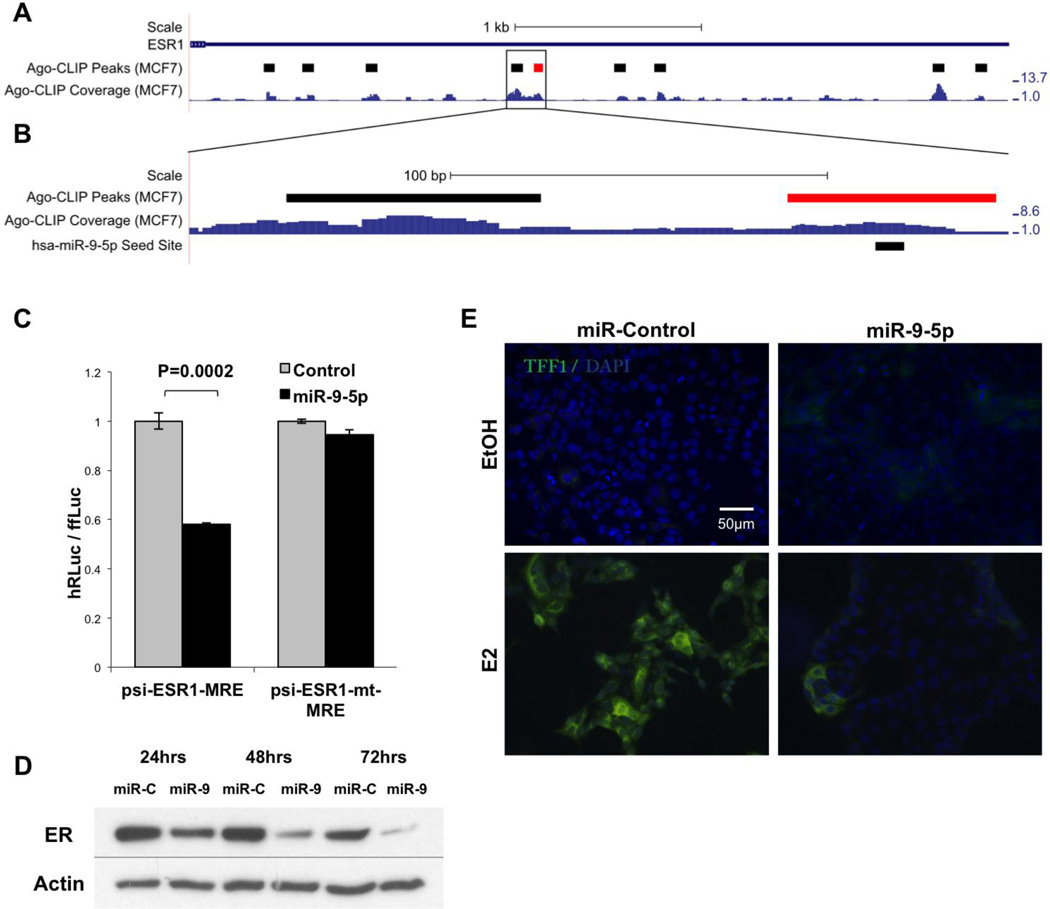
In addition to the global effects of miRNA on steroid receptor pathway, we also found that ER mRNA itself is targeted by miRNAs in ER+ breast cancer cell lines. Prior studies have reported on the role of several miRNAs in regulating the expression of ER at the individual mRNA level, but no unbiased genome-wide biochemical studies have been reported [28–30]. In our data set, statistically significant Ago footprints were present throughout the ER mRNA, including several prominent peaks in the 3’ UTR and coding regions (Figure 2A). We presumed that each of the 8 peaks in the ER 3’UTR corresponds to a functional mRNA-miRNA interaction, and that these could be narrowed down based on candidate miRNAs with potential to bind to these peaks. Initially we examined our data set for previously described miRNA-mRNA interactions, and confirmed a number of previously described pairs in our data set. These included putative binding sites for miR-17~92 cluster [29], miR-221/222 [6] and miR-26 [30] in the 3’UTR of ER (Figure 2A).
Figure 2. Mir-9-5p regulates ER in breast cancer.
A. The 3’UTR of ER, showing HITS-CLIP coverage. Statistically significant Ago footprints (peaks) are shown above the coverage (red peaks have been validated in this work). Coverage scale (right) is in Reads per Million Mapped Reads (RPMMR). B. The 3’UTR of ER, showing HITS-CLIP coverage at the miR-9-5p peak. Statistically significant Ago footprints (peaks) are shown above the coverage. Coverage scale (right) is in Reads per Million Mapped Reads (RPMMR). The sequence complementary to the miR-9-5p seed site is shown below the coverage. C. Luciferase assay in HEK293 cells showing repression of luciferase expression from the ESR1-MRE-containg luciferase plasmid upon co-transfection with a miR-9-5p mimic. Mutation of the sequence complementary to the seed site abolishes this repression. D. Western blot analysis of MCF7 cells transfected with miR-9 or control mimic at 5 nM concentration. ER protein expression was downregulated in miR-9 transfected cells starting 24 hours after transfection. Same western blots were stripped and re-probed for actin to ensure equal protein loading. E. TFF1 (green), a downstream target of activated ER, was not induced in miR-9-5p overexpressing breast cancer cells by 17-beta estradiol. Nuclei were counterstained with DAPI (blue). Cells were transfected with either Control (left column) or miR-9-5p (right column) miRNA mimics and treated with EtOH (vehicle control; top row) or E2 (bottom row). Portions of this figure based on output from the UCSC Genome Browser (http://genome.ucsc.edu).
We then investigated an Ago-mRNA peak in the ER 3’ UTR, 1759 nt downstream of the translational stop site (Figure 2A and B). Interestingly, it lies within a region that has been annotated as important for maintaining ER mRNA stability [31]. Identification of miRNA seed matches in the 60 nt Ago footprint using the predictive algorithms RNA22 and RNA hybrid [32, 33] suggested that this Ago-mRNA peak likely represents a binding site for miR-9-5p (Supplemental Figure 3). We confirmed that miR-9-5p binds to the sequence representing the Ago footprint and that mutation in the binding sequence leads to a decrease in binding effectiveness by luciferase assay (Figure 2C). Both miR-9-5p and control mimics were used and transfected into MCF7 cells and led to a reduction of ER protein expression following miRNA overexpression starting at 24 hours following transfection and remained decreased up to 72 hours later (Figure 2D).
In order to test the functional significance of miR-9-5p regulation of ER, we analyzed two downstream estrogen-regulated genes, TFF1 (Trefoil Factor 1, an estrogen regulated tumor suppressor [34]) and PR (Progesterone Receptor, another steroid receptor) [35], neither of which were predicted to bind to miR-9-5p. Immunocytochemistry for both proteins was performed on ER+ cell lines (MCF7 and BT474) with stable transfections of miR-9-5p or the control vector (Figure 2E). Both TFF1 and PR proteins were upregulated in response to estradiol treatment in control vector transfected cells. This effect was however ameliorated when miR-9-5p was over-expressed in addition to the estradiol treatment (Figure 2E, PR data not shown). Thus miR-9-5p affects downstream signaling of ER suggesting functional relevance of this interaction.
MiR-9 targets multiple steroid receptor pathways
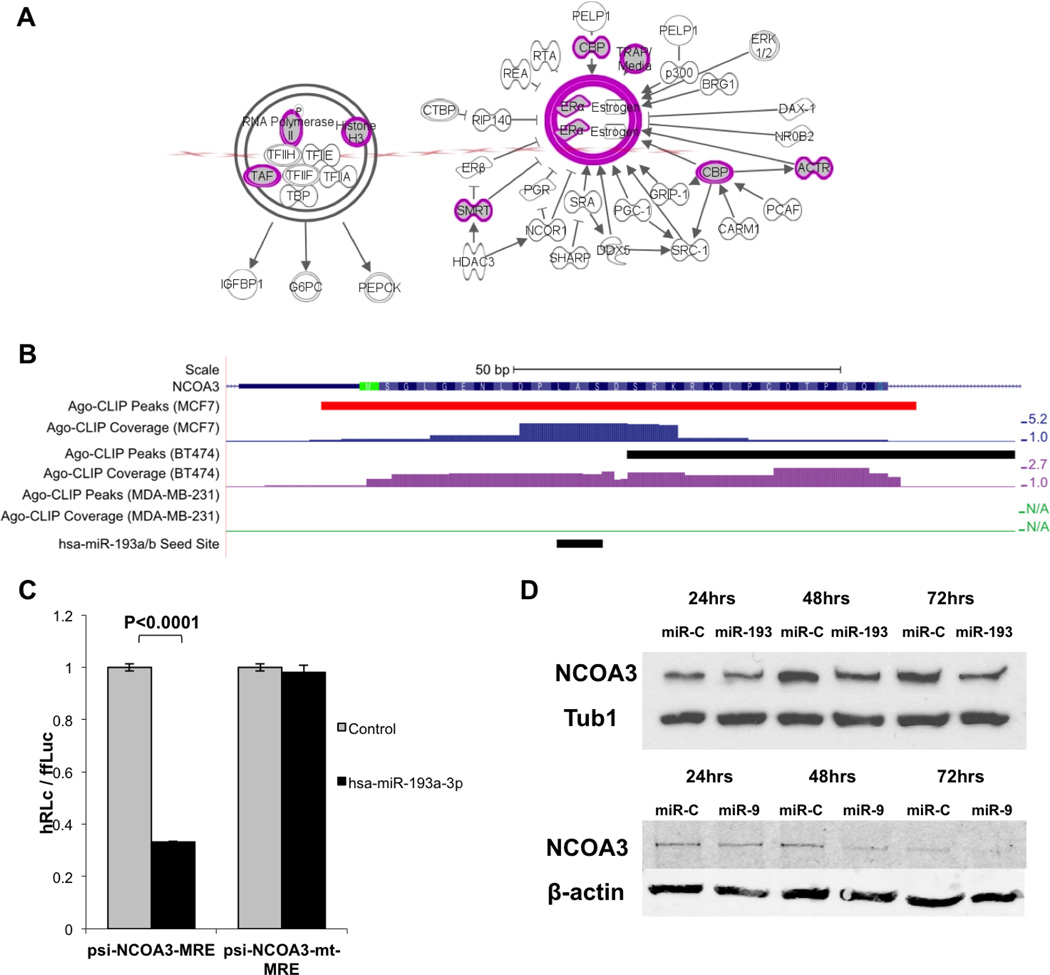
Given the involvement of miR-9-5p in ER mRNA regulation, we further hypothesized that it may be involved in direct or indirect regulation of multiple nodes of the ER signaling pathway. To test this, we used IPA to identify the pathway enriched in the Ago-mRNA peaks predicted to be bound by miR-9-5p. Consistent with our hypothesis, ER signaling pathway was the top canonical pathway targeted by miR-9-5p in ER+ cell lines, with multiple predicted gene targets. In addition, miR-9-5p acted as a regulator of common nodes of other hormone receptor signaling pathways, (androgen receptor (AR), retinoic acid receptor (RXR/RAR)) and their co-factors (i.e. NCOA2, NCOA3 and NCOA4; Figure 3A, Supplemental Figure 4A). These results support miR-9-5p as part of a control mechanism for a coherent set of signaling components at the center of cellular response to hormonal cues in ER+ breast cancer. As miR-9-5p and miR-221, which has also been reported to regulate ER, share little sequence homology, we used IPA to determine the overlap between their respective predicted targets and pathways. Among estrogen signaling pathway components, only ER itself was found to have any overlap between the predicted targets, suggesting a distinct physiologic role for both miRNAs (Supplemental Figure 3B). Additionally, there was no enrichment for other hormone receptor pathways in the miR-221 target set, suggesting that the cellular role of miR-221 is distinctly different than that of miR-9-5p. The expression level of miR-221 in ER+ disease is low [25], which may in part explain its distinct role and targets.
Figure 3. ER and NC0A3 are regulated by distinct miRNAs.
A. Predicted targets of miR-9-5p contain multiple nodes of ER signaling. These include ACTF (NCOA3) and TAF both co-regulated by miR-193a/b-3p (Figure base on output from IPA). B. The first coding exon of NCOA3, showing HITS-CLIP coverage. Statistically significant Ago footprints (peaks) are shown above the coverage (red peaks have been validated in this work). Coverage scale (right) is in Reads per Million Mapped Reads (RPMMR). The sequence complementary to the miR-193a/b-3p seed site is shown below the coverage. C. Luciferase assay in HEK293 cells showing repression of luciferase expression from the NCOA3-MRE-containg luciferase plasmid upon co-transfection with a miR-193a/b-3p mimic. Mutation of the sequence complementary to the seed site abolishes this repression. D. Overexpression of miR-193a/b-3p (Top) or miR-9-5p (Bottom) leads to a downregulation of the NC0A3 protein, with a maximum difference 72 hours after treatment.
The responsiveness of ER+ breast cancer to anti-endocrine therapies depends on the functional status of the entire ER signaling axis, including the activity of the downstream regulators and co-factors. Downregulation of ER, decoupling of downstream response elements [36],overexpression of ER co-activators (i.e. NCOA3) [37, 38] and activation of parallel pathways [39, 40] have all been implicated in the development of anti-endocrine resistance. Among the direct predicted targets of miR-9-5p, we found multiple steroid receptor co-factors including NCOA3 (Nuclear Receptor Coactivator 3; also known as amplified in breast cancer 1 or SRC-3). NCOA3 is a nuclear cofactor that is either amplified (up to 10%) or overexpressed (up to 40–50%) in breast cancer, and is correlated with resistance to anti-endocrine therapy in ER+ disease [38]. NCOA3 modulates ER activity and mediates interactions with other signaling pathways including Her2 and Akt [39, 40]. Overexpression of NCOA3 correlates with poor outcomes in patients with ER+ breast cancer. As predicted, overexpression of miR-9-5p lead to downregulation of NCOA3 at the protein level (Figure 3D). We therefore proceeded to further investigate NCOA3 regulation by miRNAs and found it to be also a predicted target of not only miR-9-5p, but also miR-193b-3p, one of the highly expressed miRNAs in ER+ cell lines (Supplemental Table 2). In addition, miR-9 and 193 were found to share another target in the ER pathway, several members of the TAF gene family which are part of the TFIID complex [41] (Figure 3A). Using seed-pair matching of NCOA3 and miR-193-3p (Supplemental Figure 3) we identified an Ago-mRNA peak in the first coding exon of NCOA3, 15 nt downstream of the start codon (Figure 3B). We used a luciferase assay to confirm the binding of miR-193-3p to the Ago-mRNA peak in the CDS (Figure 3C). Additionally, overexpression of miR-193-3p led to a significant downregulation of NCOA3 protein 24 hours after transfection (Figure 3D). Together, these data establish that NCOA3 is a direct target of miR-193-3p, and confirm that targeting occurs through the site identified by HITS-CLIP.
Modulation of estrogen response by miRNAs
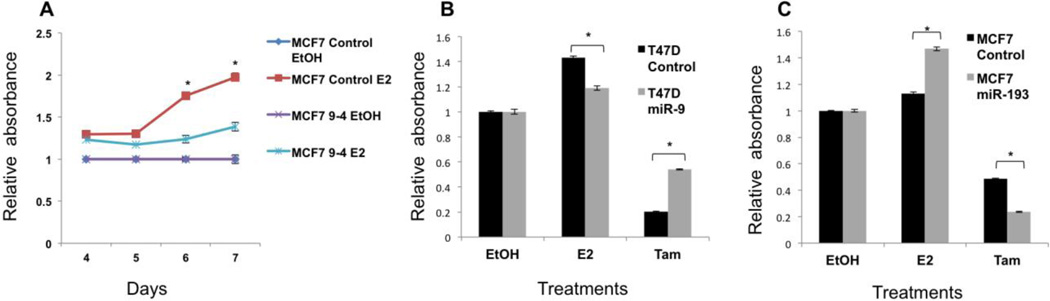
Based on the above results, we hypothesized that both miR-9-5p and miR-193-3p would modulate the response of breast cancer cells to estrogen and anti-endocrine therapy. To test this, we first determined growth rates of estrogen responsive cell lines MCF7and T47D cells that we engineered to stably express miR-9-5p or miR-193-3p. 17-beta estradiol acts as a mitogen, whereas tamoxifen inhibits growth in these cells. Based on our results, we expected miR-9-5p to mitigate the response to estradiol by decreasing the target receptor expression. Indeed, overexpression of miR-9-5p in multiple ER+ cell lines blunted the effect of 17-beta estradiol during a 7-day period (Figure 4A and B). The miR-9-5p overexpressing cells lost their rapid growth response to 17-beta estradiol, similar to the phenotype of ER- breast cancers. We also measured the effect of miR-9-5p on pharmacological inhibition of ER by addition of tamoxifen. As anticipated, increased expression of miR-9-5p caused resistance to tamoxifen (Figure 4B). Since overexpression of NCOA3 in breast cancer is correlated with resistance to anti-endocrine therapy in ER+ disease, we hypothesized that a decrease in NCOA3 would enhance the responsiveness of ER+ cancer cells to estrogen and tamoxifen. Indeed, the overexpression of miR-193-3p had an effect opposite to that of miR-9-5p: an increase in miR-193-3p levels exaggerated the effects of both estradiol and tamoxifen on ER+ breast cancer cells (Figure 4C), confirming the functional regulation of NCOA3 by miR-193-3p.
Figure 4. MiR-9-5p and miR-193a/b-3p modulate the response of ER+ breast cancer to estrogen.
A. Response of control MCF7 cells and a single cell clone (clone 4 in this example) of miR-9-5p overexpressing MCF7 cells were examined. Cells were treated with ethanol (ETOH) or 17-beta estradiol (E2). MiR-9-5p overexpression blunted the response of ER+ cells to 17-beta estradiol (statistically significant difference p<0.05 marked with asterisk) on days 6 and 7. B. Similar effect of decrease in sensitivity was observed on other ER+ cell lines. Here, miR-9-5p overexpressing T47D cells show a decrease in response both to 17-beta estradiol, as well as 4-hydroxytamoxifen (student t-test was used for statistical analysis, p<0.05). C. MiR-193a/b-3p further exaggerated the response of ER+ cells (control and miR-193a/b-3p overexpressing MCF7 cells) to both 17-beta estradiol (E2) and 4-hydroxytamoxifen (Tam).
HITS-CLIP analysis predicts patient outcomes in clinical samples from patients with ER+ breast cancer
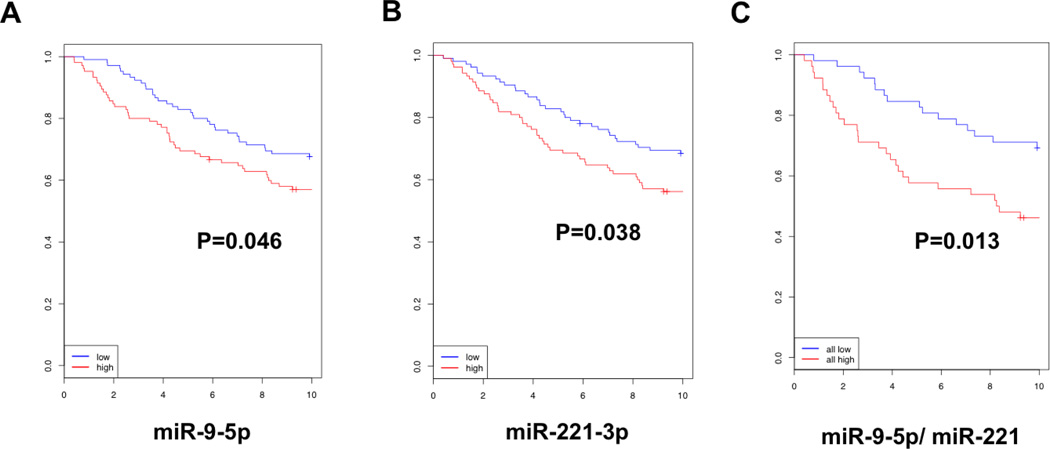
Given that miR-9-5p is a direct regulator of both ER and down-stream regulators of ER signaling, we hypothesized that expression levels of miR-9-5p in breast cancer samples could predict clinical outcome. Specifically, we hypothesized that high expression levels of miR-9-5p would decrease responsiveness of tumors to anti-endocrine therapies, thus contributing to earlier disease relapse and subsequent poor overall outcome in patients with ER+ breast cancer. To test this hypothesis, we used data from previously published cohorts of patient samples with corresponding clinical data, as well as matched miRNA expression levels measured by RNA-seq [19]. We first classified each miRNA expression as either high or low, based on the median expression for each individual miRNA within the whole data set. Then we applied the survival package within R to determine whether expression levels of a specific miRNA affected patient outcomes. Kaplan Meyer plots were generated with R software (Figure 5). We then performed a supervised analysis of miR-9-5p expression and found that high expression levels were predictive for early relapse (p=0.046, Figure 5A). In agreement with our in vitro data, high miR-9-5p expression correlated with decreased levels of ER mRNA, in ER+ patient tumors (p=0.025). MiR-221-3p has been previously described as a direct regulator of ER, and the 3’ UTR Ago peaks corresponding to the published regulatory elements were seen in our datasets as well [6]. High expression level of miR-221-3p also predicted for a worse outcome in the same complete cohort of 216 patients (p=0.038, Figure 5B). The patient outcome predictions could be further refined by combining the miR-9-5p and miR-221-3p expression levels in patients with ER+ breast cancer (Figure 5C). MiR-221-3p level inversely correlated with ER mRNA expression, similar to miR-9-5p. From these results we concluded that while miR-221-3p and miR-9-5p share ER as a target, miR-9-5p is involved in the global regulation of the endocrine axis under physiologic conditions in primary breast cancers.
Figure 5. Ago-HITS-CLIP defines functional regulatory networks and predicts patient outcomes in breast cancer.
A. MiR-9-5p expression level predicts disease recurrence in patients with breast cancer (p=0.046). B. MiR-221-3p, a known regulator of ER, similarly predicts patient outcomes and combination for both miR-9-5p and miR-221-3p further improves the predictive value (C).
MiRNA networks regulate the state of differentiation and disease severity in breast cancer
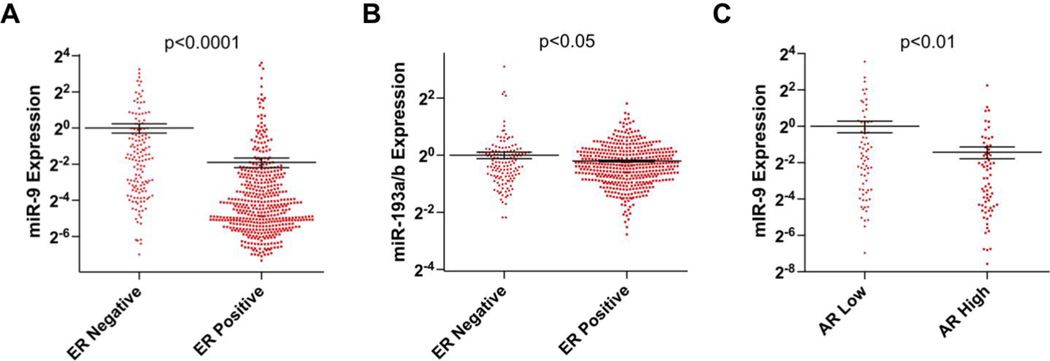
Expression of steroid receptors such as ER, PR and AR have been shown to define a more differentiated breast cancer phenotype, which correlates with lower tumor grade and better patient outcomes [42–45]. We speculated that if miR-9-5p was a global regulator of steroid receptors (ER and AR) and its co-factors (NCOA2-4), we could verify such an association in an independent data set. We used the Breast Invasive Carcinoma (BRCA) miRNA-seq and mRNA-seq data from The Cancer Genome Atlas for our secondary analysis (n=597, annotated patient samples) [20]. We again confirmed that miR-9-5p expression was inversely correlated with ER status across all breast cancers (Figure 6A). Furthermore, in ER-positive tumors, miR-9-5p expression was approximately 2-fold lower in those with 50–100% of cells staining positive for ER by immunohistochemistry than in those with <50% (data not shown). For miR-193a/b the inverse correlation with ER expression was less striking, but still statistically significant (Figure 6B). The clinically relevant functional steroid receptor in triple negative disease is AR, where its expression is inversely correlated with aggressiveness of disease. Supporting the role of miR-9-5p as a universal steroid receptor regulator in breast cancer, low AR mRNA expression correlated well with high miR-9-5p levels in patients with triple negative disease (Figure 6C) and confirmed the predictive value of a genome wide analysis.
Figure 6. MiRNA regulation of steroid receptors in breast cancer.
A. miR-9-5p expression inversely correlates with ER mRNA levels of patients with breast cancer in the TCGA data set (p<0.001). B. ER expression is inversely correlated with miR-193a/b-3p (p<0.01). C. miR-9-5p expression level is inversely correlated with AR mRNA levels in ER- tumors in the TCGA.
DISCUSSION
Here we demonstrate that an unbiased genome-wide biochemical analysis of miRNA-mRNA binding based on HITS-CLIP for the Argonaute protein can define individual miRNA-mRNA interactions and regulatory networks in breast cancer subtypes. More importantly, the results can be placed into biological and clinical context when correlated with patient outcomes. Given the importance of ER regulation in breast cancer biology, we initially focused on the regulation of the endocrine signaling axis in ER+ breast cancers by miRNAs as proof of principle for this model. Multiple prior studies have reported on the contribution of miRNAs to the biological differences between ER+ and triple negative breast cancer [6, 28, 29]. While we found our results to be generally concordant with published data, our unbiased genome-wide approach also uncovered multiple novel regulators of the hormonal axis in breast cancer including miR-193 and miR-9-5p. We show that miR-9 affects several key nodes in steroid hormone regulation of ER+ breast cancers, further affirming that miRNAs are a part of larger regulatory network, fine tuning cellular responses of complex pathways such as steroid receptor signaling.
We demonstrate that a CLIP peak in the 3’ UTR of ER is a direct target of miR-9-5p, and that overexpression of miR-9-5p reduces endogenous ER protein levels as well as that of several ER target genes. We also establish that a CLIP peak in the coding sequence of NCOA3, an ER cofactor, is a direct target of miR-193a/b-3p, and that overexpression of both miR-193a/b-3p and miR-9-5p results in reduced NCOA3 protein expression (the latter likely through a separate exonic CLIP peak). As other ER pathway components and downstream targets also contain CLIP peaks with seed matches to miR-9-5p and miR-193a/b-3p, we propose that these miRNAs cooperate to regulate global estrogen signaling in breast cancer. We further demonstrate that miR-9-5p and miR-193a/b-3p help regulate responsiveness of breast cancer cells to estrogen/tamoxifen. While simultaneous repression of both ER and NCOA3 by miR-9-5p leads to a decrease in response to estrogen, decrease of NCOA3 alone by miR-193a/b-3p sensitizes ER+ breast cancer cells to tamoxifen. Thus, miR-9-5p and miR-193a/b-3p allow for fine-grained regulation on the ER signaling pathway, through a combination of shared and discrete targets.
The present analysis of the miRNA-mRNA interactome offers a detailed look at the post-transcriptional regulation under physiological conditions. This is exemplified in the largely non-overlapping set of targets for miR-9-5p and miR-221. Both miRNAs have been shown to regulate ER, however their cellular functions are most likely different under normal conditions. This observation is supported by the generally low expression of miR-221 in ER+ breast cancers. Similarly, while many prior studies have linked expression profiles to patient outcomes, we believe that taking a functional approach to target-miRNA interactions, such as the one presented here, can add a functional dimension and help explain the biological meaning of descriptive analyses. As evidenced by our results, Ago-HITS-CLIP is a powerful tool that can be used to define novel miRNA-mRNA interactions as well as functional regulatory networks.
However, a few disadvantages were evident. First, while HITS-CLIP in its present form remains a sophisticated method for the enrichment of sequences bound by Argonaute, target validation still relies upon miRNA binding predictions and standard protein expression and binding experiments. It should also be noted that not all targets could be directly validated by such standard methods. An exciting refinement of the original HITS-CLIP protocol termed CLASH (Crosslinking, ligation and sequencing of hybrids) was recently reported [46]. CLASH relies on hybrid mRNA-mRNA molecules formed during a ligation step of the CLIP library preparation and can thus reliably identify targets of at least some of the miRNAs directly from the sequencing data. A second is that the technique without further refinements remains non-quantitative for mRNA levels. Our results show that while the Ago-miRNA peaks maintain relative quantitation, Ago-mRNA peaks do not. Incorporating quantitation during the library preparation steps (by adding degenerate primers called UMIs or Unique Molecular Identifiers) is one way to circumvent this problem [47]. Finally, UV cross-linking is very inefficient and the technique in its current form requires a large number of cells for library preparation. Although addition of photoactivable nucleoside analogues (PAR-CLIP) was reported to vastly improve cross-linking efficiencies, our experiments failed to show such benefit (data not shown) and the technique was noted to be similar in efficiency by other investigators [48]. As a result of this high cell number requirement, HITS-CLIP and related techniques remain unusable for primary clinical samples.
Despite these limitations, our study highlights the importance and utility of unbiased genome-wide biochemical approaches like HITS-CLIP that enable us to define novel clinically relevant miRNA-based regulatory pathways of endocrine responsiveness and resistance in breast cancer. In addition to identifying two miRNAs that play a key role in the regulation of estrogen signaling in breast cancer, we have also generated three valuable genome-wide miRNA-target interaction datasets. Further analysis of these data, particularly in combination with other genome-wide approaches including profiling of primary tumor samples, will undoubtedly reveal further insights into the role of miRNAs in breast cancer biology.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Dr. Zissimos Mourelatos (University of Pennsylvania, Philadelphia, PA) for generously providing the Ago antibody for initial experiments, Jill Castoe (High Throughput Sequencing Core, UC Denver) for assistance with next generation sequencing, Dr. Carol Sartorius (UC Denver) for critical reading of the manuscript and Dr. Kenneth Jones (UC Denver) for logistical support of data analysis. The work was supported in part by grants from the National Institute of Health (HL104070 and DK073701) to MMP and CA164048 to PK; March of Dimes Basil O'Connor Starter Scholar, Damon Runyon Rachleff Innovation Award and American Cancer Society Research Scholar Grant to J.R.H.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1.Desantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2013 doi: 10.3322/caac.21203. [DOI] [PubMed] [Google Scholar]
- 2.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 3.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Bian C, Liao L, Li J, Zhao RC. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat. 2010 doi: 10.1007/s10549-010-0954-4. [DOI] [PubMed] [Google Scholar]
- 6.Zhao J-J, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Thomson DW, Bracken CP, Goodall GJ. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011;39:6845–6853. doi: 10.1093/nar/gkr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;(38 Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 10.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keydar I, Chen L, Karby S, Weiss FR, Delarea J, Radu M, Chaitcik S, Brenner HJ. Establishment and characterization of a cell line of human breast carcinoma origin. Eur J Cancer. 1979;15:659–670. doi: 10.1016/0014-2964(79)90139-7. [DOI] [PubMed] [Google Scholar]
- 13.Korch C, Spillman MA, Jackson TA, Jacobsen BM, Murphy SK, Lessey BA, Jordan VC, Bradford AP. DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination. Gynecol Oncol. 2012;127:241–248. doi: 10.1016/j.ygyno.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pillai MM, Yang X, Balakrishnan I, Bemis L, Torok-Storb B. MiR-886-3p down regulates CXCL12 (SDF1) expression in human marrow stromal cells. PloS One. 2010;5:e14304. doi: 10.1371/journal.pone.0014304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 16.Balakrishnan I, Yang X, Brown J, Ramakrishnan A, Torok-Storb B, Kabos P, Hesselberth JR, Pillai MM. Genome-wide analysis of miRNA-mRNA interactions in marrow stromal cells. Stem Cells Dayt Ohio. 2013 doi: 10.1002/stem.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, Zhang S, Wang T, Sinibaldi D, Coppola D, Heller R, Ellis LM, Karras J, Bromberg J, Pardoll D, Jove R, Yu H. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 18.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu X-D, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, Taylor M, Harris AL, Ragoussis J. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe J-P, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo W-L, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurteau GJ, Carlson JA, Spivack SD, Brock GJ. Overexpression of the microRNA hsa-miR-200c leads to reduced expression of transcription factor 8 and increased expression of E-cadherin. Cancer Res. 2007;67:7972–7976. doi: 10.1158/0008-5472.CAN-07-1058. [DOI] [PubMed] [Google Scholar]
- 25.Stinson S, Lackner MR, Adai AT, Yu N, Kim H-J, O’Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T, Newman RJ, Yue P, Bourgon R, Modrusan Z, Stern HM, Warming S, de Sauvage FJ, Amler L, Yeh R-F, Dornan D. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:ra41. doi: 10.1126/scisignal.2001538. [DOI] [PubMed] [Google Scholar]
- 26.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinforma Oxf Engl. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avery-Kiejda KA, Braye SG, Forbes JF, Scott RJ. The expression of Dicer and Drosha in matched normal tissues, tumours and lymph node metastases in triple negative breast cancer. BMC Cancer. 2014;14:253. doi: 10.1186/1471-2407-14-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, lorio MV, Li M, Volinia S, Alder H, Nakamura T, Nuovo G, Liu Y, Nephew KP, Croce CM. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J, Hannon GJ, Stebbing J. The estrogen receptor-α-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci. 2009;106:15732–15737. doi: 10.1073/pnas.0906947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Zheng J, Zhang Y, Yang L, Wang J, Ni J, Cui D, Yu C, Cai Z. Tumor-specific expression of microRNA-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and -independent pathways. Mol Ther J Am Soc Gene Ther. 2011;19:1521–1528. doi: 10.1038/mt.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keaveney M, Parker MG, Gannon F. Identification of a functional role for the 3’ region of the human oestrogen receptor gene. J Mol Endocrinol. 1993;10:143–152. doi: 10.1677/jme.0.0100143. [DOI] [PubMed] [Google Scholar]
- 32.Miranda KC, Huynh T, Tay Y, Ang Y-S, Tarn W-L, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 33.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA N Y N. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buache E, Etique N, Alpy F, Stoll I, Muckensturm M, Reina-San-Martin B, Chenard MP, Tomasetto C, Rio MC. Deficiency in trefoil factor 1 (TFF1) increases tumorigenicity of human breast cancer cells and mammary tumor development in TFF1-knockout mice. Oncogene. 2011;30:3261–3273. doi: 10.1038/onc.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foekens JA, Rio MC, Seguin P, van Putten WL, Fauque J, Nap M, Klijn JG, Chambon P. Prediction of relapse and survival in breast cancer patients by pS2 protein status. Cancer Res. 1990;50:3832–3837. [PubMed] [Google Scholar]
- 36.Clark GM, McGuire WL, Hubay CA, Pearson OH, Marshall JS. Progesterone receptors as a prognostic factor in Stage II breast cancer. N Engl J Med. 1983;309:1343–1347. doi: 10.1056/nejm198312013092240. [DOI] [PubMed] [Google Scholar]
- 37.Jordan VC, O’Malley BW. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2007;25:5815–5824. doi: 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- 38.Osborne CK, Bardou V, Hopp TA, Chamness GC, Hilsenbeck SG, Fuqua SAW, Wong J, Allred DC, Clark GM, Schiff R. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95:353–361. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 39.Arpino G, Wiechmann L, Osborne CK, Schiff R. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29:217–233. doi: 10.1210/er.2006-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osborne CK, Schiff R. Mechanisms of endocrine resistance in breast cancer. Annu Rev Med. 2011;62:233–247. doi: 10.1146/annurev-med-070909-182917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papai G, Weil PA, Schultz P. New insights into the function of transcription factor TFIID from recent structural studies. Curr Opin Genet Dev. 2011;21:219–224. doi: 10.1016/j.gde.2011.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasparini P, Fassan M, Cascione L, Guler G, Balci S, Irkkan C, Paisie C, Lovat F, Morrison C, Zhang J, Scarpa A, Croce CM, Shapiro CL, Huebner K. Androgen receptor status is a prognostic marker in non-basal triple negative breast cancers and determines novel therapeutic options. PloS One. 2014;9:e88525. doi: 10.1371/journal.pone.0088525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park S, Park HS, Koo JS, Yang Wl, Kim SI, Park B-W. Higher expression of androgen receptor is a significant predictor for better endocrine-responsiveness in estrogen receptor-positive breast cancers. Breast Cancer Res Treat. 2012;133:311–320. doi: 10.1007/s10549-011-1950-z. [DOI] [PubMed] [Google Scholar]
- 44.Peters KM, Edwards SL, Nair SS, French JD, Bailey PJ, Salkield K, Stein S, Wagner S, Francis GD, Clark SJ, Brown MA. Androgen receptor expression predicts breast cancer survival: the role of genetic and epigenetic events. BMC Cancer. 2012;12:132. doi: 10.1186/1471-2407-12-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 46.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the Human miRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kivioja T, Vaharautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J. Counting absolute numbers of molecules using unique molecular identifiers. Nat Meth. 2012;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 48.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.